Abstract
The present study deployed a Mediator (MED) genes-mediated integrated genomic strategy for understanding the complex genetic architecture of grain size/weight quantitative trait in rice. The targeted multiplex amplicon resequencing of 55 MED genes annotated from whole rice genome in 384 accessions discovered 3971 SNPs, which were structurally and functionally annotated in diverse coding and non-coding sequence-components of genes. Association analysis, using the genotyping information of 3971 SNPs in a structured population of 384 accessions (with 50–100 kb linkage disequilibrium decay), detected 10 MED gene-derived SNPs significantly associated (46% combined phenotypic variation explained) with grain length, width and weight in rice. Of these, one strong grain weight-associated non-synonymous SNP (G/A)-carrying OsMED4_2 gene was validated successfully in low- and high-grain weight parental accessions and homozygous individuals of a rice mapping population. The seed-specific expression, including differential up/down-regulation of three grain size/weight-associated MED genes (including OsMED4_2) in six low and high-grain weight rice accessions was evident. Altogether, combinatorial genomic approach involving haplotype-based association analysis delineated diverse functionally relevant natural SNP-allelic variants in 10 MED genes, including three potential novel SNP haplotypes in an OsMED4_2 gene governing grain size/weight differentiation in rice. These molecular tags have potential to accelerate genomics-assisted crop improvement in rice.
Rice (Oryza sativa L.) is a vital staple food and nutritional source for over half of the human population across the globe. With decreasing farmland and increase in global population, the improvement of rice yield and productivity is imperative to sustain world food security in the present scenario of climate change. Enhancing grain yield is thus the primary objective of current rice breeding and genomics research1,2. Grain size, the major determinant of grain weight and one of the crucial components of grain yield is positively correlated with grain length, grain width, grain thickness and grain filling degree3. Therefore, grain size/weight is a known target trait for both domestication and artificial breeding for enhancing productivity in rice2,4,5. Advancements of genome mapping, sequencing, and functional genomics have provided powerful tools to uncover the molecular basis of complex quantitative grain size traits in rice. These efforts led to identify thousands of low-resolution QTLs associated with rice grain size/weight. Of these, a set of selected high-resolution major QTLs has been narrowed-down into six most promising genes (GS3, GW2, qSW5/GW5, GS5, TGW6 and GIF1) governing grain size (length, width, weight and filling degree) through map-based cloning and mutant complementation analyses in rice4,6,7,8,9,10,11,12,13,14. Detailed molecular characterization and functional validation of these six genes have inferred their negative (GS3, GW2 and qSW5/GW5) or positive (GS5, TGW6 and GIF1) regulatory mechanism during plant growth and seed development controlling grain size/weight in rice1,2. Most of these genes majorly regulate signaling pathways mediated by phytohormones, proteasomal degradation and G-proteins to control cell elongation and proliferation in the seeds of rice. Genetic association analysis of these known cloned/characterized genes with grain morphology of numerous germplasm lines reflects existence of their multiple natural allelic variants in a large rice gene pool with certain degree of pleiotropic/epistasis effects on other undesirable grain yield component traits1,2. These intricate and complex interactions of grain size genes impose major hindrances for marker-assisted genetic improvement of grain size/weight and yield in rice. Henceforth, a clear understanding of the genetic/molecular basis of grain size/weight variation, by identification of novel functionally relevant genes and alleles governing the target traits, is extremely important for rice genomics-assisted crop improvement program.
In eukaryotes, transcription of genes is regulated by various transcription factors and it is further assisted through different cofactors15. A number of studies have established Mediator complex as a crucial cofactor involved in RNA pol II mediated transcription regulation16,17. The Mediator complex encompassing more than 25 subunits in eukaryotes, acts as an interface between transcriptional activators/repressors and RNA pol II and thus can have both inducing as well as repressing effect on transcription17,18,19. The role of these Mediator subunits in transcription has been established in the formation of pre-initiation complex, transcription initiation and elongation, splicing, gene looping and transcription termination19,20,21,22,23,24,25,26,27,28,29,30. Abundance of intrinsic disorder regions (IDRs) in MED subunits and the modular arrangement consisting of Head, Middle, Tail and Kinase modules makes Mediator a versatile regulator, as it can interact with various factors and attain diverse configurations for regulating multiple processes in eukaryotes18,31,32. Plant Mediator complex composition is no different from animals except for the absence of MED1 and presence of some plant-specific MED subunits33,34. In plants, specifically in Arabidopsis, diverse functions ranging from development to biotic and abiotic stresses have been attributed to different MED subunits. For instance, MED12 and MED13, which are the parts of Kinase module, regulate timing of embryo patterning35. Similarly, STRUWELLPETER, identified as MED14, is known to regulate cell proliferation in Arabidopsis36. MED18 regulates flowering time and identity of floral organs by transcriptional regulation of floral regulators37. MED16, MED2 and MED14 are vital for providing freezing tolerance in Arabidopsis38. A single MED subunit, MED25 regulates multiple growth, development and stress tolerance traits, including plant organ size control39, root hair differentiation40, lateral root development through auxin signalling41, defense against fungal pathogens by regulating jasmonic acid42,43 and abscissic acid signalling43. MED15 and MED16 modulate response to biotic factors by regulating salicylic acid response44,45. Likewise, MED21 regulates response against necrotrophic fungi46 and MED8 controls flowering time and resistance towards necrotrophic fungi42. The aforementioned previous studies in Arabidopsis and expression profiling of MED genes in rice and Arabidopsis ascertain their definitive role in diverse useful yield component and stress tolerance traits, besides basal regulation of gene expression34,47,48,49,50. However, none of the MED genes regulating specific agronomic traits, including grain size/yield has been identified and functionally validated so far to be utilized in marker-assisted selection for genetic improvement of rice. In this context, it would be interesting to decipher the possible role of MED genes in grain size/weight regulation and seed development in rice.
An integrated approach of SNP marker-based high resolution candidate gene-based association analysis, traditional QTL mapping, differential expression profiling and molecular haplotyping is well documented as an attractive strategy for efficient dissection of complex quantitative yield component traits in multiple crop plants, including rice1,2,4,11,51,52,53,54,55,56,57. This strategy will also prove useful in rapid identification of natural allelic variants within genes associated with grain size/weight and understanding their mechanism of interaction/regulation for grain size/weight variation in rice cultivars adapted to diverse agroclimatic conditions. All these inputs obtained from the combinatorial approach could essentially expedite marker-assisted breeding for selecting cultivars with large grain size and more yield in rice.
Considering the aforesaid possibilities, genetic association analysis of grain size and weight traits was performed based on precise field phenotyping and genotyping of informative SNPs mined from 55 MED subunit genes (distributed across rice genome) in 384 diverse low and high grain weight rice accessions (association panel). This strategy was further integrated with traditional bi-parental mapping population validation, differential expression profiling and gene-based SNP haplotyping/LD (linkage disequilibrium) mapping to delineate functionally relevant natural allelic variants and haplotypes in the potential MED gene(s) regulating grain size (grain length, grain width) and 1000-grain weight in rice.
Results and Discussion
Discovery, annotation and genotyping of MED gene-derived SNPs
The implication of integrated genomic strategy (combining association analysis, QTL mapping, expression profiling and molecular haplotyping) for efficient dissection of complex quantitative traits and rapid identification of potential candidate genes especially regulating grain size/weight traits is well demonstrated in crop plants, including rice2,4,55,56,57,58. In this context, the current study integrated candidate gene-based association mapping with bi-parental mapping population validation, differential gene expression profiling and gene-based haplotyping/LD mapping to scale-down the candidate Mediator (MED) genes governing grain size, including grain length, grain width and grain weight in rice. A diverse array of MED genes is known to regulate multiple agronomic traits, including yield component and abiotic/biotic stress tolerance traits in crop plants34,49,59. Primarily, to perform candidate gene-based association mapping, the diverse coding and non-coding (introns, URRs and DRRs along with 5′ and 3′ UTRs, respectively) sequence components of 55 MED genes annotated from whole rice genome were sequenced and genotyped in low and high grain weight 384 rice accessions (belonging to an association panel) by targeted multiplex-amplicon resequencing to discover potential gene-derived SNP allelic variants.
