Abstract
We characterized the dynamics of autophagy in vitro using four different cell systems and analyzing markers widely used in this field, i.e. LC3 (microtubule-associated protein 1 light chain 3; protein recruited from the cytosol (LC3-I) to the autophagosomal membrane where it is lipidated (LC3-II)) and p62/SQSTM1 (adaptor protein that serves as a link between LC3 and ubiquitinated substrates), (Klionsky et al., 2016) [1]. Data provided include analyses of protein levels of LC3 and p62 by Western-blotting and endogenous immunofluorescence experiments, but also p62 mRNA levels obtained by quantitative PCR (qPCR). To monitor the turnover of these autophagy markers and, thus, measure the flux of this pathway, cells were under starvation conditions and/or treated with bafilomycin A1 (Baf. A1) to block fusion of autophagosomes with lysosomes.
Abbreviations: Baf. A1, Bafilomycin A1; EBSS, Earle׳s Balanced Salt Solution; FBS, Fetal bovine serum; GAPDH, Glyceraldehyde 3-phosphate dehydrogenase; HFs, Human fibroblasts; LC3, Microtubule-associated protein 1 light chain 3; MEFs, Mouse embryonic fibroblasts; PBS, Phosphate-buffered saline; qPCR, Quantitative PCR; SB, Sample buffer; SDS, Sodium dodecyl sulfate; TBST, Tris-buffered saline with Tween 20
Keywords: Autophagy, LC3, p62, Western-blot
Specifications Table
| Subject area | Biochemistry and Molecular Biology |
| More specific subject area | Autophagy |
| Type of data | Figures |
| How data was acquired | Western-blotting (SDS-gel electrophoresis and semi-dry transfer; Bio-Rad equipment and ImageJ software), immunofluorescence (inverted microscope (OLYMPUS IX-51) and ImageJ software), quantitative PCR (qPCR) (Applied Biosystems 7500 PCR real system), data statistical analysis (SPSS software). |
| Data format | Analyzed |
| Experimental factors | Four cell lines (Mouse embryonic fibroblasts (MEFs), Human fibroblasts (HFs), SH-SY5Y human neuroblastoma cells, and N27 rat dopaminergic cells). Starvation-induced autophagy by incubation with Earle׳s Balanced Salt Solution (EBSS) medium, bafilomycin A1 (Baf. A1) treatment (blocking autophagosome-lysosome fusion), and dual treatment (Baf. A1+EBSS). |
| Experimental features | Western-blotting analysis of LC3 and p62 proteins using Sample buffer (SB) 1X lysis buffer, detection of endogenous LC3 and p62 immunofluorescence and p62 mRNA levels by qPCR. |
| Data source location | Not applicable |
| Data accessibility | Data is within this article |
Value of the data
-
•
This data provides characterization of basal macroautophagy flux and response to classical inducers (EBSS) and inhibitors (Baf. A1) of this mechanism in MEFs, HFs, SH-SY5Y and N27 cell lines.
-
•
The data would be valuable for further studies of autophagy dynamics in these four cell lines.
-
•
This data could give a basis for further experiments on revealing the underlying mechanism of autophagy in these cell lines.
-
•
The data support the development of transcription analysis of autophagy markers.
1. Data
In order to understand autophagy dynamics [1] and develop a robust methodological assay, we used four cell models: MEFs, HFs, SH-SY5Y human neuroblastoma cells, and N27 rat mesencephalic dopaminergic cells. LC3 and p62 protein levels were analyzed by Western-blotting assay, using SB 1X as a lysis buffer (Fig. 1). Furthermore, we measured the endogenous immunostaining of LC3 and p62 proteins in all cell models used in this report (Fig. 2). To complete this data analysis, p62 mRNA expression was performed in all cell lines (Fig. 3).
