Abstract
Extracellular vesicles, including exosomes, are nanoscale vesicles that carry molecular information of parental cells. They are being pursued as biomarkers of cancers that are difficult to detect or serially follow. Here we present a compact sensor technology for rapid, on-site exosome screening. The sensor is based on an integrated magnetic-electrochemical assay: exosomes are immunomagnetically captured from patient samples, and profiled through electrochemical reaction. By combining magnetic enrichment and enzymatic amplification, the approach enables i) highly sensitive, cell-specific exosome detection, and ii) sensor miniaturization and scale-up for high throughput measurements. As a proof-of-concept, we implemented a portable, eight-channel device, and applied it to screen extracellular vesicles in plasma samples from ovarian cancer patients. The sensor allowed for the profiling of multiple protein markers simultaneously within an hour, outperforming conventional methods in assay sensitivity and speed.
Keywords: extracellular vesicles, electrochemical sensing, cancer, diagnostics, point-of-care
Growing evidence has positioned extracellular vesicles (EVs) as an effective readout of cancer management1-6. Exosomes, in particular, have emerged as a potent biomarker. Exosomes are nanoscale vesicles actively secreted by cells. These vesicles carry molecular constituents of their originating cells, including transmembrane and cytosolic proteins7, mRNA8, DNA9 and microRNA10, and can thus serve as cellular surrogates11. Combined with their relative abundance and ubiquitous presence in bodily fluids (e.g. serum, ascites, urine, CSF)12-14, exosomes can offer unique advantages for longitudinal monitoring15-17. Exosome analyses are minimally invasive and afford relatively unbiased readouts of the entire tumor burden, less affected by the scarcity of the samples or intra-tumoral heterogeneity5.
Analyzing extracellular vesicles in routine clinical settings, however, still remain a difficult task, mainly due to the lack of adequately sensitive and fast assay platforms, particularly for protein analyses. Flow cytometry provides high throughput detection18, yet tends to miss small vesicles (<200 nm) because of weak light scattering19. New counting approaches based on particle-tracking or dynamic light scattering can offer more accurate particle counts, >103-fold higher than those reported by flow cytometry20, but yield limited molecular information. Conversely, conventional molecular assays (e.g. Western blot, ELISA) require large amounts of samples, and become impractical for clinical research needs, notably serial analyses, large patient cohorts, or limited specimens in biorepositories.
We have previously developed miniaturized systems to facilitate EV analyses in clinical environments. These systems include microfluidic devices for sample preparation21, 22 and analytical tools for protein analyses3, 5. Ensuing clinical studies with patient samples established EVs’ clinical utility for cancer diagnosis and treatment monitoring3, 5: tumor-specific exosomes could be identified based on their unique transmembrane protein signature, and exosomal protein changes could be correlated with treatment responses. Translating these systems into routine clinical tests, however, is still limited by i) separate procedures and devices required for exosome isolation and detection; ii) technical complexities involved in device fabrication and operation; and iii) relatively high costs of analytical instruments (e.g. NMR detector, spectral microscope).
We reasoned that electrochemical sensing could be an effective detection modality that is easily applicable to clinical settings. Electrochemical sensing could achieve high sensitivity through signal amplification with redox-active reporters23-28. The readout system measures electrical currents, and can be realized as a compact and low-power portable device29, 30. Harnessing electrochemical sensing for exosome analyses, however, is challenged by both assay design and hardware development: i) combining exosome isolation and detection in a single platform; ii) providing fast, high-throughput assays for multiplexed screening; and iii) making the system affordable and easy-to-use.
We herein report on a new integrated approach, iMEX (integrated magnetic-electrochemical exosome), for fast, streamlined exosome analyses. The iMEX combines two orthogonal modalities, magnetic selection and electrochemical detection. Magnetic beads are used for exosome capture and labeling; captured exosomes are detected via electrochemical sensing. The iMEX offers distinct advantages: i) cell-specific exosomes can be isolated directly from complex media without need for extensive filtration or centrifugation; ii) the assay can achieve high detection sensitivity through magnetic enrichment and enzymatic amplification; iii) through the electrical detection scheme, sensors can be miniaturized and expanded for parallel measurements. To prove the concept, we implemented a portable iMEX platform with eight detection channels. The iMEX was able to detect exosomes at a sensitivity of <105 vesicles. Importantly, it only used 10 μL of samples and generated readouts within 1 hour. We further explored iMEX's potential clinical utility by profiling EVs collected from ovarian cancer patients. With its capacities for fast, high-throughput, and on-spot analysis, the iMEX could accelerate the transition of EV and exosome analyses towards routine clinical testing.
RESULTS AND DISCUSSION
iMEX platform
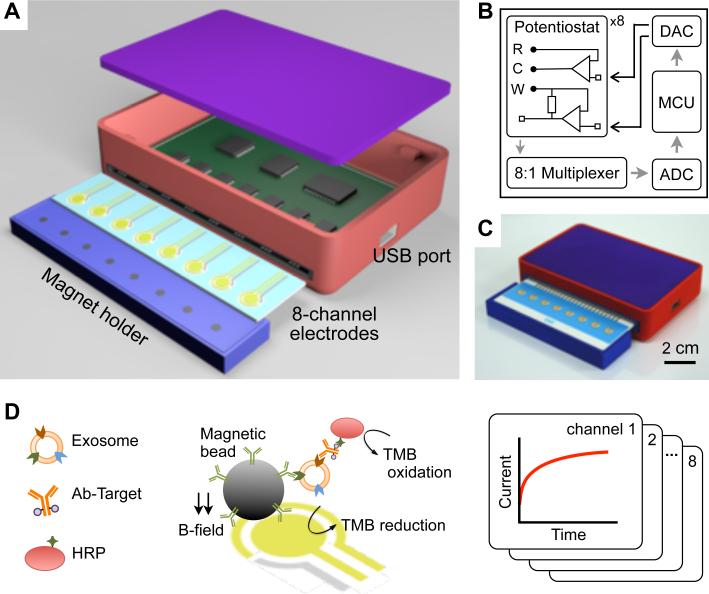
The miniaturized iMEX system had eight independent channels (Figure 1A). Each channel was equipped with a potentiostat capable of measuring a wide range of current (±7.5 μA). The input signal was conditioned by a low-pass filter (cut-off frequency, 5 Hz) to suppress high frequency noise (Supplementary Figure 1). The eight potentiostats were connected to a digital-to-analog converter for potential control, an analog-to-digital converter for signal digitization, a multiplexer for channel selection, and a micro-controller unit for system operation (Figure 1B; Supplementary Figure 2). We packaged the device as a handheld unit (Figure 1C). A card-edge connector was used for the quick attachment of the electrode cartridge. A magnet holder, containing 8 cylindrical magnets, was placed underneath the electrode cartridge. These magnets are used to concentrate magnetic beads to the sensor surface. The overall device cost was <$50. The system performance was comparable to that of commercial equipment (SP-200, Bio-Logic; Supplementary Figure 1B), however at much smaller footprint and cost. Furthermore, the iMEX effectively provided a simultaneous readout from all electrodes through rapid polling of each channel (50 msec per channel). All data were monitored and analyzed by custom-designed software (Supplementary Figure 3).