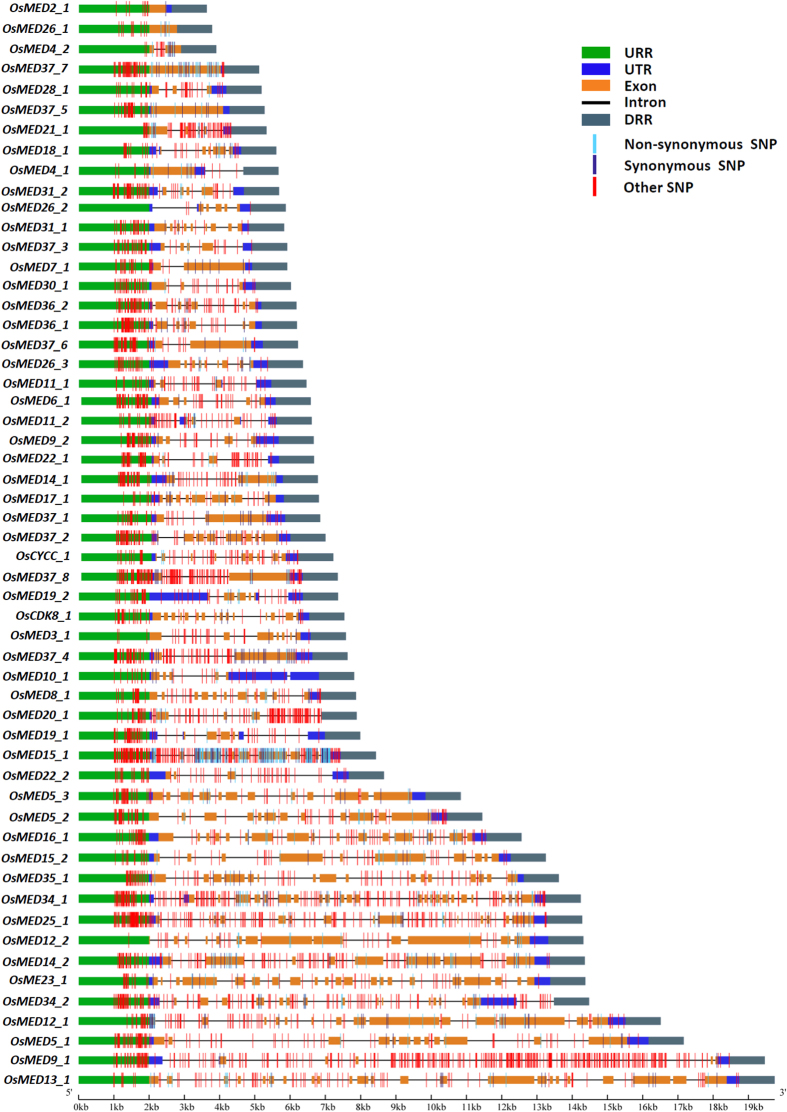
The targeted resequencing of coding and non-coding intronic and regulatory sequence components of 55 MED genes in 384 diverse low and high grain weight rice accessions (association panel) using the Illumina TruSeq Custom Amplicon strategy mined 3971 high-quality SNPs with an average frequency of 72.2 SNPs/gene (Fig. 1, Tables 1, S1). These SNPs were physically mapped across 12 chromosomes of rice with a highest (14.3%, 568 SNPs) and lowest (1.8%, 73) density on chromosomes 9 and 6, respectively (Figure S1, Table 1). The structural annotation of 3971 SNPs in the MED genes revealed the presence of 3306 (83.3%) and 665 (16.7%) SNPs in the non-coding and coding regions of genes, respectively (Fig. 1, Tables 1, S1). Among the non-coding SNPs, 1545 (46.7%) and 1761 (53.3%) SNPs were derived from the regulatory (URRs and DRRs along with 5′ and 3′ UTRs, respectively) and intronic sequence components of genes, respectively. The 665 coding SNPs included 323 (48.6%) non-synonymous (missense and nonsense) and 342 (51.4%) synonymous SNPs in the MED genes, respectively (Fig. 1, Tables 1, S1). The informative SNPs (specifically the non-synonymous and regulatory SNPs) discovered from diverse coding and non-coding sequence components of MED genes can serve as a useful genomic resource to be utilized for manifold genomics-assisted breeding applications, including genetic association analysis and targeted mapping of potential genes regulating multiple traits of agronomic importance in rice.
Figure 1. Gene structure of 55 Mediator (MED) subunit genes depicting the structural annotation of SNPs in different coding (synonymous and non-synonymous) as well as non-coding regulatory (URRs and DRRs along with 5′ and 3′ UTRs, respectively) and intronic sequence components of genes.
URR: upstream regulatory region, DRR: downstream regulatory region, UTR: 5′ or 3′ untranslated region. The sub-genic regions are indicated as per the MED gene annotation information available at Rice Genome Annotation Project (RGAP, release 7.0).
Table 1. Genomic distribution of SNPs mined from rice Mediator genes.
| Ricechromosomes | Mediator genesannotated | Number (%) ofgenic SNPsdiscovered | Regulatory-SNPs | Intronic-SNPs |
CDS-SNPs |
|
|---|---|---|---|---|---|---|
| Non-synonymous | Synonymous | |||||
| Os_chr01 | 4 | 484 (12.2) | 84 | 376 | 9 | 15 |
| Os_chr02 | 6 | 328 (8.3) | 160 | 121 | 17 | 30 |
| Os_chr03 | 5 | 286 (7.2) | 155 | 94 | 5 | 32 |
| Os_chr04 | 4 | 504 (12.7) | 166 | 185 | 92 | 61 |
| Os_chr05 | 6 | 370 (9.3) | 170 | 147 | 19 | 34 |
| Os_chr06 | 1 | 73 (1.8) | 51 | 20 | 0 | 2 |
| Os_chr07 | 4 | 263 (6.6) | 108 | 99 | 26 | 30 |
| Os_chr08 | 5 | 327 (8.2) | 129 | 131 | 36 | 31 |
| Os_chr09 | 6 | 568 (14.3) | 192 | 289 | 39 | 48 |
| Os_chr10 | 7 | 324 (8.2) | 127 | 126 | 48 | 23 |
| Os_chr11 | 4 | 332 (8.4) | 133 | 150 | 24 | 25 |
| Os_chr12 | 3 | 112 (2.8) | 70 | 23 | 8 | 11 |
| Total | 55 | 3971 | 1545 (46.7) | 1761 (53.3) | 323 (48.6) | 342 (51.4) |
Parentheses indicate the proportion of each kind of SNP mined from its respective total MED gene-derived SNPs.
