Abstract
Background
Prominence of glycolysis in glioblastomas may be non-specific or a feature of oncogene-related subgroups (i.e. amplified EGFR, etc.). Relationships between amplified oncogenes and expressions of metabolic genes associated with glycolysis, directly or indirectly via pH, were therefore investigated.
Methods
Using multiplex ligation-dependent probe amplification, copy numbers (CN) of 78 oncogenes were quantified in 24 glioblastomas. Related expressions of metabolic genes encoding lactate dehydrogenases (LDHA, LDHC), carbonic anhydrases (CA3, CA12), monocarboxylate transporters (SLC16A3 or MCT4, SLC16A4 or MCT5), ATP citrate lyase (ACLY), glycogen synthase1 (GYS1), hypoxia inducible factor-1A (HIF1A), and enolase1 (ENO1) were determined in 22 by RT-qPCR. To obtain supra-glycolytic levels and adjust for heterogeneity, concurrent ENO1 expression was used to mathematically transform the expression levels of metabolic genes already normalized with delta-delta crossing threshold methodology.
Results
Positive correlations with EGFR occurred for all metabolic genes. Significant differences (Wilcoxon Rank Sum) for oncogene CN gains in tumors of at least 2.00-fold versus less than 2.00-fold occurred for EGFR with CA3's expression (p < 0.03) and for RNF139 with CA12 (p < 0.004). Increased CN of XIAP associated negatively. Tumors with less than 2.00-fold CN gains differed from those with gains for XIAP with CA12 (p < 0.05). Male gender associated with CA12 (p < 0.05).
Conclusions
Glioblastomas with CN increases in EGFR had elevated CA3 expression. Similarly, tumors with RNF149 CN gains had elevated CA12 expression.
General significance
In larger studies, subgroups of glioblastomas may emerge according to oncogene-related effects on glycolysis, such as control of pH via effects on carbonic anhydrases, with prognostic and treatment implications.
Abbreviations: CN, copy number; DAPI, diaminephylindole; ddCt, delta-delta crossing threshold; GB, glioblastoma; GOI, gene of interest; HKG, housekeeping gene; IRES, internal ribosome entry site; MLPA, multiplex ligation-dependent probe amplification; MPNST, malignant peripheral nerve sheath tumor; MTB/GF, metabolic/growth factor; NB, normal brain; RT-qPCR, real time quantitative PCR; REMBRANDT, Repository of Molecular Brain Neoplasia Database; SLC, solute carrier; WHO, World Health Organization
Keywords: Amplified oncogenes, Glycolysis, Carbonic anhydrase, EGFR, RNF139, XIAP
Graphical abstract
Highlights
-
•
PCR of glioblastomas show oncogene copy numbers relate to metabolic gene expressions.
-
•
ENO1(ENOLASE1) transformations yielded “supra-glycolytic” metabolic gene expressions.
-
•
EGFR, RNF139, and XIAP associated with expressions of two carbonic anhydrase genes.
-
•
Male gender associated (+) with the transformed expression of carbonic anhydrase 12.
-
•
Oncogene amplifications may aid control of pH to protect glycolysis in glioblastomas.
1. Introduction
Increased reliance on glycolysis for ATP is a defining feature of malignancies, including the incurable, high-grade brain tumors known as glioblastomas. Enzymes of the glycolytic pathway in tumors are protected under conditions that are inhospitable for normal cells. In previous studies of glioblastoma cells, we identified their pseudopodia as being glycolytic subcellular domains that also contain oncogene products, including increased Met [1] and phosphorylated EGFR. Discovering increased anti-phosphotyrosine and anti-phosphoserine/threonine-reactive bands on immunoblots of pseudopodia compared to whole cells suggested to us that multiple oncogenes could be present in glycolytic regions of these cells (unpublished). In view of the functional roles and associations of the proteins encoded by oncogenes in enhancement of tumors and their malignant behavior, oncogenes that are amplified are logical candidates to investigate for aiding glycolysis. Oncogenes and their products potentially increase protection of glycolytic enzymes from the protons released by non-aerobic production of ATP involving lactic acid.
Copy numbers (CNs) of amplified oncogenes and expression levels of metabolic genes can be determined in the same tumors to detect functional associations. PCR techniques quantify gene expression and CN with impressive sensitivity. Heterogeneity within each tumor due to a malformed vasculature, etc. leads to fluctuation in metabolism. To account for heterogeneity in levels of glycolytic activity and obtain supra-glycolytic levels, the expression results for metabolic genes in tumor samples were transformed mathematically to levels indicated by expression of ENO1 that encodes enolase 1, the major isoform of enolase in glioblastomas. Enolase catalyzes the ninth of ten steps in the traditional glycolytic pathway. The expression of ENO1 in tumors, given as a value relative to normal brain expression, was used to scale the expression levels of other metabolic genes in the same tumor. The metabolic genes' expression levels were transformed to be multiples of concurrent ENO1 levels. Studies have shown that ENO1 is seldom lost in high grade brain tumors. ENO1 was retained in 98.6% of glioblastomas with some variation in expression levels noted [2]. Expressing each metabolic gene as a multiple of ENO1's expression level in the same sample led to the discovery of positive correlations between oncogene CNs and the expression of metabolic genes in this study. Possibly glioblastomas harbor functional subgroups defined by changes in CN of oncogenes.
Not surprisingly, including control of pH among the functional choices of genes to study yielded significant results. Warburg utilized large amounts of bicarbonate in his model of cancer [3]. The bicarbonate buffer system is the most important physiologic buffer. It controls the body's pH via independent regulation of its components, carbon dioxide and bicarbonate, by the lungs and kidneys, respectively, thus providing tremendous capacity [4]. Determining how tumors take advantage of the circulating 4300 mmoles of bicarbonate that the kidneys filter and mostly recycle daily [5] is urgently needed. Our study identified multiple associations between EGFR, RNF139, and XIAP oncogenes and gender with expression levels of two carbonic anhydrases that catalyze interconversions among components of the bicarbonate buffer system.
2. Materials and methods
2.1. Patients in the study
With patient and Institutional Review Board approval, frozen samples of glioblastomas resected from 25 adult patients during the time interval, 2002–2008, were obtained from the University of Pittsburgh Brain Tumor Tissue Bank. Clinical information for 24 patients included a mean age of 62.9 yr., range 19–81 yr., M:F = 11:13, and data for specific patients are listed (Table 1). Only one case, GB25, was unsatisfactory for all parts of the study.
Table 1.
Clinical information regarding glioblastoma (GB) patients.
| GB | Age (yrs) |
Sex | GB | Age (yrs) |
Sex |
|---|---|---|---|---|---|
| 01 | 36 | M | 13 | 66 | M |
| 02 | 57 | M | 14 | 53 | M |
| 03 | 77 | F | 15 | 69 | F |
| 04 | 58 | F | 16 | 19 | M |
| 05 | 43 | F | 17 | 78 | F |
| 06 | 59 | F | 18 | 65 | M |
| 07 | 74 | F | 19 | 62 | F |
| 08 | 69 | F | 20 | 69 | F |
| 09 | Unka | Unk | 21 | 80 | M |
| 10 | 46 | M | 22 | 81 | F |
| 11 | 69 | M | 23 | 55 | F |
| 12 | 64 | M | 24 | 80 | M |
| 25 | 81 | F | |||
Unknown data.
2.2. Multiplex ligation-dependent probe amplification (MLPA) analysis of oncogenes
Copy number (CN) was determined for each of seventy-eight oncogenes (AKT1, AURKA, BCAR3, BCAS1 and 2, BCL2, BCL2A1, BCL2L1, 11, 13, and 14, BCL6, BCLG, BIRC2, 3, and 5, BRAF, BRMS1, CCNA1, CCND1 and 2, CCNE1, CDK4 and 6, CENPF, CTTN, CYP27B1, EGFR, ERBB2 and 4, ESR1, EVI1, FGF3 and 4, FGFR1, GNAS, GSTP1, HMGA1, IGF1R, IGFBP2, 4 and 5, IRS2, JAK2, MDM2 and 4, MET, MMP7, MOS, MYCL, MYBL1 and 2, MYC, MYCN, NAIP, NFKBIE, NRAS, NTRK1-3, PDGFRA and B, PIK3C2B, PIK3CA, PPM1D, PSMB4, PTK2, PTP4A3, PTPN1, RELA, RNF139, RUNX1, SERBINB2, 7, and 9, TERT, TOM1L2, UCKL1, and XIAP). Multiplex ligation-dependent probe amplification (MLPA) was successfully performed in 24 tumors (GB01-GB24) to obtain CN data. Normal brain (NB) from the occipital lobe of an 82 yr. old female (Biochain, Hayward, CA) was used to normalize tumor DNA results for each oncogene's CN. All tumors were analyzed concurrently with NB in the MLPA assays. Briefly, in MLPA, DNA (200 ng) from tumor samples and NB was denatured at 98 °C and hybridized with MLPA probes (P171, P172, P173 Tumor Gains kits, MRC-Holland, The Netherlands) for genes that are known to be amplified in tumors. Hybridization occurred during 16–20 h at 60 °C. Ligations with Ligase65 (MRC-Holland) were at 54 °C for 15 min followed by 98 °C for 5 min. Ligation products were amplified with PCR primers and polymerase for 35 cycles (30 s 95 °C, 30 s 60 °C, 60 s 72 °C) and then 20 min at 72 °C at the end using a thermocycler (MasterCycler personal Eppendorf, Hamburg, GM). PCR products were separated and analyzed via capillary electrophoresis using the SS600 Fragment Analysis option with fluorescent signal detection on a CEQ8000 instrument (Beckman-Coulter, Fullerton, CA) using the manufacturer's reagents and size standards. Stuffer sequences incorporated into PCR probes of specific lengths identified the amplified products of PCR reactions in regard to each gene. Peak heights reflected initial amounts of DNA. Slightly less efficient PCR of longer amplicons accounted for minor reductions in peak heights that were corrected with NB by dividing tumor oncogene CN by NB CN for the same oncogene. These methods have been previously described more fully [6], [7]. Note that in earlier studies BCL2L14 was listed as BCLG/BCL2L14, CTTN as EMS, NAIP as BIRC1, XIAP as BIRC4, and UCKL1 as FLJ20517. Some cytogenetic loci for various genes have also changed and are given in this paper as they are currently listed on the Online Mendelian Inheritance in Man (OMIM) or HUGO Gene Nomenclature Committee (HGNC) websites, http://www.omim.org/ and http://www.genenames.org/, respectively.
2.3. RT-qPCR for metabolic and growth factor (MTB/GF) gene expression
Numerous genes encoding proteins that favor glycolysis under adverse conditions, including acidosis and intermittent disconnection from the blood supply (i.e. invasive cell migration), were considered among genes whose encoded proteins enable glycolysis during hypoxia. Using early (2007–2008) versions of the Repository of Molecular Brain Neoplasia Database (REMBRANDT) [8], ENO1 was chosen to represent the expression levels of genes encoding enzymes in the energy generation phase of glycolysis and then served to transform expressions of other metabolic genes and growth factor genes. Nine other genes (ACLY, CA3, CA12, GYS1, HIF1A, LDHA, LDHC, SLC16A3 (or MCT4), and SLC16A4) were chosen as genes of interest (GOI) from the functionally relevant metabolic groups of genes. Two growth factors, EGF and PDGFA, were also chosen for this study. For each group of genes (glucose transport, glycolysis, mitochondria, links between metabolic pathways, fat metabolism, control of pH/lactate, glycogen metabolism, gluconeogenesis, and growth factors) and for each individual gene, the prevalence of 2-fold or greater elevations in glioblastomas, the likelihood of a glioma being a glioblastoma if an elevation in expression was present, significantly poorer survival in glioblastomas with elevations (see Section 3.2), as well as functional relevance based on previous studies, were considered in compiling a list of 12 metabolic/growth factor (MTB/GF) genes to study for associations with oncogenes. Data for the MTB/GF genes in their respective groups and the controls are given in Section 3.2. Since ENO1 was used for MTB/GF gene transformations and SLC16A4's locus was potentially within an oncogene amplicon (see Section 3.1), data is reported most comprehensively for eight of the ten metabolic genes. The RNA-derived cDNAs were prepared from tumor samples and the reference pooled non-malignant brain RNA from 23 adults, mean 68.3 yr., range 23–86 yr. of age. Sufficient RNA was obtained from 22 tumors (GB01-08 and GB10-23) for analysis (M:F = 10:12, 19–80 yr. of age). Template cDNAs in each tumor and the reference were tested in triplicate for quantitative expression levels of MTB/GF genes and two housekeeping genes (HKGs), which were ACTB and GAPDH on 96 well plates (SABioSciences Corp. Qiagen, Valencia, CA), using an ABI 7500 Prism thermocycler (Applied BioSystems, ThermoFisher Scientific, Waltham, MA). The delta-delta crossing threshold (ddCt) method was used to determine fold increases in the tumors compared to reference derived cDNA with averaged HKGs used for normalization. Each dCt equaled Ct(MTB/GF)—average Ct(HKG). The ddCt for each gene equaled dCt(tumor sample) − dCt(reference non-malignant brain). The fold-change for each MTB/GF gene compared to the normal reference was equal to 2− ddCt. This methodology and the HKGs have been more fully described [9]. After normalizing with the HKGs, MTB/GF gene expression levels were transformed (division by concurrent expression of ENO1). Results for each MTB/GF gene were used for comparisons with CN of oncogenes in Fisher's Exact tests, correlations, and Wilcoxon Rank Sum comparisons.
