SUMMARY
AIDS caused by simian immunodeficiency virus (SIV) infection is associated with gastrointestinal disease, systemic immune activation and, in cross sectional studies, changes in the enteric virome. Here we performed a longitudinal study of a vaccine cohort to define the natural history of changes in the fecal metagenome in SIV-infected monkeys. Matched rhesus macaques were either uninfected or intrarectally challenged with SIV, with a subset receiving the Ad26 vaccine, an adenovirus vector expressing the viral Env/Gag/Pol antigens. Progression of SIV infection to AIDS was associated with increased detection of potentially pathogenic viruses and bacterial enteropathogens. Specifically, adenoviruses were associated with an increased incidence of gastrointestinal disease and AIDS-related mortality. Viral and bacterial enteropathogens were largely absent from animals protected by the vaccine. These data suggest that the SIV-associated gastrointestinal disease is associated with the presence of both viral and bacterial enteropathogens and protection against SIV infection by vaccination prevents enteropathogen emergence.
Keywords: virome, microbiome, SIV, AIDS, vaccine, adenovirus, picornavirus, circovirus
INTRODUCTION
Systemic immune activation is associated with progressive infection by the lentiviruses HIV and SIV and is a strong predictor of progression to AIDS (Deeks et al., 2004; Giorgi et al., 1999; Hunt et al., 2008; Liu et al., 1997). One contributor to systemic immune activation is thought to be damage to the epithelial barrier of the gastrointestinal tract, but the specific mechanisms responsible for this are incompletely defined. Gastrointestinal barrier compromise is concomitant with the detection of bacterial and viral pathogen-associated molecular patterns (PAMPS) and antigens in the blood (Brenchley et al., 2006; Handley et al., 2012; Klase et al., 2015) which likely contribute to immune activation. SIV infection is associated with depletion of lamina propria CD4 T cells and CD103+ dendritic cells resulting in limited production of interleukin (IL)-17 and IL-22, two cytokines important for epithelial barrier function (Brenchley et al., 2008; Cecchinato et al., 2008; Estes et al., 2008; Klatt et al., 2012; Raffatellu et al., 2008). In addition, expansion of the enteric virome during pathogenic SIV infection is associated with direct damage to the epithelial barrier by adenoviruses and spillage of enteric parvoviruses into the circulation (Handley et al., 2012). Although bacterial lipopolysaccharides (LPS) and LPS-binding protein can be detected in the serum of SIV infected macaques or HIV-infected persons, changes in bacterial community structure have not been detected in cross-sectional studies in monkeys (Handley et al., 2012; McKenna et al., 2008). However, recent work has indicated that HIV infection is associated with the emergence of groups of bacteria containing potential enteropathogens (Mutlu et al., 2014).
To better understand the relationship between progressive lentivirus infection and the enteric virome and bacterial microbiome, we performed a longitudinal study to determine how the fecal metagenome changed over the course of infection. Further, to define whether changes in the metagenome are due to SIV exposure itself rather than progressive SIV infection and immunodeficiency, we studied a cohort of animals vaccinated with adenovirus serotype 26 (Ad26) vectors expressing Env/Gag/Pol antigens with or without Env protein boost (Barouch et al., 2015). Vaccine protection in this cohort of animals is associated with strong antibody responses to the Env protein and decreased detection of picornavirus sequences in the feces (Barouch et al., 2015). This cohort allowed us to contrast changes in the fecal metagenome between animals that were intrarectally challenged with SIV, but which were or were not protected against establishment of chronic lentivirus infection and provided a valid control group for environmental effects on the metagenome in monkeys followed over time.
Here we show that animals not protected by vaccination were more likely to have gastrointestinal disease. Gastrointestinal adenoviruses, adeno-associated viruses and picornaviruses were more abundant in the unprotected animals and were associated with gastrointestinal disease. Similar to previous reports, gastrointestinal bacterial community structure remained almost indistinguishable between SIV unchallenged, protected and unprotected SIV challenged animals (Handley et al., 2012; McKenna et al., 2008). However, a small subset of severely ill animals exhibited altered bacterial community structure with distinguishable alterations in bacterial β-diversity and increased abundance of Enterobacteriaceae and Moraxellaceae, two bacterial families containing enteropathogens. In a related study in humans, we also found that adenoviruses and bacterial enteropathogens are more prominent in fecal samples of HIV infected patients with very low CD4 T cell numbers (<200) suggesting similar host-pathogen interactions in human and macaques (Monaco et al., in revision). Taken together, these findings indicate that both the enteric virome and bacterial microbiome change during progressive lentivirus infection concurrent with development of severe immunodeficiency. Effective vaccination against SIV prevents these changes and the emergence of potentially pathogenic bacteria and viruses that might damage the intestinal epithelial barrier.
RESULTS
Rhesus monkey vaccine cohort
This study utilizes a well-characterized Rhesus monkey vaccine cohort from a previously published vaccine study (Barouch et al., 2015). Barouch et al. reported increased protection against SIV challenge in Ad26 vaccine primed animals boosted with Env protein when compared to vaccine alone (Figure 1, S1) (Barouch et al., 2015). This cohort included 36 animals. Five animals were neither vaccinated nor challenged with SIV (‘SIV unchallenged’ herein) (Figure 1A and S1). Seven animals received a sham vaccine, 12 received Ad26 vaccine alone, and 12 received Ad26 vaccine followed by an Env protein boost protocol. Of the SIV-challenged animals a total of 23 became at least transiently viremic while eight animals were protected against SIV infection. Of the 23 infected animals, two animals receiving both Ad26 vaccine and Env boost controlled viremia to below the limit of detection. Therefore, studies of the virome and bacterial microbiome contained 21 animals with SIV viremia (‘unprotected’ herein), 8 protected animals (‘protected’ herein), and 2 animals able to control infection. When comparing Ad26 vaccine alone to Ad26 + Env boost, we did not observe any significant alterations to the composition of the enteric bacterial microbiome or virome (data not shown). Throughout all analyses, these controller animals behaved similarly to protected animals and treated as protected animals in all analysis. The cohort was monitored for 52-weeks. Animals that died prior to the 52-week time point were considered to have died of AIDS related illness (‘AIDS related death’ herein). On average, animals succumbed to AIDS 45 +/− 14 weeks post-challenge. Serum levels of LPS-binding protein, percentage of CD4 T cells, and SIV load were measured and fecal samples collected at 5 and 32 weeks post-challenge or at necropsy in animals that succumbed to AIDS related illness. Animals were designated as having gastrointestinal disease following autopsy performed after sacrifice of AIDS-related death if they had villus blunting or fusion, intestinal abnormal numbers of either eosinophils or basophils, lymphocytic infiltrates or cytoplasmic inclusion bodies.
Figure 1.
