Abstract
RNA-targeting approaches are emerging as viable therapeutics that offer an alternative method to modulate traditionally ‘undrugable’ targets. In the case of dominantly inherited neurodegenerative diseases, gene suppression strategies can target the underlying cause of these intractable disorders. Polyglutamine diseases are caused by CAG expansions in discrete genes, making them ideal candidates for gene suppression therapies. Here, we discuss the current state of gene suppression approaches for Huntington's disease and the spinocerebellar ataxias, including the use of antisense oligonucleotides, short-interfering RNAs, as well as viral vector-mediated delivery of short hairpin RNAs and artificial microRNAs. We focus on lessons learned from preclinical studies investigating gene suppression therapies for these disorders, particularly in rodent models of disease and in non-human primates. In animal models, recent advances in gene suppression technologies have not only prevented disease progression in a number of cases, but have also reversed existing disease, providing evidence that reducing the expression of disease-causing genes may be of benefit in symptomatic patients. Both allele- and non-allele-specific approaches to gene suppression have made great strides over the past decade, showing efficacy and safety in both small and large animal models. Advances in delivery techniques allow for broad and durable suppression of target genes, have been validated in non-human primates and in some cases, are currently being evaluated in human patients. Finally, we discuss the challenges of developing and delivering gene suppression constructs into the CNS and recent advances of potential therapeutics into the clinic.
Introduction
Gene suppression approaches for dominantly inherited neurodegenerative diseases have made great strides over the past decade, especially in the cases of the polyglutamine-repeat disorders. In particular, gene suppression approaches for Huntington's disease (HD) and the spinocerebellar ataxias (SCAs) have shown great promise and are quickly moving from testing in rodent models of disease into large animal models and, in the case of HD, into the first clinical trial in human patients. Polyglutamine-repeat disorders stem from abnormally long CAG expansions, which confer a toxic gain of function on the expressed protein, resulting in symptoms that are different for each disease. In this review, we will address the progress made in the fields of HD and the SCAs, with a specific focus on the use of RNA interference (RNAi) and antisense oligonucleotide (ASO) approaches. Additionally, we will discuss the hurdles to successful gene suppression approaches and clinical trial development for these disorders.
Therapeutic Approaches for Co-Opting Endogenous mRNA Degradation Pathways
RNA interference
RNA interference is an evolutionarily conserved process of post-transcriptional gene suppression that utilizes native non-coding double-stranded RNA sequences to reduce mRNA expression. MicroRNAs (miRNAs) are transcribed within the nucleus by pol II and pol III promoters to form stem-loop structures known as primary-miRNAs (pri-miRNAs). Pri-miRNAs are cleaved by the Drosha-DGCR8 microprocessor complex to form a precursor-miRNA (pre-miRNA) hairpin-like structure. Pre-miRNAs are then exported to the cytoplasm by Exportin-5 whereupon they are further processed by Dicer to create a mature miRNA duplex. The antisense, or ‘guide’ strand is loaded into the RNA-induced silencing complex (RISC), while the sense or ‘passenger’ strand is degraded. Upon entering RISC, the guide strand silences its target based on the degree of complementarity of the seed sequence to the target. Perfect complementarity, in the case of short-interfering RNAs (siRNAs), leads to Ago-2-mediated cleavage, whereas imperfect base pairing, in the case of miRNAs, leads to destabilization or translational repression (1).
Over the years, the endogenous cellular RNAi pathway has been co-opted to suppress specific genes of interest. RNAi effectors are generated in the laboratory and introduced into the cell as siRNAs, short hairpin RNAs (shRNAs) or artificial miRNAs (artificial sequences cloned into the context of an endogenous miRNA sequence, such as miR-30). siRNAs are 21 nucleotide-long duplexes that are processed by Dicer and silence target mRNAs through Ago-2-mediated cleavage. These can be delivered in a variety of flavors including chemically modified single-stranded siRNAs (2,3), liposome-formulated siRNAs (4) or nanoparticles (5). Early generation RNAi triggers were in the form of shRNAs embedded in a pri-miRNA scaffold, or miRNA backbone, and delivered by viral vectors (6–8). These shRNAs were extremely potent and achieved high levels of knockdown in vivo. However, the shRNAs were expressed at a high level and have been shown in certain cases to saturate endogenous RNAi machinery (9,10). Newer techniques use siRNAs embedded into artificial miRNA backbones to achieve stable and efficient mRNA suppression without the buildup of antisense precursor species in the cell.
The delivery and stable expression of RNAi suppression constructs remains a major challenge for gene therapy applications. siRNAs do not readily cross the blood–brain barrier (BBB) and when injected directly into the brain do not typically penetrate the plasma membrane without modification. To circumvent this issue, many studies have used shRNAs and artificial miRNAs expressed from viral vectors to achieve stable expression in particular brain regions of interest. Recombinant adeno-associated virus (AAV) and lentivirus (LV) are the most common gene therapy vectors for directed shRNA and miRNA delivery into the brain. They provide stable expression of RNAi triggers while being minimally immunogenic, non-pathogenic and replication incompetent (11). Recombinant LV can be pseudotyped with different glycoproteins to alter their cellular tropism (12), transduces both dividing and non-dividing cells and integrates into the host's genome unless the integrase is inactivated (13). However, LV-mediated transduction often results in low copy number, making it a weaker vehicle compared with AAV. AAVs provide strong and stable gene expression without integrating into the host genome, remaining as nuclear episomes. The numerous, variable capsid serotypes of AAV provide specific cell and tissue tropisms, making it a desirable choice for directed gene therapy (14,15). AAV serotypes bind to distinct cellular receptors, display patterns of retrograde and anterograde transport following injection and newer generation serotypes are being engineered with increased tropism to distinct tissues types and brain regions.
It should be noted that viral vector-mediated delivery can induce a neutralizing antibody response against the viral capsid, diminishing transgene expression in target tissues (16,17). Several animals used in preclinical research, as well as humans, have varying levels of pre-existing antibodies to many of the AAV serotypes, with highest levels against AAVs 1 and 2 compared with AAVs 5, 6, 8 and 9 (18,19). Research animals and human patients can be pre-screened for the presence of anti-AAV neutralizing antibodies and current research is investigating novel engineered AAV capsids that may be able to evade neutralizing antibody responses. In the case of AAVs, sustained expression has been seen over 6 years in canine and non-human primates (20), making the need for re-delivery unlikely. An outstanding example of AAV tolerability and re-delivery, should it be necessary, is the work by Bennett et al. (21) which demonstrated that re-administration of AAV to the eye of patients was extremely well tolerated several years after the original delivery.
Antisense oligonucleotides
ASOs are single-stranded nucleotides, typically 12–22 bases in length, that bind complementary target RNA through Watson and Crick hybridization resulting in modulation of the target RNA. ASOs are primarily used to modulate RNA either through the recruitment of RNase H, an endogenous enzyme that recognizes RNA/DNA heteroduplexes and degrades the target RNA (22), or by obstructing interactors of the target RNA to inhibit translation or modulate splicing. The antisense mechanism invoked depends on both the ASO's binding location on the target RNA and the chemical modifications of the ASO (23).
More than 30 years of optimization of oligonucleotide modifications have generated molecules with characteristics amenable to the treatment of neurodegenerative diseases. Most, if not all, ASOs in development or in the clinic today contain a substitution of sulfur for non-bridging oxygen atom in the phosphate backbone, transforming the phosphodiester linkage to a phosphorothioate (24). This improves pharmacokinetics by increasing resistance to nucleases and improves distribution and pharmacodynamics by increasing protein binding (25). Another common modification is a 2′ alteration to the sugar moiety. This includes 2′-O-methoxyethyl (MOE) modifications, as well as introduction of bicyclic nucleosides such as locked nucleic acids (LNAs) and constrained 2′-O-ethyl nucleic acids (cEts). Sugar modifications increase binding affinity to the target RNA, increase resistance to nucleases and can decrease non-specific toxicities, each to a different degree (26,27). For the higher affinity modifications, like LNAs, increased potency can come at the cost of tolerability (28). Importantly, most sugar modifications render the modified nucleotide resistant to RNase H activity.
Each modification, with its unique properties, can be used as a building block to assemble an ASO with a desired set of characteristics. If RNA degradation is required, a gapmer design can be used. This is done by incorporating sugar modifications into the 5′ and 3′ residues to improve binding affinity and resistance to nucleases (the wing) while leaving a stretch of uninterrupted, unmodified deoxynucleic acids in the center (the gap) to engage RNase H (27). Alternatively, ASOs with fully modified sugars are used to bind RNA with high affinity without degrading the target RNA. This is typically done to obstruct binding of other interactors, such as hnRNPs to alter splicing, or inhibit translation by blocking ribosomes (29–31). Higher affinity modifications, like cEts, can be employed to target tissues or sites on a gene that are normally less amenable to ASO activity (32,33). Thus, each ASO has a common set of class properties that can be partially predicted based on the modifications used as well as the unique set of properties determined by the sequence of the ASO.