2.4. Detection of MTB gene relationships with the presence of any amplified oncogenes
Tumors were initially stratified according to whether they had oncogene(s) amplified at 3.00-fold or greater levels (Yes or No) to facilitate analysis with Fisher Exact tests. The results for each MTB/GF gene were used to determine tumor membership in Group (+) if the MTB/GF gene had ≥ 1.10 × (times) ENO1 expression and Group (−) if the MTB/GF gene had < 1.10 × ENO1 expression levels. The cut-off of 1.10 × expression of ENO1 was chosen to form the groups with no other cut-offs attempted to avoid selection bias. The 1.10 × threshold was used only for the Fisher Exact tests reported in Section 3.4.
2.5. EGFR fluorescence in situ hybridization (FISH)
A frozen tissue sample available on GB06 was evaluated for EGFR signals with fluorescence in situ hybridization (FISH). The FISH probes for the EGFR band region, 7p11.2–7p12 and a control locus, 7p11.1–q11.1, D7Z1, (Vysis Locus Specific Identifier EGFR SpectrumOrange and CEP 7 SpectrumGreen, respectively, Abbott Molecular Inc., Des Plaines, IL) were hybridized to interphase nuclei in a 5 μm section overnight. Un-hybridized probes were washed away. Diaminophylindole (DAPI) fluorescent blue (Abbott Molecular) stained the nuclei. Slides were scanned on a fluorescent microscope (Leica DMR, Wetzlar, GM) for analysis with images captured using a digital camera (Applied Imaging, San Jose, CA) and CytoVision v4.02 imaging software (Applied Imaging). The methodology was described in a previous study [6].
2.6. Statistical tests
The Fisher's Exact tests were described in Section 2.4. Pearson's correlation coefficients were calculated to detect positive and negative correlations between expression values of MTB/GF genes and CN of oncogenes. Correlations were also performed for 3 genes in the 8q amplicon with each other. Wilcoxon Rank Sum comparisons were performed to detect associations with metabolic gene expression in the group of oncogenes defined by (1) exhibiting at least one 3.00-fold CN gain among all the tumors and (2) whose data yielded two groups of comparable sizes (10 and 12, 9 and 13, or 8 and 14 tumors) when oncogene CN gains of 2.00-fold or greater were considered. Associations with metabolic gene expressions have p-values stated for trends and significance levels less than 0.05. Also, chi square analyses were performed on tumors in the REMBRANDT database during the selection of genes for expression analysis, as described in the Results section (end of Section 3.2).
3. Results
3.1. Oncogene amplifications detected with MLPA
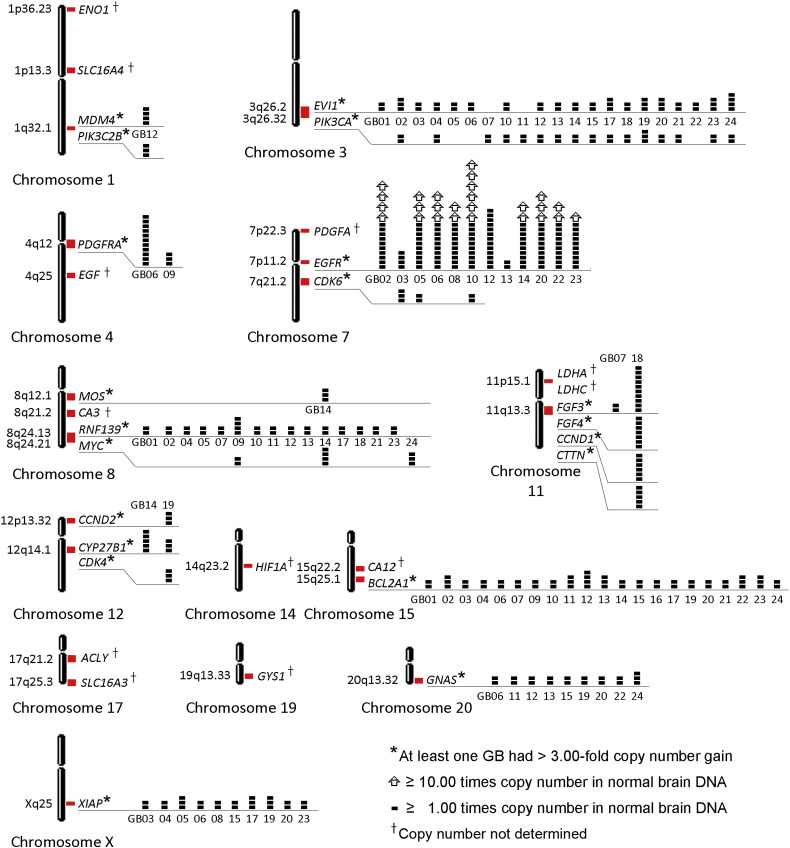
Twenty-four glioblastomas (GB1–GB24) assayed with MLPA for the CN levels of 78 oncogenes yielded amplifications, 3.00-fold or greater, in 18 glioblastomas for one or more of twenty oncogenes, listed as follows: BCL2A1, CCND1, CCND2, CDK4, CDK6, CTTN, CYP27B1, EGFR, EVl1, FGF3, FGF4, GNAS, MDM4, MOS, MYC, PDGFRA, PIK3C2B, PIK3CA, RNF139, and XIAP. Low level gains in CN that did not achieve a 3.00-fold threshold also occurred frequently. For the twenty oncogenes with at least one 3.00-fold amplification in the tumors, all of the CN gains for each one that were 2.00-fold or greater are shown at their chromosomal loci (Fig. 1). Locations of MTB/GF genes for which only RNA expression data were determined in the study are also included. Each of the 24 glioblastomas tested had at least one of the CN gains shown. Genes not amplified at or beyond the 3.00-fold threshold in at least one tumor are not shown. Tumors with CN gains of EGFR also had CN gains of one to seven other oncogenes among those shown (Fig. 1). Twelve genes, whose gains were always below 3.00-fold among the glioblastomas, included NRAS (1p13.2), BCAS2 (1p13.2), ERBB4 (2q34), BCL6 (3q27.3), NAIP (5q13.2), MET (7q31.2), BRAF (7q34), MDM2 (12q15), PPM1D (17q23.2), BCL2L1 (20q11.21), AURKA (20q13.2), and RUNX1 (21q22.12), in at least one of 23 tumors. Of these, MDM2 was co-amplified along with CYP27B1 and CDK4 nearby on chromosome 12 in GB19, and with only CYP27B1 in GB14. These findings are consistent with the complexity of the 12q13-15 amplicon described in glioblastomas [10]. Also, AURKA was co-amplified with nearby GNAS on chromosome 20 in GB06.
Fig. 1.
Loci of amplified oncogenes and metabolic/growth factor genes. The chromosomal loci (red rectangles) of twenty oncogenes amplified at least 3.00-fold and metabolic/growth factor (MTB/GF) genes are shown. The copy number (CN) amplification of oncogenes in each glioblastoma (GB) are depicted with small solid black rectangles (1.00 times normal per rectangle) and open arrows (10.00 times normal per arrow) as indicated in comparison to normal brain DNA. The highest CN gains occurred for EGFR. The superscript (†) indicates the MTB/GF genes with expression data but no CN data.
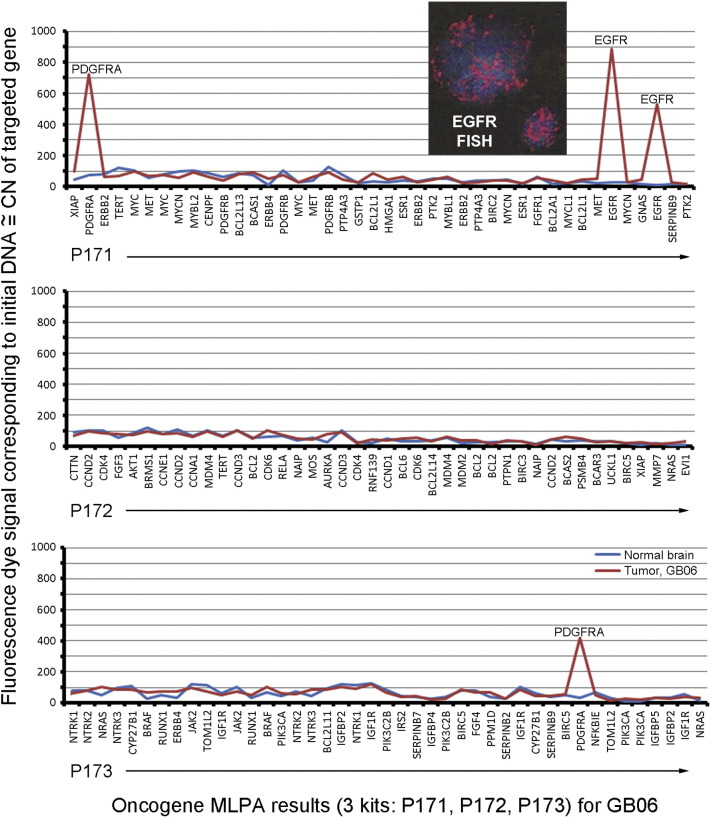
It was noted that near or at the locus for SLC16A4 (1p13.3), CN gains occurred for oncogenes, NRAS (1p13.2) and BCAS2 (1p13.3). Although results for SLC16A4 are included when those for all the MTB/GF genes are listed in tables, they are not shown when results of metabolic genes are highlighted as a group (eight genes remaining) in the following figures due to the possibility that SLC16A4 was a bystander in an amplicon containing the oncogenes, NRAS and BCAS2. Also, deletions of oncogenes detected are not reported in this study. An example of MLPA amplification results for PDGFRA and EGFR are shown for GB06 with FISH signals for EGFR in the frozen sample included as an insert (Fig. 2).
Fig. 2.
Quantified oncogenes in GB06. An example of multiplex ligation-dependent probe amplification (MLPA) results for 78 oncogenes in one glioblastoma. Its high level amplifications of PDGFRA and EGFR were readily detected as fluorescence signals from each of the 2 sets of probes used for each oncogene. The two-fold elevations in tumor versus normal brain were detected using mathematical comparisons described in Section 2.2. The red and blue lines represent tumor and normal DNA, respectively. Also, the inset at the top shows fluorescence in situ hybridization (FISH) signals for EGFR (red) in two cells of the frozen tissue sample used for MLPA. The nuclei stained blue with diaminophylindole (DAPI).
3.2. Expression of metabolic/growth factor (MTB/GF) genes in REMBRANDT
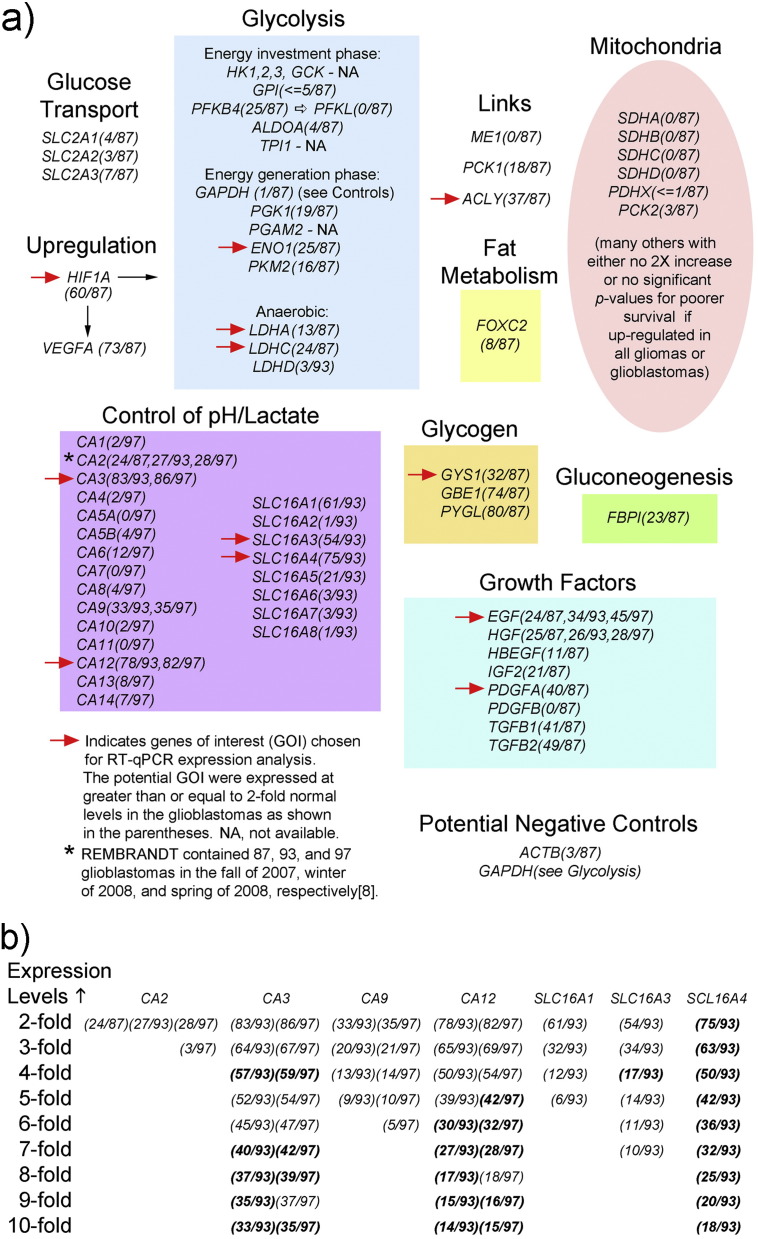
Specific members of functional groups and gene families were chosen for study of their expression levels in samples of glioblastomas based on queries of the REMBRANDT database of brain tumors [8] and their potential support of glycolysis (Fig. 3a).