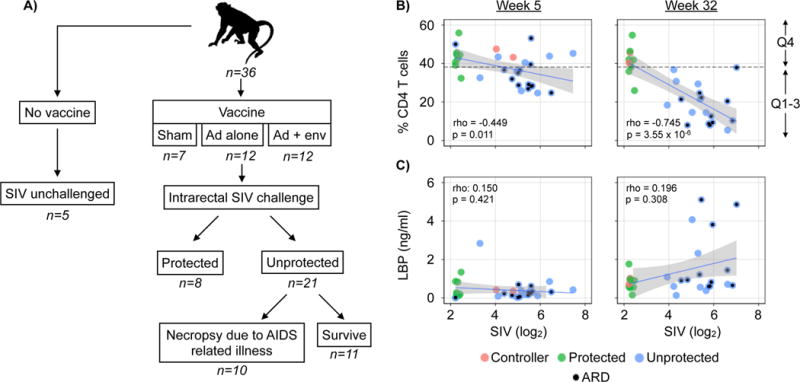
Study design and the relationships between CD4 T cells, LPS-binding protein and SIV loads. (A) Animal cohort and study design (B) Relationship between the percentage of blood CD4 T cells and SIV load (log2) 5 and 32 weeks post-challenge. (C) Relationship between LPS-binding protein (ng/ml) and SIV load (log2) and the percentage of blood CD4 T cells 32 weeks post-challenge. (D) Association between vaccine protection to SIV and the presence of gastrointestinal disease (p = 0.002, odds ratio: 30.99). (E) Association between the detection of elevated levels of LPS-binding protein and gastrointestinal disease p-value: 0.038, odds ratio: 7.50). Animals that succumbed to AIDS related death (ARD) are indicated with a black circle. Fitted LOESS lines (blue) and confidence intervals (gray). Spearman’s rho and p-values were calculated using Spearman’s rank correlation test. Significance in Panels D&E determined using Fisher’s exact test. p ≤ 0.01 = **. For more data on individual measurements of CD4 T cells, LPS-binding protein and SIV loads see Figure S1.
Associations between serum SIV load, CD4 T cells and LPS-binding protein and gastrointestinal disease and survival
To define associations between SIV-mediated immunocompromise, gastrointestinal leakage as measured by the presence of serum LPS-binding protein and gastrointestinal disease, we compared the percentage of blood CD4 T cells, serum SIV load and serum LPS-binding protein levels in animals from a previously published vaccine study (Barouch et al., 2015). As expected, increased serum SIV load correlated with a decrease in the percentage of blood CD4 T cells (Figure 1B and S1B). The strength of this correlation increased between weeks 5 and 32 post-challenge. Gastrointestinal disease was identified in 13 animals and was also more likely to be found in unprotected animals (Figure 1D) (p = 0.002, odds ratio: 30.99).
Serum LPS-binding protein was not consistently detected until 32 weeks post-challenge (Figure 1C and S1C). There was no significant correlation between SIV viral load or the percentage of CD4 T cells and LPS-binding protein levels (Figure 1C). Ignoring the protected animals and analyzing only the unprotected animals to assess correlations between LPS-binding protein levels and SIV load also did not reveal a statistically significant correlation (week 5: rho: −0.196, p-value: 0.394, Week 32: rho: 0.234, p-value: 0.334). However, it was notable that LPS-binding protein levels above were only detected in animals with SIV loads above the median (p-value: 0.014, odds ratio: 14.00). Detection of serum LPS-binding protein was also significantly associated with animals with gastrointestinal disease (Figure 1E) (p-value: 0.038, odds ratio: 7.50). Therefore, detection of serum LPS-binding protein was not significantly correlated with either SIV load or the percentage of CD4 T cells, but was more likely detected in unprotected animals with gastrointestinal disease.
The fecal sequenome in SIV challenged and unchallenged macaques
To determine the relative abundances of fecal sequences originating from food, macaque, bacteria or viruses, we mapped sequences to a collection of reference genomes (Figure 2). Total nucleic acid (DNA and cDNA to enable detection of both RNA and DNA viruses) was extracted from fecal samples. Using the MiSeq 2×250 paired-end protocol we obtained 1.97 +/− 0.78 million sequences per sample of which 93 +/− 3% were of high-quality and used for all subsequent analyses (Table S1). Only 15 +/− 11% of all sequences mapped to the collection of reference genomes. The largest percentage of sequences mapped to bacterial genomes, with the second most frequently mapped genome belonging to wheat, a primary protein source in the monkey diet. Fewer sequences mapped to other dietary components, such as banana and peanut. A few animals had sequences that mapped to the Drosophila melanogaster genome. This is likely due to the ingestion of insects. Low numbers of sequences also mapped to the Candida albicans genome, suggesting the presence of yeast species.
Figure 2. Sequence mapping to reference genome databases.
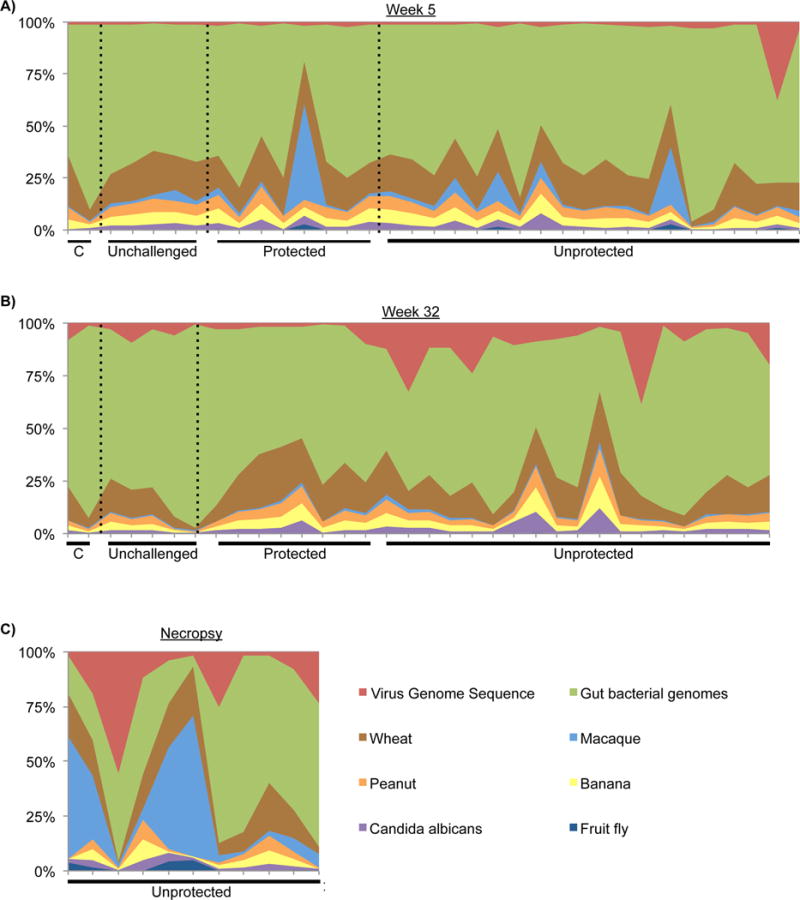
Relative abundance of sequences that map to genome databases from samples collected at (A) week 5, (B) week 32 or (C) necropsy in animals that die due to AIDS related death. Abbreviation: C = controller. Statistical analysis comparing the number of food, microbial and host sequences in SIV unchallenged and SIV challenged unprotected and protected macaques can be viewed in Figure S2.