Like siRNAs, ASOs do not cross an intact BBB. However, ASOs are soluble in artificial cerebrospinal fluid (CSF) and can be delivered directly into the CSF surrounding the brain and spinal cord. Once introduced into the CSF, modified ASOs have sufficient stability and uptake to distribute to the brain parenchyma and enter cells, both in preclinical models and in human patients (34–37). MOE-modified gapmers have long half-lives (35,37,38); therefore, it is likely that only intermittent and infrequent delivery of ASOs to the CSF will be required. Finally, ASOs behave in a dose-dependent manner, making it possible to moderate the degree of target modulation to a level that is safe yet still effective (35).
Gene Suppression for Dominantly Inherited Diseases
Huntington's Disease
HD is an autosomal dominantly inherited neurodegenerative disorder that results from an expanded CAG repeat in the HTT gene (mHTT) on chromosome 4 (39). The encoded protein, mutant huntingtin (mHTT), contains an expanded polyglutamine (PolyQ) tract at the N-terminus, which leads to aberrant folding and the inability to be processed appropriately by the cell. mHTT takes on a toxic gain of function leading to cellular dysfunction, inclusion body formation, gliosis and brain atrophy (40). In general, non-affected individuals have fewer than 36 glutamine repeats in their huntingtin gene (HTT), and 36–39 repeats results in a reduced-penetrance of the disease, with a later onset and slower progression of symptoms. Repeats of 40 or higher result in full expression of disease phenotypes. The number of CAG repeats is directly related to the age of onset of symptoms, and patients with exceptionally high repeat lengths have the juvenile form of HD (41). While numerous brain regions are affected, the striatum and overlaying cortex are the most-heavily affected regions. HD symptoms have a devastating impact on the patient's quality of life. Motor symptoms include involuntary hyperkinetic movements, an abnormal gait and difficulty with speech and swallowing (42,43). Personality changes are defined by wide mood swings, aggression, anxiety and clinical depression (44–46). Cognitive decline is characterized by both long- and short-term memory dysfunction and loss of executive function (46–49). Eventually, patients require help with all activities of daily living and end up wheelchair-bound and bedridden. HD is invariably fatal and patients typically succumb to the disease 10–15 years following diagnosis. Currently, all treatments for HD are palliative and thus, finding a therapeutic that ameliorates neuropathology and related behavioral dysfunction would be of tremendous benefit to patients. Because the genetic mutation that causes HD is known, gene suppression strategies have garnered a great deal of interest as a potential interventional therapeutic.
RNAi studies for HD in rodents and non-human primates
Over the past decade, numerous potential therapeutic approaches have emerged to reduce the expression of mHTT in the brain, including the use of siRNAs, shRNAs, artificial miRNAs, zinc finger transcriptional repressors and ASOs (Table 1). It has been more difficult than first anticipated to specifically target the mutant allele, while preserving expression of the unaffected allele. Several early proof-of-concept studies targeted only the human mHTT transgene in both transgenic and AAV-mediated rodent models of HD. These studies demonstrated that solely reducing expression of the mutant transgene, while leaving the endogenous rodent huntingtin (Hdh) alleles unaffected, was efficacious in reducing neuropathology and behavioral readouts germane to each disease model, including striatal inclusion formation, neuronal atrophy and motor dysfunction (7,50,52,53). These initial studies were incredibly important in highlighting the overall efficacy of an RNAi approach in preventing HD disease symptomology. In an attempt to specifically target the mHTT allele, other groups have identified particular single-nucleotide polymorphism (SNP) variants that reside only on the mutant allele using DNA collected from HD patient lymphocytes and fibroblasts (75,76). In 2009, Pfister et al. (77) identified a set of five allele-specific siRNAs corresponding to three SNP sites that could potentially be used to treat a large portion of HD patients in the US and Europe. More recently, Monteys et al. (59) demonstrated partial allele specificity in vivo using an artificial miRNA designed to target an SNP in a double transgenic engineered HD mouse model. Aside from targeting SNPs in the HTT gene itself, Becanovic et al. (78) have shown that targeting an SNP in the HTT promoter effectively reduces NF-kB binding and reduces huntingtin protein (HTT) expression in HD patient cells and in transgenic mouse striatum.
Table 1.
In vivo gene reduction strategies for HD and SCAs
| Disease | Target gene | Therapeutic construct | Injection site | Animal model | Synopsis of study results | Ref. |
|---|---|---|---|---|---|---|
| HD | Human mHTT | AAV2/1-shRNA | Striatum | N171–82Q Tg mouse | Reduction in human mHTT prevented inclusions, gait deficits and rotorod dysfunction | (50) |
| HD | Human mHTT | AAV2/5-shRNA | Striatum | R6/1 Tg mouse | Reduction in human mHTT reduced inclusion burden, normalized striatal transcripts and prevented clasping phenotype | (7) |
| HD | Human mHTT | siRNA | ICV | R6/2 Tg mouse | Reduction in human mHTT reduced inclusion burden, prolonged longevity, improved motor dysfunction and slowed weight loss | (51) |
| HD | Human mHTT | siRNA | Striatum | AAV-HTT-injected mouse | Reduction in human mHTT prolonged striatal neuron survival, reduced aggregates and prevented motor dysfunction | (52) |
| HD | Human mHTT | AAV1/2-shRNA | Striatum | AAV-HTT-injected rat | Reduction in human mHTT prevented neuronal loss and forelimb impairment | (53) |
| HD | Human mHTT/Mouse Hdh | AAV2/1-shRNA/AAV2/1-miRNA | Striatum | CAG140 KI mouse | Placing an siRNA construct in the construct of an artificial miRNA compared with an shRNA alleviated toxicity due to high expression of antisense RNAs | (8) |
| HD | Human mHTT/Mouse Hdh | AAV2/1-miRNA | Striatum | N171–82Q Tg mouse | Reducing expression of both human mHTT and endogenous mouse Hdh reduced inclusion formation, prevented rotorod deficits and extended lifespan | (54) |
| HD | Human mHTT/Rat Hdh | LV-shRNA | Striatum | LV-HTT-injected rat | Reducing expression of both human mHTT and endogenous rat Hdh lowered inclusion formation, prevented rotorod deficits and was well tolerated out to 9 months post-injection | (55) |
| HD | Human mHTT | AAV2/5-Cre | Hypothalamus | BACHD Tg mouse | Reducing human mHTT expression prevented weight gain and impaired glucose metabolism | (56) |
| HD | Human mHTT | AAV2/9-miRNA | Jugular vein | BACHD, N171–82Q Tg mouse | Vascular delivery of an miRNA reduced mHTT in several brain regions, reduced aggregates, prevented regional cortical and striatal atrophy and prevented weight loss | (57) |
| HD | Human mHTT/Mouse Hdh | AAV2/5-miRNA | Striatum | YAC128 Tg mouse | Reducing expression of both human mHTT and endogenous mouse Hdh after symptom development lowered inclusion burden, normalized striatal transcripts and prevented motor phenotypes | (58) |
| HD | Human mHTT | AAV2/1-miRNA | Striatum | HTT double Tg mice | Partial allele selectivity was achieved by targeting an SNP that resides on the mHTT transgene | (59) |
| HD | Human mHTT | AAV2/1-miRNA | Hypothalamus | N171–82Q Tg mouse | Reducing human mHTT expression prevented inclusion formation, neuropepetide dysregulation, glucose dysregulation and weight loss | a |
| HD | Rhesus HTT | AAV2/1-miRNA | Putamen | NHP | Reduction in rhesus HTT did not induce neuronal loss, gliosis, an immune response, motor dysfunction or weight loss | (60) |
| HD | Rhesus HTT | AAV2/2-miRNA | Putamen | NHP | Reduction in rhesus HTT did not induce neuronal loss, gliosis, an immune response, motor dysfunction or weight loss | (61) |
| HD | Sheep HTT | AAV2/9-miRNA | Striatum | Sheep | Reduction in sheep HTT did not induce neuronal loss, gliosis, an immune response, motor dysfunction or weight loss | b |
| HD | Rhesus HTT | siRNA | Putamen | NHP | siRNA treatment for 28 days reduces rhesus HTT expression throughout the putamen and was well tolerated | (62) |
| HD | Rhesus HTT | siRNA | Putamen | NHP | siRNA treatment for 4 days resulted in sustained HTT suppression out to an estimated 27–39 days post-infusion | (63) |
| HD | Human mHTT | ASO | ICV | BACHD, YAC128 R6/2 Tg mouse | Reduction in human mHTT in young and aged mice reversed rotarod deficits in YAC128 and BACHD mice, ameliorated hypoactivity and anxiety in aged BACHD mice and prevented loss of brain mass in R6/2 | (35) |