Fig. 3.
Searches for MTB/GF genes of interest (GOI) in the REMBRANDT database. Genes were chosen based on functional relevance and REMBRANDT [8] expression data. Section 3.2 explains the selection of GOI indicated with red arrows in detail. (a) Functional groupings. (b) Utilization of expression levels above two-fold in selection of GOI in the carbonic anhydrase and solute carrier 16 families of genes. Expression levels that had significant p-values (< 0.05) for poorer survival (Kaplan Meier) are bolded. The full gene names are as follows: ACLY (ATP citrate lyase), ACTB (actin B), ALDOA (aldolase A), CA (carbonic anhydrase), EGF (epidermal growth factor), ENO1 (enolase 1), FBP1 (fructose 1,6 bisphosphatase), FOXC2 (forkhead box C2), GAPDH (glyceraldehyde-3-phosphate dehydrogenase), GBE1 (glycogen binding enzyme), GCK (glucokinase), GPI (glucophosphoisomerase), GYS1 (glycogen synthase 1), HBEGF (heparin-binding EGF like growth factor), HGF (hepatocyte growth factor), HIF1A (hypoxia-inducible factor 1, alpha subunit), HK (hexokinase), IGF2 (insulin-like growth factor II), LDH (lactate dehydrogenase), ME1 (malic enzyme 1), PCK (phosphoenolpyruvate carboxykinase), PDGF (platelet-derived growth factor), PDHX (pyruvate dehydrogenase complex, component X), PFKFB4 (6-phosphofructo-2-kinase/fructose-2,6 bisphosphatase 4), PGAM2 (phosphoglycerate mutase 2), PGK1 (phosphoglycerate kinase 1), PFKL (phosphofructokinase L), PKM2 (pyruvate kinase, muscle, 2), PYGL (glycogen phosphorylase, liver), SDH (succinate dehydrogenase complex), SLC2 (solute carrier family 2), SLC16 (solute carrier family 16), TGFB (transforming growth factor, beta), and TPI1 (triosephosphate isomerase 1). Note that SCL16A3 is also known as MCT4 (monocarboxylate transporter 4).
The highest proportions of 2-fold increased gene expressions occurred among the following functional groups: the energy generation phase of glycolysis, anaerobic glycolysis, upregulation of glycolysis, links between metabolic pathways, control of pH/lactate, glycogen metabolism, and growth factors. Within four of these groups, ENO1, LDHC, CA3, and ACLY had the greatest numbers of elevations for expression.
Additionally, in regard to the anaerobic glycolysis group, elevations of LDHA expression levels at least 2-fold were present in 14 of all the gliomas so that if it was elevated in a glioma, there was a 92.9% chance that the tumor was a glioblastoma. The chances of a glioma with 2-fold or greater elevations of LDHC or ENO1 for being a glioblastoma were 85.7% and 89.3%, respectively. Therefore LDHA was also included. Results for CA12 and SCL16A4 (Fig. 3b) were comparable to CA3's results, along with impressive significantly poorer survival results at high expression levels, therefore they were also included. The well-known function of the protein encoded by SLC16A3 (better known as MCT4) and occurrence of its results within the range of the other chosen members in this functional group warranted its inclusion.
Within the glycogen metabolism group, GYS1, GBE, and PYGL had at least 2-fold elevations in 36, 101, and 114 gliomas, respectively, with poorer Kaplan Meier survival in all gliomas for each of the three genes at such significant levels that their p-values were all given as 0.0. The chances of the elevations occurring in a glioblastoma, when present in a glioma, were 88.9%, 73.3%, and 70.2%, respectively, for GYS1, GBE1, and PYGL. With this data, along with functional data reported previously [11], GYS1 was chosen to study. Elevations of at least 2-fold expression among the growth factors, EGF, PDGFA, TGFB1, and TGFB2 occurred in 32, 52, 58, and 66, respectively, of all gliomas so the tumors with elevations were also glioblastomas at rates of 75.0%, 76.9%, 70.7%, and 74.2%, respectively. Considering this data along with the abundance of literature supporting their roles in brain tumors, EGF and PDGFA, were chosen. Also, later searches showed that increased expression of EGF was present in a higher percentage of glioblastomas than the 2007 REMBRANDT data indicated. The well-known regulator of glycolysis in hypoxia, HIF1A, was also included. Functions of the proteins encoded by the MTB/GF and control genes are given (Table 2).
Table 2.
Functions of proteins encoded by metabolic and growth factor (MTB/GF) genes selected for expression analysis and studies for associations with oncogenes.
| 1. ACLY, ATP citrate lyase | Cleaves citric acid, synthesizes acetyl CoA |
| 2. CA3, Carbonic anhydrase III | Interconversions of CO2/HCO3− in cytoplasm |
| 3. CA12, Carbonic anhydrase XII | Interconversions of CO2/HCO3− at cell surface |
| 4. ENO1, Enolase 1* | Catalyzes ninth of ten steps in glycolysis |
| 5. GYS1, Glycogen synthase 1 | Synthesis of glycogen from simpler carbohydrates |
| 6. HIF1A, Hypoxia-inducible factor 1, alpha subunit | Transcription factor, upregulates genes in hypoxia |
| 7. LDHA, Lactate Dehydrogenase A | Interconversion of lactic acid and pyruvate |
| 8. LDHC, Lactate Dehydrogenase Cǂ | Interconversion of lactic acid and pyruvate |
| 9. SLC16A3, Solute carrier family 16 (monocarboxylic acid transporter), member 3. See protein name |
Transports lactic acid and pyruvate across membranes, protein called monocarboxylate transporter 4 (MCT4) |
| 10. SLC16A4, Solute carrier family 16 (monocarboxylic acid transporter), member 4. See protein name |
Transports lactic acid and pyruvate across membranes, proteincalled monocarboxylate transporter 5 (MCT5) |
| 11. EGF, Epidermal Growth Factor | Differentiation, etc. |
| 12. PDGFA,Platelet-derived growth factor, alpha polypeptide |
Mesenchymal cell mitogenesis, etc. |
| 13. ACTB, Actin B | Structural, Housekeeping gene (HKG) for RT-qPCR |
| 14. GAPDH, Glyceraldehyde-3- phosphate dehydrogenase |
Glycolysis, Housekeeping gene (HKG) for RT-qPCR |
ENO1 was used to transform the expression levels of the other MTB/GF genes.
LDHC gene expression was originally thought to be limited to testes but has been found in various types of malignancy.
In support of these choices, significantly poorer Kaplan Meier survival in all gliomas was reported in the database for ACLY, GYS1, ENO1, and HIF1A, at 2 or 3-fold levels of increased gene expression. The other chosen metabolic genes, CA3, CA12, SLC16A3 (or MCT4), SCL16A4, LDHA, and LDHC, had significantly poorer survival in glioblastomas with increased expression at levels that varied from 2 to 10-fold above normal. Also, EGF and PDGFA had significantly poorer survival associated with elevated expression levels in all gliomas and glioblastomas and in all gliomas, respectively. Additionally, by chi square analyses the likelihood for elevated levels of the genes to occur in a glioblastoma that also contained a 2-fold elevation in expression of ENO1 (our putative indicator of glycolytic activity) was p < 0.05 for ACLY, GYS1, SLC16A4, LDHA, and HIF1A. Trends for PDGFA, LDHC, SLC16A3 (or MCT4), and EGF occurred with p-values of 0.05, 0.05, 0.14, and 0.16, respectively. Specific p-values for the expression of CA3 at 3-fold, 7-fold, and 10-fold elevations to occur in tumors with 2-fold elevations in ENO1's expression were 0.48, 0.27, and 0.25, respectively. The downward trend in p-values for fold elevations of CA3 extending to the highest limit in the database was valued. The p-values for expression of CA12 at 2-fold, 3-fold, and 5-fold elevations to occur in tumors with 2-fold elevations in ENO1's expression were 0.04, 0.002, and 0.0001, respectively.
3.3. MTB/GF gene expression mathematically transformed by concurrent ENO1 levels
RNA from 22 glioblastomas in this study yielded results that could be analyzed with RT-qPCR. One of the unsatisfactory specimens (GB25) was also unsatisfactory for MLPA when determining CN of oncogenes. The concurrent values for expression of ENO1, ranging from 0.222 to 1.972 times normal, were used to transform the expression of each MTB/GF gene to multiples of ENO1 to reflect their levels relative to glycolytic pathway activity to obtain the supra-glycolytic levels in adjusting for tumor heterogeneity. The range of gene expression for pathway activity was assumed to range from 0.222 to 1.972 times normal according to the values of ENO1. In a previous study the expression of ACLY in this group of tumors was shown to correlate with concurrent expression of ENO1 with no transformation performed and minus one outlier [9]. The mean expressions of MTB/GF genes with 95% confidence intervals (CI) are shown for all 22 glioblastomas (Fig. 4a).
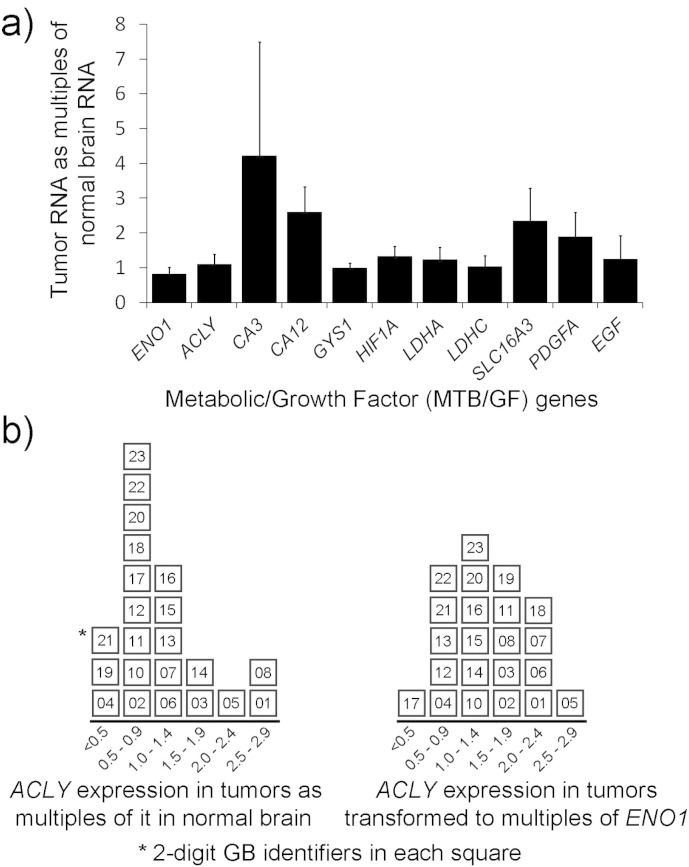
Fig. 4.
Expressions of MTB/GF genes and the effect of ENO1 transformation on ACLY in considering ENO1 as a candidate for MTB/GF transformations. (a) Comparisons of MTB/GF gene expressions, including ENO1, prior to transformations in 22 glioblastomas (GBs). Normal brain RNA was from pooled non-malignant brain tissue. Each gene's fold change was obtained with the ddCt method described in Section 2.3. Means with 95% confidence intervals are shown for each gene. (b) Effects of ENO1 when used to transform a metabolic gene's expression data. On the left, the distribution of GBs, each identified with a 2-digit number, is displayed according to ACLY's expression in each as a fold-change compared to normal brain's ACLY RNA. The distribution shown on the right is according to each tumor's expression of ACLY following transformation (division by concurrent ENO1 expression).