MED gene-based association mapping of rice grain size
For candidate gene-based association mapping, the genotyping data of 3971 informative MED gene-derived SNPs (with 5% minor allele frequency) exhibiting polymorphism among 384 rice accessions was utilized. The use of these SNPs in determination of population genetic structure and PCA (principal component analysis) differentiated all 384 rice accessions from each other, which clustered into two distinct population groups- POP I and POP II. The determination of LD patterns in a population of 384 accessions using 3971 SNPs (physically mapped on 12 chromosomes) exhibited a broader LD estimate (r2: 0.32–0.78) and faster LD decay (r2 decreased to half of its maximum value) nearly at 50–100 kb physical distance of rice chromosomes. This estimate is comparable with the chromosomal LD decay documented in previous candidate gene-based and genome-wide association mapping studies of rice1,51,52,53,54. Therefore, the LD decay documented in the present study using the genotyping information of MED gene-derived SNPs mapped on 12 rice chromosomes is adequate enough for efficient trait association mapping to identify potential genic loci governing useful agronomic traits, including grain size/weight in rice.
The normal frequency distribution along with a broader phenotypic variation and higher heritability for grain size, including grain length (6.6 to 11.2 mm, mean ± SD: 8.5 ± 0.71, mean CV: 8% and mean H2: 75%), grain width (1.9 to 3.5 mm, 2.8 ± 0.31, 11% and 73%) and 1000-grain weight (15.4 to 39.2 g, 26.5 ± 4.8, 18% and 82%), in 384 rice accessions were observed across two diverse geographical locations/years based on ANOVA (Figure S2, Table S2). The ANOVA outcomes inferred a highly significant difference (P < 0.0001) among rice accessions for grain size/weight trait variation despite significant environmental (years and geographical locations) and block replication effects on these traits (Table S3). A significant interaction between genotypes (G)/accessions and environments (E) for grain size/weight traits was evident. These observations infer complex quantitative genetic inheritance pattern of grain size (grain length, grain width) and grain weight traits in rice and thus require an efficient integrated genomics-assisted breeding strategy (like association/genetic mapping and molecular haplotyping) for genetic dissection of these target traits in rice. Further, consistent phenotypic expression of grain size traits, based on high heritability across diverse geographical locations/years in 384 accessions of an association panel, implicates the robustness of grain size/weight phenotypic data generated in the present study for trait association mapping in rice. Therefore, the mean phenotyping data of accessions, revealing consistent phenotypic expression for grain size/weight traits across geographical locations/years, was utilized for subsequent SNP marker-trait association study.
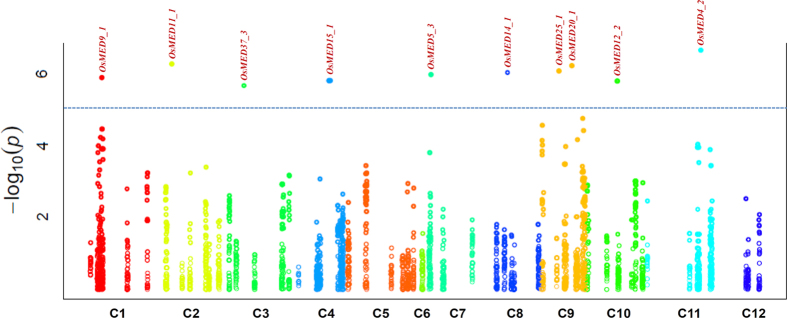
The use of CMLM and P3D/EMMAX model-based approaches (at a FDR cut-off ≤0.05) in genetic association analysis identified 10 SNPs in 10 MED genes exhibiting significant association (at a P value ≤ 10−5) with grain length, grain width and grain weight in rice (Fig. 2, Table 2). These grain size-associated MED gene-derived SNPs were mapped on nine chromosomes (excluding chromosomes 5, 6 and 12) of rice. Of these, a maximum of two trait-associated SNPs were represented from rice chromosome 9. Six and four grain size trait-associated genomic SNP loci were derived from coding (six non-synonymous SNPs) and non-coding [URR (three SNPs) and intronic (one SNP)] sequence components of 10 MED genes, respectively (Table 2). The estimated minor allele frequency (MAF) for 10 grain size/weight-associated MED genes in a constituted association panel varied from 15–26% with an average of 21%. The proportion of phenotypic variation for grain length, grain width and grain weight explained by maximum effect 10 SNP loci in 10 MED genes (OsMED9_1, OsMED11_1, OsMED37_3, OsMED15_1, OsMED5_3, OsMED14_1, OsMED25_1, OsMED20_1, OsMED12_2 and OsMED4_2) among 384 rice accessions varied from 15 to 33% R2. The percentage of combined PVE (phenotypic variation explained) revealed by all significant 10 MED gene-derived SNPs was 46%. Interestingly, seven, four and 10 MED-gene based SNPs associated with grain length, grain width and grain weight revealing combined PVE of 43% (varied from 15–33%), 41% (18–33%) and 48% (15–33%) were identified, respectively (Table 2). Five (grain length and grain weight), one (grain width and grain weight) and two (grain length, grain width and grain weight) MED gene-derived SNPs exhibited significant association with multiple grain size traits in rice. A strong association of one non-synonymous SNP (G/A) scanned in OsMED4_2 gene with grain length, grain width and grain weight (33% PVE with P value 0.3 × 10−8) followed by one regulatory (URR) SNP (T/A) identified in OsMED25_1 gene with grain length and grain weight (28% PVE with P value 1.3 × 10−6) was evident (Table 2). The added-advantage of CMLM and P3D/EMMAX strategies based on their efficacy towards scanning of non-spurious SNP marker-trait association with maximal statistical power and high prediction accuracy over other association model-based approaches hitherto has been well-documented in crop plants57,60,61. In this perspective, the potential MED gene-derived SNP loci associated with grain length, grain width and grain weight scanned in this study deploying CMLM and P3D/EMMAX-based association mapping strategy is relevant and thus can be applied for deciphering the complex gene regulatory networks underlying grain size/weight trait variation in rice. Notably, six (OsMED15_1, OsMED14_1, OsMED12_2, OsMED25_1, OsMED5_3 and OsMED4_2) of the 10 high grain size/weight-associated SNPs-containing MED genes identified in our study are known to govern diverse growth and developmental processes in plants34,35,36,39,49,59,62,63,64. Especially, the role of OsMED15 gene in controlling seed development as well as its significant association potential for high and low grain weight differentiation is well-demonstrated in rice49,59. Similarly, the involvement of another gene MED12 in controlling embryo patterning during seed development has been deciphered in Arabidopsis35. MED25 of Arabidopsis has been found to be involved in the regulation of timing and process of flowering, which though not demonstrated experimentally, may further affect timing and process of seed setting and development42. The heterozygote mutant lines of Atmed14 are dwarf with abnormal architecture including abnormal floral structure suggesting a probable influence on seed setting and maturation36. As the med14 mutant of Arabidopsis shows reduced cell numbers in all the aerial organs36, there is a possibility that MED14 can directly or indirectly affects overall seed yield. In rice, MED4 interacts with SAD1 to regulate tiller number which can affect the overall grain yield64. The essential role of a MED5 gene in repressing phenyl propanoid biosynthesis62 as well as in regulating proper plant growth/development, including cell wall lignification has been demonstrated in Arabidopsis63. Therefore, grain size/weight trait-associated 10 SNP loci identified from diverse non-synonymous coding and regulatory sequence components of 10 MED genes are assumed to be functionally relevant. Such non-synonymous and regulatory SNPs are known to regulate diverse grain size and weight traits during seed development in crop plants, including rice2,4,55,56,57,58. Henceforth, the trait-associated novel natural SNP allelic variants-containing MED genes identified by candidate gene-based association mapping can essentially be utilized for establishing rapid marker-trait linkages and efficient identification/mapping of genes governing grain size/weight trait in rice.
Figure 2. Manhattan plot illustrating the significance of SNP loci-containing MED genes for grain weight trait association in rice.
X-axis represents the relative density of SNPs mined from MED subunit genes distributed over 12 rice chromosomes. Y-axis indicates the-log10 (P) value to scan the significant trait-associated SNP loci at a cut-off P ≤ 10−5.