| HD | Human mHTT/Mouse Hdh | ASO | ICV | BACHD Tg mouse, non-Tg mouse | Simultaneous suppression of human mHTT and normal mouse Hdh reversed rotarod and open-field deficits for 8 months post-treatment. No difference from mutant selective suppression. No behavioral changes in ASO treated non-transgenic littermates | (35) |
| HD | Monkey HTT | ASO | IT | NHP | Monkey HTT was reduced in the cord and cortex for >8 weeks post-treatment termination | (35) |
| HD | Human mHTT | ASO | ICV | YAC128 Tg mouse | Reduced mHTT improved motor deficits and this correlated with improvements in several striatal dysregulated transcripts | (64) |
| HD | Human mMTT/Mouse Hdh | ASO | ICV | BACHD, YAC128 Tg mouse | Single bolus injection of non-allele-selective huntingtin ASO decreased mHTT and mouse huntingtin protein levels >80% and did not induce gliosis or astrocytosis | (65) |
| HD | Human HTT SNP targeting | ASO | ICV | BACHD Tg mouse | Single bolus injection of SNP targeting ASO decreased mHTT protein levels 50% and did not induce gliosis or astrocytosis | (65) |
| HD | Human HTT SNP targeting | ASO | ICV | Hu97/18 double Tg mouse | Single bolus achieved 75% reduction in human mHTT protein 4 weeks post-dose, with no change in normal human HTT protein | (66) |
| HD | Human HTT SNP targeting | ASO | IT | Rat | No gliosis, astrocytosis, change in body weight, grip strength or open field | (58) |
| HD | Human HTT SNP targeting | ASO | ICV | Hu97/18 double Tg | Selective human mHTT suppression >90%. mHTT protein suppression maintained at >50% for 36 weeks after single ICV bolus injection | (38) |
| HD | Human HTT SNP targeting | ASO | ICV/IT | Mouse, rat | Lead SNP ASOs did not induce gliosis, astrocytosis, weight loss or alteration in neurological exams in mice or rats | (38) |
| HD | CAG | ss-siRNA | ICV | Q150 KI mouse | Selectively decreased CAG expanded huntingtin protein 50–75% in various regions throughout the mouse brain | (2) |
| SCA1 | Human mATXN1 | AAV2/1-shRNA | CBL CTX | SCA1 Tg mouse (B05) | Suppression of cerebellar mATXN1 prevented cerebellar degeneration and motor phenotypes | (6) |
| SCA1 | Human mATXN1/ mouse Atxn1 | AAV2/1-miRNA | DCN | SCA1 Tg mouse (B05) | Suppression of cerebellar human mATXN1 and mouse Atxn1 levels improved behavioral and molecular phenotypes | (67) |
| SCA1 | Mouse Atxn1 | AAV2/5-miRNA | DCN | SCA1 KI mouse | Suppression of cerebellar Atxn1 levels improved behavioral and neuropathological phenotypes for out to 1 year post-injection | (68) |
| SCA3 | Human mATXN3 SNP targeting | LV-shRNA | Striatum | SCA3 rat | Allele-specific targeting of a SNP on human mATXN3 prevented inclusion formation and neuron loss | (69) |
| SCA3 | Human mATXN3/Rat Atxn3 | LV-shRNA | Striatum | SCA3 rat | Non-allele-specific targeting of human mATXN3 and rat Atxn3 reduced neuropathology | (70) |
| SCA3 | Human mATXN3 | AAV2/1-miRNA | DCN | SCA3/MJD84.2 mouse | Allele-specific suppression of ATXN3 resolved molecular phenotypes | (71) |
| SCA3 | Human mATXN3 | AAV2/1-miRNA | DCN | SCA3 rat | Lifelong suppression of mATXN3 was well tolerated but did not prevent motor impairment or prolong lifespan | (72) |
| SCA7 | Human mATXN7/Mouse Atxn7 | AAV2/1-miRNA | Retina | SCA7 Tg | Suppression of mATXN7 and mouse Atxn7 was well tolerated, did not induce neuropathology and normal retinal function was preserved | (73) |
| SCA7 | Human mATXN7/Mouse Atxn7 | AAV2/1-miRNA | DCN | SCA7 Tg | Suppression of human mATXN7 and mouse Atxn7 improved behavioral and molecular phenotypes | (74) |
Tg, transgenic; inj., injected; LV, lentivirus.
aMcBride, oral presentation, Gordon Research Conference on CAG Repeat Disorders, 2015.
bAronin, poster presentation, CHDI conference, 2015.
An alternative gene suppression strategy for HD that has made substantial progress over the last decade is the partial reduction in both mutant and native HTT alleles (known as non-allele-specific RNAi). While it is clear that normal HTT protein plays an important functional role in the neuron and is necessary during development, the goal of non-allele specific RNAi is to establish the lowest level of knockdown that leads to amelioration of neuropathology and behavioral deficits while maintaining a positive safety and tolerability profile. Numerous reports have shown that a partial reduction in endogenous huntingtin in the adult brain of multiple species is well tolerated and this approach is now being pursued by many teams as a viable clinical approach to treat HD. Reducing both mutant and endogenous huntingtin in both transgenic HD mouse and rat striatum by 40–60% prevents motor deficits, extends lifespan and does not lead to toxicity in both fragment and full-length mouse HD models in both short- and long-term studies (8,54,55). Encouragingly, reducing both HTT alleles in post-symptomatic transgenic HD mice was well tolerated and resulted in reduced inclusion formation and rotorod dysfunction, suggesting that RNAi may be beneficial in HD patients that already present with disease phenotypes (55,58).
Following success in rodent models of HD, the safety of partially reducing endogenous HTT has been evaluated in non-human primate studies following the delivery of AAV vectors expressing artificial miRNAs and shRNAs. McBride et al. (60) demonstrated that a 45% reduction in rhesus HTT did not induce neuronal loss, gliosis, behavioral dysfunction or weight loss and these results were further extended when Grondin et al. (61) showed that sustained HTT suppression out to 6 months post-infusion was well tolerated. Two follow-up studies found that siRNA infusion into the rhesus putamen reduced HTT to similar degrees and that partial HTT suppression lasted for up to 39 days after the infusion of siRNA was terminated (62,63). Similar tolerability has recently been shown following a 50% reduction in HTT in the sheep putamen by Neil Aronin's laboratory (poster presentation, CHDI conference, 2015). Together, these results show that a partial reduction in both mutant and wild-type HTT alleles may be a viable therapeutic strategy to treat HD.
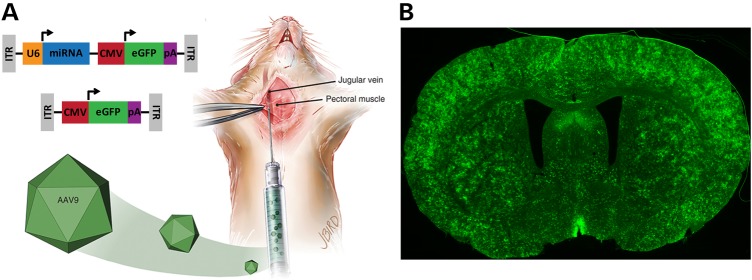
Historically, RNAi constructs evaluated in animal models of HD have been administered via a direct injection into the striatum, the primary site of degeneration in HD, or into the adjacent lateral ventricle (Fig. 1). However, many extra-striatal brain regions are also affected in HD and have become recent targets for gene suppression therapy. Hult et al. (56) lowered mHTT expression in the BACHD transgenic HD mouse hypothalamus using an AAV-Cre-based approach and effectively prevented the characteristic metabolic phenotype. More recently, McBride et al. found that a partial reduction in both human mHTT and mouse Hdh using an AAV-miRNA approach in the N171–82Q transgenic mouse hypothalamus partially prevented neuropeptide dysregulation, glucose homeostasis and altered energy metabolism (McBride, oral presentation, Gordon Research Conference on CAG Repeat Disorders, 2015). Because neurodegeneration is seen throughout many regions of the brain, a global reduction in mHTT expression throughout the brain would be expected to provide a greater benefit compared with targeting individual affected regions. In 2014, Dufour et al. (57) showed that AAV serotype 9 expressing an artificial miRNA targeting mHTT crossed the BBB following injection into the jugular vein of transgenic HD mice and significantly reduced mHTT expression in the cortex, striatum, hypothalamus, hippocampus and thalamus (Fig. 2). This widespread mHTT knockdown was associated with prevention of cortical and striatal atrophy as well as reduced inclusion body formation. This study was the first to show a reduction in mHTT expression in brain following vascular administration of a gene suppression construct and highlights the possibility of sustained mHTT knockdown from a single, systemic injection.
Figure 1.
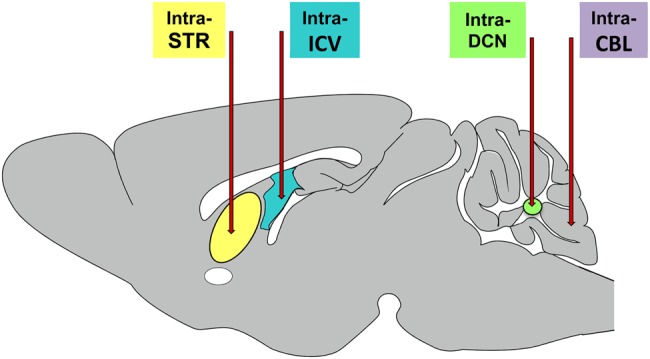
Cartoon of common delivery sites for gene suppression constructs evaluated in mouse models of HD and the SCAs. Primary delivery sites include the striatum, lateral ventricle, DCN and the cerebellar cortex. Delivery into the striatum (STR) (7,8,50,53,54,58,59,79), DCN (67,68,71,74,80) and cerebellar cortex (CBL) (6) result largely in focal expression of the injected construct, with evidence if anterograde and retrograde transport when AAVs are used as the delivery vehicle. In contrast, injection into the CSF via injections into the lateral ventricle (ICV) (2,35,38,51,64,66) or the cisterna magna (CM) result in more widespread delivery throughout the brain.