The number of sequences mapping to reference genomes was compared between SIV unchallenged, protected and unprotected animals at 5 and 32 weeks post-challenge and at the time of necropsy due to AIDS related death (Figure S2). The number of sequences mapping to the macaque genome was increased in samples collected at necropsy in comparison to SIV unchallenged and unprotected samples collected at 32 weeks post-challenge (Figure S2B). This is potentially due to the necropsy procedure contaminating fecal samples with host cells, or, alternatively, to increases in host cell shedding as a result of AIDS related gastrointestinal disease.
The number of sequences mapping to viral genomes was significantly increased in samples collected at necropsy when compared to SIV unchallenged and protected animals (Figure S2D). There was no correlation between sequences mapping to the macaque genome and sequences mapping to viral genomes in samples collected at necropsy, suggesting that the increases in sequences mapping to viral genomes at necropsy can not be attributed to increases in host cell contamination (Spearman’s rho: −0.21, p-value = 0.543).
Together, these data suggest that protection is not associated with changes in the numbers of sequences that map to genomes associated with common food or bacterial genomes. In addition, gastrointestinal virome expansion is not detectable during the early phase of disease and is most significantly associated with animals that succumb to AIDS related illness.
The enteric virome in SIV challenged and unchallenged macaques
To define associations between SIV-mediated immunocompromise and specific types of viruses, we classified and counted the number of viral sequences between experimental groups across different time points. Viral sequences were classified using a two-staged approach. In the first stage, all dereplicated (removing >95% identical sequences) high-quality sequences were queried against a virus protein database using blastx. Sequences were most frequently assigned to families of bacteriophages (Caudovirales, Microviridae), several eukaryotic viruses (Picornaviridae, Adenoviridae, Parvoviridae, Circoviridae, Picobirnaviridae and Poxviridae), and several large dsDNA viruses that are known to infect algae and insect cells (Phycodnaviridae, Mimiviridae, Iridoviridae, Marseilleviridae, and Nudiviridae) as well as other viral families that are defined as “unclassified” in the NCBI Taxonomy Database (Figure S3A). At this stage, all sequences that were assigned to specific viral taxa were binned as being “potentially viral”. These potentially viral sequences were passed through a second set of blastx and blastn queries encapsulated in the VirusSeeker pipeline (Zhao et al., in preparation). In brief, all potentially viral sequences were queried against the NCBI non-redundant nucleic acid and protein databases using a combination of blastn and blastx sequentially. Potential virus sequences assigned to non-viral taxonomic lineages by VirusSeeker were considered to be non-viral or ambiguous in origin and were excluded from subsequent analysis. Comparison of the number of viral sequences identified prior to, and after VirusSeeker analysis revealed that sequences assigned to a number of viral families during search of a virus-only database, many putatively belonging to large dsDNA viruses, are likely non-viral in origin (Figure S3B). The remaining viral families after this conservative analysis included the Picornaviridae, Adenoviridae, Phycodnaviridae, Parvoviridae, Circoviridae and Picobirnaviridae. All of the sequences assigned to one of these families were used in subsequent analyses. Sequences assigned to the Phyconaviridae did not show any differences when comparisons were made between all experimental groups and time points (data not shown).
The diversity of genera identified within each selected viral family varied greatly. The Picobirnaviridae were represented by a single genus, the human picobirnavirus. In contrast, sequences were assigned to 19 different species of Circoviridae, 14 genera of Picornaviridae, 6 genera of Adenoviridae and 7 genera of Parvoviridae (Figure S4A–D). Sequences assigned to the Picornaviridae were assigned to two dominant genera, the enteroviruses and sapeloviruses. Similarly, the Parvoviridae were dominated by sequences assigned to the adeno-associated viruses.
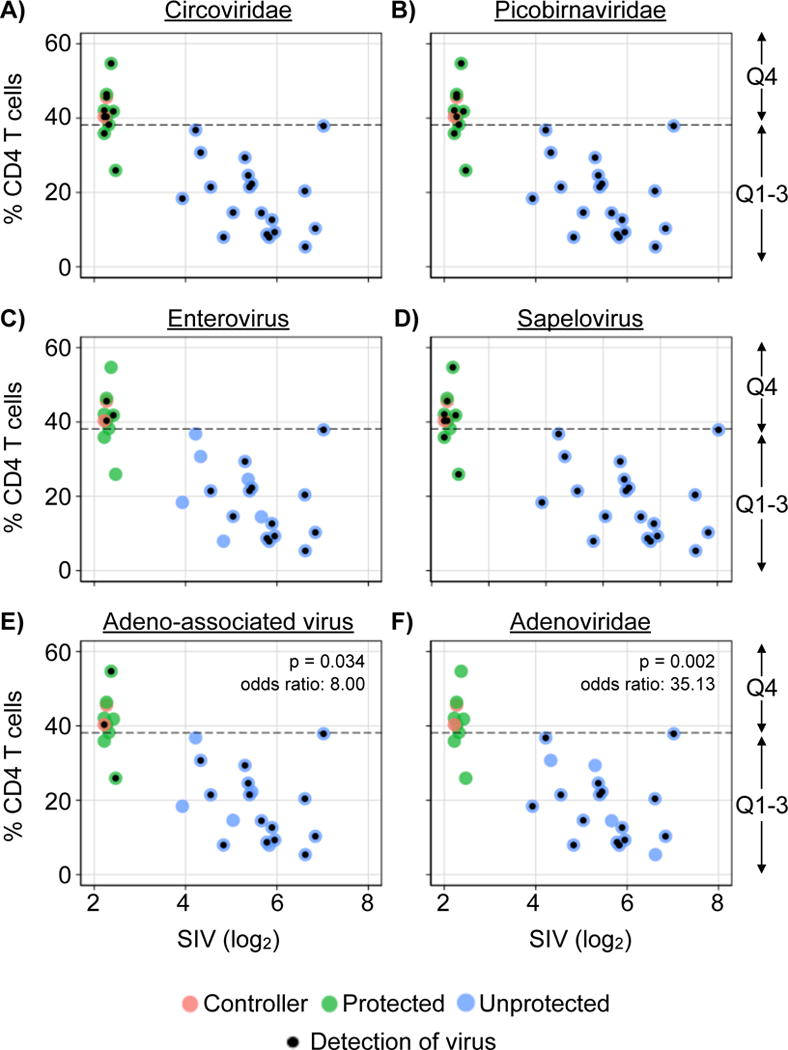
Associations between viruses and SIV-associated immunodeficiency and gastrointestinal disease
To define potential associations between viruses and SIV infection or SIV-induced immunodeficiency we compared the numbers of viral sequences in samples from SIV unchallenged animals with that of samples from SIV challenged animals that were either protected or unprotected from SIV challenge (Figure 3). Circoviridae, Picobirnaviridae, sapelovirus, adeno-associated virus and Adenoviridae sequences were all detected by 5 weeks post-challenge. Sequences assigned to the Circoviridae and Picobirnaviridae were the most frequently detected, being found in 94 and 81% of all samples respectively. However, there were no statistically significant differences in the numbers of Circoviridae or Picobirnaviridae sequences when comparing experimental groups (Figure 3A & B). This indicates that SIV-effects on the fecal virome are not seen for all types of viruses.