The medians were within the lower halves of the 95% CI shown, except for GYS1 and SLC16A3 (or MCT4). GYS1's median was in the upper half of the 95% CI shown. SLC16A3's median was not in the gene's 95% CI and its 1st, 2nd (median), and 3rd quartiles were 0.668, 1.378, and 3.340, respectively. CA3 with the largest degree of variability shown had 1st, 2nd, and 3rd quartiles of 0.822, 1.019, and 2.568, respectively. The distributions of 22 individual glioblastomas according to their expression of ACLY, before and after transformation with concurrent ENO1, are shown (Fig. 4b). The transformation produced results in the same range with GBs more evenly distributed. However, only 5 tumors retained the same level of expression and 7 shifted more than 1 level of expression that is indicated on the x-axis. Distributions of the glioblastomas according to their expressions of eight metabolic genes, after transformation with concurrent ENO1 expressions can be compared to ACLY's distribution (Fig. 5).
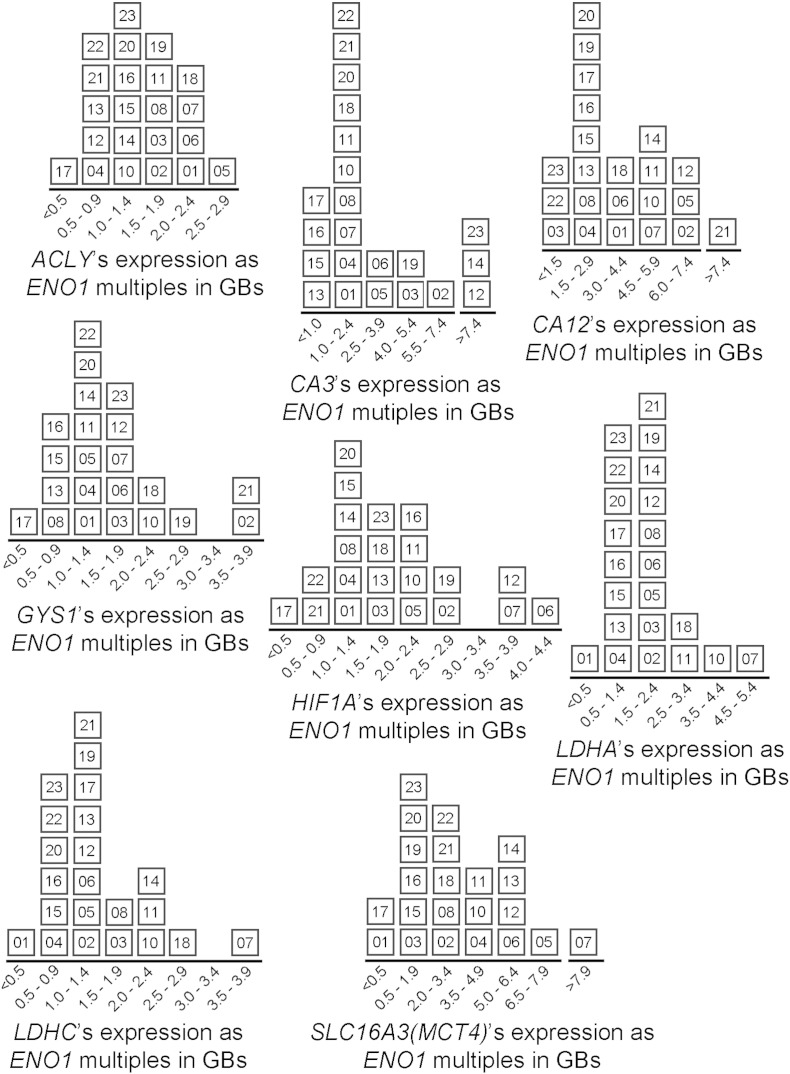
Fig. 5.
Distributions of glioblastomas according their expressions of each metabolic gene following mathematical transformation by ENO1. This was described for ACLY (Fig. 4b). Each tumor is identified by a 2-digit number. The highest transformed values occurred for CA3, CA12, and SCL16A3 (or MCT4).
3.4. Fisher's exact tests of MTB/GF gene expression with oncogene copy number gains
Expression levels of the MTB/GF genes were transformed so that levels above those attributed to ongoing glycolysis in a heterogeneous tumor environment could be analyzed. After transforming their levels to represent multiples of ENO1's concurrent expression, Groups (+) and (−) were obtained for each MTB/GF gene based on whether its expression level was equal to or greater than 1.10 times the expression of ENO1 or not, respectively. Statistical significance, p = 0.041 (Fisher's Exact), for a difference was found for LDHC's Group (+) as shown (Table 3, bottom row).
Table 3.
Stratification of each glioblastoma(GB) by the presence of amplified oncogenes and by expression levels of transformed metabolic/growth factor (MTB/GF) genes listed in the top row. Each MTB/GF level was divided by concurrent expression of ENO1 to become a multiple of ENO1 expression. If the result was ≥ 1.10 (arbitrary threshold), then the GB is included in Group (+) and otherwise is in Group (−). A significant (*p < 0.05) association for LDHC with the presence of amplified oncogenes and a trend for LDHA were detected in the GBs.
| GB | Amplified oncogenes | LDHC | LDHA | CA3 | ACLY | GYS1 |
SLC16A3 (MCT4) |
SLC16A4 (MCT5) |
HIF1A | PDGFA | EGF | CA12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | No | − | − | − | + | + | − | + | + | − | − | + |
| 02 | Yes | + | + | + | + | + | + | + | + | + | + | + |
| 03 | Yes | + | + | + | + | + | − | − | + | + | + | − |
| 04 | No | − | − | + | − | − | + | − | − | + | + | + |
| 05 | Yes | + | + | + | + | + | + | + | + | + | + | + |
| 06 | Yes | + | + | + | + | + | + | + | + | + | + | + |
| 07 | No | + | + | + | + | + | + | + | + | + | + | + |
| 08 | Yes | + | + | + | + | − | + | − | − | + | − | + |
| 10 | Yes | + | + | + | + | + | + | + | + | + | − | + |
| 11 | Yes | + | + | + | + | + | + | + | + | + | − | + |
| 12 | Yes | + | + | + | − | + | + | + | + | − | − | + |
| 13 | Yes | − | + | − | − | − | + | − | + | − | − | + |
| 14 | Yes | + | + | + | + | − | + | + | + | − | + | + |
| 15 | No | − | − | − | − | − | − | − | + | + | + | + |
| 16 | No | − | − | − | + | − | − | − | + | − | − | + |
| 17 | Yes | − | − | − | − | − | − | + | − | − | − | + |
| 18 | Yes | + | + | + | + | + | + | + | + | + | + | + |
| 19 | Yes | + | + | + | + | + | + | − | + | − | − | + |
| 20 | Yes | − | − | − | + | + | − | − | + | + | − | + |
| 21 | No | − | + | + | − | + | + | + | − | + | − | + |
| 22 | Yes | − | − | − | − | + | + | + | − | − | − | − |
| 23 | Yes | − | − | + | + | + | − | + | + | + | + | + |
| Total in Group (−) | 10 | 8 | 7 | 7 | 7 | 7 | 8 | 5 | 8 | 12 | 2 | |
| Total in Group (+) | 12 | 14 | 15 | 15 | 15 | 15 | 14 | 17 | 14 | 10 | 20 | |
| Numerators = Number of GBs with at least one amplified oncogene (3-fold or greater) in Group(−) or Group(+) | ||||||||||||
| Group (−) | 5/10 | 4/8 | 4/7 | 4/7 | 4/7 | 4/7 | 5/8 | 3/5 | 6/8 | 9/12 | 2/2 | |
| Group (+) | 11/12 | 12/14 | 12/15 | 12/15 | 12/15 | 12/15 | 11/14 | 13/17 | 10/14 | 7/10 | 14/20 | |
| Fisher's Exact (p) | 0.041* | 0.085 | 0.213 | 0.213 | 0.213 | 0.213 | 0.273 | 0.319 | 0.376 | 0.354 | 0.519 | |
A tumor with elevated LDHC levels had a greater likelihood of having any 3.00-fold amplified oncogenes compared to Group(−) that was comprised of tumors with LDHC levels below the cut-off. In Group(+) for LDHC, 92% (11/12) of the tumors had amplified oncogenes versus 50% (5/10) in Group(−). The amplified oncogenes included EGFR, EVI1, BCL2A1, and PIK3CA in 8, 7, 6, and 3 tumors, respectively, with EGFR frequently being co-amplified. A strong tendency, p = 0.085 (Fisher's Exact), was also seen for the higher expression levels of LDHA to have amplified oncogenes, with 86% of its Group(+) having them versus 50% of its Group(−) tumors. Having only two glioblastomas in CA12's Group(−) defined by this threshold limited its analysis.
3.5. Correlations of CN gains for oncogenes with expression of MTB/GF genes
Each of the twenty oncogenes with at least one 3.00-fold elevation among the glioblastomas exhibited positive correlations of their CN gains with increased expression levels of at least two MTB/GF genes (Table 4). All MTB/GF genes had positive correlations with CN of EGFR and also for sums of CN for all 20 oncogenes (not shown). Among the oncogenes, CCND1 had the next highest frequency of positive correlations with MTB/GF genes and PIK3C2B, PIK3CA, MOS, RNF139, CYP27B1, and BCL2A1, each correlated positively with seven MTB/GF genes.
Table 4.
Correlations between individual MTB/GF genes and copy numbers (CN) of oncogenes, listed horizontally, in 22 glioblastomas. Expression values of MTB/GF genes after ddCt normalization were divided by concurrent expression levels of normalized ENO1 to obtain MTB/GF values as multiples of ENO1's expression prior to calculating correlations. Positive correlations less than 0.2 are indicated by ‘+’ and the negative ones are indicated by ‘-’. Genes are listed in the order of their chromosomal loci. Similar patterns of correlations for potential amplicons comprised of oncogenes are indicated in the bottom row. Note that SLC16A3 is also known as MCT4.
| MTB/GF genes listed below | 1q32.1 |
3q26.2 |
3q26.32 |
4q12 |
7p11.2 |
7q21.2 |
8q12.1 |
8q24.13 |
8q24.21 |
11q13.3 |
12p13.32 |
12q14.1 |
15q25.1 |
20q13.32 |
Xq25 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MDM4 | PIK3C2B | EVI1 | PIK3CA | PDGFRA | EGFR | CDK6 | MOS | RNF139 | MYC | CCND1 | CTTN | FGF3 | FGF4 | CCND2 | CYP27B1 | CDK4 | BCL2A1 | GNAS | XIAP | |
| 1p13.3 SLC16A4 |
||||||||||||||||||||
| − | − | − | + | 0.28 | + | − | + | + | − | + | − | − | − | − | − | − | 0.24 | 0.22 | + | |
| 4q25 EGF |
||||||||||||||||||||
| − | − | + | − | 0.39 | 0.25 | 0.37 | − | − | − | − | − | − | − | − | − | − | − | − | + | |
| 7p22.3 PDGFA |
||||||||||||||||||||
| − | − | − | − | 0.63 | 0.40 | + | − | − | − | − | − | − | − | − | − | − | + | 0.27 | − | |
| 8q21.2 CA3 |
||||||||||||||||||||
| 0.33 | 0.34 | 0.24 | + | − | + | + | 0.46 | 0.35 | 0.46 | − | − | − | − | − | 0.41 | 0.44 | 0.36 | + | 0.00 | |
| 11p15.1 LDHA LDHC |
||||||||||||||||||||
| − | + | − | 0.28 | − | + | + | + | + | + | 0.28 | + | 0.21 | + | + | + | + | − | − | − | |
| − | + | − | 0.27 | − | + | + | 0.26 | + | + | 0.42 | 0.31 | 0.37 | 0.30 | + | + | + | − | − | − | |
| 14q23.2 HIF1A |
||||||||||||||||||||
| 0.34 | 0.46 | − | + | 0.52 | + | − | − | − | − | + | − | − | − | 0.21 | − | − | 0.34 | 0.38 | − | |
| 15q22.2 CA12 |
||||||||||||||||||||
| 0.31 | 0.33 | + | + | − | + | − | + | 0.39 | + | + | − | − | − | − | + | + | 0.19 | − | − | |
| 17q21.2 ACLY |
||||||||||||||||||||
| − | − | − | − | 0.31 | + | + | + | − | − | 0.36 | 0.33 | 0.34 | 0.30 | + | + | − | − | − | − | |
| 17q25.3 SLC16A3 |
||||||||||||||||||||
| + | 0.21 | − | − | 0.20 | + | − | 0.27 | 0.20 | 0.23 | + | − | − | − | − | + | + | + | + | − | |
| 19q13.33 GYS1 |
||||||||||||||||||||
| + | + | 0.30 | 0.33 | + | 0.21 | − | − | + | − | + | + | + | + | 0.26 | + | − | + | − | − | |
| Similar patterns for the potential amplicons | 9 of 11 MTB/GF genes | 8 of 11 MTB/GF genes | 7 of 11 MTB/GF genes | 9 of 11 MTB/GF genes | ||||||||||||||||
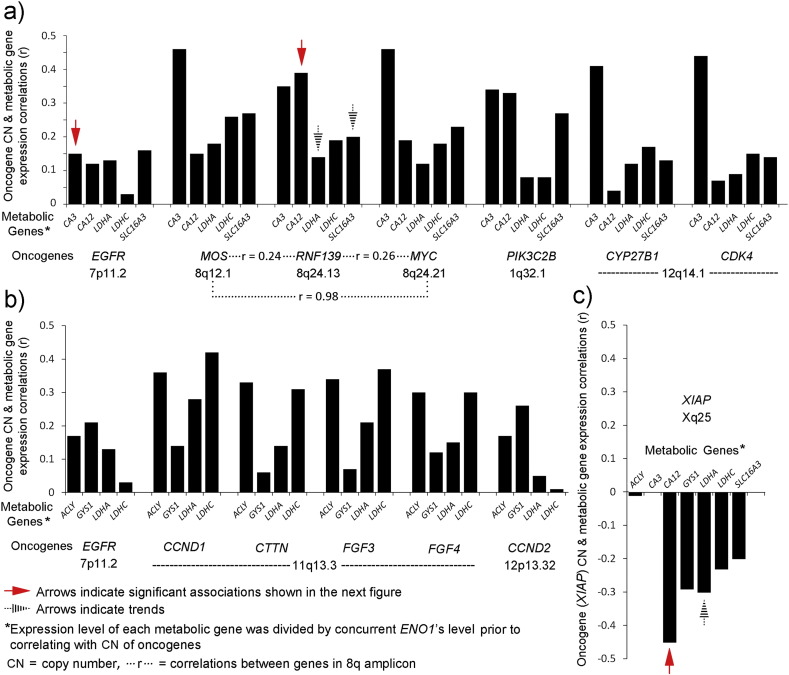
In contrast, XIAP exhibited predominantly negative correlations and correlated positively with only two MTB/GF genes. Among the MTB/GF genes, LCHC, LDHA, and GYS1 each had expression levels that correlated positively with CN of fourteen oncogenes that are listed. All of the metabolic genes had at least nine positive oncogene correlations each whereas the growth factor genes each had five. The highest individual correlation of a MTB/GF gene's expression with CN of an oncogene, r = 0.63, occurred for PDGFA with CN of PDGFRA, the gene for one of its receptors. The correlation of HIF1A's expression with CN of PDGFRA, r = 0.52, was the next highest. The correlations of CA3's expression with CN of MOS and MYC, r = 0.46 for each, were noted with the realization that all 3 genes are located on 8q (Fig. 1). Patterns for positive correlations in potential amplicons derived from 1q, 8q, 11q, and 12q occurred (Table 4, bottom row).