Table 2. SNPs identified in the MED genes associated with grain size traits in rice.
| OsMEDgenes | MSU locusIDs | Chromosomes | AssociatedSNPs | Physicalpositions(bp) | Minor allelefrequency(MAF %) | Functionalsignificanceof SNPs | Encoded aminoacid | P-value | PVE(R2%) | Associated grainsize traits |
|---|---|---|---|---|---|---|---|---|---|---|
| OsMED9_1 | LOC_Os01g31629 | Oschr01 | (C/G) | 17320769 | 17 | Intron | – | 1.7 × 10−5 | 21 | GWg |
| OsMED11_1 | LOC_Os02g09600 | Oschr02 | (G/A) | 4940189 | 15 | Non-Synonymous coding | Threonine (ACG)/Methionine (ATG) | 1.7 × 10−6 | 25 | GL and GWg |
| OsMED37_3 | LOC_Os03g16860 | Oschr03 | (A/G) | 9369204 | 16 | URR | – | 1.1 × 10−5 | 18 | GWi and GWg |
| OsMED15_1 | LOC_Os04g03860 | Oschr04 | (G/A) | 1754571 | 18 | Non-Synonymous coding | Glycine(GGG)/Arginine (AGG) | 0.8 × 10−5 | 17 | GL and GWg |
| OsMED5_3 | LOC_Os07g48350 | Oschr07 | (G/A) | 28899486 | 20 | Non-Synonymous coding | Alanine (GCG)/Threonine (ACG) | 1.5 × 10−5 | 20 | GWg and GWi |
| OsMED14_1 | LOC_Os08g24400 | Oschr08 | (G/T) | 14736149 | 18 | URR | – | 2.1 × 10−5 | 15 | GL and GWg |
| OsMED25_1 | LOC_Os09g13610 | Oschr09 | (T/A) | 7912591 | 26 | URR | – | 1.3 × 10−6 | 28 | GL and GWg |
| OsMED20_1 | LOC_Os09g27140 | Oschr09 | (A/G) | 16507513 | 21 | Non-Synonymous coding | Phenylalanine (TTC)/Leucine (CTC) | 0.9 × 10−7 | 26 | GL, GWi and GWg |
| OsMED12_2 | LOC_Os10g40260 | Oschr 10 | (G/C) | 21503285 | 22 | Non-Synonymous coding | Valine (GTT)/Leucine (CTT) | 1.1 × 10−5 | 24 | GL and GWg |
| OsMED4_2 | LOC_Os11g05150 | Oschr11 | (C/T) | 2260577 | 24 | Non-Synonymous coding | Glutamic acid (GAG)/Lysine(AAG) | 0.3 × 10−8 | 33 | GL, GWi and GWg |
GL: Grain Length, GWi: Grain Width and GWg: 1000-Grain Weight, PVE: Phenotypic Variation Explained and URR: Upstream Regulatory Region.
Validation of grain size-associated MED genes in a mapping population
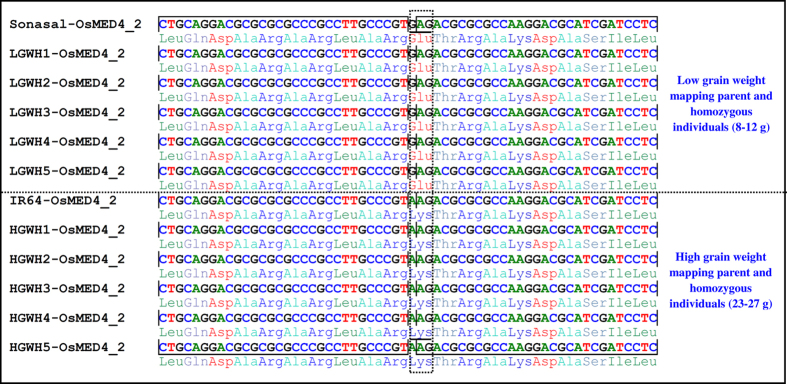
To validate 10 MED gene-derived SNPs exhibiting significant association with grain length, grain width and grain weight in rice, the SNPs exhibiting parental polymorphism (between IR 64 and Sonasal) were genotyped in 10 of each low and high grain weight homozygous individuals of a F4 mapping population (IR 64 × Sonasal). One non-synonymous SNP (G/A)-containing OsMED4_2 gene showing strong association with grain size, grain width and grain weight (based on trait association analysis), was validated in a mapping population (Fig. 3). All low (8–12 g) and high (23–27 g) grain weight parental accessions and homozygous individuals of a mapping population contained the identical high (A) and low (G) grain size-associated SNP alleles identified from an OsMED4_2 gene (Fig. 3). Henceforth, a stronger SNP allele effect of OsMED4_2 gene with high and low grain weight differentiation in rice was apparent. In contrast, SNP alleles mined from nine other MED genes revealing association with high and low grain weight differentiation could not correspond to the phenotypes of the low and high grain weight mapping parents and homozygous individuals. However, large-scale validation and genotyping of all 10 grain size/weight-associated MED gene-derived SNPs in the numerous bi-parental mapping populations contrasting for grain size/weight are required to ascertain the definitive association potential of these identified functionally relevant molecular tags in grain size/weight trait regulation in rice. Altogether, 10 grain size/weight-associated MED genes, including one non-synonymous SNP-containing OsMED4_2 (validated by both trait association analysis and in bi-parental mapping population) were selected as target candidates for grain weight/size trait regulation by their further validation through differential expression profiling in rice.
Figure 3. SNP (G to A) depicting the missense non-synonymous amino acid substitution [Glutamic acid (GAG) to Lysine (AAG)] in an OsMED4_2 gene differentiated the low (8–12 g) and high (23–27 g) grain weight parental accessions and representative homozygous individuals of a F4 mapping population (IR 64 × Sonasal).
The MED gene sequence region-carrying a non-synonymous SNP allelic variant is highlighted with a dotted box.