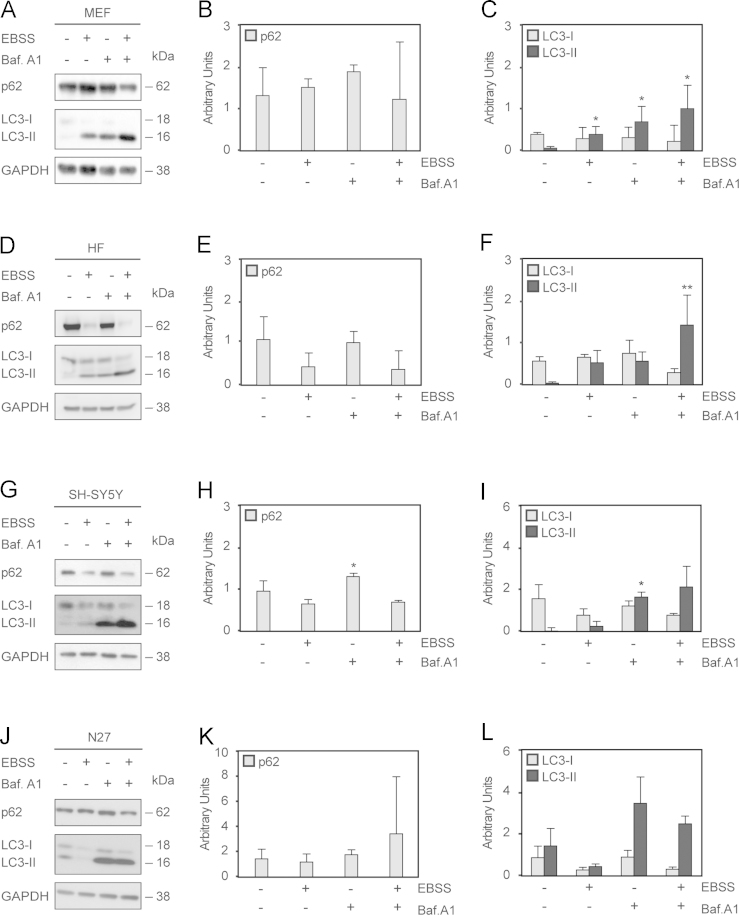
Fig. 1.
LC3 and p62 modulation after EBSS±Baf. A1 treatment in four cell models. Cells were treated with EBSS, Baf. A1 and dual treatment for 4 h, as described in Section 2. Representative blots of 3 independent experiments of MEFs (A), HFs (D) SH-SY5Y (G) and N27 rat dopaminergic cells (J) are shown. GAPDH was used as a loading control. p62 densitometry expressed in arbitrary units is shown in panels B (MEFs), E (HFs), H (SH-SY5Y) and K (N27). LC3 (isoforms I and II) densitometry expressed in arbitrary units is shown in panels C (MEFs), F (HFs), I (SH-SY5Y) and L (N27). Data represent the mean±SEM; n=3 (*p≤0.05, **p≤0.01, ***p≤0.001, related to the corresponding non-treated condition).