Figure 2.
Global suppression of HTT in the transgenic HD mouse brain. (A) Cartoon depicting the delivery of an AAV9 vector expressing an mHTT-specific miRNA and a GFP control gene (top construct, AAV9-mi2.1-GFP) or AAV9 vector expressing GFP alone (bottom construct, AAV9-GFP) into the mouse jugular vein. (B) Vascular delivery of AAV9-mi2.1-GFP resulted in widespread expression of both the therapeutic miRNA and GFP throughout the N171–82Q transgenic mouse brain and a concomitant reduction in mHTT in several brain regions including the cortex, striatum, hypothalamus, hippocampus and thalamus (57).
ASO strategies for HD
Since the identification of the single causative mutation in HD, multiple ASO approaches using different antisense mechanisms to modulate mHTT have been tested. The most advanced of these approaches, total HTT suppression using traditional MOE gapmer ASO designs, is currently being tested in the clinic. Here, an MOE gapmer ASO complementary to the human huntingtin RNA is introduced directly into the CSF. This is predicted to result in partial suppression of both the mutant and normal HTT alleles. As ASOs behave in a dose-dependent manner, HTT suppression can be targeted from 0 to 75% (35). In preclinical models, huntingtin targeting MOE gapmer ASOs have both prevented and reversed disease (35,64). Suppression of human mHTT in the YAC128 and BACHD models of HD reversed existing behavioral phenotypes and prevented progressive loss of brain mass in R6/2 mice (35). Similarly, ASO-mediated human mHTT suppression in YAC128 mice ameliorated key striatal gene expression changes (64). Interestingly, the earlier in disease the treatment was initiated, the more robust the disease reversal. Perhaps the finding of greatest significance from these rodent studies was that transient ASO suppression of mHTT (∼4 months) was sufficient to obtain a long-term benefit (>9 months). One can imagine a treatment where intermittent suppression of HTT (‘a huntingtin holiday’) is all that is required to provide continuous benefit (81).
ASOs can be introduced into the CNS by direct injection in the CSF either by intrathecal (IT) injection in non-human primates and human patients or (due to surgical limitations) intracerebral ventricular (ICV) injection in rodents (Fig. 1). Human huntingtin ASOs delivered ICV in rodents suppress human HTT mRNA >75% throughout the brain and spinal cord (35). In larger non-human primate brains, huntingtin ASOs complementary to monkey HTT RNA delivered IT are present in the spinal cord, cortex and to a lesser degree deeper brain structures (35). Tissue adjacent to the CSF has the highest total tissue levels of ASO, suggesting most ASO reaches the tissue by passive diffusion. ASOs are also found in and diffusing radially from the Virchow–Robins space, a fluid-filled area that surrounds veins and arteries in the brain parenchyma and is likely another route of ASO distribution. In areas of low bulk ASO levels, populations of neuronal cell bodies that contain high levels of ASO are often found, making it unlikely that ASOs arrived to these neurons by passive diffusion. One possibility is that ASOs are actively transported to these cell bodies by anterograde or retrograde transport. However, the exact mechanisms of ASO uptake and transport in the CNS have not been fully elucidated and warrant further study. Regardless of mechanism, what is clear is that delivery of huntingtin ASOs to the CSF results in distribution of active ASO to much of the CNS.
Modified ASOs have long half-lives in the CNS. In BACHD mice expressing the human mHTT gene, human mHTT mRNA is suppressed up to 12 weeks following termination of treatment (35). Similar longevity of suppression is achieved in non-human primates treated with a huntingtin ASO. This is further supported by clinical data, as ASO was detectable in the brain and spinal cord of an amyotrophic lateral sclerosis (ALS) patient 3 months after an IT delivery of ASO (34). The long half-lives of ASOs in the CNS are particularly beneficial for slowly progressing diseases, where patients will likely require treatment for years, if not decades. In cases where targeting inherited disease causing mutations is possible, as in HD, infrequent dosing becomes even more valuable, as ideally patients could be identified and treatment initiated before disease manifests, but would likely require nearly a lifetime of treatment.
As with siRNAs, one primary question for any gene suppression approach is the necessity for allele selectivity. Also, like siRNA and miRNA approaches, in preclinical models, ASO-mediated huntingtin suppression in normal mice and non-human primates does not induce a phenotype (35). In disease models, simultaneous reduction in endogenous mouse Hdh along with human mHTT yielded the same phenotypic reversal as reduction in mHTT alone (35). However, despite lack of evidence that partial suppression of normal huntingtin will be detrimental, an allele-selective approach is appealing and remains tractable.
An alternative approach is to use RNase H ASOs to target SNPs linked to the CAG expanded HTT allele and not present in the normal HTT allele. More than 90% of mHTT linked SNPs are present in intronic regions, making targeting of these sites with siRNAs unfeasible. ASOs, however, are active against pre-mRNAs as well as mature mRNAs and can target intronic sequences. Using population genetics, researchers have identified heterozygous SNPs that are linked to the CAG allele in nearly half of the HD population (65). Strategic incorporation of high affinity modifications (cEts) into MOE gapmers and targeting SNP sites, yielded >50-fold selectivity of the mHTT allele over the normal allele containing a one base mismatch at the SNP site (38,65,66). Although promising, one limitation is that 3–5 ASOs at unique sites would be required to target 80% of the HD population allele selectively (82).
To allele selectively target all HD patients, fully 2′ sugar modified ASOs complementary to the CAG repeat can preferentially obstruct the translation of the expanded CAG containing transcript, resulting in 6-fold selectivity over unexpanded HTT (83–86). Selectivity is likely achieved through steric hindrance, where longer transcripts allow for binding of multiple CAG targeting ASOs. A similar approach targeting the CAG repeat, but employing the miRNA pathway utilizes a single-stranded siRNA (ss-siRNA) that contains a mismatch to the CAG results in the recruitment of Ago2 to the expanded transcript, and suppression of translation without altering mRNA levels (2). The CAG targeting ss-siRNAs were validated both in vitro in patient fibroblasts and in vivo in a knock-in (KI) HD mouse model (2). One consideration when developing these approaches is to determine the potential and safety of unwanted suppression of other CAG-containing transcripts. Another consideration is that the current CAG targeting ss-siRNAs are not as potent as the other siRNA and ASO approaches. Despite the challenges, CAG targeting is particularly attractive as a single drug has the potential to treat all polyglutamine diseases.
RNAi studies for the SCAs in rodents and non-human primates
Like HD, many of the SCAs are part of a subset of disorders known collectively as polyglutamine-repeat diseases. In particular, SCA 1, 2, 3, 6, 7 and 17 are categorized as autosomal-dominant polyglutamine diseases.
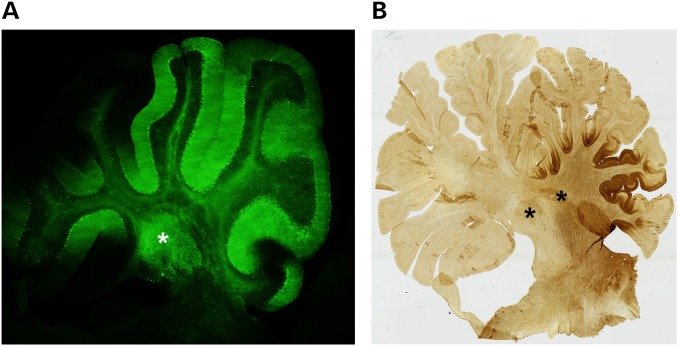
Spinocerebellar ataxia type 1 (SCA1) is a late onset neurodegenerative disease caused by a CAG expansion in the coding region of ataxin-1 (ATXN1) which affects 1–2 in 100 000 people worldwide (87,88). Symptoms include loss of coordination, slurred speech, difficulty swallowing and mild cognitive impairment. Neurodegeneration occurs predominantly in cerebellar Purkinje cells, deep cerebellar nuclei (DCN), brainstem nuclei and spinocerebellar tracts (Fig. 1) (89–92). Reliable mouse models have been integral in understanding disease pathology (93–95). RNAi as a potential therapeutic was first demonstrated using AAV expressing shRNAs targeting ATXN1 in the transgenic mouse model of SCA1. Directed injections to the medial cerebellar lobules led to significant knockdown of ataxin-1 and an improvement of behavioral and neuropathological phenotypes (6). It has since been shown that delivery to the DCN, versus the cerebellar cortex itself, leads to a wider biodistribution of therapeutic load by trafficking retrograde up the axons to the Purkinje cell soma (67,68,72). More recently, artificial miRNAs targeting human ATXN1 (in a transgenic model) or mouse Atxn1 (in a knock-in model) were delivered using AAV serotypes 2/1 and 2/5, respectively (Fig. 3A) (67,68). Directed stereotaxic injections of RNAi constructs to the DCN of pre-symptomatic SCA1 mice prevented motor deficits and improved cellular and molecular phenotypes. Significant knockdown of ataxin-1 was achieved 35 weeks (67) and 60 weeks (68) post-delivery with no overt glial response in either case. Following success in mouse models, the feasibility and safety was evaluated in non-human primates. AAV2/1 expressing the same artificial miRNA used in transgenic SCA1 mice was stereotaxically injected to the DCN of adult, naive rhesus macaques using magnetic resonance imaging-guided techniques (Fig. 3B). The DCN, cerebellar cortex, brainstem nuclei and projections to the ventral lateral thalamus were safely transduced with significant reduction in endogenous ATXN1 mRNA levels ≥30% in transduced areas (96).