Differential expression profiling of grain size-associated MED genes
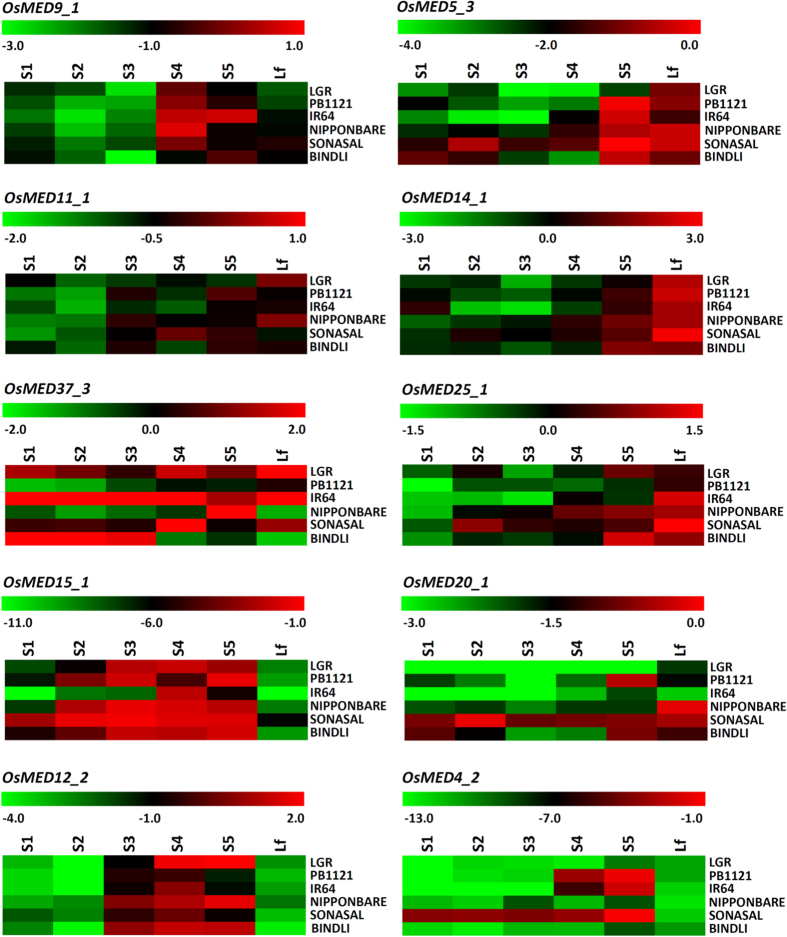
The grain size-associated ten SNPs-containing MED genes (identified by candidate gene-based association analysis), including one validated in bi-parental mapping population, were assayed for differential expression profiling to access the functional regulatory pattern of these genes in controlling grain size/weight of rice. The flag leaves and five seed developmental stages (S1 to S5) of two low (Sonasal and Bindli) and four high/medium (Pusa Basmati 1121, IR 64, Nipponbare and LGR) grain weight rice accessions were utilized for quantitative RT-PCR assay (Fig. 4). Of these, regulatory, intronic and non-synonymous SNPs-containing nine genes (except OsMED11_1) were ≥2 fold differentially regulated in at least one of the five seed developmental stages as compared to flag leaf in all the six rice accessions under study (Figure S3). Out of these nine genes, non-synonymous SNPs-containing four genes (OsMED4_2, OsMED12_2, OsMED15_1 and OsMED37_3) exhibited very high expression in seed stages (>7 fold upregulation in at least one of the five seed development stages as compared to flag leaf) of at least two varieties and among these, OsMED4_2 showed seed-specific expression (Fig. 4, S3, Table S4). Remarkably, one non-synonymous SNP (G/A)-carrying seed-specific OsMED4_2 validated by both genetic association analysis and in mapping population revealed almost an inversely correlated differential expression pattern in seed developmental stages of some of the selected low and high grain weight rice accessions (Fig. 4, S3, Table S4). A decreased expression of OsMED4_2 gene in the initial three seed developmental stages (S1, S2 and S3) of high (Pusa Basmati 1121, IR 64 and LGR) and increased expression in low (Sonasal) grain weight rice accessions than that of flag leaves of respective accessions was observed (Figs 4 and 5D, S3, Table S4). A pronounced higher expression of OsMED4_2 in S4 and S5 seed developmental stages of both high (IR 64 and Pusa Basmati 1121) and low (Sonasal) grain weight rice accessions was also apparent (Figs 4 and 5D, S3, Table S4). Interestingly, OsMED4_2 with non-synonymous SNPs validated in high and low grain weight parental accessions of a mapping population (IR 64 × Sonasal), exhibited differential regulation pattern in these accessions during seed development, implicating functional significance of this gene in grain weight regulation of rice. It would be thus interesting to constitute gene-specific haplotypes by targeting/combining other novel coding and non-coding SNP allelic variants mined from this OsMED4_2 gene and determine trait association potential of the gene haplotypes with grain size/weight variation in naturally occurring rice accessions.
Figure 4. Heat maps depicting the differential expression profiles of 10 grain size/weight-associated MED genes in five seed developmental stages (S1–S5) of six contrasting low (Sonasal and Bindli) and high (LGR, Pusa Basmati 1121, Nipponbare and IR 64) grain weight rice accessions as compared to flag leaves.
Color scale at the top of each map represents –ΔCt (Ct ubiquitin - Ct gene) values. Green, black and red color shows low, medium and high expression, respectively. Lf- flag leaf and PB1121- Pusa Basmati 1121.
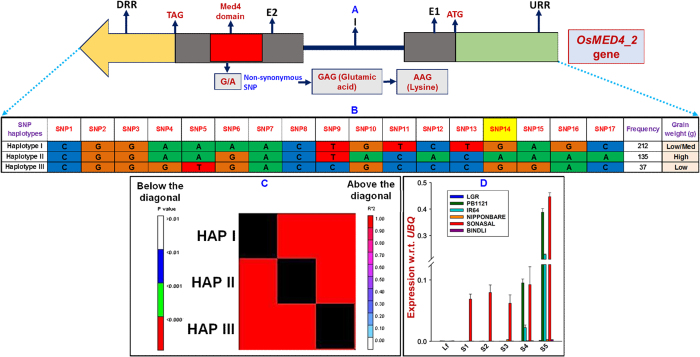
Figure 5. The molecular haplotyping and SNP haplotype-specific association analysis/LD mapping in an OsMED4_2 gene (A) validating its strong association potential for grain weight/size differentiation in rice. The genotyping of 17 SNPs, including one missense non-synonymous SNP (G/A, shaded with yellow colour) [encoding for Glutamic acid (GAG) to Lysine (AAG)] among 384 rice accessions (association panel) constituted three haplotypes (B). (C) Three SNP haplotype marker-based genotyping information produced a higher LD estimate and resolution covering the entire gene. (D) The differential expression profiling of OsMED4_2 gene in five seed developmental stages (S1–S5) and flag leaves (Lf) of six contrasting low (Sonasal and Bindli) and high (LGR, PusaBasmati 1121, Nipponbare and IR 64) grain weight rice accessions.
E1: Exon1, E2: Exon2, I: Intron, URR: upstream regulatory region and DRR: downstream regulatory region.
Molecular haplotyping in a grain size-associated MED gene
For molecular haplotyping of a strong grain size-associated OsMED4_2 gene (validated by association analysis, expression profiling and in mapping population), the cloned PCR amplicon sequencing and Illumina targeted multiplex-gene amplicon resequencing of entire 2 kb URR, exons, 1 kb DRR and intronic region of target gene in 384 rice accessions were performed (Fig. 5A). This discovered 17 SNPs from diverse coding and non-coding (including three non-synonymous, seven intronic and two URR SNPs) sequence components of the gene. The haplotype analysis in OsMED4_2 gene, by deploying the genotyping data of 17 SNPs among 384 rice accessions, constituted overall three haplotypes (Fig. 5B). The three SNP haplotype-based LD mapping in OsMED4_2 gene exhibited a higher degree of LD (r2 > 0.90 with P < 1.0 × 10−7) resolution in this gene (Fig. 5C). The association analysis using OsMED4_2 gene-derived SNP haplotypes demonstrated its strong association potential (PVE: 44% with P: 1.1 × 10−10) for grain size/weight trait variation. Remarkably, two major haplotypes of OsMED4_2 gene differentiated by a functional non-synonymous coding SNP (G/A) revealed strong association potential for low/medium (haplotypes I and III) and high grain weight (haplotype II) differentiation in rice (Fig. 5B). Nevertheless, novel haplotypes (with diverse allelic recombination) in an OsMED4_2 gene exhibiting differential trait association potential for rice grain size/weight were identified by SNP-based high-resolution molecular haplotyping. Altogether, a higher association potential of OsMED4_2 gene with grain size/weight trait variation in rice was ascertained by their combined validation through candidate gene-based association analysis, in mapping population, differential expression profiling and high-resolution molecular haplotyping/LD mapping. The grain size/weight is a complex quantitative trait and controlled by a complex regulatory networks involving a diverse arrays of genes in rice1,2. A number of known genes underlying QTLs governing grain length, grain width and grain weight have been cloned and characterized so far in rice4,6,7,8,9,10,11,12,13,14. In spite of several major efforts, no such potential robust genes/QTLs (validated in multiple genetic backgrounds/environments) have been identified till date to be deployed in marker-assisted breeding for selecting accessions with high grain weight and yield in rice. In the present study, efforts have been made to integrate candidate gene-based association analysis with mapping population validation, differential gene expression profiling and gene-based molecular haplotyping/LD mapping effectively, which enabled to delineate diverse natural SNP allelic variants in 10 MED genes, including three novel haplotypes in OsMED4_2 gene regulating grain weight/size differentiation in rice (Figure S4).