Figure 1. Integrated magnetic-electrochemical exosome (iMEX) platform.
(A) Sensor schematic. The sensor can simultaneously measure signals from eight electrodes. Small cylindrical magnets are located below the electrodes to concentrate immunomagnetically captured exosomes. (B) Circuit diagram. The sensor system has eight potentiostats, an 8-to-1 multiplexer, an analog-to-digital converter (ADC), a digital-to-analog converter (DAC), and a micro-controller unit (MCU). Each potentiostat has three electrodes: reference (R), counter (C), and working (W). (C) A packaged device. The device has a small form factor (9 × 6 × 2 cm3). (D) Schematic of iMEX assay. Exosomes are captured on magnetic beads directly in plasma and labeled with HRP enzyme for electrochemical detection. The magnetic beads were coated with antibodies against CD63, an enriched surface marker in exosomes. The working (W) and the counter (C) electrodes were made of gold (Au), and the reference electrode (R) was made of silver/silver chloride (Ag/AgCl). Eight channels are simultaneously monitored for high-throughput analysis. HRP, horseradish peroxidase; TMB, 3,3’,5,5’-tetramethylbenzidine.
Figure 1D summarizes the iMEX assay scheme. Exosomes are first captured onto immunomagnetic beads. Secondary antibodies with an oxidizing enzyme (horseradish peroxidase; HRP) are then used, followed by mixing the beads with chromogenic electron-mediators (3,3’,5,5’-tetramethylbenzidine; TMB) which generate electrical current when HRP is encountered (Supplementary Fig. 4; see Methods and Experimental section for details). Using magnetic beads significantly simplifies the assay procedures: excess agents (e.g. antibodies, enzymes) can be removed via magnetic washing, and captured exosomes can be magnetically concentrated on the electrodes to improve the detection sensitivity.
Assay optimization
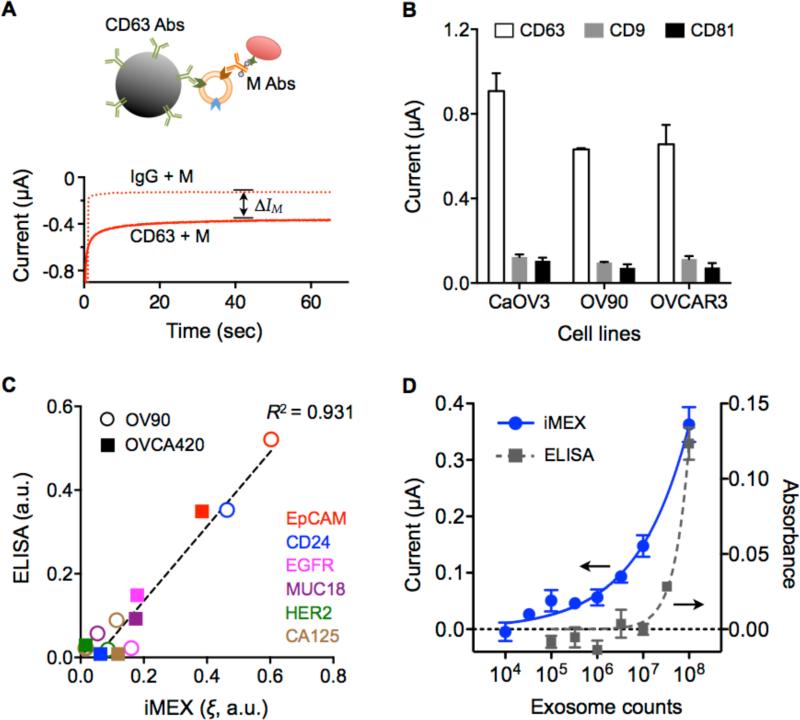
We optimized the iMEX assay protocol. We applied the chronoamperometry method for signal detection: the electrical current generated from TMB reduction was monitored while a reduction potential (−100 mV versus Ag/AgCl reference electrode) was applied to a working electrode. The current level (I) reached a plateau within 1 minute after the reduction potential was applied (Figure 2A). We averaged the current level (I) from 40 to 45 seconds as a representative value.
Figure 2. Characteristics of the iMEX diagnostic platform.
(A) Schematic of the electrochemical measurement in the iMEX assay. With –100 mV reduction potential, the current level was reached a plateau within 1 minute. The current difference between the CD63-bead and IgG-bead samples (ΔIM) was used as a representative value of a target protein marker. Abs, antibodies. M, marker. (B) Signal comparison of three tetraspanin markers (CD63, CD9, and CD81) in cancer exosomes. Signals from CD63 were much higher than those from other markers in exosomes collected from ovarian cancer cell lines (CaOV3, OV90, and OVCAR3). (C) Comparison between iMEX and ELISA. Six surface proteins were profiled in two ovarian cancer cell lines (OV90 and OVCA420). The results showed high correlation (R2 = 0.931). a.u., arbitrary unit. (D) Varying number of exosomes were spiked into human plasma and assayed by iMEX and ELISA. The detection limits were 3×104 (iMEX) and 3×107 (ELISA). All measurements were performed in triplicate, and the data are displayed as mean ± SD.