Figure 3. Quantification of eukaryotic virus sequences in fecal samples.
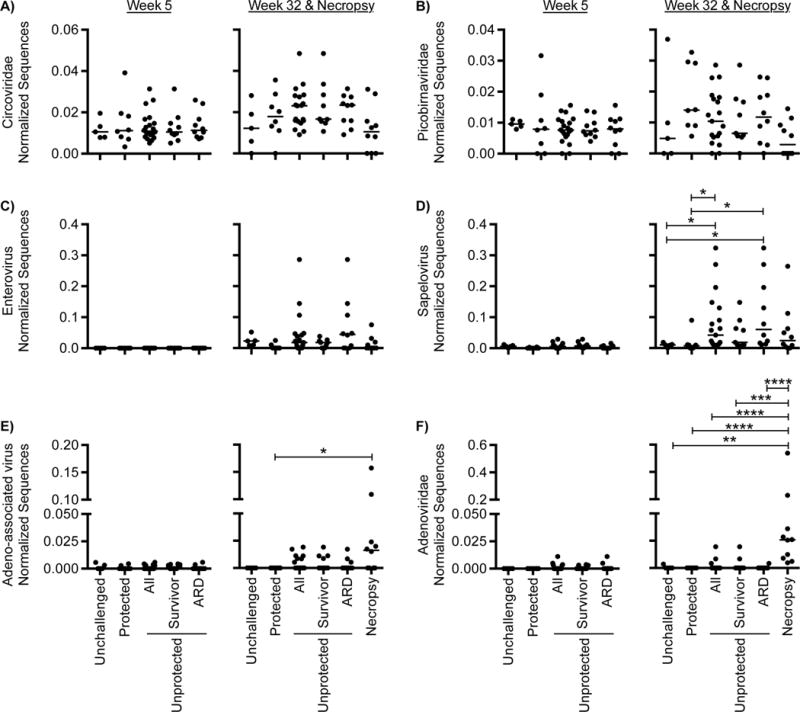
Abundance of eukaryotic virus sequences in samples collected from SIV unchallenged and SIV challenged unprotected and protected macaques collected at 5 and 32 weeks post-challenge. (A) Circoviridae (B) Picobirnaviridae, (C) enterovirus (D) sapelovirus (E) adeno-associated virus and (F) Adenoviridae. Sequences were normalized by dividing by the number of dereplicated (<95% identical), high-quality sequences. The square root of the normalized value is used to standardize the y-axis. Differences between groups were considered statistically significant if p < 0.05 using the nonparametric Kruskal-Wallis Test correcting for multiple comparisons with Dunn’s procedure. p ≤ 0.05 = *, p ≤ 0.01 = **, p ≤ 0.001 = ***, p ≤ 0.0001 = ****. The median for each group is indicated with a horizontal line with error bars indicating interquartile range. More data on viruses detected in fecal samples can be found in Figure S3.
Notably absent at the 5 week time-point were sequences from the enteroviruses, which only became detectable at 32 weeks (Figure 3C). Of the two Picornaviridae genera examined, only the sapeloviruses were associated with lack of vaccine protection against SIV 32 weeks post-challenge with SIV (Figure 3C & D).
Sequences from Circoviridae and both enterovirus and sapelovirus increased between 5 and 32 weeks post-challenge in the unprotected but not in the protected animals indicating that their expansion is associated with SIV pathogenesis (Figure S3C). While there was a significant change in enterovirus sequences in unprotected animals, we also observed that these sequences increased significantly between 5 and 32 weeks even in unchallenged animals, underling the importance of matched controls for longitudinal studies of the enteric virome (Figure S3D).
Sequences assigned to the Adenoviridae were significantly increased in samples collected at necropsy from animals that succumbed to AIDS related illness (Figure 4). Sequences from Circoviridae, Picobirnaviridae and enterovirus and sapelovirus were also common in samples taken at necropsy, however, the detection of adeno-associated virus and adenoviridae sequences was unique in that they were less prominent at both earlier time points (Figure 4E&F). Several animals with abundant enteroviral or sapeloviral sequences at the week 32 time-point had dramatically decreased levels in their necropsy collected samples (Figure 4C&D). However, the overall number of animals with detectable enterovirus or sapelovirus sequences remained largely the same.
Figure 4. Longitudinal analysis of viruses in animals succumbing to AIDS related illness.
(A–F) Quantification of specific virus sequences at 5 and 32 weeks post-infection and in samples collected at necropsy in animals that died to AIDS related illness. Bar charts indicate the % of animals wherein the specific virus was detected. (G–H) Association between specific viruses and gastrointestinal disease. Significance for longitudinal changes (A–F) was determined using nonparametric, repeated measures ANOVA correcting for multiple comparisons with Dunn’s procedure. Significance in Panels G–J determined using Fisher’s exact test. p ≤ 0.05 = *, p ≤ 0.01 = **, p ≤ 0.001 = ***, p ≤ 0.0001 = ****.
Adenovirus sequences were significantly associated with animals that succumbed early to AIDS related symptoms (10/10, 100%) compared to the animals that survived (3/21 13.6%), Fisher’s exact p-value < 0.001, odds ratio = 135.0). Adenovirus (Fisher’s exact p = 0.0253, odds ratio = 8.00), Adeno-associated virus (Fisher’s exact p = 0.026, odds ratio = 7.86) and enterovirus (Fisher’s exact p-value: 0.026, odds ratio: 7.86), were significantly associated with gastrointestinal disease, while sapelovirus sequences were detected equivalently between animals with or without gastrointestinal disease (Figure 4G&H). Of note, none of the viruses detected in this study correlated with the detection of serum LPS-binding protein suggesting that gastrointestinal virome expansion is significantly associated with gastrointestinal disease but not, at least with the sensitivity of this analysis, with a specific virus infection. There were no associations between specific gastrointestinal disease pathologies and the detection of specific types of viral taxa.