Figure 3.
Transgene expression in the mouse and NHP cerebellum following AAV-mediated expression in the DCN. (A) Sagittal section of mouse cerebellum following direct delivery of AAV2/5 expressing an miRNA targeting Atxn1 and eGFP to the DCN. Green staining demonstrates biodistribution to multiple lobules from single injection site. (B) Sagittal section of NHP cerebellum following direct delivery of AAV2/1 expressing and an miRNA targeting ATXN1 and eGFP to the DCN. Brown staining shows biodistribution throughout multiple lobules. *DCN.
Spinocerebellar ataxia type 3 (SCA3), also known as Machado–Joseph disease, is the most common of the SCAs. SCA3 is a late onset, progressive neurodegenerative disease caused by a CAG expansion in the ataxin-3 (ATXN3) gene. Symptoms are characterized by ataxia as well as dystonia and peripheral neuropathy (97). Previously, LV expressing both allele-specific and non-allele-specific shRNAs targeting ATXN3 in rats and mice were both beneficial well tolerated (69,70,98). More recently, two complementary papers studied the effects of artificial siRNAs targeting ATXN3 in transgenic mice. Both studies used stereotaxic injections of AAV2/1 to target the DCN of SCA3 mice. Combined, the studies demonstrated that directed delivery of siRNAs achieved prolonged knockdown of ATXN3 and cleared nuclear aggregates characteristic in the mouse model (71). However, despite significant knockdown of cerebellar ATXN3 and apparent tolerability of the constructs, the mice still developed motor phenotypes (72).
Spinocerebellar ataxia type 7 (SCA7) is caused by a CAG expansion in ataxin-7. In addition to the characteristic cerebellar neurodegeneration and ataxic symptoms, patients also lose their vision (99). Recently, non-allele-specific artificial miRNAs targeting ataxin-7 (ATXN7) were injected into either the eye or the DCN of SCA7 Tg mice. AAV2/1 delivery to the DCN resulted in improvement of motor phenotypes as well as improvement of cerebellar Purkinje cell dendrites, known to be affected in the model. In addition, long-term knockdown of ATXN7 was achieved in the cerebellum and brainstem without toxicity (74). Artificial miRNA expressed from AAV2/1 injected to the retina of SCA7 mice achieved ≥50% suppression of ATXN7 in the retina with no adverse effects in retinal functioning (73).
Ongoing and future clinical trials
The success of the last decade of work has led to the first in man gene suppression therapy in HD patients. ASOs have previously been delivered IT to both ALS (34) and spinal muscular atrophy patients (100). Recently, a phase I clinical trial delivering a huntingtin ASO has been initiated in early-stage HD patients. This is the first potentially disease-modifying therapy to enter trials for HD, and certainly the first to target HTT, the underlying cause of the disease. Additional Phase I clinical studies, utilizing AAV-delivered miRNAs that have made substantial progress in rodent and NHP studies, are likely to follow in the next few years. The first HD gene suppression trial is focused on safety and tolerability of an IT-delivered RNase H MOE-modified ASO targeting human HTT. In addition to safety endpoints, neuroimaging assessments, as well as exploratory endpoints quantifying cognitive, motor, and neuropsychiatric aspects of disease will be performed.
One key challenge in the clinic for centrally delivered gene suppression approaches is identification of a target engagement biomarker. Blood, urine and other easily accessible matrices are not likely to reflect changes limited to the CNS, as is the case for most CNS gene suppression approaches. Numerous groups have been working to determine if HTT is detectible in the CSF. This year, two groups have successfully detected mHTT in the CSF (101,102); mutant HTT increases with disease burden and tracks with tau and neurofilament light chain load in the CSF (102). Southwell et al. (101) has demonstrated, in a humanized mouse model, that mHTT levels in the CSF decrease after ASO-mediated suppression of HTT in the CNS, suggesting that CSF HTT can be used to track changes in HTT levels in tissues. Analysis of HTT levels in CSF will allow for confirmation of target engagement in the CNS, and aid in dose selection. Similar attempts to detect and quantify other polyQ disease proteins in the CSF are also underway.
As discussed, there are multiple methods for delivery of RNA therapeutics to the CNS. In diseases where pathology is restricted to a distinct brain region or tissue, or if global suppression is unfavorable for a given target, then directed intracranial injections can be employed (7,8,50,53,54,58,59,67,68,71,74,79,80). If broader suppression is needed, injections directly into the CSF (2,35,38,51,64,66) or the vasculature (57,103) can provide a more global distribution to the brain and spinal cord. These advances are encouraging and future studies are warranted to identify alternative methods to deliver RNA therapeutics across the BBB.
This is an exciting time for gene suppression therapies in neurodegenerative diseases. It is clear from the preclinical data that suppression of the underlying cause of disease, particularly in dominantly inherited diseases like polyQ diseases, can have a dramatic effect on disease course in animal models. We are entering a time where technologies have advanced and delivery techniques have been refined to a point where treatment of human patients is tractable. With human clinical trials ongoing, and more imminent, it is becoming increasingly likely that gene suppression approaches will have a real chance at altering the course of these devastating diseases.
Conflict of Interest statement. H.B.K. is an employee of Isis Pharmaceuticals.
Funding
Support for M.S.K. comes from NIH NS045667, the University of Iowa Roy J. Carver Trust and the National Ataxia Foundation; support for J.L.M comes from NIH NS069798, The Hereditary Disease Foundation, The Medical Research Foundation and ONPRC Core Grant P51OD011092.