To capture exosomes, we used magnetic beads that were coated with antibodies against tetraspanin, transmembrane proteins enriched in exosomes5, 31, 32. We first compared signal levels with differently sized beads (diameters, 2.7 μm and 8.8 μm). When the total surface area of beads was matched to capture similar amount of exosomes, the measured signal levels were nearly identical (Supplementary Figure 5A). This result can be explained with diffusivity in porous media: The effective diffusivity (De) for stacked beads can be expressed as , where D0 is the diffusivity in free media, and ε is the porosity of the structure48. In case of uniformly sized beads, both ε (≥ 0.47) and m (= 3/2) are bead-size independent; the iMEX signals are thus expected to remain constant. We opted to use 2.7-μm beads; bigger beads tended to sediment, requiring frequent shaking of samples. Compared to the no-enrichment scheme, magnetic enrichment led to ~72% increase in the analytical signal (Supplementary Figure 5B).
We next tested three representative tetraspanin proteins (CD63, CD9, CD81) as a target; these markers are reportedly enriched in exosomes13, 33-39. We prepared 2.7-μm magnetic beads specific to each marker (see Methods and Experimental section for details). When applied to exosomes from different cell lines, CD63-based capture showed consistently high signal (Figure 2B); we thus opted to use CD63 as a marker for exosome enrichment.
For each target marker (M), we prepared a pair of magnetic beads: one conjugated with antibodies against CD63 (CD63-beads) and the other with antibodies against iso-type matched IgG (IgG-beads). Exosomes were mixed with each bead-type and subsequently labeled with antibodies against a target marker; the net signal difference ΔIM (= ICD63+M – IIgG+M; Figure 2A) was then obtained. We used ΔICD63 to estimate the total exosome load, and defined a normalized metric ξM (= ΔIM/ΔICD63) as the expression level of a target marker (M). Note that such scaling would compensate for variations in exosome numbers among samples.
iMEX validation
We applied the developed iMEX protocol to profile exosomes for transmembrane proteins. For this validation study, we harvested exosomes from cell culture (OV90, OVCA420) through a conventional method (see Methods and Experimental section for details), and spiked them into phosphate buffered saline (PBS) solution (~109 exosomes/mL). Samples were aliquoted, and processed by iMEX and enzyme-linked immunosorbent assays (ELISA). Comparative analysis showed high correlation between two methods (Figure 2C; R2 = 0.931), confirming iMEX's analytical capacity. The iMEX assay, however, was faster (1 hr) and consumed smaller amounts of samples (10 μL) than ELISA (5 hr, 100 μL).
We further tested iMEX for detecting exosomes in biofluids. Cancer exosomes were collected from cell culture (OV90), and varying numbers of exosomes were spiked into undiluted human plasma. Titration experiments established the limit of detection (LOD) of 3×104 exosomes, with the dynamic ranges spanning four orders of magnitude (Figure 2D). Similar measurements with ELISA required more than 107 exosomes for reliable detection. Using matched controls (IgG-beads) was important to compensate for background signals from sample-dependent, non-specific exosome binding.
Profiling of protein markers in cell derived exosomes
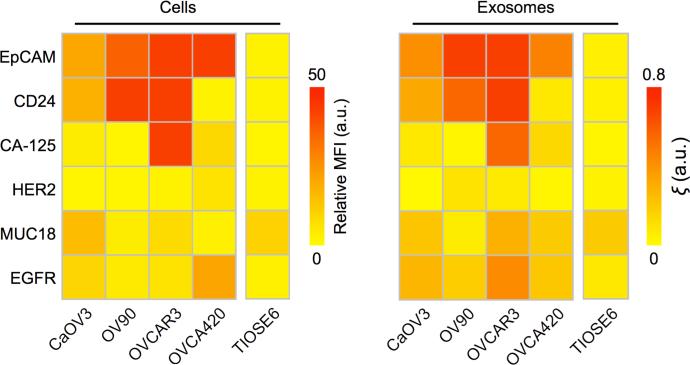
We applied the iMEX to screen exosomal surface markers from a panel of ovarian cancer cell lines. Because iMEX enriches CD63-positive (CD63+) exosomes and labels them for target proteins, we were able to examine how closely CD63+ exosomes reflect their cells of origin. We chose six representative surface markers based on prior studies: epithelial cell adhesion molecule (EpCAM)40, CD2441, cancer antigen 125 (CA125)42, 43, human epidermal growth factor 2 (HER2)44, 45, mucin 18 (MUC18)46, and epidermal growth factor receptor 2 (EGFR)47. The cellular expression levels of these markers were measured with flow cytometry; exosomes were harvested from the conditioned cell-culture media, and profiled with iMEX. The molecular profiles of cells and CD63+ exosomes were highly correlated (Figure 3), which supported the use of exosomes as cellular surrogates.
Figure 3. Profiling of surface proteins in ovarian cancer cells and their secreting exosomes.
Four ovarian cancer cell lines (CaOV3, OV90, OVCAR3, and OVCA420) and one normal cell line (TIOSE6) were screened for six putative cancer markers (via flow cytometry, left panel). Cell-derived exosomes were immunomagnetically captured (CD63-specific) and assayed by iMEX (right panel). The profiling data showed a good match between cells and CD63-positive exosomes. The iMEX assay was in duplicate, and the mean values are displayed.