The involvement of OsMED4 gene in transcriptional regulation by its effective interaction with other protein-coding genes and signalling pathways underlying various aspects of plant development and growth has been deciphered recently64. MED4 is a subunit in the Middle module of the complex. Just like yeast and mammalian MED4, Arabidopsis MED4 interacts with MED9 and thus appears to be an important component for integrity of Middle module structure32. On the basis of very high sequence homology between Arabidopsis and rice MED4, it can be postulated that OsMED4 might be interacting with OsMED9. MED4 has two IDRs, one at each terminal, separated by a region which is predicted to be helical in nature. In yeast, a fragment harbouring this helical region and the C-terminal IDR was found to be important in the interaction of MED4 with MED7, MED9, MED10 and MED2165. The non-synonymous SNP (G/A) was found to be present in the CDS sequence corresponding to this helical region of OsMED4_2. Interestingly, in one of the earlier study, this region emerged as a signature motif for MED4 suggesting its importance in MED4 functioning66. This part of MED4 might thus be important in rice for maintaining the integrity of the Middle module. MED4 is a very disordered protein with a strong tendency to interact with other proteins32. In Arabidopsis, AtMED4 interacts with more than hundred proteins, including a couple of transcription factors like WOX13 and UNE12 that play important role in seed development and maturation32. WOX13 controls medio-lateral patterning of the fruit, which is the basis for seed maturation and dispersal67. On the other hand, mutation in UNE12 shows defect in embryo sac functions such as pollen tube guidance or fertilization68. There is a possibility that in rice also, MED4 is targeted by orthologs of WOX13 and UNE12 for their function. So any variation in the important residues of MED4 that disrupts its interaction with such transcription factors (WOX13 or UNE12) or other Mediator subunits (MED7, MED9, MED10 or MED21) can exhibit effect on the process of fertilization, seed setting, development and maturation. Such possible transcriptional mechanism of trait regulation due to non-synonymous SNP substitutions in the CDS of genes encoding variable amino acid residues and altered secondary structure of proteins has already been demonstrated in multiple known cloned grain size genes of rice2. It will be interesting to expand the SNP analysis in a larger set of diverse rice varieties to the whole genome level to decipher the genetic network significantly associated with rice grain size/weight and then see if OsMED4_2 is a part of the network. Thus, the grain size/weight-associated functionally relevant molecular tags (alleles and haplotypes) identified in the MED genes using a combinatorial genomic approach can be useful for rapid quantitative dissection of complex grain size/weight trait and eventually in marker-assisted breeding to develop improved rice cultivars with high grain weight and yield.
Methods
Targeted multiplex-gene amplicon resequencing
The genomic DNA was isolated from the young leaves of 384 low and high grain weight diverse rice accessions using QIAGEN DNeasy96 Plant Kit (QIAGEN, USA) according to the manufacturer’s protocol. For mining and genotyping of gene-based SNPs, a set of 55 MED genes structurally and functionally annotated from whole rice genome34 were utilized. These selected genes were resequenced using the genomic DNA of 384 rice accessions employing the multiplexed amplicon resequencing method (TruSeq Custom Amplicon v1.5) of Illumina MiSeq next-generation sequencer (Illumina, USA). The CDS (coding sequences)/exons, introns, 2000-bp URRs (upstream regulatory regions) and 1000-bp DRRs (downstream regulatory regions) of 55 MED genes were targeted for designing and synthesizing the custom oligo probes using Design Studio software (Illumina, USA). All the probes were pooled into a custom amplicon tube to produce amplicons with an average size of 400 bp per reaction and template library was made using TruSeq Custom Amplicon Assay kit v1.5. The sample-specific indices were added to each library by PCR using common primers from the TruSeq Amplicon Index kit. The normalization of the uniquely tagged pooled amplicon libraries was performed and the generated clusters were sequenced by Illumina MiSeq platform. Illumina Amplicon Viewer was used to visualize the sequenced amplicons and sequence variants. The high-quality gene amplicons sequence reads of each accession were mapped to the pseudomolecules of reference Nipponbare rice genome (MSU, http://rice.plantbiology.msu.edu, Release 7.0) and non-erroneous high-quality SNPs were detected among accessions following methods of Saxena et al.57 and Kujur et al.69.
To ascertain the reliability and accuracy of identified SNPs, the genomic DNA of 24 rice accessions (selected from 384 low and high grain weight accessions) were PCR amplified with 55 MED gene-specific primers. The amplified PCR products were sequenced by automated 96 capillary ABI 3730xl DNA Analyzer (Applied Biosystems, USA). Subsequently, the high-quality gene sequences were aligned and compared to discover SNPs among accessions as per Saxena et al.70.
Association mapping
For phenotyping, 384 diverse rice accessions belonging to an association panel were grown in the field (as per randomised complete block design with two replications) for two consecutive years (2012 and 2013) during crop growing season at two diverse geographical locations (New Delhi-latitude 28°4′ N and longitude 77.1′ E and Tamil Nadu-11° N and 78 °E) of India. The accessions were phenotyped with replications for grain length (mm), grain width (mm) and grain weight (g) by measuring the weight of 1000 mature dried grains (at 10% moisture content) selected from 10–15 representative plants of each accession. The diverse statistical parameters, including frequency distribution, coefficient of variation (CV) and broad-sense heritability (H2) of grain size (grain length, width and weight) traits among accessions were estimated using SPSSv17.0 as per Bajaj et al.71. The determination of population genetic structure, PCA and LD decay among accessions using MED gene-derived SNPs was performed following Kujur et al.56.
For association mapping, the grain length, grain width and 1000-grain weight phenotypic and MED gene-based SNP genotyping information (5% MAF) as well as population structure ancestry coefficient (Q matrix), kinship matrix (K) and PCA (P) data of 384 rice accessions were integrated. MAF using the SNP genotyping data was measured using TASSEL v5.0 (http://www.maizegenetics.net/#!tassel/c17q9). Association analysis was performed using CMLM (compressed mixed linear model) and P3D (population parameters previously determined)/EMMAX (efficient mixed model association eXpedited) model-based approach of GAPIT as per Kujur et al.56 and Kumar et al.61. To ensure the accuracy of association outcomes, the relative distribution of observed and expected -log10(P)-value of each SNP marker-trait association was compared individually with their quantile-quantile plots. According to false discovery rate (FDR cut-off ≤0.05), the adjusted P-value threshold of significance was corrected for multiple comparisons. The potential SNP loci in the diverse coding and non-coding sequence components of MED genes revealing significant association with grain length, grain width and grain weight trait at a highest R2 (degree of SNP marker-trait association) and lowest FDR adjusted P-values (threshold P ≤ 10−5) were selected.
Validation of associated SNPs in a mapping population
To ascertain the potential of MED gene-derived SNPs for grain length, grain width and grain weight association, the trait-associated SNPs were selected to validate in a traditional bi-parental mapping population. For this, 10 of each low (Sonasal with 1000-grain weight: 10 g) and high (IR 64: 25 g) grain weight homozygous individuals derived from a F4 mapping population (IR 64 × Sonasal) along with parental accessions were selected for DNA isolation. The grain size/weight-associated SNPs exhibiting polymorphism between the mapping parents were genotyped in the selected 20 homozygous low and high grain weight mapping individuals using MALDI-TOF mass array SNP genotyping assay following Saxena et al.57,70. The correspondence of low and high grain size/weight-associated SNPs with their presence in the low and high grain weight homozygous mapping individuals was determined to validate the grain size/weight trait association potential of MED gene-derived SNPs.