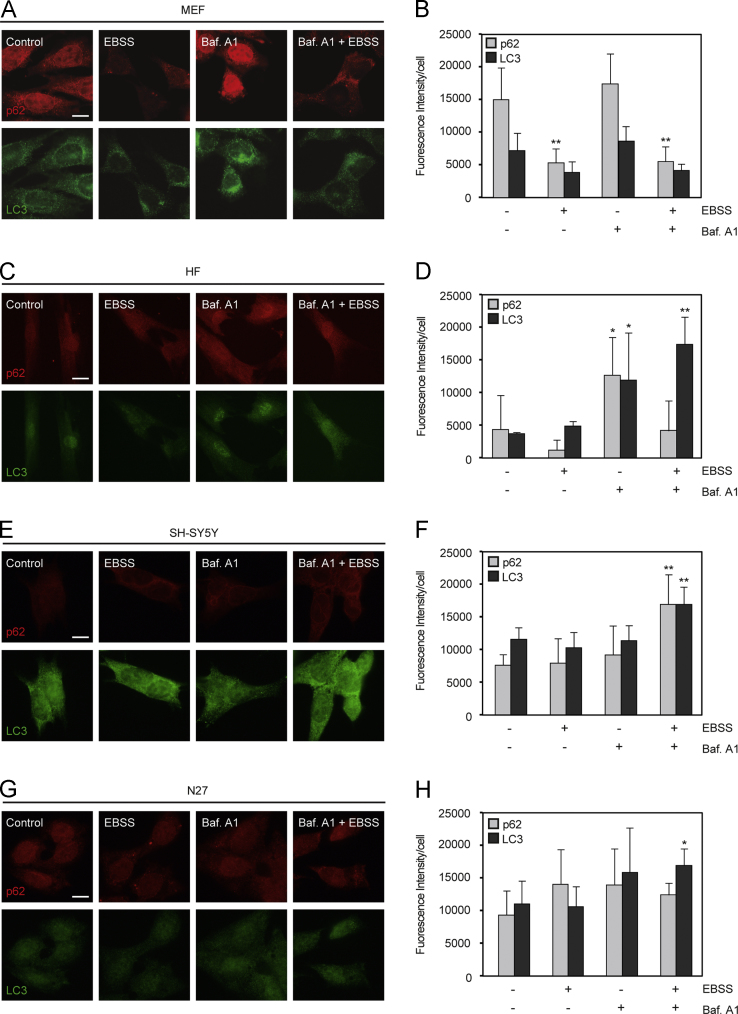
Fig. 2.
Endogenous immunofluorescence of LC3 and p62 after EBSS ± Baf. A1 treatment in four cell models. Cells were treated with EBSS, Baf. A1 and dual treatment for 4 h, as described in Section 2. Representative images from MEFs (A), HFs (C), SH-SY5Y (E) and N27 cells (G) are shown. Scale bars equate to 10 µm. Histograms show quantification of fluorescence intensity per cell from MEFs (B), HFs (D), SH-SY5Y (F) and N27 cells (H). Data represent the mean±SEM; n=3 (*p≤0.05, **p≤0.01, related to the corresponding non-treated condition).
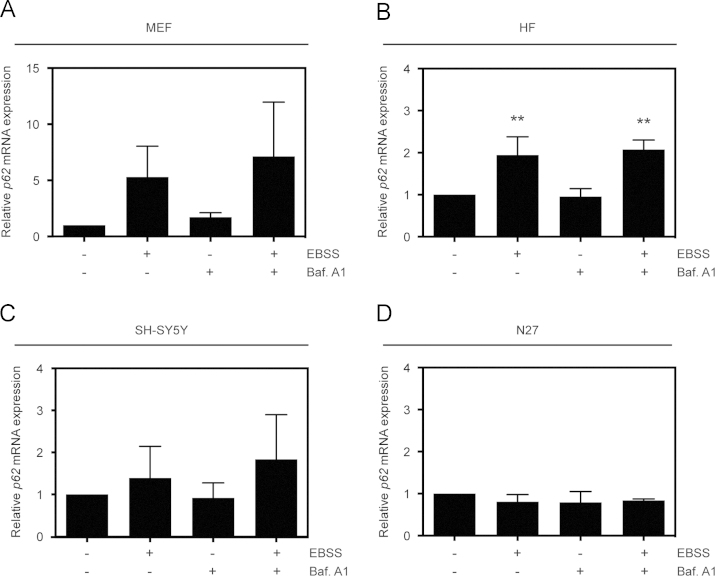
Fig. 3.
p62 mRNA analysis after EBSS±Baf. A1 treatment in four cell models. Cells were treated with EBSS, Baf. A1 and dual treatment for 4 h, as described below. Quantification of p62 mRNA expression levels from all lines studied is shown: MEFs (A), HFs (B), SH-SY5Y (C) and N27 cells (D). Relative expression was determined using GAPDH as a housekeeping gene. Data represent the mean±SEM; n=3 (**p≤0.01, related to the corresponding non-treated condition).