Clinical: Direct analyses of plasma from patients with ovarian cancer
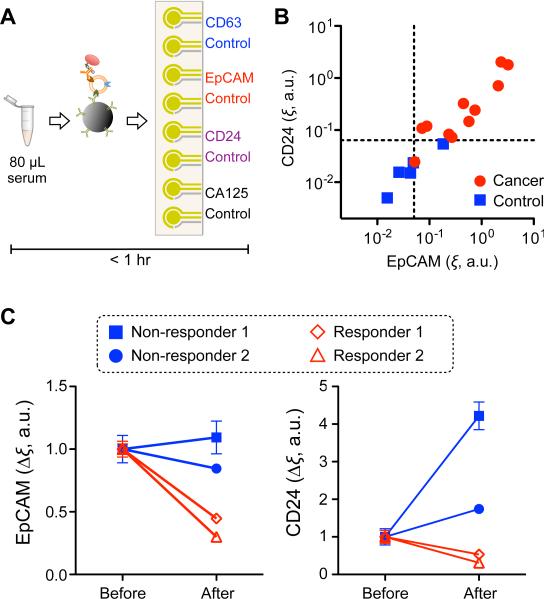
The iMEX assay isolates EVs directly from plasma or serum, and allows profiling in a rapid, high-throughput manner — key for successful integration into the clinical workflow. To demonstrate clinical feasibility, we customized the iMEX assay for ovarian cancer EV detection in blood (Figure 4A). Clinical plasma samples were aliquoted without any purification, and each aliquot (10 μL per marker) was incubated with magnetic beads for EV capture (15 min), followed by magnetic washing. The bead-bound EVs were consecutively labeled for target markers (15 min) and HRP (15 min), and loaded onto the device. With the 8-electrodes independently operated, we were able to simultaneously measure four different markers (CD63, EpCAM, CD24, and CA125) along with their respective IgG-controls. The IgG-controls were beneficial to specific detection of target molecules among non-purified clinical samples.
Figure 4. iMEX for clinical applications.
(A) iMEX assay for clinical sample analysis. 10 μL of plasma per marker was used, and the entire assay was completed within 1 hour without filtration and centrifugation processes. (B) Plasma samples from ovarian cancer patients (n = 11) and healthy controls (n = 5) were analyzed with the iMEX assay. EpCAM and CD24 levels were much higher in cancer patients. The EpCAM and CD24 expression levels (ξEpCAM vs. ξCD24) were highly correlated (R2 = 0.870). (C) Longitudinal monitoring of drug treatment responses. Plasma samples from four ovarian cancer patients were analyzed with the iMEX assay. EpCAM and CD24 levels in responders were decreased significantly, but their levels in non-responders were stable (EpCAM) or increased (CD24) after treatment. All measurements were in duplicate. a.u., arbitrary unit.
We tested single-time point plasma samples from 11 ovarian cancer patients and five healthy controls. The expression levels of EpCAM and CD24 in EVs were much higher in ovarian cancer patients than healthy controls, and both metrics showed high correlation (Figure 4B, Supplementary Figure 6). We next examined iMEX's potential for serial EV testing by measuring EpCAM and CD24 in plasma collected at two time points (2 months apart) from four ovarian cancer patients undergoing drug treatment. The iMEX assays were conducted blinded to treatment response. For ‘non-responding’ patients, expression levels of EpCAM and CD24 increased while ‘responding’ patients displayed a significant decrease in both markers (Figure 4C). The level of CD24 showed steeper increases than that of EpCAM for non-responders.
CONCLUSION
We developed the iMEX technology to aid the translation of exosome analysis into clinical settings. A unique feature of iMEX is the integration of vesicle isolation and detection into a single platform. The use of magnetic actuation simplifies vesicle isolation and subsequent assay steps, and the electrochemical sensing facilitates high-throughput screening and sensor miniaturization. The current study validated these concepts: i) a portable detection system was implemented with the capacity for parallel measurements; ii) the iMEX enriched exosomes directly from blood, and profiled them for molecular information; iii) the entire iMEX assay (i.e., exosome isolation, labeling, detection) was completed within 1 hour while consuming only 10 μL of plasma per marker. We also demonstrated iMEX's clinical potential by profiling EVs within blood collected from ovarian cancer patients.
The bead-based magnetic enrichment brings several advantages in the iMEX measurement. First, the method provides a convenient way of concentrating signal sources on the electrodes, which enhances the detection sensitivity. Second, compared to the surface-based capture wherein antibodies are immobilized on the chip surface, the bead-based method is amenable to reliable and simpler conjugation chemistry, and benefits from faster binding kinetics between antibodies and exosomes. Third, the bead-bound vesicles could be readily recovered for downstream molecular analyses in tandem with iMEX. For instance, bead-bound EVs can be eluted31 or lysed3 to profile their nucleic acid contents.
The iMEX system can complement the other exosome analysis platforms we have developed (Supplementary Table 1), particularly the nano-plasmonic exosome (nPLEX) sensor3. The nPELX is well suited to conduct massively parallel exosome screening, potentially for central clinical laboratories; it has high detection sensitivity, down to ~103 exosomes, and is equipped with >1000 detection sites3. The system complexity and the requirement for nanofabrication, however, is a limiting factor for nPLEX's routine clinical applications. The iMEX system has lower sensitivity and throughput than nPLEX, but is affordable and miniaturized for on-site exosome detection.
In the current work, we focused on profiling CD63+ EV population (exosomes), which was motivated by two factors: i) the signal from CD63 capture was the highest (Figure 2B) among the tetraspanin markers tested; and ii) we and others have previously shown that ovarian cancer exosomes are enriched with CD633, 35. The iMEX profiling found a high correlation in protein expression between CD63-positive exosomes and their parent cells (Figure 3); this result validated the potential use of CD63-positive exosomes as cellular surrogates. However, we note the need to extend the exosome-capture strategy, considering that diverse EV types (e.g. CD63-negative) may exist in patient samples. Examining these populations could yield more precise information to capture tumor heterogeneity. The iMEX method can be readily adopted for such purposes by changing capture antibodies.
We envision multiple directions to further advance the iMEX technology. First, the assay throughput can be improved by increasing the number of detection sites. Electrochemical sensing is ideally suited for such a scale-up: the sensing elements (electrodes) can be readily microfabricated into a large array format, and signals (electrical currents) can be read out by compact electronics with high-speed multiplexing. Second, the detection sensitivity could be improved by exploring new designs for electrochemical signal detection. The signal level is correlated with the surface area of a sensor and the amount of enzyme bound to target entities; thus, higher sensitivity can be achieved by using a nanostructured sensor surface or multi-label nanoparticles24, 49, 50. Third, detection targets can be expanded to include other exosomal constituents. For example, exosomes carry various nucleic acids (e.g. mRNA, microRNA); analyzing nucleic acids along with exosomal proteins would provide more accurate snapshot of tumor states6, 8, 9, 51. Electrochemical sensing has been applied to detect a trace amount of nucleic acids (<1 pM) without PCR amplification23, 28, 52. We expect similar approaches could be adopted to profile exosomal nucleic acids. The resulting iMEX could be a powerful clinical tool for affordable, scalable, and comprehensive exosome analyses, thereby deepening our insights into tumor biology and accelerating effective cancer management.