Viral associations with Immune Correlates
To identify how the abundance of viruses correlated with the protective immune response, we combined the abundance data for all viruses with the immune parameters collected as part of the vaccine study and performed a principal-components analysis (Barouch et al., 2015). The PCA plot separated the SIV challenged unprotected and the protected samples across principal-components axis 1 (Figure 5A). Samples from unprotected animals were clustered to the left of 0.0 on axis 1, while protected samples clustered to the right. Singular value decomposition was used to identify variables that most strongly contribute to this clustering (Figure 5B). As previously reported, samples in the protected cluster were defined by having antibody responses to the vaccine antigens SIV Gag, Env and Pol as well as abundant CD4 and CD8 T cells (Barouch et al., 2015). Variables contributing to the unprotected cluster were high serum SIV loads, serum LBP and detectable gastrointestinal viruses at both 5 and 32 weeks post SIV challenge.
Figure 5. Associations between vaccine immune correlates and the detection of enteric virus.
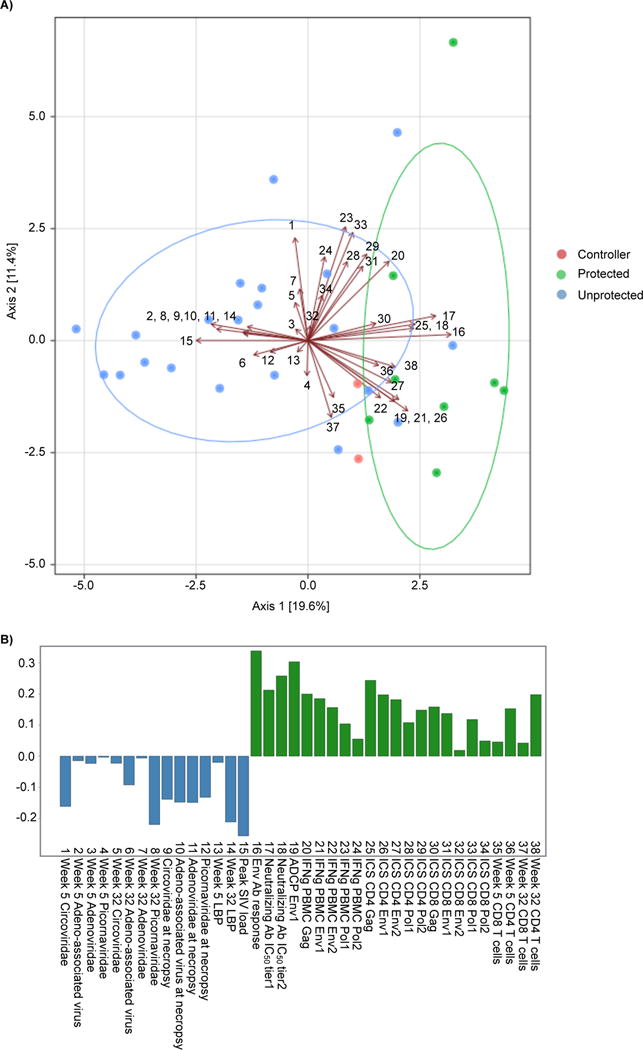
(A) Principal components analysis of immune correlates and enteric virus abundance. Points represent individual samples. Arrows and associated numbers are the input variables that can be viewed in (B). The major groups of viremic and protected animals are circled with a median elliptical centroid. (B) Singular value decomposition of the variables used for principal components analysis. Abbreviations: Ab = antibody, ICS=intracellular cytokine staining, IFNg = interferon gamma, ADCP=antibody-dependent cellular phagocytosis, IC50=inhibitory concentration 50, PBMC=peripheral blood mononuclear cell.
Analysis of the bacterial component of the metagenome
16S rRNA gene profiling was used to identify associations between gastrointestinal bacterial community structure, vaccine mediated protection, and overall animal survival. There were no statistically significant differences in the richness and diversity of bacterial communities when comparing SIV unchallenged, unprotected or protected animals at the 5 and 32 week time-point (Figure 6A & B). There were statistically significant increases in the richness of bacterial communities within all experimental groups between the 5 and 32 week time-points, however diversity remained the same throughout the study (Figure S5). Similarly, with the exception of a few of the samples collected at necropsy from animals succumbing to AIDS related illness, there were no distinguishable differences in bacterial beta-diversity when comparing across sampling times or between outcomes using PCoA analysis of UniFrac distances (Figure 6C & D). Therefore, other than in a few severely ill animals that succumb to AIDS related illness, the overall bacterial community structure appears to be relatively stable after SIV challenge regardless of the presence or absence of vaccine-mediated protection.
Figure 6. Bacterial community profiling.
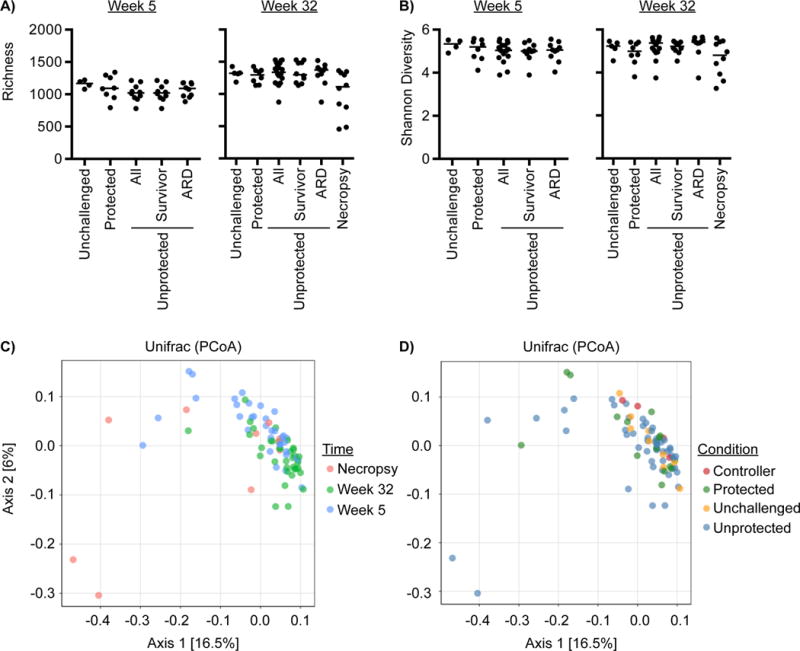
Comparison of (A) bacterial richness and (B) Shannon diversity in SIV unchallenged and SIV challenged unprotected and protected macaques collected at 5 and 32 weeks post-challenge or at necropsy due to AIDS related death (ARD). Principal Coordinate Analysis (PCoA) plots of the weighted UniFrac distances colored by (C) sampling time or (D) animal condition. Differences in richness or Shannon diversity between groups were considered significant if p < 0.05 using the nonparametric Kruskal-Wallis Test correcting for multiple comparisons with Dunn’s procedure. The median for each group is indicated with a horizontal line. For longitudinal comparisons of richness and Shannon diversity within groups between 5 and 32 weeks post-challenge see Figure S5.