Also, two patterns of possible functional relevance emerged between metabolic genes and oncogenes. In one pattern, LDHA, LDHC, CA3, CA12, and SLC16A3 (or MCT4), encoding proteins responsible for generation of ATP in anaerobic glycolysis and buffering of the resulting hydrogen ions, shared positive correlations with MOS, RNF139, MYC, PIK3C2B, CYP27B1, and CDK4 as well as with EGFR (Table 4 and Fig. 6a). In a search for oncogenes that associate with glycolytic tumor cell migration [12], a second oncogene pattern was characterized by positive correlations of the lactate dehydrogenases and putative metabolic adaptors, ACLY and GYS1 [9], [11], for short-term or intermittent dependence on glycolysis. This group consisted of ACLY, GYS1, LDHA, and LDHC, positively correlating with the oncogenes, CCND1, CTTN, FGF3, FGF4, and CCND2, as well as with EGFR, (Table 4 and Fig. 6b). Strong negative correlations for five of the seven metabolic genes in these two groups were found for CN of XIAP (Table 4 and Fig. 6c).
Fig. 6.
Patterns of functional relevance in the correlations between metabolic genes and oncogenes. (a) A pattern attributed to genes whose expressions protect glycolysis in acidic conditions, possibly to supra-physiologic limits. Positive correlations of the ENO1 transformed expressions of metabolic genes (CA3, CA12, LDHA, LDHC, and SLC16A3(or MCT4)) occurred with CN of oncogenes (EGFR, MOS, RNF139, MYC, PIK3C2B, CYP27B1, and CDK4). Those supported by significant associations (Wilcoxon Rank Sum, Section 3.6) are indicated by solid red arrows and trends by black striped arrows. Complexity within a potential 8q amplicon is indicated by strong correlation (r = 0.98) between MOS and MYC but weaker correlations between each of them and intervening RNF139. (b) A pattern attributed to non-aerobic glycolysis operating in intermittent avascular conditions based on previous studies of glycolytic-dependent glioblastoma cell migration [9], [11], involving ENO1 transformed expressions of metabolic genes, ACLY, GYS1, LDHA, and LDHA, was identified by shared positive correlations with CN of oncogenes, EGFR, CCND1, CTTN, FGF3, FGF4, and CCND2. (c) The CN gain of XIAP revealed an opposing pattern for metabolic genes' expressions. The significant association (Wilcoxon Rank Sum, Section 3.6) and a trend are indicated by the solid red and striped black arrows, respectively.
3.6. Associations of metabolic gene expressions with CN of oncogenes and gender
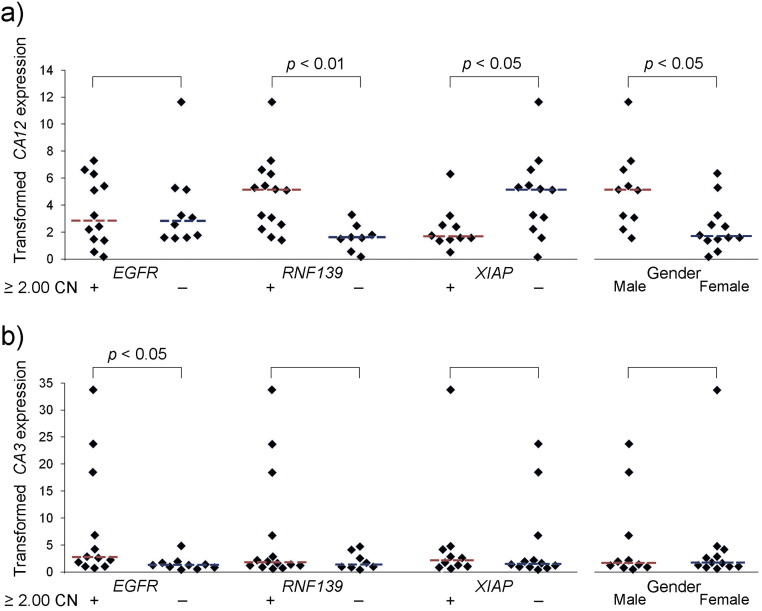
Associations with CN of several oncogenes and gender were searched for among eight metabolic genes, ACLY, CA3, CA12, GYS1, HIF1A, LDHA, LDHC, and SLC16A3 using Wilcoxon Rank Sum comparisons. There were 4 oncogenes, EGFR, RNF139, GNAS, and XIAP, whose CN gains were sufficient in number among the glioblastomas (Fig. 1) so that comparisons could be made using 2.00-fold CN gains as the threshold for inclusion into one of two comparable-size groups for each of these four oncogenes. Additionally, stratification by gender (Table 1) also yielded comparable size groups. Due to XIAP's location on the X chromosome, gender is a contributing factor to XIAP's CN [13]. The gender ratio for those without XIAP CN gains of at least 2.00-fold was M:F = 10:2 and only females had XIAP CN gains that were 2.00-fold or more when standardized by normal DNA from a female for the CN studies. The largest number of significant associations occurred for CA12. Its associations with RNF139's CN gains, XIAP's lack of CN gains, and male gender, had p-values of 0.0037, 0.0426, and 0.0169, respectively, (Fig. 7a). Medians for RNF139's CN gains, XIAP's lack of gains, and male gender, all 5.1424 (level of CA12 given as a multiple of concurrent ENO1's expression) are shown as red dashed lines. The medians for CA12's transformed expression levels in the opposing groups were 1.6024, 1.7213, and 1.7213, respectively, and are shown by blue dashed lines. A significant association occurred for CA3 with 2.00-fold or greater CN gains of EGFR, p = 0.0249, with medians of 2.7444 and 1.2811 for CA3's transformed expression levels in groups, with and without at least 2.00-fold CN gains of EGFR, respectively, (Fig. 7b). Strong trends for associations were found for transformed LDHA's expression levels with RNF139's 2.00-fold or greater CN gains, lack of XIAP's CN gains, and male gender with p-values of 0.0950, 0.0804, and 0.1229, respectively, and a trend was also detected for an association between transformed SLC16A3(or MCT4)’s expression levels and RNF139's 2.00-fold or greater CN gains, with a p-value of 0.11 (not shown). No significant associations or trends were found for GNAS using the 2.00-fold threshold for its CN gains with metabolic genes (not shown).
Fig. 7.
Expressions of carbonic anhydrase genes, CA3 and CA12, transformed by ENO1, associated significantly in Wilcoxon Rank Sum analyses with CN changes and gender in glioblastomas. (a) Transformed CA12 expression associated significantly with CN gains of RNF139 that were 2.00-fold or greater, lack of a 2.00-fold or greater CN gain in XIAP, and with male gender. (b) Transformed CA3 expression associated significantly with CN gains of EGFR that were 2.00-fold or greater. The red and blue dashed lines among the data points indicate the medians for each data group as indicated.
4. Discussion
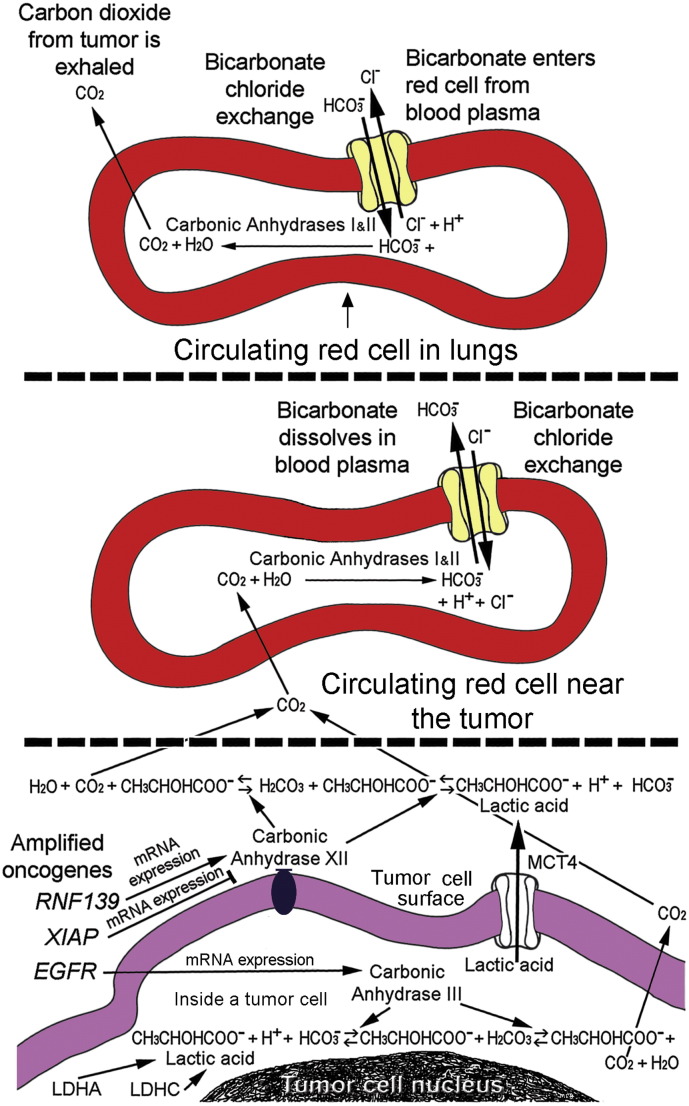
When respiration is inhibited, such as during hypoxia, the glycolytic pathway lacks NAD+ from mitochondria. To compensate, large amounts of the organic acid, lactate, are produced from pyruvate via lactate dehydrogenases to maintain the NAD+ levels needed for continuous cycling of non-aerobic glycolysis. The capacity for buffering hydrogen ions becomes critical in glycolytic conditions due to susceptibility of phosphofructokinase and other glycolytic enzymes to inhibition by acidity [14], [15], [16], [17], in addition to the sensitivity of many cellular proteins to pH changes. To control pH, there are alterations in carbonic anhydrase family members, solute carrier (SLC) family members that transport monocarboxylates, vacuolar type H+-ATPases, membrane sodium/hydrogen exchangers, chloride/bicarbonate and sodium/bicarbonate exchangers, etc. that occur in a cooperative manner. Some of the regulation is known, such as the effect of HIF-1α on carbonic anhydrase IX and other proteins, but the complete picture is not known. The heightened activity of tumor cells to manipulate their pH for compensatory purposes is a hallmark of malignancy. This malignant feature has been implicated in drug resistance and provides new treatment targets to consider clinically [18], [19], [20], [21], [22], [23], [24], [25].