METHODS AND EXPERIMENTAL
Fabrication of the iMEX system
The device consists of a micro-controller (Atmega328, Atmel Corporation), a digital-to-analog converter (DAC8552, Texas Instruments), an analog-to-digital converter (ADC161S626, Texas Instruments), a multiplexer (ADG708, Analog Devices), and eight potentiostats. Each potentiostat consists of two operational amplifiers (AD8606, Analog Devices): one amplifier maintains the potential difference between a working electrode and a reference electrode, and the other one works as a transimpedance amplifier to convert a current to a voltage signal. The current measuring range of the transimpedance amplifier was ±7.5 μA. The eight-channel electrodes are commercially available (DropSens, Spain).
Preparation of immunomagnetic beads
5 mg of magnetic beads coated with epoxy groups (Dynabeads M-270 Epoxy, Invitrogen) were suspended in 1 mL of 0.1M sodium phosphate solution at room temperature for 10 minutes. The magnetic beads were separated from the solution with a permanent magnet and re-suspended in 100 μL of the same solution. 100 μg of antibodies against CD63 (Ancell) or respective IgG (Ancell) were added and mixed thoroughly. 100 μL of 3M ammonium sulfate solution was added, and the whole mixture was incubated overnight at 4°C with slow tilt rotation. The beads were washed twice with phosphate buffer saline (PBS) solution and finally re-suspended in 2 mL of PBS with 1% bovine serum albumin (BSA). More details can be found in the manual provided by the manufacturer of the magnetic beads.
Biotinylation of labeling antibodies
10 mM Sulfo-NHS-Biotin (Pierce) solution in PBS was incubated with antibodies for two hours at room temperature. Unreacted Sulfo-NHS-Biotin was removed using Zeba spin desalting column, 7K MWCO (Thermo Scientific). Antibodies were kept at 4 °C until use.
iMEX assay
10 μL of exosomes-spiked PBS solution (or plasma) was mixed with 50 μL of the immunomagnetic bead solution for 15 minutes at room temperature. The bead concentration was determined according to the following criterion: , where Cb and Ce are the bead and the exosome concentrations, respectively; Vb and Ve are the volume of the bead solution and the exosome-spiked solution (or plasma), respectively; Rb and Re are the mean radius of beads and exosomes, respectively. This requirement ensured that sufficient bead surface was available for exosome capture. In our experiment condition, Re ~ 50 nm, Rb = 1.4 μm, and Ce ~ 1010 /mL. Therefore, we adjusted the bead concentration to ~108 /mL. The magnetic beads were separated from the solution with a permanent magnet and re-suspended in 80 μL of PBS (1% BSA). After 5 seconds of vortexing, the beads were separated and re-suspended in 80 μL of PBS (1% BSA). 10 μL of antibodies of interest (20 μg/mL in PBS) were mixed with the beads for 15 minutes at room temperature. The magnetic beads were separated and washed as described before, and they were re-suspended in 50 μL of PBS (1% BSA). 5 μL of streptavidin-conjugated HRP enzymes (1:100 diluted in PBS) were mixed with the beads for 15 minutes at room temperature. The magnetic beads were separated and washed as described before, and they were re-suspended in 7 μL of PBS. The prepared bead solution and 20 μL of UltraTMB solution (ThermoFisher Scientific) were loaded on top of the screen-printed electrode. After 3 minutes, chronoamperometry measurement was started with the electrochemical sensor. The current levels in the range of 40-45 seconds were averaged.
Enzyme-linked immunosorbent assay (ELISA)
CD63 antibody (Ancell) and IgG1 antibody (Ancell) were dilute to 5 μg/mL concentration in PBS and added to the Maxisorp 96 well plate (Nunc) for overnight incubation at 4°C. After washing with PBS, 2% BSA in PBS blocking solution was added to the plate for 1 hour incubation at room temperature. Subsequently, 108 exosomes in 100 μL PBS were added to each well for 1 hour incubation at room temperature. After discarding the blocking solution, antibodies (1 μg/mL) against various markers were added to each well and incubated at room temperature for 1 hour. Unbounded antibodies were washed with PBS three times. Streptavidin-HRP molecules were added to the each well for 1 hour at room temperature. After washout with PBS, the chemiluminescence signal was measured.
Flow cytometry
5×105 cells per antibody were used for flow cytometry experiments. Cells were fixed with 4% paraformaldehyde for 10 minutes at room temperature, and then washed with PBS (0.5% BSA). Subsequently, cells were blocked with BSA (0.5% in PBS) and then incubated with primary antibodies (4 μg/mL). After primary antibody incubation, cells were washed, incubated with fluorophore conjugated secondary antibody (2 μg/mL; Abcam) and washed. The fluorescence signals from the labeled cells were measured using BD LSRII Flow Cytometer (BD Biosciences). Mean fluorescent intensities (MFIs) recorded were normalized using the following formula [(signal-IgG isotype control)/secondary]. Blocking and incubation with antibodies (primary and secondary) were preformed for 30 min each at room temperature. Every washing step comprised of three 5-min washes at 300 g with PBS (0.5% BSA).
Cell culture
OV90, OVCAR3, OCVA420, and TIOSE6 cells were grown in RPMI-1640 medium (Cellgro). CaOV3 were cultured in Dulbecco's modified essential medium (DMEM, Cellgro). All media were supplemented with 10% FBS and penicillin-streptomycin (Cellgro). All cell lines were tested and were free of mycoplasma contamination (MycoAlert Mycoplasma Detection Kit, Lonza, LT07-418).
Exosome isolation from cultured cells
We used a conventional method to harvest exosomes from cell culture media. Cells at passages 1–15 were cultured in vesicle-depleted medium (with 5% depleted FBS) for 48 hours. Conditioned medium from ~107 cells was collected, centrifuged at 300g for five minutes. Supernatant was filtered through a 0.2-μm membrane filter (Millipore) and concentrated by 100,000g for one hour. After the supernatant was removed, exosome pellet was washed with PBS and centrifuged at 100,000g for 1 hour. Exosome pellet was resuspended in PBS.