Differentially abundant bacterial taxa in SIV challenged macaques
While there were no major changes in the overall bacterial communities detected by 16S rRNA sequencing, others have reported emergence of specific groups of bacteria in association with HIV infection (Mutlu et al., 2014), a finding that we have verified (Monaco et al, In revision). Variance stabilization techniques based on mixture models of microbiome data permits detailed and robust comparisons of differential abundances while substantially improving both power and accuracy (McMurdie and Holmes, 2014). Using these techniques, we identified bacterial families differentially abundant between experimental groups. SIV-challenged unprotected versus protected animals had relatively few differentially abundant bacterial families at both the 5 and 32-weeks time-points (Figure 7A&B, Table S2). Several members of the Bacteroidetes were associated with the lack of protection, while Clostridial bacteria were normally associated with protection. Ruminococcaceae switched from being more abundant in the unprotected animals at week 5 to being in the protected animals at week 32. There were also relatively few changes in specific bacterial taxa when comparing animals with gastrointestinal disease to those without (Figure 7C).
Figure 7. Differentially abundant bacterial taxa.
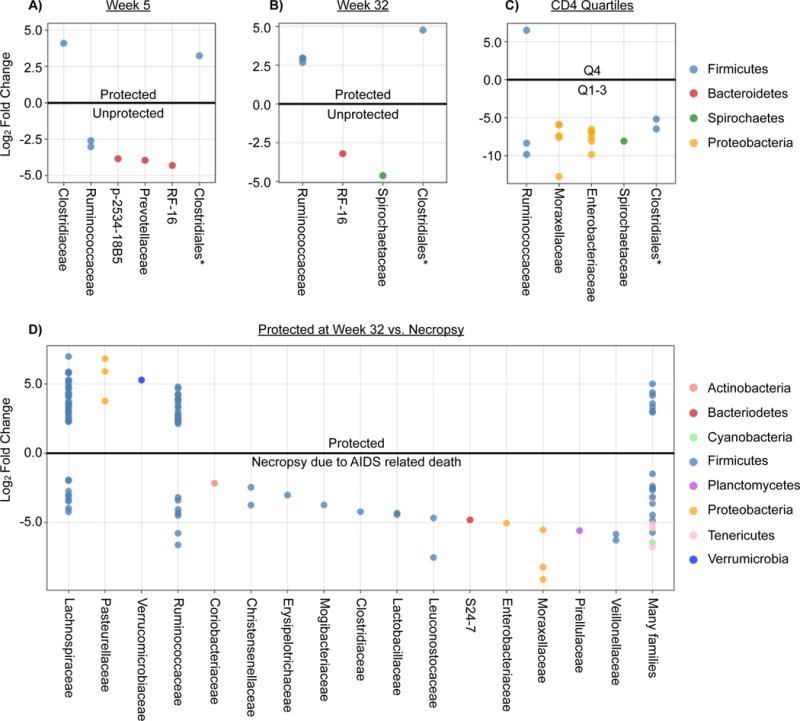
Differentially abundant taxa in SIV-challenged unprotected and protected animals (A) 5 weeks (B) 32 weeks post-challenge. (C) Bacterial taxa differentially abundant in animals with or without gastrointestinal disease. (D) Bacterial taxa differentially abundant in animals within the top (4th quartile) or bottom three quartiles of percentage of blood CD4 T cells.
Enterobacteriaceae and Moraxellaceae were more abundant in animals with a low percentage of blood CD4 T cells (lower 3 quartiles) when compared to animals in the top quartile of CD4 T cells (Figure 7D). These data suggest that bacterial families that frequently harbor enteropathogens are more abundant in animals that succumb to AIDS related illness or that have lower levels of CD4 T cells.
Associations between bacteriophages and bacteria
To define the relationships between bacteriophage populations and SIV infection we compared the numbers of bacteriophage sequences in fecal samples from SIV unchallenged, unprotected, protected animals and those that succumbed to AIDS related illness. There was no significant difference in bacteriophage richness or diversity when comparing any of these groups (Figure S6A–D). There were also no statistically significant correlations between bacteriophage and bacteria richness and Shannon diversity (Table S3 & S4). Taken together these data indicate that SIV infection and vaccine-mediated protection were not associated with major alterations in bacteriophage populations.
DISCUSSION
In this study we analyzed a vaccination cohort to better understand changes in the fecal metagenome during chronic lentivirus infection leading to AIDS (Barouch et al., 2015). This study design allowed us to assess both longitudinal changes in the metagenome and to define the relationship between changes we observed and the level of progressive immunodeficiency. We compared events in animals without chronic SIV infection despite challenge with the virus (protected), SIV-infected animals that survived to the end of the study period (unprotected), and animals that succumb to AIDS. In animals with the most severe immunocompromise, and those dying of AIDS, we detected an expansion of sequences from groups of organisms, both viral and bacterial, which might include pathogens that damage the intestinal epithelium. Specifically, we detected increased Adenoviridae sequences in samples collected at necropsy from animals that succumbed to AIDS related disease and an association between three types of viruses, the enteroviruses, adeno-associated viruses and adenoviruses, and the presence of gastrointestinal disease.
The bacterial microbiome was markedly similar between protected and unprotected animals with only a few taxa differentially abundant between protected and unprotected animals or animals with or without gastrointestinal disease. The only notable differences in bacterial taxa were the emergence of bacterial taxa containing known enteropathogens in animals with diminished CD4 T cells. The emergence of potential bacterial enteropathogens and Adenoviridae mirrors what is observed in humans who progress to AIDS (Monaco, et al., in revision).
Taken together, we found that successful vaccination and specific vaccine-elicited immune responses were associated with prevention of expansion of the fecal virome, and the emergence of specific bacterial taxa that may contain enteropathogens. Gastrointestinal disease was strongly associated with the detection of enteropathogenic viruses. While bacterial enteropathogens emerged in animals with SIV-associated immunodeficiency, they were not associated with gastrointestinal disease. These data suggest that the enteropathy often associated with AIDS may be due to pathogen infection.
Enteric virome expansion in SIV unchallenged and challenged macaques
Expansion of the enteric virome has previously been linked to pathogenic SIV infection in macaques (Handley et al., 2012). This previous cross-sectional study linked expansion of Picornaviridae, Adenoviridae and Parvoviridae with SIV pathogenesis. The current study confirms these findings and expands on them by providing higher taxonomic resolution and longitudinal monitoring of specific enteric virus acquisition over time. Cross-sectional analysis revealed no SIV-associated changes in the number of Circoviridae sequences (Handley et al., 2012). However, longitudinal analysis shows that there is a significant increase in Circoviridae sequences between the 5 and 32 week time-points in unprotected animals (Figure S3C).