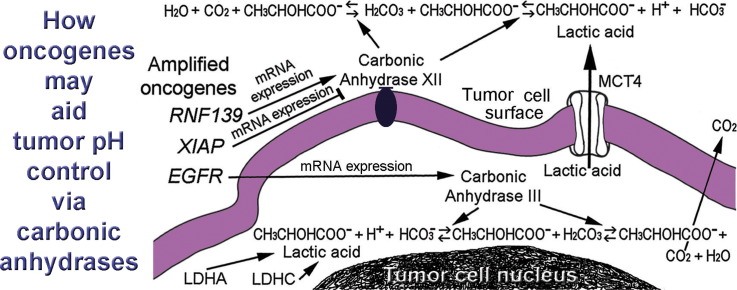
The carbonic anhydrase family members are prominent mediators of pH control in tumors. The amounts of bicarbonate, HCO3 −, needed to buffer protons would be depleted without replenishment by carbonic anhydrases that catalyze interconversions of carbon dioxide and water to bicarbonate and protons and vice versa via carbonic acid. The carbonic anhydrases possess impressively rapid catalytic rates, up to a million-fold maximal turnover rate per second for the carbonic anhydrase II isoform in red cells [26]. The catalytic rate of CAIII (encoded by CA3) in nucleated cells is slower but its activity can be enhanced by phosphates [27]. Also, isozyme-specific residues in the active site of CAIII, when replaced by their counterparts in CAII, permit CAIII to achieve the impressive kinetics of CAII [26], [28], [29]. Carbonic anhydrase III is a major protein in muscle and myoepithelial cells [30], [31], [32]. Expression of CA3 and other genes, including HIF1A, were significantly augmented in muscle of endurance runners exercising in hypoxia (14.5% oxygen) [33]. In a separate study, repeated sprints by runners in hypoxia also led to increased expression levels of CA3 [34]. Levels of CAIII also increase with age in human muscle [35]. In the central nervous system, anoxic stress for 4 h led to increased levels of CAIII, among other isoforms, in the cerebral cortex, hippocampus, cerebellum, and retina of piglets [36]. Interestingly, in hepatoma cells overexpression of CA3 by transfection resulted in increased extracellular acidification, anchorage-independent growth and invasiveness with elevated focal adhesion kinase (FAK) and Src activity [37]. Increased EGFRvIII expression has been associated with protein levels of CAIII and poor survival in glioblastomas [38]. Previously, we detected four phosphorylated tyrosine (pY) residues (Y845, Y992, Y1045, and Y1068) in EGFR within pseudopodia of migrating glioblastoma cells [39]. The pseudopodia constitute a relatively glycolytic subcellular domain compared to whole cells [1]. Others have proposed that EGFR's “Y/pY fingerprints” defined in time courses may “encode” acid–base disturbances [40]. This study of glioblastomas showed that CN gains in EGFR of 2.00-fold or greater significantly associated with expression levels of CA3, transformed by concurrent levels of ENO1. Thus, the metabolic role played by carbonic anhydrase III in glioblastomas is possibly influenced by effects due to gains in CN of EGFR and its signaling on CA3's expression levels.
The other significant associations found in this study were for CN of oncogenes and gender with CA12 that encodes the transmembrane carbonic anhydrase, CAXII. Gains in CN of RNF139, lack of gains in XIAP, and male gender associated significantly with expression of CA12 in this study's glioblastomas. Interestingly, the two amplified oncogenes, RNF139 and XIAP, that had opposing associations with expression of CA12, both encode RING proteins that are E3 ubiquitin ligases [41], [42], [43], [44].
Previously CAXII has been found in multiple types of tumors with expression results in some suggesting a malignant role. Immunohistochemistry demonstrated CAXII in oncocytomas and clear-cell carcinomas of the kidneys [45] and ovarian carcinomas [46]. In assays of hypoxic colon adenocarcinoma cells, silencing of CA9 alone led to a 40% reduction in xenograft tumor volume with accompanying up-regulation of CA12 mRNA levels, whereas silencing of both CA9 and CA12 led to a greater (85%) reduction in tumor volume. Also, when CA9 was silenced in vivo, there was an increase in the amount of CAXII shown with immunostaining [47]. Gene knockdown studies of CA12 in breast carcinoma cells decreased their invasiveness that was restored by overexpressing CA12 [48]. Protein expression of CAXII in oral squamous carcinoma has been associated with more advanced clinical stages, larger tumor size, recurrence, and poorer prognosis [49]. Both CA9/CAIX and CA12/CAXII have been studied in lung adenocarcinoma cell lines [50]. In colon carcinoma, CAXII was upregulated on the surfaces of chemoresistant cells [51]. In diffuse astrocytomas, two forms of CAXII occur derived from alternative splicing. Immunoreactivity for CAXII was found in 98% of 363 astrocytomas and its increased expression correlated with higher World Health Organization (WHO) grade, older age, and poorer prognosis independent of patient age and WHO grade [52]. Our selection of CA12 as a gene from the large carbonic anhydrase family to study was based partly on significantly poorer patient survival at high levels of expression in glioblastomas found in the REMBRANDT database during 2007–2008 [8].
Statistically significant associations found in our study included greater expression of CA12 (transformed by ENO1) occurring with 2.00-fold or greater CN of RNF139, previously known as TRC8 for “translocation in renal carcinoma”. RNF139 occurs as a chromosomal translocation, t(3;8)(p14.2;q24.1) in some familial renal cancers [53], [54], [55] and another RNF139 translocation, t(8;22)(q24.13;q11.21) has been reported in a dysgerminoma [56]. In clear cell renal carcinomas, RNF139 has been thought to act as a tumor suppressor [41], [42], [57], [58], [59]. However, RNF139 is expressed in non-renal tissues, including brain [60], and gains in the expression and CN of RNF139 have recently been noted in studies that include non-renal tumors. In a refractory cancer gene set interaction network (113 cancer patient samples) annotated with tissue type specificity, RNF139 was overexpressed in 9 of 16 pancreas samples, p = 0.0085, compared to normal tissue samples [61]. Also, RNF139 was one of three predicted driver genes in cancer shared among breast, melanoma, and liver cancers based on a computational method that analyzed CN aberrations in human cancer genomes [62]. A study on primary oral tumor samples and nearby tumor-free tissues identified RNF139 among a group of 14 genes exhibiting CN gains out of 133 cancer-related genes [63]. Increases in RNF139 gene expression were found in comparisons of Barrett esophagus (precancerous) and esophageal adenocarcinomas with normal esophagus [64]. Our study suggests that CN gains in RNF139 found in glioblastomas are associated with the increased expression of CA12.
Another statistically significant association for an oncogene in our study occurred for the greater expression of CA12 (transformed by ENO1) when CN of XIAP (X-linked inhibitor of apoptosis, previously known as BIRC4) was less than 2.00-fold. In other words, there was a negative correlation with CN gains in XIAP for increased expression of CA12. Others have found XIAP and two other inhibitors of apoptosis in twelve malignant glioma cell lines [65]. Although XIAP is known as an endogenous repressor of the terminal caspase cascade in apoptosis, it may also be involved in other activities, including modulation of signal transduction and protein ubiquitination. XIAP is synthesized during conditions of stress that inhibit protein synthesis [66]. When XIAP is translated by a mechanism involving an internal ribosome entry site (IRES), interactions of XIAP IRES with MDM2 may affect stabilization of itself and MDM2. Interestingly, because XIAP can induce autophagy, it can have an antitumor effect in early tumorigenesis in contrast to being an aid for tumor survival in stressful conditions. Therefore, XIAP's role in tumors has been proposed to be contextual [67]. Mutational loss of XIAP is responsible for X-linked lymphoproliferative syndrome type 2 [68], [69].
Although XIAP's CN could potentially be influenced by X chromosomal inactivation, instead, its CN is normally determined by the number of X chromosomes. One copy of XIAP in males and two in females, with no evidence of a CN variation, were found in 400 individuals tested with quantitative PCR in a recent asthma study [13]. Also, differences in response to cerebral ischemia in stroke according to gender have been attributed to XIAP's correspondence to chromosomal dosage [70]. In regard to tumors, increased XIAP demonstrated with immunohistochemistry in hepatocellular carcinoma specimens was associated with small tumor size, early tumor stage, and better relapse-free and overall survival [71]. Also, in a mouse model that demonstrates progression to malignant peripheral nerve sheath tumors (MPNST) from neurofibromas, a loss in CN for XIAP was found in early passage cell lines established from the MPNSTs [72]. Accordingly, the negative correlation and significant association for lack of XIAP gains with expression of CA12 in our study suggests that it may regulate expression of CA12 to control acidity in tumors. However, this relationship is complex in that there are also reports of several types of tumors, including gliomas, where targeting XIAP has been suggested for treatment purposes [73], [74], [75], [76].
Regulation of pH is an essential requirement for life in that protein interactions are very sensitive to pH, such as those of phosphofructokinase. Vulnerabilities of phosphofructokinase and other glycolytic enzymes are being considered as important treatment targets [77]. The access of tumors to the bicarbonate buffer system in the bloodstream is possibly optimized by oncogenes (Fig. 8). This study identified CN gains of EGFR and RNF139, male gender, and lack of XIAP's CN gains as having significant associations with expressions of carbonic anhydrases that catalyze interconversions of the bicarbonate buffer's components within and around tumor cells. Gains in CN for EGFR significantly associated with expression of CA3, that encodes an intracellular carbonic anhydrase, CAIII. Gains in RNF139 significantly associated with CA12's expression that encodes CAXII, whose activity is extracellular. The proposed effects of gains in CN of the three oncogenes are illustrated (Fig. 8, bottom panel). Gender and lack of CN gains for XIAP may be factors in tumor biology. The gains in CN for EGFR and RNF139 within tumors may aid tumors metabolically and potentially can serve as therapeutic targets to counteract non-physiologic pH changes in tumors.
Fig. 8.
Roles for amplified oncogenes, EGFR, RNF139, and XIAP, to control pH via access to the systemic bicarbonate buffer system in aiding glioblastoma cells. Red cells and plasma in the blood stream (upper panels) remove carbons via CO2 that is exhaled in the lungs. Transport of CO2/HCO3− occurs in a relay fashion within the circulation. Large amounts of bicarbonate in the bloodstream buffer protons and levels are maintained via recycling in the kidneys where the protons carried by bicarbonate are expelled. Amplified EGFR and RNF139 may enhance expression of CA3 and CA12, respectively, to increase encoded tumor carbonic anhydrases (lower panel). In contrast, XIAP may reduce expression of CA12. Lactic acid membrane transporter, MCT4, is encoded by SLC16A3. The edge of a tumor cell is in the lowest panel. Red cells are depicted with reactions derived from other diagrams [78], [79], [80]. H+ ions (protons) from ATP breakdown during anaerobic metabolism involving lactic acid is a process explained elsewhere [81].
Also, in tumor cells there is a functional relationship between lactate dehydrogenases and carbonic anhydrases in that with increasing ATP via lactate production, there is higher demand for pH regulation mediated by carbonic anhydrases. Accordingly, in this study there was a trend for CN gains of RNF139 to associate with expression of LDHA. CN gains of RNF139 also showed a trend for associating with expression of SLC16A3 (also known as MCT4) that extrudes lactic acid from cells. Additionally, male gender and the lack of CN gains for XIAP trended positively with expression of LDHA. For LDHC, a Fisher's Exact test revealed a significant association for its increased expression with any 3.00-fold or greater oncogene CN gain in this study.
5. Conclusions
The sensitivity of PCR methodologies, selection of metabolic genes using functional relevance and REMBRANDT data, inclusion of many known oncogenes to screen, and using ENO1 transformation to adjust for tumor heterogeneity and obtain supra-glycolytic metabolic gene expression levels were contributing factors that led to the identification of significant associations between oncogene CN changes and metabolic gene expression levels in this study. Regulation of pH emerged as a potential oncogene-mediated advantage for tumor cells so that roles for oncogenes in controlling tumor pH are compelling to consider in future treatment regimens. Slight increases in expression of carbonic anhydrases with rapid catalytic rates due to oncogene CN changes may provide dramatic advantages for tumors. Our findings suggest that oncogenes in glioblastomas increase utilization of the body's vast, systemic bicarbonate buffering system to aberrantly control pH. Identification of participating oncogenes may identify functional subgroups among glioblastomas in larger studies. These oncogenes, when amplified, may be culprits that enable malignant metabolism to robustly energize tumor cells that outcompete normal cells with clinically important prognostic and therapeutic implications.
Disclosure summary
The authors declare no conflict of interest.
Transparency document
Transparency document.
Acknowledgments
We thank The Pittsburgh Foundation's Walter L. Copeland Fund for Cranial Research (D2006-0379) and Louisiana State University Health Sciences Center in Shreveport Grant-in-Aid for funding. Technical assistance, including usage of equipment, was graciously provided as needed by Molecular Diagnostic Laboratories in the Departments of Pathology at Louisiana State University Health, Shreveport, LA, and at the University of Pittsburgh Medical Center, Pittsburgh, PA. We are especially grateful for the contributions of Jeffrey A. Kant, MD, PhD, former director of the Molecular Diagnostics Laboratory, Department of Pathology, University of Pittsburgh, Pittsburgh, PA, now deceased, to many of the early steps taken in determining how this study progressed and evolved.
Footnotes
The Transparency document associated with this article can be found, in online version.