Clinical samples preparation
The study was approved by the Institutional Review Board at the Dana-Farber/Harvard Cancer Center (PI: Castro), and the procedures followed were in accordance with institutional guidelines. Informed consent was obtained from all subjects (n = 11). Peripheral blood was withdrawn (~15 mL) from patients with ovarian cancer and centrifuged at 400 g for 15 minutes to separate plasma from red blood cells and buffy coat. 10 μL of plasma was used for each surface marker analysis.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Dr. Breakefield (Massachusetts General Hospital) for many helpful discussions and granting the use of laboratory resources; and Dr. Birrer (Massachusetts General Hospital) for the gift of the OVCA420 cells. This work was supported in part by US NIH Grants R01-HL113156 (to H.L.), R01-EB00462605 (to R.W.), P01CA069246 (to R.W.), T32CA79443 (to R.W.), K12CA087723-11A1 (to C.M.C.), Ovarian Cancer Research Fund Liz Tilberis Early Career Award (to C.M.C.), DOD Ovarian Cancer Research Program Award W81XWH-14-1-0279 (to H.L. and C.M.C.), and Basic Science Research Program (NRF-2014R1A6A3A03060030 to J.P.) funded by the Ministry of Education, South Korea.
Footnotes
AUTHOR CONTRIBUTIONS S.J., J.P., D.P., C.M.C, R.W., and H.L. designed the study, analyzed data, prepared figures and wrote the manuscript. S.J., J.P., and D.P. performed the research. All authors contributed to the manuscript.
Conflict of Interest
The authors declare no competing financial interest.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publication website at DOI:
REFERENCES
- 1.Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK, Becker A, Hoshino A, Mark MT, Molina H, Xiang J, Zhang T, Theilen TM, Garcia- Santos G, Williams C, Ararso Y, Huang Y, Rodrigues G, Shen TL, Labori KJ, et al. Pancreatic Cancer Exosomes Initiate Pre-Metastatic Niche Formation in the Liver. Nat. Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, Singh S, Williams C, Soplop N, Uryu K, Pharmer L, King T, Bojmar L, Davies AE, Ararso Y, Zhang T, et al. Tumour Exosome Integrins Determine Organotropic Metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Im H, Shao H, Park YI, Peterson VM, Castro CM, Weissleder R, Lee H. Label-free Detection and Molecular Profiling of Exosomes With a Nano-Plasmonic Sensor. Nat. Biotechnol. 2014;32:490–495. doi: 10.1038/nbt.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N, Reissfelder C, Pilarsky C, Fraga MF, Piwnica-Worms D, Kalluri R. Glypican-1 Identifies Cancer Exosomes and Detects Early Pancreatic Cancer. Nature. 2015;523:177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shao H, Chung J, Balaj L, Charest A, Bigner DD, Carter BS, Hochberg FH, Breakefield XO, Weissleder R, Lee H. Protein Typing of Circulating Microvesicles Allows Real-time Monitoring of Glioblastoma Therapy. Nat. Med. 2012;18:1835–1840. doi: 10.1038/nm.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shao H, Chung J, Lee K, Balaj L, Min C, Carter BS, Hochberg FH, Breakefield XO, Lee H, Weissleder R. Chip-based Analysis of Exosomal mRNA Mediating Drug Resistance in Glioblastoma. Nat. Commun. 2015;6:6999. doi: 10.1038/ncomms7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graner MW, Alzate O, Dechkovskaia AM, Keene JD, Sampson JH, Mitchell DA, Bigner DD. Proteomic and Immunologic Analyses of Brain Tumor Exosomes. FASEB J. 2009;23:1541–1557. doi: 10.1096/fj.08-122184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WTJ, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma Microvesicles Transport RNA and Proteins That Promote Tumour Growth and Provide Diagnostic Biomarkers. Nat. Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO, Skog J. Tumour Microvesicles Contain Retrotransposon Elements and Amplified Oncogene Sequences. Nat. Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosomemediated Transfer of Mrnas and Micrornas is a Novel Mechanism of Genetic Exchange Between Cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 11.Kalra H, Simpson RJ, Ji H, Aikawa E, Altevogt P, Askenase P, Bond VC, Borras FE, Breakefield X, Budnik V, Buzas E, Camussi G, Clayton A, Cocucci E, Falcon-Perez JM, Gabrielsson S, Gho YS, Gupta D, Harsha HC, Hendrix A, et al. Vesiclepedia: A Compendium for Extracellular Vesicles With Continuous Community Annotation. PLoS Biol. 2012;10:e1001450. doi: 10.1371/journal.pbio.1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raimondo F, Morosi L, Chinello C, Magni F, Pitto M. Advances in Membranous Vesicle and Exosome Proteomics Improving Biological Understanding and Biomarker Discovery. Proteomics. 2011;11:709–720. doi: 10.1002/pmic.201000422. [DOI] [PubMed] [Google Scholar]
- 13.Théry C, Zitvogel L, Amigorena S. Exosomes: Composition, Biogenesis and Function. Nat. Rev. Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 14.Théry C, Ostrowski M, Segura E. Membrane Vesicles as Conveyors of Immune Responses. Nat. Rev. Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 15.Andre F, Schartz NE, Movassagh M, Flament C, Pautier P, Morice P, Pomel C, Lhomme C, Escudier B, Le Chevalier T, Tursz T, Amigorena S, Raposo G, Angevin E, Zitvogel L. Malignant Effusions and Immunogenic Tumour-derived Exosomes. Lancet. 2002;360:295–305. doi: 10.1016/S0140-6736(02)09552-1. [DOI] [PubMed] [Google Scholar]