References
- 1.Ha M., Kim V.N. (2014) Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol., 15, 509–524. [DOI] [PubMed] [Google Scholar]
- 2.Yu D., Pendergraff H., Liu J., Kordasiewicz H.B., Cleveland D.W., Swayze E.E., Lima W.F., Crooke S.T., Prakash T.P., Corey D.R. (2012) Single-stranded RNAs use RNAi to potently and allele-selectively inhibit mutant huntingtin expression. Cell, 150, 895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lima W.F., Prakash T.P., Murray H.M., Kinberger G.A., Li W., Chappell A.E., Li C.S., Murray S.F., Gaus H., Seth P.P. et al. (2012) Single-stranded siRNAs activate RNAi in animals. Cell, 150, 883–894. [DOI] [PubMed] [Google Scholar]
- 4.de Fougerolles A.R. (2008) Delivery vehicles for small interfering RNA in vivo. Hum. Gene Ther., 19, 125–132. [DOI] [PubMed] [Google Scholar]
- 5.Lecaros R.L., Huang L., Lee T.C., Hsu Y.C. (2015) Nanoparticle delivered VEGF-A siRNA enhances photodynamic therapy for head and neck cancer treatment. Mol. Ther., doi:10.1038/mt.2015.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xia H., Mao Q., Eliason S.L., Harper S.Q., Martins I.H., Orr H.T., Paulson H.L., Yang L., Kotin R.M., Davidson B.L. (2004) RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat. Med., 10, 816–820. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez-Lebron E., Denovan-Wright E.M., Nash K., Lewin A.S., Mandel R.J. (2005) Intrastriatal rAAV-mediated delivery of anti-huntingtin shRNAs induces partial reversal of disease progression in R6/1 Huntington's disease transgenic mice. Mol. Ther., 12, 618–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McBride J.L., Boudreau R.L., Harper S.Q., Staber P.D., Monteys A.M., Martins I., Gilmore B.L., Burstein H., Peluso R.W., Polisky B. et al. (2008) Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc. Natl Acad. Sci. USA, 105, 5868–5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boudreau R.L., Spengler R.M., Davidson B.L. (2011) Rational design of therapeutic siRNAs: minimizing off-targeting potential to improve the safety of RNAi therapy for Huntington's disease. Mol. Ther., 19, 2169–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boudreau R.L., Martins I., Davidson B.L. (2009) Artificial microRNAs as siRNA shuttles: improved safety as compared to shRNAs in vitro and in vivo. Mol. Ther., 17, 169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dissen G.A., McBride J, Lomniczi A., Matagne V., Dorfman M., Neff T.L., Galimi F., Ojeda S.R. (2012) Using lentiviral vectors as delivery vehicles for gene therapy. Controlled Genetic Manipulations, 65, 69–96. [Google Scholar]
- 12.Cronin J., Zhang X.Y., Reiser J. (2005) Altering the tropism of lentiviral vectors through pseudotyping. Curr. Gene Ther., 5, 387–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parr-Brownlie L.C., Bosch-Bouju C., Schoderboeck L., Sizemore R.J., Abraham W.C., Hughes S.M. (2015) Lentiviral vectors as tools to understand central nervous system biology in mammalian model organisms. Front. Mol. Neurosci., 8, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murlidharan G., Samulski R.J., Asokan A. (2014) Biology of adeno-associated viral vectors in the central nervous system. Front. Mol. Neurosci., 7, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kantor B., McCown T., Leone P., Gray S.J. (2014) Clinical applications involving CNS gene transfer. Adv. Genet., 87, 71–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Z., Asokan A., Samulski R.J. (2006) Adeno-associated virus serotypes: vector toolkit for human gene therapy. Mol. Ther., 14, 316–327. [DOI] [PubMed] [Google Scholar]
- 17.Louis Jeune V., Joergensen J.A., Hajjar R.J., Weber T. (2013) Pre-existing anti-adeno-associated virus antibodies as a challenge in AAV gene therapy. Hum. Gene Ther. Methods, 24, 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boutin S., Monteilhet V., Veron P., Leborgne C., Benveniste O., Montus M.F., Masurier C. (2010) Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum. Gene Ther., 21, 704–712. [DOI] [PubMed] [Google Scholar]
- 19.Rapti K., Louis-Jeune V., Kohlbrenner E., Ishikawa K., Ladage D., Zolotukhin S., Hajjar R.J., Weber T. (2012) Neutralizing antibodies against AAV serotypes 1, 2, 6, and 9 in sera of commonly used animal models. Mol. Ther., 20, 73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stieger K., Schroeder J., Provost N., Mendes-Madeira A., Belbellaa B., Le Meur G., Weber M., Deschamps J.Y., Lorenz B., Moullier P. et al. (2009) Detection of intact rAAV particles up to 6 years after successful gene transfer in the retina of dogs and primates. Mol. Ther., 17, 516–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bennett J., Ashtari M., Wellman J., Marshall K.A., Cyckowski L.L., Chung D.C., McCague S., Pierce E.A., Chen Y., Bennicelli J.L. et al. (2012) AAV2 gene therapy readministration in three adults with congenital blindness. Sci. Transl. Med., 4, 120ra115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerritelli S.M., Crouch R.J. (2009) Ribonuclease H: the enzymes in eukaryotes. FEBS J., 276, 1494–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bennett C.F., Swayze E.E. (2010) RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol., 50, 259–293. [DOI] [PubMed] [Google Scholar]
- 24.Eckstein F. (2000) Phosphorothioate oligodeoxynucleotides: what is their origin and what is unique about them? Antisense Nucleic Acid Drug Dev., 10, 117–121. [DOI] [PubMed] [Google Scholar]
- 25.Stein C.A., Subasinghe C., Shinozuka K., Cohen J.S. (1988) Physicochemical properties of phosphorothioate oligodeoxynucleotides. Nucleic Acids Res., 16, 3209–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henry S.P., Geary R.S., Yu R., Levin A.A. (2001) Drug properties of second-generation antisense oligonucleotides: how do they measure up to their predecessors? Curr. Opin. Investig. Drugs, 2, 1444–1449. [PubMed] [Google Scholar]
- 27.Monia B.P., Lesnik E.A., Gonzalez C., Lima W.F., McGee D., Guinosso C.J., Kawasaki A.M., Cook P.D., Freier S.M. (1993) Evaluation of 2′-modified oligonucleotides containing 2′-deoxy gaps as antisense inhibitors of gene expression. J. Biol. Chem., 268, 14514–14522. [PubMed] [Google Scholar]
- 28.Swayze E.E., Siwkowski A.M., Wancewicz E.V., Migawa M.T., Wyrzykiewicz T.K., Hung G., Monia B.P., Bennett C.F. (2007) Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res., 35, 687–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hua Y., Vickers T.A., Okunola H.L., Bennett C.F., Krainer A.R. (2008) Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet., 82, 834–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh N.N., Shishimorova M., Cao L.C., Gangwani L., Singh R.N. (2009) A short antisense oligonucleotide masking a unique intronic motif prevents skipping of a critical exon in spinal muscular atrophy. RNA Biol., 6, 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baker B.F., Lot S.S., Condon T.P., Cheng-Flournoy S., Lesnik E.A., Sasmor H.M., Bennett C.F. (1997) 2′-O-(2-Methoxy)ethyl-modified anti-intercellular adhesion molecule 1 (ICAM-1) oligonucleotides selectively increase the ICAM-1 mRNA level and inhibit formation of the ICAM-1 translation initiation complex in human umbilical vein endothelial cells. J. Biol. Chem., 272, 11994–12000. [DOI] [PubMed] [Google Scholar]
- 32.Seth P.P., Siwkowski A., Allerson C.R., Vasquez G., Lee S., Prakash T.P., Kinberger G., Migawa M.T., Gaus H., Bhat B. et al. (2008) Design, synthesis and evaluation of constrained methoxyethyl (cMOE) and constrained ethyl (cEt) nucleoside analogs. Nucleic Acids Symp. Ser., 553–554. [DOI] [PubMed] [Google Scholar]
- 33.Pandey S.K., Wheeler T.M., Justice S.L., Kim A., Younis H., Gattis D., Jauvin D., Puymirat J., Swayze E.E., Freier S.M. et al. (2015) Identification and characterization of modified antisense oligonucleotides targeting DMPK in mice and nonhuman primates for the treatment of myotonic dystrophy type 1. J. Pharmacol. Exp. Ther., 355, 310–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller T.M., Pestronk A., David W., Rothstein J., Simpson E., Appel S.H., Andres P.L., Mahoney K., Allred P., Alexander K. et al. (2013) An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: a phase 1, randomised, first-in-man study. Lancet Neurol., 12, 435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kordasiewicz H.B., Stanek L.M., Wancewicz E.V., Mazur C., McAlonis M.M., Pytel K.A., Artates J.W., Weiss A., Cheng S.H., Shihabuddin L.S. et al. (2012) Sustained therapeutic reversal of Huntington's disease by transient repression of huntingtin synthesis. Neuron, 74, 1031–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith R.A., Miller T.M., Yamanaka K., Monia B.P., Condon T.P., Hung G., Lobsiger C.S., Ward C.M., McAlonis-Downes M., Wei H. et al. (2006) Antisense oligonucleotide therapy for neurodegenerative disease. J. Clin. Invest., 116, 2290–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rigo F., Chun S.J., Norris D.A., Hung G., Lee S., Matson J., Fey R.A., Gaus H., Hua Y., Grundy J.S. et al. (2014) Pharmacology of a central nervous system delivered 2′-O-methoxyethyl-modified survival of motor neuron splicing oligonucleotide in mice and nonhuman primates. J. Pharmacol. Exp. Ther., 350, 46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Southwell A.L., Skotte N.H., Kordasiewicz H.B., Ostergaard M.E., Watt A.T., Carroll J.B., Doty C.N., Villanueva E.B., Petoukhov E., Vaid K. et al. (2014) In vivo evaluation of candidate allele-specific mutant huntingtin gene silencing antisense oligonucleotides. Mol. Ther., 22, 2093–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.(1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell, 72, 971–983. [DOI] [PubMed] [Google Scholar]