Differential expression profiling
To determine the regulatory pattern of genes associated (validated by association analysis and in mapping population) with grain size/weight, the differential expression profiling of these genes was performed using the quantitative RT-PCR assay. The total RNA was isolated from three biological replicates of flag leaf (considered as control) and five different seed developmental stages (defined as per Agarwal et al.72 and Sharma et al.73) of four high/medium (Pusa Basmati 1121, IR 64, Nipponbare and LGR) and two low (Sonasal and Bindli) grain weight rice accessions as previously described74. The purified RNA was tested for quality by denaturing agarose gel electrophoresis and NANODROP 2000 Spectrophotometer (Thermo Scientific, NanoDrop products, USA). One μg of high quality total RNA was used for cDNA synthesis using first strand cDNA synthesis kit (Applied Biosystems, USA). The cDNA (1:100 dilution) along with 1X Fast SYBR Green Master Mix (Applied Biosystems) and 200 nM of forward and reverse gene-specific primers (Table S5) in a total reaction volume of 10 μl was amplified in quantitative RT-PCR assay by ViiA™ 7 Real-Time PCR system (Applied Biosystems). The normalization and differential expression were calculated as reported previously75.
Molecular haplotyping
For gene-based SNP haplotyping, the 2 kb URR, exon, intron and 1 kb DRR of grain weight-regulating candidate MED gene (validated by association analysis, in mapping population and expression profiling), amplified from 384 rice accessions (association panel) were cloned and sequenced as per Kujur et al.55 and Saxena et al.70. The high-quality MED gene sequences were aligned among accessions using the CLUSTALW multiple sequence alignment tool of MEGA v6.076 and SNPs in the genes were discovered. The genotyping data of MED gene-derived SNPs generated by cloned PCR amplicon sequencing and aforesaid Illumina targeted multiplex-gene amplicon resequencing among accessions, was used to constitute haplotypes within the gene. For gene haplotype-based association analysis, the SNP haplotype genotyping information in the MED gene was further correlated with 1000-grain weight phenotyping data of 384 rice accessions using aforementioned genetic association analysis strategy.
Additional Information
How to cite this article: Malik, N. et al. An Integrated Genomic Strategy Delineates Candidate Mediator Genes Regulating Grain Size and Weight in Rice. Sci. Rep. 6, 23253; doi: 10.1038/srep23253 (2016).
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the financial support for this study provided by a research grant (102/IFD/SAN/2161/2013-14) from the Department of Biotechnology (DBT), Government of India. NM and ND acknowledge the CSIR (Council of Scientific and Industrial Research) for Senior Research Fellowship awards.
Footnotes
Author Contributions N.M. and N.D. conducted all experiments and drafted the manuscript. A.K.S. actively participated in phenotyping of germplasm lines and mapping population of rice. P.A. and S.K.P. involved in SNP discovery, gene expression profiling and data analysis, and also assisted in manuscript writing. J.K.T. and A.K.T. conceived and designed the study, guided data analysis and interpretation, participated in drafting and correcting the manuscript critically and gave the final approval of the version to be published. All authors have read and approved the final manuscript.
References
- Huang R. et al. Genetic bases of rice grain shape: so many genes, so little known. Trends Plant Sci. 18, 218–226 (2013). [DOI] [PubMed] [Google Scholar]
- Zuo J. & Li J. Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu. Rev. Genet. 48, 99–118 (2014). [DOI] [PubMed] [Google Scholar]
- Xu J. L., Xue Q. Z., Luo L. J. & Li Z. K. Genetic dissection of grain weight and its related traits in rice (Oryza sativa L.). Chin. J. Rice Sci. 16, 6–10 (2002). [Google Scholar]
- Li Y. et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43, 1266–1269 (2011). [DOI] [PubMed] [Google Scholar]
- Meyer R. S. & Purugganan M. D. Evolution of crop species: genetics of domestication and diversification. Nat. Rev. Genet. 14, 840–852 (2013). [DOI] [PubMed] [Google Scholar]
- Fan C. et al. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112, 1164–1171 (2006). [DOI] [PubMed] [Google Scholar]
- Fan C., Yu S., Wang C. & Xing Y. A causal C-A mutation in the second exon of GS3 highly associated with rice grain length and validated as a functional marker. Theor. Appl. Genet. 118, 465–472 (2009). [DOI] [PubMed] [Google Scholar]
- Song X. J., Huang W., Shi M., Zhu M. Z. & Lin H. X. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39, 623–630 (2007). [DOI] [PubMed] [Google Scholar]
- Shomura A. et al. Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 40, 1023–1028 (2008). [DOI] [PubMed] [Google Scholar]
- Weng J. et al. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 18, 1199–1209 (2008). [DOI] [PubMed] [Google Scholar]
- Mao H. et al. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. USA 107, 19579–19584 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44, 950–954 (2012). [DOI] [PubMed] [Google Scholar]
- Zhang X. et al. Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. USA 109, 21534–21539 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru K. et al. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 45, 707–711 (2013). [DOI] [PubMed] [Google Scholar]
- Roeder R. G. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 579, 909–915 (2005). [DOI] [PubMed] [Google Scholar]
- Conaway R. C., Sato S., Tomomori-Sato C., Yao T. & Conaway J. W. The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem. Sci. 30, 250–255 (2005). [DOI] [PubMed] [Google Scholar]
- Malik S. & Roeder R. G. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 11, 761–772 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss Z. C., Ebmeier C. C. & Taatjes D. J. The Mediator complex and transcription regulation. Crit. Rev. Biochem. Mol. Biol. 48, 575–608 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway R. C. & Conaway J. W. The Mediator complex and transcription elongation. Biochim. Biophys. Acta 1829, 69–75 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. et al. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol. Cell 17, 683–694 (2005). [DOI] [PubMed] [Google Scholar]
- Malik S., Barrero M. J. & Jones T. Identification of a regulator of transcription elongation as an accessory factor for the human Mediator coactivator. Proc. Natl. Acad. Sci. USA 104, 6182–6187 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. J. et al. Mediator coordinates PIC assembly with recruitment of CHD1. Genes Dev. 25, 2198–2209 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H. et al. Human Mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 146, 92–104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B. et al. Functional association of Gdown1 with RNA polymerase II poised on human genes. Mol. Cell 45, 38–50 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. et al. Mediator complex regulates alternative mRNA processing via the MED23 subunit. Mol. Cell 45, 459–469 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nock A., Ascano J. M., Barrero M. J. & Malik S. Mediator-regulated transcription through the +1 nucleosome. Mol. Cell 48, 837–848 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jishage M. et al. Transcriptional regulation by Pol II(G) involving mediator and competitive interactions of Gdown1 and TFIIF with Pol II. Mol. Cell 45, 51–63 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukundan B. & Ansari A. Srb5/Med18-mediated termination of transcription is dependent on gene looping. J. Biol. Chem. 288, 11384–11394 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oya E. et al. Mediator directs co-transcriptional heterochromatin assembly by RNA interference-dependent and -independent pathways. PLoS Genet. 9, e1003677 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaschka C. et al. Architecture of the RNA polymerase II-Mediator core initiation complex. Nature 518, 376–380 (2015). [DOI] [PubMed] [Google Scholar]
- Ebmeier C. C. & Taatjes D. J. Activator-Mediator binding regulates Mediator-cofactor interactions. Proc. Natl. Acad. Sci. USA 107, 11283–11288 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagulapalli M., Maji S., Dwivedi N., Dahiya P. & Thakur J. K. Evolution of disorder in Mediator complex and its functional relevance. Nucleic Acids Res. 44, 1591–1612 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backstrom S., Elfving N., Nilsson R., Wingsle G. & Bjorklund S. Purification of a plant Mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol. Cell 26, 717–729 (2007). [DOI] [PubMed] [Google Scholar]