- 16.Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, Garcia-Santos G, Ghajar C, Nitadori-Hoshino A, Hoffman C, Badal K, Garcia BA, Callahan MK, Yuan J, Martins VR, Skog J, Kaplan RN, Brady MS, et al. Melanoma Exosomes Educate Bone Marrow Progenitor Cells Toward a Pro-metastatic Phenotype Through Met. Nat. Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grange C, Tapparo M, Collino F, Vitillo L, Damasco C, Deregibus MC, Tetta C, Bussolati B, Camussi G. Microvesicles Released From Human Renal Cancer Stem Cells Stimulate Angiogenesis and Formation of Lung Premetastatic Niche. Cancer Res. 2011;71:5346–5356. doi: 10.1158/0008-5472.CAN-11-0241. [DOI] [PubMed] [Google Scholar]
- 18.Rubin O, Crettaz D, Canellini G, Tissot JD, Lion N. Microparticles in Stored Red Blood Cells: an Approach Using Flow Cytometry and Proteomic Tools. Vox Sang. 2008;95:288–297. doi: 10.1111/j.1423-0410.2008.01101.x. [DOI] [PubMed] [Google Scholar]
- 19.van der Pol E, van Gemert MJ, Sturk A, Nieuwland R, van Leeuwen TG. Single vs Swarm Detection of Microparticles and Exosomes By Flow Cytometry. J. Thromb. Haemost. 2012;10:919–930. doi: 10.1111/j.1538-7836.2012.04683.x. [DOI] [PubMed] [Google Scholar]
- 20.Lawrie AS, Albanyan A, Cardigan RA, Mackie IJ, Harrison P. Microparticle Sizing By Dynamic Light Scattering in Fresh-frozen Plasma. Vox Sang. 2009;96:206–212. doi: 10.1111/j.1423-0410.2008.01151.x. [DOI] [PubMed] [Google Scholar]
- 21.Rho J, Chung J, Im H, Liong M, Shao H, Castro CM, Weissleder R, Lee H. Magnetic Nanosensor for Detection and Profiling of Erythrocyte-derived Microvesicles. ACS Nano. 2013;7:11227–11233. doi: 10.1021/nn405016y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee K, Shao H, Weissleder R, Lee H. Acoustic Purification of Extracellular Microvesicles. ACS Nano. 2015;9:2321–2327. doi: 10.1021/nn506538f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei F, Patel P, Liao W, Chaudhry K, Zhang L, Arellano-Garcia M, Hu S, Elashoff D, Zhou H, Shukla S, Shah F, Ho CM, Wong DT. Electrochemical Sensor for Multiplex Biomarkers Detection. Clin. Cancer Res. 2009;15:4446–4452. doi: 10.1158/1078-0432.CCR-09-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munge BS, Coffey AL, Doucette JM, Somba BK, Malhotra R, Patel V, Gutkind JS, Rusling JF. Nanostructured Immunosensor for Attomolar Detection of Cancer Biomarker Interleukin-8 Using Massively Labeled Superparamagnetic Particles. Angew. Chem. Int. Ed. Engl. 2011;50:7915–7918. doi: 10.1002/anie.201102941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wen Y, Pei H, Shen Y, Xi J, Lin M, Lu N, Shen X, Li J, Fan C. DNA Nanostructure-based Interfacial Engineering for PCR-free Ultrasensitive Electrochemical Analysis of microRNA. Sci. Rep. 2012;2:867. doi: 10.1038/srep00867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsieh K, Patterson AS, Ferguson BS, Plaxco KW, Soh HT. Rapid, Sensitive, and Quantitative Detection of Pathogenic DNA at the Point of Care Through Microfluidic Electrochemical Quantitative Loop-mediated Isothermal Amplification. Angew. Chem. Int. Ed. Engl. 2012;51:4896–4900. doi: 10.1002/anie.201109115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campuzano S, Torrente-Rodriguez RM, Lopez-Hernandez E, Conzuelo F, Granados R, Sanchez-Puelles JM, Pingarron JM. Magnetobiosensors Based on Viral Protein P19 for Microrna Determination in Cancer Cells and Tissues. Angew. Chem. Int. Ed. Engl. 2014;53:6168–6171. doi: 10.1002/anie.201403270. [DOI] [PubMed] [Google Scholar]
- 28.Das J, Ivanov I, Montermini L, Rak J, Sargent EH, Kelley SO. An Electrochemical Clamp Assay for Direct, Rapid Analysis of Circulating Nucleic Acids in Serum. Nat. Chem. 2015;7:569–575. doi: 10.1038/nchem.2270. [DOI] [PubMed] [Google Scholar]
- 29.Nemiroski A, Christodouleas DC, Hennek JW, Kumar AA, Maxwell EJ, Fernandez-Abedul MT, Whitesides GM. Universal Mobile Electrochemical Detector Designed for Use in Resource-limited Applications. Proc. Natl. Acad. Sci. U.S.A. 2014;111:11984–11989. doi: 10.1073/pnas.1405679111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rowe AA, Bonham AJ, White RJ, Zimmer MP, Yadgar RJ, Hobza TM, Honea JW, Ben-Yaacov I, Plaxco KW. Cheapstat: an Open-source, “do-it-yourself” Potentiostat for Analytical and Educational Applications. PLoS One. 2011;6:e23783. doi: 10.1371/journal.pone.0023783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jorgensen M, Baek R, Pedersen S, Sondergaard EK, Kristensen SR, Varming K. Extracellular Vesicle (EV) Array: Microarray Capturing of Exosomes and Other Extracellular Vesicles for Multiplexed Phenotyping. J. Extracell. Vesicles. 2013;2:20920. doi: 10.3402/jev.v2i0.20920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J, Wei F, Schafer C, Wong DT. Detection of Tumor Cell-specific miRNA and Protein in Exosome-like Microvesicles From Blood and Saliva. PLoS One. 2014;9:e110641. doi: 10.1371/journal.pone.0110641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen C, Skog J, Hsu CH, Lessard RT, Balaj L, Wurdinger T, Carter BS, Breakefield XO, Toner M, Irimia D. Microfluidic Isolation and Transcriptome Analysis of Serum Microvesicles. Lab Chip. 2010;10:505–511. doi: 10.1039/b916199f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gercel-Taylor C, Atay S, Tullis RH, Kesimer M, Taylor DD. Nanoparticle Analysis of Circulating Cell-derived Vesicles in Ovarian Cancer Patients. Anal. Biochem. 