- 40.Vonsattel J.P., Myers R.H., Stevens T.J., Ferrante R.J., Bird E.D., Richardson E.P. Jr (1985) Neuropathological classification of Huntington's disease. J. Neuropathol. Exp. Neurol., 44, 559–577. [DOI] [PubMed] [Google Scholar]
- 41.Andrew S.E., Goldberg Y.P., Kremer B., Telenius H., Theilmann J., Adam S., Starr E., Squitieri F., Lin B., Kalchman M.A. et al. (1993) The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington's disease. Nat. Genet., 4, 398–403. [DOI] [PubMed] [Google Scholar]
- 42.Heinsen H., Strik M., Bauer M., Luther K., Ulmar G., Gangnus D., Jungkunz G., Eisenmenger W., Gotz M. (1994) Cortical and striatal neurone number in Huntington's disease. Acta Neuropathol., 88, 320–333. [DOI] [PubMed] [Google Scholar]
- 43.Yanagisawa N. (1992) The spectrum of motor disorders in Huntington's disease. Clin. Neurol. Neurosurg., 94 Suppl, S182–S184. [DOI] [PubMed] [Google Scholar]
- 44.Epping E.A., Mills J.A., Beglinger L.J., Fiedorowicz J.G., Craufurd D., Smith M.M., Groves M., Bijanki K.R., Downing N., Williams J.K. et al. (2013) Characterization of depression in prodromal Huntington disease in the neurobiological predictors of HD (PREDICT-HD) study. J. Psychiatr. Res., 47, 1423–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jason G.W., Pajurkova E.M., Suchowersky O., Hewitt J., Hilbert C., Reed J., Hayden M.R. (1988) Presymptomatic neuropsychological impairment in Huntington's disease. Arch. Neurol., 45, 769–773. [DOI] [PubMed] [Google Scholar]
- 46.Tabrizi S.J., Langbehn D.R., Leavitt B.R., Roos R.A., Durr A., Craufurd D., Kennard C., Hicks S.L., Fox N.C., Scahill R.I. et al. (2009) Biological and clinical manifestations of Huntington's disease in the longitudinal TRACK-HD study: cross-sectional analysis of baseline data. Lancet Neurol., 8, 791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bonner-Jackson A., Long J.D., Westervelt H., Tremont G., Aylward E., Paulsen J.S., Investigators P.-H. and Coordinators of the Huntington Study, G. (2013) Cognitive reserve and brain reserve in prodromal Huntington's disease. J. Int. Neuropsychol. Soc., 19, 739–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harrington D.L., Liu D., Smith M.M., Mills J.A., Long J.D., Aylward E.H., Paulsen J.S. (2014) Neuroanatomical correlates of cognitive functioning in prodromal Huntington disease. Brain Behav., 4, 29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harrington D.L., Smith M.M., Zhang Y., Carlozzi N.E., Paulsen J.S. and PREDICT-HD Investigators of the Huntington Study Group. (2012) Cognitive domains that predict time to diagnosis in prodromal Huntington disease. J. Neurol. Neurosurg. Psychiatry, 83, 612–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harper S.Q., Staber P.D., He X., Eliason S.L., Martins I.H., Mao Q., Yang L., Kotin R.M., Paulson H.L., Davidson B.L. (2005) RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc. Natl Acad. Sci. USA, 102, 5820–5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y.L., Liu W., Wada E., Murata M., Wada K., Kanazawa I. (2005) Clinico-pathological rescue of a model mouse of Huntington's disease by siRNA. Neurosci. Res., 53, 241–249. [DOI] [PubMed] [Google Scholar]
- 52.DiFiglia M., Sena-Esteves M., Chase K., Sapp E., Pfister E., Sass M., Yoder J., Reeves P., Pandey R.K., Rajeev K.G. et al. (2007) Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc. Natl Acad. Sci. USA, 104, 17204–17209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Franich N.R., Fitzsimons H.L., Fong D.M., Klugmann M., During M.J., Young D. (2008) AAV vector-mediated RNAi of mutant huntingtin expression is neuroprotective in a novel genetic rat model of Huntington's disease. Mol. Ther., 16, 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boudreau R.L., McBride J.L., Martins I., Shen S., Xing Y., Carter B.J., Davidson B.L. (2009) Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington's disease mice. Mol. Ther., 17, 1053–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Drouet V., Perrin V., Hassig R., Dufour N., Auregan G., Alves S., Bonvento G., Brouillet E., Luthi-Carter R., Hantraye P. et al. (2009) Sustained effects of nonallele-specific Huntingtin silencing. Ann. Neurol., 65, 276–285. [DOI] [PubMed] [Google Scholar]
- 56.Hult S., Soylu R., Bjorklund T., Belgardt B.F., Mauer J., Bruning J.C., Kirik D., Petersen A. (2011) Mutant huntingtin causes metabolic imbalance by disruption of hypothalamic neurocircuits. Cell Metab., 13, 428–439. [DOI] [PubMed] [Google Scholar]
- 57.Dufour B.D., Smith C.A., Clark R.L., Walker T.R., McBride J.L. (2014) Intrajugular vein delivery of AAV9-RNAi prevents neuropathological changes and weight loss in Huntington's disease mice. Mol. Ther., 22, 797–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stanek L.M., Sardi S.P., Mastis B., Richards A.R., Treleaven C.M., Taksir T., Misra K., Cheng S.H., Shihabuddin L.S. (2014) Silencing mutant huntingtin by adeno-associated virus-mediated RNA interference ameliorates disease manifestations in the YAC128 mouse model of Huntington's disease. Hum. Gene Ther., 25, 461–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Monteys A.M., Wilson M.J., Boudreau R.L., Spengler R.M., Davidson B.L. (2015) Artificial miRNAs targeting mutant huntingtin show preferential silencing in vitro and in vivo. Mol. Ther. Nucleic Acids, 4, e234. [DOI] [PubMed] [Google Scholar]
- 60.McBride J.L., Pitzer M.R., Boudreau R.L., Dufour B., Hobbs T., Ojeda S.R., Davidson B.L. (2011) Preclinical safety of RNAi-mediated HTT suppression in the rhesus macaque as a potential therapy for Huntington's disease. Mol. Ther., 19, 2152–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grondin R., Kaytor M.D., Ai Y., Nelson P.T., Thakker D.R., Heisel J., Weatherspoon M.R., Blum J.L., Burright E.N., Zhang Z. et al. (2012) Six-month partial suppression of Huntingtin is well tolerated in the adult rhesus striatum. Brain, 135, 1197–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stiles D.K., Zhang Z., Ge P., Nelson B., Grondin R., Ai Y., Hardy P., Nelson P.T., Guzaev A.P., Butt M.T. et al. (2012) Widespread suppression of huntingtin with convection-enhanced delivery of siRNA. Exp. Neurol., 233, 463–471. [DOI] [PubMed] [Google Scholar]
- 63.Grondin R., Ge P., Chen Q., Sutherland J.E., Zhang Z., Gash D.M., Stiles D.K., Stewart G.R., Sah D.W., Kaemmerer W.F. (2015) Onset time and durability of huntingtin suppression in rhesus putamen after direct infusion of antihuntingtin siRNA. Mol. Ther. Nucleic Acids, 4, e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stanek L.M., Yang W., Angus S., Sardi P.S., Hayden M.R., Hung G.H., Bennett C.F., Cheng S.H., Shihabuddin L.S. (2013) Antisense oligonucleotide-mediated correction of transcriptional dysregulation is correlated with behavioral benefits in the YAC128 mouse model of Huntington's disease. J. Huntington‘s Dis., 2, 217–228. [DOI] [PubMed] [Google Scholar]
- 65.Carroll J.B., Warby S.C., Southwell A.L., Doty C.N., Greenlee S., Skotte N., Hung G., Bennett C.F., Freier S.M., Hayden M.R. (2011) Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the Huntington disease gene/allele-specific silencing of mutant huntingtin. Mol. Ther., 19, 2178–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ostergaard M.E., Southwell A.L., Kordasiewicz H., Watt A.T., Skotte N.H., Doty C.N., Vaid K., Villanueva E.B., Swayze E.E., Bennett C.F. et al. (2013) Rational design of antisense oligonucleotides targeting single nucleotide polymorphisms for potent and allele selective suppression of mutant Huntingtin in the CNS. Nucleic Acids Res., 41, 9634–9650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Keiser M.S., Geoghegan J.C., Boudreau R.L., Lennox K.A., Davidson B.L. (2013) RNAi or overexpression: alternative therapies for Spinocerebellar Ataxia Type 1. Neurobiol. Dis., 56, 6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Keiser M.S., Boudreau R.L., Davidson B.L. (2014) Broad therapeutic benefit after RNAi expression vector delivery to deep cerebellar nuclei: implications for spinocerebellar ataxia type 1 therapy. Mol. Ther., 22, 588–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alves S., Nascimento-Ferreira I., Auregan G., Hassig R., Dufour N., Brouillet E., Pedroso de Lima M.C., Hantraye P., Pereira de Almeida L., Deglon N. (2008) Allele-specific RNA silencing of mutant ataxin-3 mediates neuroprotection in a rat model of Machado–Joseph disease. PloS One, 3, e3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Alves S., Nascimento-Ferreira I., Dufour N., Hassig R., Auregan G., Nobrega C., Brouillet E., Hantraye P., Pedroso de Lima M.C., Deglon N. et al. (2010) Silencing ataxin-3 mitigates degeneration in a rat model of Machado–Joseph disease: no role for wild-type ataxin-3? Hum. Mol. Genet., 19, 2380–2394. [DOI] [PubMed] [Google Scholar]