Contributor Information
Marie E. Beckner, Email: mbeckne1@kent.edu, mbeckner@sprynet.com.
Ian F. Pollack, Email: pollaci@upmc.edu, ian.pollack@chp.edu.
Mary L. Nordberg, Email: mary.nordberg@deltamdx.com.
References
- 1.Beckner M.E., Chen X., An J., Day B.W., Pollack I.F. Proteomic characterization of harvested pseudopodia with differential gel electrophoresis and specific antibodies. Lab. Invest. 2005;85:316–327. doi: 10.1038/labinvest.3700239. [DOI] [PubMed] [Google Scholar]
- 2.Muller F.L., Colla S., Aquilanti E., Manzo V., Genovese G., Lee J., Eisenson D., Narurkar R., Deng P., Nezi L., Lee M., Hu B., Hu J. Passenger deletions generate therapeutic vulnerabilities in cancer. Nature. 2012;488:337–342. doi: 10.1038/nature11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 4.Laiken N., Fanestil D.D. 12th ed. West JB ed., Williams & Wilkins; Baltimore, MD: 1990. Acid–base Balance and Regulation of H+ Excretion in Best and Taylor's Physiological Basis of Medical Practice; pp. 486–493. Chapter 32. [Google Scholar]
- 5.Skelton L.A., Boron W.F., Zhou Y. Acid–base transport by the renal proximal tubule. J. Nephrol. 2010;23(Suppl. 16):S4–S18. [PMC free article] [PubMed] [Google Scholar]
- 6.Truong L.N., Patil S., Martin S.S., LeBlanc J.F., Nanda A., Nordberg M.L., Beckner M.E. Rapid detection of high-level oncogene amplifications in ultrasonic surgical aspirations of brain tumors. Diagn. Pathol. 2012;7:66. doi: 10.1186/1746-1596-7-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beckner M.E., Sampath R., Flowers A.B., Katira K., D'Souza D., Patil S., Patel R.B., Nordberg M.L., Nanda A. Low-level amplification of oncogenes correlates inversely with age for patients with nontypical meningiomas. World Neurosurg. 2013;79:313–319. doi: 10.1016/j.wneu.2011.08.023. [DOI] [PubMed] [Google Scholar]
- 8.Repository of Molecular Brain Neoplasia Database (REMBRANDT) v1.52 and earlier versions, generated and maintained by the NCI and NINDS at NIH. February, 2008. http://rembrandt.nci.nih.gov Available at: (data (93 glioblastomas available))
- 9.Beckner M.E., Fellows-Mayle W., Zhang Z., Agostino N.R., Kant J.A., Day B.W., Pollack I.F. Identification of ATP citrate lyase as a positive regulator of glycolytic function in glioblastomas. Int. J. Cancer. 2010;126:2282–2295. doi: 10.1002/ijc.24918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beckner M.E. Co-amplified oncogenes in glioblastomas. In: Bezerra M.F., Alves C.R., editors. Glioblastoma: Risk Factors, Diagnosis and Treatment Options. Nova Science Publishers, Inc.; Hauppauge, NY: 2012. pp. 101–115. Chapter 4. [Google Scholar]
- 11.Beckner M.E., Gobbel G.T., Abounader R., Burovic F., Agostino N.R., Laterra J., Pollack I.F. Glycolytic glioma cells with active glycogen synthase are sensitive to PTEN and inhibitors of PI3K and gluconeogenesis. Lab. Invest. 2005;85:1457–1470. doi: 10.1038/labinvest.3700355. [DOI] [PubMed] [Google Scholar]
- 12.Beckner M.E., Stracke M.L., Liotta L.A., Schiffmann E. Glycolysis as primary energy source in tumor cell chemotaxis. J. Natl. Cancer Inst. 1990;82:1836–1840. doi: 10.1093/jnci/82.23.1836. [DOI] [PubMed] [Google Scholar]
- 13.Roscioli E., Hamon R., Ruffin R.E., Zalewski P., Grant J., Lester S. X-linked inhibitor of apoptosis single nucleotide polymorphisms and copy number variation are not risk factors for asthma. Respirology. 2013;18:697–703. doi: 10.1111/resp.12065. [DOI] [PubMed] [Google Scholar]
- 14.Schindler U., Betz E. Influence of severe hypercapnia upon cerebral cortical metabolism, CSF electrolyte concentrations and EEG in the cat. Bull. Eur. Physiopathol. Respir. 1976;12:277–284. [PubMed] [Google Scholar]
- 15.Achs M.J., Garfinkel D. Computer simulation of energy metabolism in acidotic cardiac ischemia. Am. J. Physiol. 1982;242:R533–R544. doi: 10.1152/ajpregu.1982.242.5.R533. [DOI] [PubMed] [Google Scholar]
- 16.Mulquiney P.J., Kuchel P.W. Model of the pH-dependence of the concentrations of complexes involving metabolites, haemoglobin and magnesium ions in the human erythrocyte. Eur. J. Biochem. 1997;245:71–83. doi: 10.1111/j.1432-1033.1997.00071.x. [DOI] [PubMed] [Google Scholar]
- 17.Costa Leite T., Da Silva D., Guimaraes Coelho R., Zancan P., Sola-Penna M. Lactate favours the dissociation of skeletal muscle 6-phosphofructo-1-kinase tetramers down-regulating the enzyme and muscle glycolysis. Biochem. J. 2007;408:123–130. doi: 10.1042/BJ20070687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Milito A., Fais S. Proton pump inhibitors may reduce tumour resistance. Expert. Opin. Pharmacother. 2005;6:1049–1054. doi: 10.1517/14656566.6.7.1049. [DOI] [PubMed] [Google Scholar]
- 19.Fais S., De Milito A., You H., Qin W. Targeting vacuolar H +-ATPases as a new strategy against cancer. Cancer Res. 2007;67:10627–10630. doi: 10.1158/0008-5472.CAN-07-1805. [DOI] [PubMed] [Google Scholar]
- 20.Hulikova A. Harris Al, Vaughan-Jones RD, and Swietach P. Acid-extrusion from tissue: the interplay between membrane transporters and pH buffers. Curr. Pharm. Des. 2012;18:1331–1337. doi: 10.2174/138161212799504920. [DOI] [PubMed] [Google Scholar]
- 21.Daniel C., Bell C., Burton C., Harguindey S., Reshkin S.J., Rauch C. The role of proton dynamics in the development and maintenance of multidrug resistance in cancer. Biochim. Biophys. Acta. 1832;2013:606–617. doi: 10.1016/j.bbadis.2013.01.020. [DOI] [PubMed] [Google Scholar]
- 22.Becker H.M., Klier M., Deitmer J.W. Carbonic anhydrases and their interplay with acid/base-coupled membrane transporters. Subcell. Biochem. 2014;75:105–134. doi: 10.1007/978-94-007-7359-2_7. [DOI] [PubMed] [Google Scholar]
- 23.Shiozaki A., Ichikawa D., Otsuji E., Marunaka Y. Cellular physiological approach for treatment of gastric cancer. World J. Gastroenterol. 2014;20:11560–11566. doi: 10.3748/wjg.v20.i33.11560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spugnini E.P., Sonveaux P., Stock C., Perez-Sayans M., De Milito A., Avnet S., Garcia A.G., Harguindey S., Fais S. Proton channels and exchangers in cancer. Biochim. Biophys. Acta. Oct 20 2014 doi: 10.1016/j.bbamem.2014.10.015. (pii: S0005-2736(14)00350–2. [Epub ahead of print]) [DOI] [PubMed] [Google Scholar]
- 25.Swietach P., Vaughan-Jones R.D., Harris A.L., Hulikova A. The chemistry, physiology and pathology of pH in cancer. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2014;369:20130099. doi: 10.1098/rstb.2013.0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindskog S. Structure and mechanism of carbonic anhydrase. Pharmacol. Ther. 1997;74:1–20. doi: 10.1016/s0163-7258(96)00198-2. [DOI] [PubMed] [Google Scholar]
- 27.Paranawithana S.R., Tu C., Laipis P.J., Silverman D.N. Enhancement of the catalytic activity of carbonic anhydrase III by phosphates. J. Biol. Chem. 1990;265:22270–22274. [PubMed] [Google Scholar]
- 28.Jewell D.A., Tu C., Paranawithana S.R., Tanhauser S.M., LoGrasso P.V., Laipis P.J., Silverman D.N. Enhancement of the catalytic properties of human carbonic anhydrase III by site-directed mutagenesis. Biochemistry. 1991;30:1484–1490. doi: 10.1021/bi00220a006. [DOI] [PubMed] [Google Scholar]
- 29.LoGrasso P.V., Tu C., Jewell D.A., Wynns G.C., Laipis P.J., Silverman D.N. Catalytic enhancement of human carbonic anhydrase III by replacement of phenylalanine-198 with leucine. Biochemistry. 1991;30:8463–8470. doi: 10.1021/bi00098a025. [DOI] [PubMed] [Google Scholar]
- 30.Vaananen H.K., Autio-Harmainen H. Carbonic anhydrase III: a new histochemical marker for myoepithelial cells. J. Histochem. Cytochem. 1987;35:683–686. doi: 10.1177/35.6.3106467. [DOI] [PubMed] [Google Scholar]
- 31.Tweedie S., Morrison K., Charlton J., Edwards Y.H. CAIII a marker for early myogenesis: analysis of expression in cultured myogenic cells. Somat. Cell Mol. Genet. 1991;17:215–228. doi: 10.1007/BF01232818. [DOI] [PubMed] [Google Scholar]
- 32.Edwards Y.H., Tweedie S., Lowe N., Lyons G. Carbonic anhydrase 3 (CA3), a mesodermal marker. Symp. Soc. Exp. Biol. 1992;46:273–283. [PubMed] [Google Scholar]
- 33.Zoll J., Ponsot E., Dufour S., Doutreleau S., Ventura-Clapier R., Vogt M., Hoppeler H., Richard R., Fluck M. Exercise training in normobaric hypoxia in endurance runners. III. Muscular adjustments of selected gene transcripts. J. Appl. Physiol. 2006;100:1258–1266. doi: 10.1152/japplphysiol.00359.2005. [DOI] [PubMed] [Google Scholar]
- 34.Faiss R., Leger B., Vesin J.-M., Fournier P.-E., Eggel Y., Deriaz O., Millet G.P. Significant molecular and systemic adaptations after repeated sprint training in hypoxia. PLoS One. 2013;8 doi: 10.1371/journal.pone.0056522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Staunton L., Zweyer M., Swandulla D., Ohlendieck K. Mass spectrometry-based proteomic analysis of middle-aged vs. aged vastus lateralis reveals increased levels of carbonic anhydrase isoform 3 in senescent human skeletal muscle. Int. J. Mol. Med. 2012;30:723–733. doi: 10.3892/ijmm.2012.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nogradi A., Domoki F., Degi R., Borda S., Pakaski M., Szabo A., Bari F. Up-regulation of cerebral carbonic anhydrase by anoxic stress in piglets. J. Neurochem. 2003;85:843–850. doi: 10.1046/j.1471-4159.2003.01721.x. [DOI] [PubMed] [Google Scholar]
- 37.Dai H.Y., Hong C.C., Liang S.C., Yan M.D., Lai G.M., Cheng A.L., Chuang S.E. Carbonic anhydrase III promotes transformation and invasion capability in hepatoma cells through FAK signaling pathway. Mol. Carcinog. 2008;47:956–963. doi: 10.1002/mc.20448. [DOI] [PubMed] [Google Scholar]
- 38.Johnson H., Del Rosario A.M., Bryson B.D., Schroeder M.A., Sarkaria J.N., White F.M. Molecular characterization of EGFR and EGFRvIII signaling networks in human glioblastoma tumor xenografts. Mol. Cell. Proteomics. 2012;11:1724–1740. doi: 10.1074/mcp.M112.019984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beckner M.E., Aznavoorian-Cheshire S.A., Pollack I.F. Reactivity for activated epidermal growth factor receptor is demonstrated in the pseudopodia of migratory glioma cells. Proc. Am. Assoc. Cancer Res. 2005;46:1327. [Google Scholar]
- 40.Skelton L.A., Boron W.F. Effect of acute acid–base disturbances on ErbB1/2 tyrosine phosphorylation in rabbit renal proximal tubules. Am. J. Physiol. Ren. Physiol. 2013;305:F1747–F1764. doi: 10.1152/ajprenal.00307.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brauweiler A., Lorick K.L., Lee J.P., Tsai Y.C., Chan D., Weissman A.M., Drabkin H.A., Gemmill R.M. RING-dependent tumor suppression and G2/M arrest induced by the TRC8 hereditary kidney cancer gene. Oncogene. 2007;26:2263–2271. doi: 10.1038/sj.onc.1210017. [DOI] [PubMed] [Google Scholar]
- 42.Lee J.P., Brauweiler A., Rudolph M., Hooper J.E., Drabkin H.A., Gemmill R.M. The TRC8 ubiquitin ligase is sterol regulated and interacts with lipid and protein biosynthetic pathways. Mol. Cancer Res. 2010;8:93–106. doi: 10.1158/1541-7786.MCR-08-0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galban S., Duckett C.S. XIAP as a ubiquitin ligase in cellular signaling. Cell Death Differ. 2010;17:54–60. doi: 10.1038/cdd.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakatani Y., Kleffmann T., Linke K., Condon S.M., Hinds M.G., Day C.L. Regulation of ubiquitin transfer by XIAP, a dimeric RING E3 ligase. Biochem. J. 2013;450:629–638. doi: 10.1042/BJ20121702. [DOI] [PubMed] [Google Scholar]