2. Experimental design, materials and methods
2.1. Cell culture and treatments
To perform this data analysis, we used four cell lines: MEFs, HFs, SH-SY5Y and N27 rat mesencephalic dopaminergic cells. The culture media for MEFs, HFs and SH-SY5Y were Dulbecco׳s Modified Eagle Medium-High Glucose (Sigma-Aldrich, D6546) supplemented with 10% fetal bovine serum (FBS) (Sigma-Aldrich, F7524), 1% L-glutamine (Sigma-Aldrich, G7513) and penicillin-streptomycin (Hyclone, SV30010). For N27 cell line, the culture medium was made of RPMI 1640 medium (1X) (Hyclone, SH30096.01) supplemented with 10% FBS, L-glutamine and penicillin-streptomycin.
Cells were seeded at densities of 3×105 (MEFs), 1×106 (HFs), 2×106 (SH-SY5Y) and 4×105 (N27) in 75-cm2 tissue culture flasks and incubated at 37 °C under saturating humidity in 5% CO2/95% air. Confluent cells (~80%) were trypsinized and plated into a 6 or 24-well plates at densities of 3×104 cells/mL (MEFs and HFs), 1×105 cells/mL (SH-SY5Y) and 3.5×104 cells/mL (N27).
After 24 h , the culture medium was replaced with different treatments (Control, EBSS, Baf. A1 and Baf. A1+EBSS). To induce autophagy by starvation conditions, the culture medium was replaced with EBSS (Sigma-Aldrich, E2888). To block fusion between autophagosomes and lysosomes, cells were incubated with 100 nM Baf. A1 (LC Laboratories, B-1080). For combined treatment, cells were preincubated with 100 nM Baf. A1 for 1 h and then they were washed with phosphate-buffered saline (PBS) 1X and treated with 100 nM Baf. A1 and EBSS. All treatments lasted 4 h.
2.2. Western-blotting
Following treatments, cells were processed as described previously [2]. Basically, cells were washed with PBS 1X and lysed in sample buffer (SB) 1X (2% (v/v) sodium dodecyl sulfate (SDS), 10% (v/v) glycerol, and 50 mM Tris–HCl, pH 6.8, in distilled water) by pipetting until homogenization. Protein concentration was measured based on the bicinchoninic acid assay, using bovine serum albumin as a standard. Samples were heated at 95 °C for 10 min before their quantification.
Equal amounts of protein (25–40 µg/condition) were resolved by 12% SDS-gel electrophoresis and transferred to polyvinylidene fluoride membranes, according to a partially modified conventional protocol [3]. Immunodetection included transferring (15 V during 15 min, per each membrane) and blocking of the membrane with WB blocking solution (10% w/v fat free milk in Tris-buffered saline with Tween 20 (TBST)) for 1 h at room temperature. After washing the membranes 2 times with TBST 1X, blots were incubated with the corresponding primary antibody: p62/SQSTM1 (1:5000) (BD Transduction Laboratories, 610498), LC3-B (1:5000) (Sigma–Aldrich, L7543) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (1:5000) (Millipore, MAB374) at 4 °C overnight, GAPDH (1 h at room temperature). The membranes were washed 2 times with TBST 1X and subsequently incubated with their respective HRP-conjugated secondary antibodies (1:10000) (Bio-Rad, 170–6515 and 170-5047 for rabbit and mouse antibodies, respectively) for 1 h at room temperature. Detection of bound antibodies was visualized by chemiluminescence using ECL substrate (Thermo Scientific, 32106). Quantification data analysis was performed using ImageJ software (NIH), establishing GAPDH protein levels as a loading control.