In both the previous and the current study, sapeloviruses were the dominant type of Picornaviridae sequences detected, with sequences from the related enteroviruses being less abundant. We found that enterovirus and sapelovirus expansion exhibited distinct patterns. At the earliest sampled time-point (5 weeks post SIV challenge), enterovirus sequences were completely undetectable in all animals, regardless of SIV challenge or pathogenesis. However, by 32 weeks, the number of enterovirus sequences increased significantly in both SIV unchallenged and challenged macaques. This expansion of enterovirus sequences in SIV challenged macaques was more severe in unprotected animals. This pattern is consistent with the idea that the enteroviruses infected both SIV infected and uninfected animals, but were less well controlled in the unprotected animals.
In contrast, adeno-associated virus and adenoviridae sequences were strongly associated with samples collected at necropsy from animals that succumbed to AIDS related illness. The distinction in emergence patterns between picornaviruses and adeno-associated viruses and adenoviruses suggests that they are under different types of immune control wherein the adeno-associated viruses and adenovirus only emerge in severely ill animals with gastrointestinal disease. While picornaviruses emerge earlier and in animals with longer survival times and in animals with or without gastrointestinal disease.
Associations between gastrointestinal viruses and serum LBP
Previous studies of HIV infection have identified correlations between the bacterial microbiome, microbial translocation, CD8 T cell activation and CD4 T cell abundance (Gori et al., 2008) (Pérez-Brocal et al., 2013). In addition, previous studies have demonstrated direct damage to the epithelial barrier by adenoviruses and spillage of enteric parvoviruses into the circulation during progressive SIV infection (Handley et al., 2012). Each of these studies suggests a connection between the composition of the enteric microbiome and leakage of enteric bacterial and viral products into the blood. The data presented here were unable to identify a statistically significant association between the detection of specific enteric viruses and the detection of serum LBP. However, we were able to demonstrate an association between the detection of three enteric viruses, the enterovirus, adeno-associated virus and adenoviridae, with gastrointestinal disease and that levels of of LPS-binding protein are above the intraquartile range more frequently in animals with gastrointestinal disease. Principal component analysis also suggests that enteric virus detection is correlated with the detection of LBP in the serum in unprotected, but not protected animals.
SIV and potential bacterial enteropathogens
In parallel to the virome studies, we also characterized the bacterial microbiome using 16S rRNA gene profiling. Fecal sampling provides an accurate representation of the both the colonic lumen and the mucosa without requiring invasive biopsy procedures and facilitating longitudinal sample collection (Yasuda et al., 2015) Our data support previous studies demonstrating that overall bacterial community structure is largely indistinguishable between SIV unchallenged and unprotected SIV challenged animals. Bacteriophage communities were also indistinguishable suggesting that the overall bacterial community structure is stable and is not undergoing ecological shifts driven by bacteriophage-bacteria predator-prey dynamics. We were able to identify specific bacterial families that were associated with protection against SIV challenge. There were relatively few changes in bacterial families when comparing protected to unprotected animals at the 5 and 32 week time points, and most of these changes occurred in bacterial families considered to be commensals. However, we observed an increased abundance in bacterial families that harbor enteropathogenic bacteria (Enterobacteriaceae and Moraxellaceae) in samples from animals with low CD4 T cells. These data suggest that the bacterial microbiome is relatively resistant to change during SIV mediated disease, and is only detectable when disease is severe. However, these changes are associated with an increased abundance of bacterial families that harbor known enteropathogenic bacteria, which may contribute to gastrointestinal disease during AIDS.
Conclusion
Herein we observed a striking, SIV induced immunodeficiency-associated, expansion of the fecal virome. This expansion was specific to a subset of enteric viruses, several of which are associated with gastrointestinal disease. In addition, we observed a rise in the abundance of bacterial families known to harbor enteropathogenic bacterial taxa. Given the emergence of known or potential enteric pathogens in association with SIV-induced immunodeficiency and death, we speculate that immunity to, or treatment of, enteric pathogens might prevent AIDS progression by limiting gastrointestinal disease, epithelial cell damage and release of inflammatory PAMPs and antigens into the body. We hypothesize that this might minimize systemic immune activation, which is linked to AIDS progression. Further, the expansion of the enteric virome may be a biomarker for severe deficiency in the capacity of the intestinal immune system to control pathogens. Similar findings from a study of HIV-infected patients in Uganda (Monaco et al. in revision), argue that these findings are general to AIDS pathogenesis in primates and humans. Together, these studies indicate that a likely mechanism for changes in the fecal microbiome, including bacteria and viruses, is severe immunodeficiency.
EXPERIMENTAL PROCEDURES
Animals
The animal cohort described in this study was previously published.(Barouch et al., 2015) In brief, 31 adult rhesus macaques (Macaca mulatta) that do not express the protective MHC class I alleles were primed with adenovirus serotype 26 (Ad26) vectors (Abbink et al., 2007) expressing SIVsmE543 Env/Gag/Pol antigens (Barouch et al., 2012) followed by either SIV mac32H Env gp140 protein (Malhotra et al., 2003) (Ad/Env; N=12) or Ad35 vectors (Nilsson et al.) expressing Env/Gag/Pol antigens (Ad Alone; N=12), and a control group receiving sham vaccines (Sham; N=8). The following immune correlate data were also published as part of the originating vaccine study and obtained with permission for reuse: serum SIV load, SIV ENV antibody responses, tier-1 and tier-2 neutralizing antibody IC50, antibody-dependent cellular phagocytosis, IFN-γ ELISPOT assays of peripheral blood mononuclear cells (PBMC) and intracellular cytokine staining (ICS) of CD4 T cells responses to Env, Pol and Gag peptides (Barouch et al., 2015). Animals were designated as having gastrointestinal disease by following autopsy if they had villus blunting or fusion, eosinophilia or basophilia, lymphocytic infiltrates or cytoplasmic inclusion bodies.
Additional animal assays
Serum levels of LPS-binding protein (LBP) were quantitated by ELISA per the manufactures instructions (Antibodies Online). Frequencies of CD3+CD4+ T cells were determined by staining lymphocytes with DAPI, anti-CD3 (eBioscience) and anti-CD4 (eBioscience) followed by flow cytometry analysis using a FACSCanto-II flow cytometer and FlowJo software.