- Mathur S., Vyas S., Kapoor S. & Tyagi A. K. The Mediator complex in plants: structure, phylogeny, and expression profiling of representative genes in a dicot (Arabidopsis) and a monocot (rice) during reproduction and abiotic stress. Plant Physiol. 157, 1609–1627 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillmor C. S. et al. The MED12-MED13 module of Mediator regulates the timing of embryo patterning in Arabidopsis. Development 137, 113–122 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autran D. et al. Cell numbers and leaf development in Arabidopsis: a functional analysis of the STRUWWELPETER gene. EMBO J. 21, 6036–6049 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Guan H., Leal F., Grey P. H. & Oppenheimer D. G. Mediator subunit18 controls flowering time and floral organ identity in Arabidopsis. PLoS One 8, e53924 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsley P. A. et al. The Arabidopsis Mediator complex subunits MED16, MED14, and MED2 regulate Mediator and RNA polymerase II recruitment to CBF-responsive cold-regulated genes. Plant Cell 26, 465–484 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R. & Li Y. Control of final organ size by Mediator complex subunit 25 in Arabidopsis thaliana. Development 138, 4545–4554 (2011). [DOI] [PubMed] [Google Scholar]
- Sundaravelpandian K., Chandrika N. N. & Schmidt W. PFT1, a transcriptional Mediator complex subunit, controls root hair differentiation through reactive oxygen species (ROS) distribution in Arabidopsis. New Phytol. 197, 151–161 (2013). [DOI] [PubMed] [Google Scholar]
- Raya-Gonzalez J., Ortiz-Castro R., Ruiz-Herrera L. F., Kazan K. & Lopez-Bucio J. PHYTOCHROME AND FLOWERING TIME1/MEDIATOR25 regulates lateral root formation via auxin signaling in Arabidopsis. Plant Physiol. 165, 880–894 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd B. N. et al. The Mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21, 2237–2252 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R. et al. The Arabidopsis Mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24, 2898–2916 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canet J. V., Dobon A. & Tornero P. Non-recognition-of-BTH4, an Arabidopsis Mediator subunit homolog, is necessary for development and response to salicylic acid. Plant Cell 24, 4220–4235 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wang C., Zhang Y., Sun Y. & Mou Z. The Arabidopsis mediator complex subunit16 positively regulates salicylate-mediated systemic acquired resistance and jasmonate/ethylene-induced defense pathways. Plant Cell 24, 4294–4309 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan R. et al. HISTONE MONOUBIQUITINATION1 interacts with a subunit of the Mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 21, 1000–1019 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd B. N., Cahill D. M., Manners J. M., Schenk P. M. & Kazan K. Diverse roles of the Mediator complex in plants. Semin. Cell Dev. Biol. 22, 741–748 (2011). [DOI] [PubMed] [Google Scholar]
- Pasrija R. & Thakur J. K. Analysis of differential expression of Mediator subunit genes in Arabidopsis. Plant Signal. Behav. 7, 1676–1686 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta S. & Thakur J. K. Importance of Mediator complex in the regulation and integration of diverse signaling pathways in plants. Front. Plant Sci. 6, 757 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Li L. & Qu L. J. Plant Mediator complex and its critical functions in transcription regulation. J. Integr. Plant Biol. 58, 106–118 (2016). [DOI] [PubMed] [Google Scholar]
- Huang X. et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 42, 961–967 (2010). [DOI] [PubMed] [Google Scholar]
- Huang X. et al. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat. Genet. 44, 32–39 (2012). [DOI] [PubMed] [Google Scholar]
- Huang X. et al. A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K. et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat. Commun. 2, 467 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujur A. et al. Functionally relevant microsatellite markers from chickpea transcription factor genes for efficient genotyping applications and trait association mapping. DNA Res. 20, 355–374 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujur A. et al. A genome-wide SNP scan accelerates trait-regulatory genomic loci identification in chickpea. Sci. Rep. 5, 11166 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena M. S. et al. An integrated genomic approach for rapid delineation of candidate genes regulating agro-morphological traits in chickpea. DNA Res. 21, 695–710 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj D. et al. Genome-wide conserved non-coding microsatellite (CNMS) marker-based integrative genetical genomics for quantitative dissection of seed weight in chickpea. J. Exp. Bot. 66, 1271–1290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur J. K., Agarwal P., Parida S., Bajaj D. & Pasrija R. Sequence and expression analyses of KIX domain proteins suggest their importance in seed development and determination of seed size in rice, and genome stability in Arabidopsis. Mol. Genet. Genomics 288, 329–346 (2013). [DOI] [PubMed] [Google Scholar]
- Lipka A. E. et al. GAPIT: genome association and prediction integrated tool. Bioinformatics 28, 2397–2399 (2012). [DOI] [PubMed] [Google Scholar]
- Kumar V. et al. Genome-wide association mapping of salinity tolerance in rice (Oryza sativa). DNA Res. 22, 133–145 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz N. D. et al. REF4 and RFR1, subunits of the transcriptional coregulatory complex Mediator, are required for phenylpropanoid homeostasis in Arabidopsis. J. Biol. Chem. 287, 5434–5445 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz N. D. et al. Disruption of Mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature 509, 376–380 (2014). [DOI] [PubMed] [Google Scholar]
- Li W. et al. SAD1, an RNA polymerase I subunit A34.5 of rice, interacts with Mediator and controls various aspects of plant development. Plant J. 81, 282–291 (2015). [DOI] [PubMed] [Google Scholar]
- Guglielmi B. et al. A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res. 32, 5379–5391 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbon H. M. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 36, 3993–4008 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romera-Branchat M., Ripoll J. J., Yanofsky M. F. & Pelaz S. The WOX13 homeobox gene promotes replum formation in the Arabidopsis thaliana fruit. Plant J. 73, 37–49 (2013). [DOI] [PubMed] [Google Scholar]
- Pagnussat G. C. et al. Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132, 603–614 (2005). [DOI] [PubMed] [Google Scholar]
- Kujur A. et al. Employing genome-wide SNP discovery and genotyping strategy to extrapolate the natural allelic diversity and domestication patterns in chickpea. Front. Plant Sci. 6, 162 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena M. S. et al. Natural allelic diversity, genetic structure and linkage disequilibrium pattern in wild chickpea. PLoS One 9, e107484 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj D. et al. A combinatorial approach of comprehensive QTL-based comparative genome mapping and transcript profiling identified a seed weight-regulating candidate gene in chickpea. Sci. Rep. 5, 9264 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal P., Kapoor S. & Tyagi A. K. Transcription factors regulating the progression of monocot and dicot seed development. Bioessays 33, 189–202 (2011). [DOI] [PubMed] [Google Scholar]
- Sharma R. et al. Expression dynamics of metabolic and regulatory components across stages of panicle and seed development in indica rice. Funct. Integr. Genomics 12, 229–248 (2012). [DOI] [PubMed] [Google Scholar]
- Agarwal P. et al. Genome-wide identification of C2H2 zinc-finger gene family in rice and their phylogeny and expression analysis. Plant Mol. Biol. 65, 467–485 (2007). [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay P. & Tyagi A. K. OsTCP19 influences developmental and abiotic stress signaling by modulating ABI4-mediated pathways. Sci. Rep. 5, 9998 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.