2.3. Immunofluorescence
For the detection of endogenous p62 and LC3B, cells were seeded on coverslips, fixed with paraformaldehyde (4% in PBS 1X) and permeabilized with Triton X-100 solution (0.1% in PBS 1X) for 10 min. To block non-specific binding, cells were incubated with 10% FBS in PBS 1X for 20 min, followed by incubation with primary antibodies anti-p62 (1:500) and anti-LC3B (1:500) for 1 h at room temperature. After that, cells were incubated with Alexa Fluor 488 anti-rabbit (1:1000) (Molecular Probes, A-11034) and 568 anti-mouse (1:1000) (Molecular Probes, A-11004) secondary antibodies for LC3 and p62, respectively. Finally, coverslips were mounted on microscope slides, by using fluoromount-G (SouthernBiotech, 0100–01) medium. Images were taken by using an inverted fluorescence microscope (Olympus, IX-51) equipped with a camera (Olympus, DP70). The quantitative measurement of the fluorescence signal was performed using ImageJ software analyzing at least 200 cells per condition. Immunofluorescence procedure was developed as previously described [2].
2.4. Quantitative PCR
Total RNA was extracted by RNeasy Mini Kit (Qiagen, 74104); 500 ng of total RNA were reverse-transcribed into complementary DNA by using a QuantiTect Reverse Transcription Kit (Qiagen, 205311), both according to the manufacturer׳s protocol. p62 mRNA expression was measured by qPCR with KAPA SYBR Fast reagents (KK4601), by using the primers described above. GAPDH gene expression was used as an endogenous control, and the expression level was calculated by using the (2-∆∆Ct) ratio [4].
2.5. Statistical analyses
Each experiment was repeated at least three times. The data were analyzed by two-tailed unpaired Student׳s t-test and ANOVA test where applicable, and all comparisons with a p value less than 0.05 (p<0.05) were considered statistically significant (***p<0.001, **p<0.01, *p<0.05). Non-significant results are not indicated in the figures. The data are expressed as the mean±the standard error of the mean (SEM).
Acknowledgments
We thank George Auburger (Experimental Neurology, Goethe University Medical School, Frankfurt am Main, Germany) for the MEFs, Adolfo López de Munaín (Neurology service, Instituto BioDonostia, Hospital Donostia, San Sebastian, Spain) for the HFs, and Anumantha G. Kanthasamy (Iowa State University, Ames, IA) for the N27 cells. M. R.-A. was supported by a FPU predoctoral fellowship (FPU13/01237) from Ministerio de Educación, Cultura y Deporte, Spain. R. G.-S. was supported by a Marie Sklodowska-Curie Individual Fellowship (IF-EF) from the European Commission. J. M. F. received research support from the Ministerio de Economia y Competitividad, Spain, CIBERNED (CB06/05/004 and PI12/02280). R. A. G.-P. was supported by a "Contrato destinado a la retención y atracción del talento investigador, TA130009" from Junta de Extremadura, Spain, and she received research support from Ministerio de Economía y Competitividad, Spain (PI14/00170). This work is supported also by “Fondo Europeo de Desarrollo Regional” (FEDER), from European Union. The authors also thank FUNDESALUD for helpful assistance.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.dib.2016.02.085.
Contributor Information
José M. Fuentes, Email: jfuentes@unex.es.
Rosa A. González-Polo, Email: rosapolo@unex.es.
Appendix A. Supplementary material
Supplementary material
References
- 1.Klionsky D.J., Abdelmohsen K., Abe A. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2016;12(1):1–222. doi: 10.1080/15548627.2015.1100356. (3rd ed.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gomez-Sanchez R., Pizarro-Estrella E., Yakhine-Diop S.M. Routine Western blot to check autophagic flux: cautions and recommendations. Anal. Biochem. 2015;477:13–20. doi: 10.1016/j.ab.2015.02.020. [DOI] [PubMed] [Google Scholar]
- 3.Fuentes J.M., Lompre A.M., Moller J.V. Clean Western blots of membrane proteins after yeast heterologous expression following a shortened version of the method of Perini et al. Anal. Biochem. 2000;285(2):276–278. doi: 10.1006/abio.2000.4784. [DOI] [PubMed] [Google Scholar]
- 4.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material