- 45.Parkkila S., Parkkila A.K., Saarnio J., Kivela J., Karttunen T.J., Kaunisto K., Waheed A., Sly W.S., Tureci O., Virtanen I., Rajaniemi H. Expression of the membrane-associated carbonic anhydrase isozyme XII in the human kidney and renal tumors. J. Histochem. Cytochem. 2000;48:1601–1608. doi: 10.1177/002215540004801203. [DOI] [PubMed] [Google Scholar]
- 46.Hynninen P., Vaskivuo L., Saarnio J., Haapasalo H., Kivela J., Pastorekova S., Pastorek J., Waheed A., Sly W.S., Puistola U., Parkkila S. Expression of transmembrane carbonic anhydrase IX and XII in ovarian tumours. Histopathology. 2006;49:594–602. doi: 10.1111/j.1365-2559.2006.02523.x. [DOI] [PubMed] [Google Scholar]
- 47.Chiche J., Ilc K., Laferriere J., Trottier E., Dayan F., Mazure N.M., Brahimi-Horn M.C., Pouyssegur J. Hypoxia-inducible carbonic anhydrase IX and XII promote tumor cell growth by counteracting acidosis through the regulation of the intracellular pH. Cancer Res. 2009;69:358–368. doi: 10.1158/0008-5472.CAN-08-2470. [DOI] [PubMed] [Google Scholar]
- 48.Hsieh M.J., Chen K.S., Chiou H.L., Hsieh Y.S. Carbonic anhydrase XII promotes invasion and migration ability of MDA-MB-231 breast cancer cells through the p38 MAPK signaling pathway. Eur. J. Cell Biol. 2010;89:598–606. doi: 10.1016/j.ejcb.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 49.Chien M.H., Ying T.H., Hsieh Y.H., Lin C.H., Shih C.H., Wei L.H., Yang S.F. Tumor-associated carbonic anhydrase XII is linked to the growth of primary oral squamous cell carcinoma and its poor prognosis. Oral Oncol. 2011;48:417–423. doi: 10.1016/j.oraloncology.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 50.Ilie M., Hofman V., Zangari J., Chiche J., Mouroux J., Mazure N.M., Pouyssegur J., Brest P., Hofman P. Response of CAIX and CAXII to in vitro re-oxygenation and clinical significance of the combined expression in NSCLC patients. Lung Cancer. 2013;82:16–23. doi: 10.1016/j.lungcan.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 51.Kopecka J., Campia I., Jacobs A., Frei A.P., Ghigo D., Wollscheid B., Riganti C. Carbonic anhydrase XII is a new therapeutic target to overcome chemoresistance in cancer cells. Oncotarget. 2015;6:6776–6793. doi: 10.18632/oncotarget.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haapasalo J., Hilvo M., Nordfors K., Haapasalo H., Parkkila S., Hyrskyluoto A., Rantala I., Waheed A., Sly W.S., Pastorekova S., Pastorek J., Parkkila A.-K. Identification of an alternatively spliced isoform of carbonic anhydrase XII in diffusely infiltrating astrocytic gliomas. Neuro-Oncology. 2008;10:131–138. doi: 10.1215/15228517-2007-065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gemmill R.M., West J.D., Boldog F., Tanaka N., Robinson L.J., Smith D.I., Li F., Drabkin H.A. The hereditary renal cell carcinoma 3;8 translocation fuses FHIT to a patched-related gene, TRC8. Proc. Natl. Acad. Sci. U. S. A. 1998;95:9572–9577. doi: 10.1073/pnas.95.16.9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poland K.S., Azim M., Folsom M., Goldfarb R., Naeem R., Korch C., Drabkin H.A., Gemmill R.M., Plon S.E. A constitutional balanced t(3;8)(p14;q24.1) translocation results in disruption of the TRC8 gene and predisposition to clear cell renal cell carcinoma. Genes Chromosomes Cancer. 2007;46:805–812. doi: 10.1002/gcc.20466. [DOI] [PubMed] [Google Scholar]
- 55.Kato T., Franconi C.P., Sheridan M.B., Hacker A.M., Inagakai H., Glover T.W., Arlt M.F., Drabkin H.A., Gemmill R.M., Kurahashi H., Emanuel B.S. Analysis of the t(3;8) of hereditary renal cell carcinoma: a palindrome-mediated translocation. Cancer Genet. 2014;207:133–140. doi: 10.1016/j.cancergen.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gimelli S., Beri S., Drabkin H.A., Gambini C., Gregorio A., Fiorio P., Zuffardi O., Gemmill R.M., Giorda R., Gimelli G. The tumor suppressor gene TRC8/RNF139 is disrupted by a constitutional balanced translocation t(8;22)(q24.13;q11.21) in a young girl with dysgerminoma. Mol. Cancer. 2009;8:52. doi: 10.1186/1476-4598-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Drabkin H.A., Gemmill R.M. Obesity, cholesterol, and clear-cell renal cell carcinoma (RCC) Adv. Cancer Res. 2010;107:39–56. doi: 10.1016/S0065-230X(10)07002-8. [DOI] [PubMed] [Google Scholar]
- 58.Drabkin H.A., Gemmill R.M. Cholesterol and the development of clear-cell renal carcinoma. Curr. Opin. Pharmacol. 2012;12:742–750. doi: 10.1016/j.coph.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 59.Lin P.H., Lan W.M., Chau L.Y. TRC8 suppresses tumorigenesis through targeting heme oxygenase-1 for ubiquitination and degradation. Oncogene. 2013;32:2325–2334. doi: 10.1038/onc.2012.244. [DOI] [PubMed] [Google Scholar]
- 60.Charytoniuk D., Porcel B., Rodriguez Gomez J., Faure H., Ruat M., Traiffort E. Sonic hedgehog signaling in the developing and adult brain. J. Physiol. Paris. 2002;96:9–16. doi: 10.1016/s0928-4257(01)00075-4. [DOI] [PubMed] [Google Scholar]
- 61.Jung S., Verdicchio M., Kiefer J., Von Hoff D., Berens M., Bittner M., Kim S. Learning contextual gene set interaction networks of cancer with condition specificity. BMC Genomics. 2013;14:110. doi: 10.1186/1471-2164-14-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen Y., Hao J., Jiang W., He T., Zhang X., Jiang T., Jiang R. Identifying potential cancer driver genes by genomic data integration. Sci. Rep. 2013;3:3538. doi: 10.1038/srep03538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ribeiro I.P., Marques F., Caramelo F., Pereira J., Patricio M., Prazeres H., Ferrao J., Juliao M.J., Castelo-Branco M., de Melo J.B., Baptista I.P., Carreira I.M. Genetic gains and losses in oral squamous cell carcinoma: impact on clinical management. Cell. Oncol. (Dordr) 2014;37:29–39. doi: 10.1007/s13402-013-0161-5. [DOI] [PubMed] [Google Scholar]
- 64.Wang X.W., Wei W., Wang W.Q., Zhao X.Y., Guo H., Fand D.C. RING finger proteins are involved in the progression of Barrett esophagus to esophageal adenocarcinoma: a preliminary study. Gut Liver. 2014;8:487–494. doi: 10.5009/gnl13133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wagenknecht B., Glaser T., Naumann U., Kugler S., Isenmann S., Bahr M., Korneluk R., Liston P., Weller M. Expression and biological activity of X-linked inhibitor of apoptosis (XIAP) in human malignant glioma. Cell Death Differ. 1999;6:370–376. doi: 10.1038/sj.cdd.4400503. [DOI] [PubMed] [Google Scholar]
- 66.Holcik M., Gibson H., Korneluk R.G. XIAP: apoptotic brake and promising therapeutic target. Apoptosis. 2001;6:253–261. doi: 10.1023/a:1011379307472. [DOI] [PubMed] [Google Scholar]
- 67.Liu T., Zhang H., Xiong J., Yi S., Gu L., Zhou M. Inhibition of MDM2 homodimerization by XIAP IRES stabilizes MDM2, influencing cancer cell survival. Mol. Cancer. 2015;14:65. doi: 10.1186/s12943-015-0334-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aguilar C., Latour S. X-linked inhibitor of apoptosis protein deficiency: more than an X-linked lymphoproliferative syndrome. J. Clin. Immunol. 2015;35:331–338. doi: 10.1007/s10875-015-0141-9. [DOI] [PubMed] [Google Scholar]
- 69.Dziadzio M., Ammann S., Canning C., Boyle F., Hassan A., Cale C., Elawad M., Fiil B.K., Gyrd-Hansen M., Salzer U. Speckmann c, and Grimbacher B. Symptomatic males and female carriers in a large Caucasian kindred with XIAP deficiency. J. Clin. Immunol. 2015;35:439–444. doi: 10.1007/s10875-015-0166-0. [DOI] [PubMed] [Google Scholar]
- 70.Siegel C., Li J., Liu F., Benashski S.E., McCullough L.D. miR-23a regulation of X-linked inhibitor of apoptosis (XIAP) contributes to sex differences in the response to cerebral ischemia. Proc. Natl. Acad. Sci. U. S. A. 2011;108:11662–11667. doi: 10.1073/pnas.1102635108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu W.Y., Kim H., Zhang C.L., Meng X.L., Wu Z.S. Clinical significance of autophagic protein LC3 levels and its correlation with XIAP expression in hepatocellular carcinoma. Med. Oncol. 2014;31:108. doi: 10.1007/s12032-014-0108-3. [DOI] [PubMed] [Google Scholar]
- 72.Kazmi S.J., Byer S.J., Eckert J.M., Turk A.N., Huijbregts R.P.H., Brossier N.M., Grizzle W.E., Mikhail F.M., Roth K.A., Carroll S.L. Transgenic mice overexpression neuregulin-1 model neurofibroma-malignant peripheral nerve sheath tumor progression and implicate chromosomal copy number variations in tumorigenesis. Am. J. Pathol. 2013;182:646–667. doi: 10.1016/j.ajpath.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hosni-Ahmed A., Sims M., Jones T.S., Patil R., Patil S., Abdelsamed H., Yates C.R., Miller D.D., Pfeffer L.M. J. Cancer Sci. Ther. 2014;6:370–377. doi: 10.4172/1948-5956.1000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bagulkar B.B., Gawande M., Chaudhary M., Gadbail A.R., Patil S., Bagulkar S. XIAP and Ki-67: a correlation between antiapoptotic and proliferative marker expression in benign and malignant tumours of salivary gland: an immunohistochemical study. J. Clin. Diagn Res. 2015;9 doi: 10.7860/JCDR/2015/11690.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yi X.P., Han T., Li Y.X., Long X.Y., Li W.Z. Simultaneous silencing of XIAP and survivin causes partial mesenchymal-epithelial transition of human pancreatic cancer cells via the PTEN/PI3K/Akt pathway. Mol. Med. Rep. 2015;12:601–608. doi: 10.3892/mmr.2015.3380. [DOI] [PubMed] [Google Scholar]
- 76.Lin F., Ghislat G., Luo S., Renna M., Siddiqi F., Rubinsztein D.C. XIAP and cIAP1 amplifications induce beclin 1-dependent autophagy through NFkB activation. Hum. Mol. Genet. 2015;24:2899–2913. doi: 10.1093/hmg/ddv052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sanzey M., Rahim S.A.A., Oudin A., Dirkse A., Kaoma T., Vallar L., Herold-Mende C., Bjerkvig R., Golebiewska A., Niclou S.P. Comprehensive analysis of glycolytic enzymes as therapeutic targets in the treatment of glioblastoma. PLoS One. 2015;10 doi: 10.1371/journal.pone.0123544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nelson D.L., Cox M.M. 3rd ed. Worth Publishers; New York, NY: 2000. Chapter 12, Biological Membranes and Transport in Lehninger Principles of Biochemistry; p. 413. [Google Scholar]
- 79.Casey J.R. Why bicarbonate? Biochem. Cell Biol. 2006;84:930–939. doi: 10.1139/o06-184. [DOI] [PubMed] [Google Scholar]
- 80.Vince J.W., Reithmeier R.A.F. Carbonic anhydrase II binds to the carboxyl terminus of human band 3, the erythrocyte Cl −/HCO3- exchanger. J. Biol. Chem. 1998;273:28430–28437. doi: 10.1074/jbc.273.43.28430. [DOI] [PubMed] [Google Scholar]
- 81.Swietach P., Vaughan-Jones R.D., Harris A.L., Hulikova A. The chemistry, physiology and pathology of pH in cancer. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2014;369:20130099. doi: 10.1098/rstb.2013.0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document.