- 71.Rodriguez-Lebron E., Costa Mdo C., Luna-Cancalon K., Peron T.M., Fischer S., Boudreau R.L., Davidson B.L., Paulson H.L. (2013) Silencing mutant ATXN3 expression resolves molecular phenotypes in SCA3 transgenic mice. Mol. Ther., 21, 1909–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Costa Mdo C., Luna-Cancalon K., Fischer S., Ashraf N.S., Ouyang M., Dharia R.M., Martin-Fishman L., Yang Y., Shakkottai V.G., Davidson B.L. et al. (2013) Toward RNAi therapy for the polyglutamine disease Machado–Joseph disease. Mol. Ther., 21, 1898–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ramachandran P.S., Bhattarai S., Singh P., Boudreau R.L., Thompson S., Laspada A.R., Drack A.V., Davidson B.L. (2014) RNA interference-based therapy for spinocerebellar ataxia type 7 retinal degeneration. PloS One, 9, e95362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ramachandran P.S., Boudreau R.L., Schaefer K.A., La Spada A.R., Davidson B.L. (2014) Nonallele specific silencing of ataxin-7 improves disease phenotypes in a mouse model of SCA7. Mol. Ther., 22, 1635–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lombardi M.S., Jaspers L., Spronkmans C., Gellera C., Taroni F., Di Maria E., Donato S.D., Kaemmerer W.F. (2009) A majority of Huntington's disease patients may be treatable by individualized allele-specific RNA interference. Exp. Neurol., 217, 312–319. [DOI] [PubMed] [Google Scholar]
- 76.van Bilsen P.H., Jaspers L., Lombardi M.S., Odekerken J.C., Burright E.N., Kaemmerer W.F. (2008) Identification and allele-specific silencing of the mutant huntingtin allele in Huntington's disease patient-derived fibroblasts. Hum. Gene Ther., 19, 710–719. [DOI] [PubMed] [Google Scholar]
- 77.Pfister E.L., Kennington L., Straubhaar J., Wagh S., Liu W., DiFiglia M., Landwehrmeyer B., Vonsattel J.P., Zamore P.D., Aronin N. (2009) Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington's disease patients. Curr. Biol., 19, 774–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Becanovic K., Norremolle A., Neal S.J., Kay C., Collins J.A., Arenillas D., Lilja T., Gaudenzi G., Manoharan S., Doty C.N. et al. (2015) A SNP in the HTT promoter alters NF-kappaB binding and is a bidirectional genetic modifier of Huntington disease. Nat. Neurosci., 18, 807–816. [DOI] [PubMed] [Google Scholar]
- 79.Garriga-Canut M., Agustin-Pavon C., Herrmann F., Sanchez A., Dierssen M., Fillat C., Isalan M. (2012) Synthetic zinc finger repressors reduce mutant huntingtin expression in the brain of R6/2 mice. Proc. Natl Acad. Sci. USA, 109, E3136–E3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ramachandran P.S., Keiser M.S., Davidson B.L. (2013) Recent advances in RNA interference therapeutics for CNS diseases. Neurotherapeutics, 10, 473–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu X.H., Yang X.W. (2012) ‘Huntingtin holiday’: progress toward an antisense therapy for Huntington's disease. Neuron, 74, 964–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Skotte N.H., Southwell A.L., Ostergaard M.E., Carroll J.B., Warby S.C., Doty C.N., Petoukhov E., Vaid K., Kordasiewicz H., Watt A.T. et al. (2014) Allele-specific suppression of mutant huntingtin using antisense oligonucleotides: providing a therapeutic option for all Huntington disease patients. PloS One, 9, e107434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gagnon K.T., Pendergraff H.M., Deleavey G.F., Swayze E.E., Potier P., Randolph J., Roesch E.B., Chattopadhyaya J., Damha M.J., Bennett C.F. et al. (2010) Allele-selective inhibition of mutant huntingtin expression with antisense oligonucleotides targeting the expanded CAG repeat. Biochemistry, 49, 10166–10178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hu J., Dodd D.W., Hudson R.H., Corey D.R. (2009) Cellular localization and allele-selective inhibition of mutant huntingtin protein by peptide nucleic acid oligomers containing the fluorescent nucleobase [bis-o-(aminoethoxy)phenyl]pyrrolocytosine. Bioorg. Med. Chem. Lett., 19, 6181–6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hu J., Matsui M., Corey D.R. (2009) Allele-selective inhibition of mutant huntingtin by peptide nucleic acid-peptide conjugates, locked nucleic acid, and small interfering RNA. Ann. N Y Acad. Sci., 1175, 24–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hu J., Matsui M., Gagnon K.T., Schwartz J.C., Gabillet S., Arar K., Wu J., Bezprozvanny I., Corey D.R. (2009) Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nat. Biotechnol., 27, 478–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Orr H.T., Chung M.Y., Banfi S., Kwiatkowski T.J. Jr, Servadio A., Beaudet A.L., McCall A.E., Duvick L.A., Ranum L.P., Zoghbi H.Y. (1993) Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat. Genet., 4, 221–226. [DOI] [PubMed] [Google Scholar]
- 88.Subramony S.H., Ashizawa T. (1993) In Pagon R.A., Adam M.P., Ardinger H.H., Wallace S.E., Amemiya A., Bean L.J.H., Bird T.D., Dolan C.R., Fong C.T., Smith R.J.H., Stephens K. (eds), GeneReviews®, Seattle: (WA: ), in press. [Google Scholar]
- 89.Robitaille Y., Schut L., Kish S.J. (1995) Structural and immunocytochemical features of olivopontocerebellar atrophy caused by the spinocerebellar ataxia type 1 (SCA-1) mutation define a unique phenotype. Acta Neuropathol., 90, 572–581. [DOI] [PubMed] [Google Scholar]
- 90.Robitaille Y., Lopes-Cendes I., Becher M., Rouleau G., Clark A.W. (1997) The neuropathology of CAG repeat diseases: review and update of genetic and molecular features. Brain Pathol., 7, 901–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Donato S.D., Mariotti C., Taroni F. (2012) Spinocerebellar ataxia type 1. Handb. Clin. Neurol., 103, 399–421. [DOI] [PubMed] [Google Scholar]
- 92.Rub U., Schols L., Paulson H., Auburger G., Kermer P., Jen J.C., Seidel K., Korf H.W., Deller T. (2013) Clinical features, neurogenetics and neuropathology of the polyglutamine spinocerebellar ataxias type 1, 2, 3, 6 and 7. Prog. Neurobiol., 104, 38–66. [DOI] [PubMed] [Google Scholar]
- 93.Burright E.N., Clark H.B., Servadio A., Matilla T., Feddersen R.M., Yunis W.S., Duvick L.A., Zoghbi H.Y., Orr H.T. (1995) SCA1 transgenic mice: a model for neurodegeneration caused by an expanded CAG trinucleotide repeat. Cell, 82, 937–948. [DOI] [PubMed] [Google Scholar]
- 94.Matilla A., Roberson E.D., Banfi S., Morales J., Armstrong D.L., Burright E.N., Orr H.T., Sweatt J.D., Zoghbi H.Y., Matzuk M.M. (1998) Mice lacking ataxin-1 display learning deficits and decreased hippocampal paired-pulse facilitation. J. Neurosci., 18, 5508–5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Watase K., Weeber E.J., Xu B., Antalffy B., Yuva-Paylor L., Hashimoto K., Kano M., Atkinson R., Sun Y., Armstrong D.L. et al. (2002) A long CAG repeat in the mouse Sca1 locus replicates SCA1 features and reveals the impact of protein solubility on selective neurodegeneration. Neuron, 34, 905–919. [DOI] [PubMed] [Google Scholar]
- 96.Keiser M.S., Kordower J.H., , Gonzalez-Alegre P., Davidson B.L (2015) Broad distribution of ataxin 1 silencing in rhesus cerebella for spinocerebellar ataxia type 1 therapy. Brain, [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Paulson H. (1993) In Pagon R.A., Adam M.P., Ardinger H.H., Wallace S.E., Amemiya A., Bean L.J.H., Bird T.D., Fong C.T., Smith R.J.H., Stephens K. (eds.), GeneReviews®, Seattle: (WA: ), in press. [Google Scholar]
- 98.Nobrega C., Nascimento-Ferreira I., Onofre I., Albuquerque D., Hirai H., Deglon N., de Almeida L.P. (2013) Silencing mutant ataxin-3 rescues motor deficits and neuropathology in Machado–Joseph disease transgenic mice. PloS One, 8, e52396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Garden G.A., La Spada A.R. (2008) Molecular pathogenesis and cellular pathology of spinocerebellar ataxia type 7 neurodegeneration. Cerebellum, 7, 138–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chiriboga C.A., Swoboda K.J., Darras B.T., Iannaccone S.T., Montes J., Vivo D.C.D., Norris D.A., Bennett C.F., Bishop K.M. (2015) Results from a Phase 1 study of ISIS-SMNRX in children with spinal muscular atrophy. Neurology, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Southwell A.L., Smith S.E., Davis T.R., Caron N.S., Villanueva E.B., Xie Y., Collins J.A., Ye M.L., Sturrock A., Leavitt B.R. et al. (2015) Ultrasensitive measurement of huntingtin protein in cerebrospinal fluid demonstrates increase with Huntington disease stage and decrease following brain huntingtin suppression. Sci. Rep., 5, 12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wild E.J., Boggio R., Langbehn D., Robertson N., Haider S., Miller J.R., Zetterberg H., Leavitt B.R., Kuhn R., Tabrizi S.J. et al. (2015) Quantification of mutant huntingtin protein in cerebrospinal fluid from Huntington's disease patients. J. Clin. Invest., 125, 1979–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dayton R.D., Wang D.B., Klein R.L. (2012) The advent of AAV9 expands applications for brain and spinal cord gene delivery. Expert Opin. Biol. Ther., 12, 757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]