2012;428:44–53. doi: 10.1016/j.ab.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi M, Salomon C, Tapia J, Illanes SE, Mitchell MD, Rice GE. Ovarian Cancer Cell Invasiveness is Associated With Discordant Exosomal Sequestration of Let-7 miRNA and miR-200. J. Transl. Med. 2014;12:4. doi: 10.1186/1479-5876-12-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Logozzi M, De Milito A, Lugini L, Borghi M, Calabro L, Spada M, Perdicchio M, Marino ML, Federici C, Iessi E, Brambilla D, Venturi G, Lozupone F, Santinami M, Huber V, Maio M, Rivoltini L, Fais S. High Levels of Exosomes Expressing CD63 and Caveolin-1 in Plasma of Melanoma Patients. PLoS One. 2009;4:e5219. doi: 10.1371/journal.pone.0005219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lotvall J, Hill AF, Hochberg F, Buzas EI, Di Vizio D, Gardiner C, Gho YS, Kurochkin IV, Mathivanan S, Quesenberry P, Sahoo S, Tahara H, Wauben MH, Witwer KW, Thery C. Minimal Experimental Requirements for Definition of Extracellular Vesicles and Their Functions: a Position Statement From the International Society for Extracellular Vesicles. J. Extracell. Vesicles. 2014;3:26913. doi: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melo SA, Sugimoto H, O'Connell JT, Kato N, Villanueva A, Vidal A, Qiu L, Vitkin E, Perelman LT, Melo CA, Lucci A, Ivan C, Calin GA, Kalluri R. Cancer Exosomes Perform Cell-Independent microRNA Biogenesis and Promote Tumorigenesis. Cancer Cell. 2014;26:707–721. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paggetti J, Haderk F, Seiffert M, Janji B, Distler U, Ammerlaan W, Kim YJ, Adam J, Lichter P, Solary E, Berchem G, Moussay E. Exosomes Released By Chronic Lymphocytic Leukemia Cells Induce the Transition of Stromal Cells Into Cancerassociated Fibroblasts. Blood. 2015;126:1106–1117. doi: 10.1182/blood-2014-12-618025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Runz S, Keller S, Rupp C, Stoeck A, Issa Y, Koensgen D, Mustea A, Sehouli J, Kristiansen G, Altevogt P. Malignant Ascites-derived Exosomes of Ovarian Carcinoma Patients Contain CD24 and EpCAM. Gynecol. Oncol. 2007;107:563–571. doi: 10.1016/j.ygyno.2007.08.064. [DOI] [PubMed] [Google Scholar]
- 41.Kristiansen G, Denkert C, Schluns K, Dahl E, Pilarsky C, Hauptmann S. CD24 is Expressed in Ovarian Cancer and is a New Independent Prognostic Marker of Patient Survival. Am. J. Pathol. 2002;161:1215–1221. doi: 10.1016/S0002-9440(10)64398-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bast RCJ, Xu FJ, Yu YH, Barnhill S, Zhang Z, Mills GB. CA 125: the Past and the Future. Int. J. Biol. Markers. 1998;13:179–187. doi: 10.1177/172460089801300402. [DOI] [PubMed] [Google Scholar]
- 43.Mandeville H, Rustin GJ. The Role of CA 125 in Epithelial Ovarian Carcinoma. J. BUON. 2002;7:13–17. [PubMed] [Google Scholar]
- 44.Meden H, Kuhn W. Overexpression of the Oncogene c-erbB-2 (HER2/neu) in Ovarian Cancer: a New Prognostic Factor. Eur. J. Obstet. Gynecol. Reprod. Biol. 1997;71:173–179. doi: 10.1016/s0301-2115(96)02630-9. [DOI] [PubMed] [Google Scholar]
- 45.Verri E, Guglielmini P, Puntoni M, Perdelli L, Papadia A, Lorenzi P, Rubagotti A, Ragni N, Boccardo F. HER2/neu Oncoprotein Overexpression in Epithelial Ovarian Cancer: Evaluation of Its Prevalence and Prognostic Significance. Clinical Study. Oncology. 2005;68:154–161. doi: 10.1159/000086958. [DOI] [PubMed] [Google Scholar]
- 46.Aldovini D, Demichelis F, Doglioni C, Di Vizio D, Galligioni E, Brugnara S, Zeni B, Griso C, Pegoraro C, Zannoni M, Gariboldi M, Balladore E, Mezzanzanica D, Canevari S, Barbareschi M. M-CAM Expression as Marker of Poor Prognosis in Epithelial Ovarian Cancer. Int. J. Cancer. 2006;119:1920–1926. doi: 10.1002/ijc.22082. [DOI] [PubMed] [Google Scholar]
- 47.Psyrri A, Kassar M, Yu Z, Bamias A, Weinberger PM, Markakis S, Kowalski D, Camp RL, Rimm DL, Dimopoulos MA. Effect of Epidermal Growth Factor Receptor Expression Level on Survival in Patients With Epithelial Ovarian Cancer. Clin. Cancer Res. 2005;11:8637–8643. doi: 10.1158/1078-0432.CCR-05-1436. [DOI] [PubMed] [Google Scholar]
- 48.Coutelieris FA, Delgado JMPQ. Transport processes in porous media. Springer; Berlin Heidelberg: 2012. [Google Scholar]
- 49.Ambrosi A, Airo F, Merkoci A. Enhanced Gold Nanoparticle Based Elisa for a Breast Cancer Biomarker. Anal. Chem. 2010;82:1151–1156. doi: 10.1021/ac902492c. [DOI] [PubMed] [Google Scholar]
- 50.Das J, Kelley SO. Tuning the Bacterial Detection Sensitivity of Nanostructured Microelectrodes. Anal. Chem. 2013;85:7333–7338. doi: 10.1021/ac401221f. [DOI] [PubMed] [Google Scholar]
- 51.Wei F, Lin CC, Joon A, Feng Z, Troche G, Lira ME, Chia D, Mao M, Ho CL, Su WC, Wong DT. Noninvasive Saliva-based EGFR Gene Mutation Detection in Patients With Lung Cancer. Am. J. Respir. Crit. Care Med. 2014;190:1117–1126. doi: 10.1164/rccm.201406-1003OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li XM, Wang LL, Luo J, Wei QL. A Dual-amplified Electrochemical Detection of mRNA based on Duplex-specific Nuclease and Bio-Bar-Code Conjugates. Biosens. Bioelectron. 2014;65:245–250. doi: 10.1016/j.bios.2014.10.051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.