Virome sequencing
For each sample, 100–200mg of frozen feces was resuspended in 6× volume of phosphate-buffered saline (PBS) and centrifuged (Finkbeiner et al., 2009). The fecal supernatant was passed through a 0.45-μm-pore-size membrane and total RNA plus DNA was extracted on a COBAS Ampliprep instrument (Roche) according to the manufacturers recommendation. Purified nucleic acid was reverse transcribed and PCR amplified using barcoded primers consisting of a base-balanced 16 nucleotide specific sequence upstream of a random 15-mer as previously described (Finkbeiner et al., 2009) and used for NEBNext library construction (New England BioLabs). Libraries were multiplexed (12 samples per flow-cell) on an Illumina MiSeq instrument (Washington University Center for Genome Sciences) using the paired-end 2×250 protocol. All sequences were uploaded to the ENA and are associated with project PRJEB9503.
Identification and analysis of Viral-like Sequences
Low-quality sequences and contaminants were removed prior to taxonomic assignment (Supplementary Experimental Procedures). Detection of potentially ambiguous or false-positive viral sequences was done using VirusSeeker, a custom bioinformatics pipeline designed to detect sequences sharing nucleotide and protein level sequence similarity to known viruses (Zhao et al., submitted). Briefly, potential unique viral reads were queried against the NCBI nt/nr databases, and only reads matching exclusively to viral sequences were kept for further analysis. All sequences aligning to viruses were further classified into viral families based on the NCBI taxonomic identity of the best hit.
Absolute read counts were normalized by dividing individual taxon sequence counts by the total assigned sequence count in a sample using the decostand function of the vegan R package (Oksanen et al.). Richness and diversity were calculated using the diversity function of the vegan package. Heat maps were generated using the pheatmap R package (Kolde, 2013).
Fecal sequence mapping to reference genomes and databases
Quality-controlled metagenomic sequences were aligned to the following reference genomes using BWA-MEM with default settings. (http://arxiv.org/abs/1303.3997). Reference genomes and accession numbers are listed in Supplemental Experimental Procedures. The number of sequences mapping to each individual reference genome or database was normalized by dividing by the total number high-quality sequences per sample.
16S rRNA Gene Profiling
Fecal nucleic acid was extracted from 100–200mg frozen feces as previously described (Norman et al., 2015). Primer selection and polymerase chain reaction was performed following previously described methods (Caporaso et al., 2011). Amplicons were pooled and purified using 0.6× Agencourt Ampure XP beads (Beckman-Coulter) according to the manufacturer’s instructions. The final pooled samples were sequenced on the Illumina MiSeq platform (Washington University Center for Genome Sciences; 2×250 standard run). 16S rRNA gene sequences were uploaded to the EMBL-EBI European Nucleotide Archive under the study accession PRJEB9503.
Demultiplexing and operational taxonomic unit (OTU) picking of 16S rRNA genes sequences was done in QIIME (Quantitative Insights Into Microbial Ecology, v.1.8.0) (Caporaso et al., 2010). Closed reference OTUs sharing 97% identity were clustered using the UCLUST algorithm (Edgar, 2010) and assigned taxonomy according to the Greengenes database (v.13.8) (McDonald et al., 2012). Rarified (25,000 sequences) and non-rarified Chao1 and Faith’s phylogenetic diversity were calculated in QIIME. All subsequent analysis was performed using PhyloSeq (v1.10.0) (McMurdie and Holmes, 2012). Samples were normalized to the relative abundance per sample for downstream analysis in PhyloSeq which included calculations of observed species richness, per sample Shannon diversity and UniFrac distances between samples or bacterial families (Lozupone and Knight, 2005). Differential abundance of bacterial taxa between experimental groups was determined using the PhyloSeq DESeq2 extension using the Wald significance test and a parametric fit type (v.1.6.3) (Anders and Huber, 2010; McMurdie and Holmes, 2012).
Statistical Analysis
Associations between groups were determined using two-by-two contingency tables. Significance was determined using Fisher’s exact test. Spearman rank correlations were calculated and significance tested using the cor and cor.test functions in R. Univariate and multivariate statistical tests were performed in GraphPad Prism version 6 (GraphPad Software). Standard deviations are represented throughout as +/−. All p-values < 0.05 were considered significant.
Supplementary Material
Table 1.
Association between the detection of a specific virus type and the lack of protection
| Virus | Fisher’s exact p-value | Odds Ratio |
|---|---|---|
| Picobirnaviridae | 0.534 | 2.333 |
| Circoviridae | 1.000 | 2.048 |
| Enterovirus | 0.121 | 4.667 |
| Sapelovirus | 0.237 | 5.000 |
| Picornaviridae | 0.237 | 5.000 |
| Adeno-associated virus | 0.045 | 7.000 |
| Adenoviridae | 0.0003 | 46.850 |
| Adeno-associated virus – Adenoviridae co-infection | 0.2465 | 3.273 |
Table 2.
Association between the detection of a specific virus type and dying to AIDS related illness.
| Virus | Fisher’s exact p-value | Odds Ratio |
|---|---|---|
| Picobirnaviridae | 1.000 | 1.537 |
| Circoviridae | 1.000 | 0.488 |
| Enterovirus | 0.068 | 5.333 |
| Sapelovirus | 0.533 | 3.973 |
| Picornaviridae | 0.533 | 3.973 |
| Adeno-associated virus | 0.129 | 4.400 |
| Adenoviridae | < 0.0001 | 63.000 |
| Adeno-associated virus – Adenoviridae co-infection | 0.4253 | 2.333 |
Acknowledgments
This work was supported by NIH projects OD011170, AI078526, AI096040, AI111918 and AI101354. We would like to acknowledge Elizabeth Curran, Jennifer Shields and Erica N. Borducchi for their technical assistance. We would also like to thank Megan Baldridge, Rachel Presti, Brian Keller and Jason Norman for generous advice and assistance in preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
S.A.H. completed all components of the data analysis, figure generation and writing of the text of the manuscript with the assistance of C.D., C.M., and G.Z., L.D., A.S. and M.N. assisted in the isolation of fecal nucleic acid and sequence data generation and analysis. A.D.M. coordinated all aspects of necropsy and tissue analysis from the research macaques. D.W., D.H.B., and H.W.V. designed and implemented the study.
References
- Abbink P, Lemckert AAC, Ewald BA, Lynch DM, Denholtz M, Smits S, Holterman L, Damen I, Vogels R, Thorner AR, et al. Comparative seroprevalence and immunogenicity of six rare serotype recombinant adenovirus vaccine vectors from subgroups B and D. J Virol. 2007;81:4654–4663. doi: 10.1128/JVI.02696-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biology. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch DH, Alter G, Broge T, Linde C, Ackerman ME, Brown EP, Borducchi EN, Smith KM, Nkolola JP, Liu J, et al. Protective efficacy of adenovirus/protein vaccines against SIV challenges in rhesus monkeys. Science. 2015;349:320–324. doi: 10.1126/science.aab3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch DH, Liu J, Li H, Maxfield LF, Abbink P, Lynch DM, Iampietro MJ, SanMiguel A, Seaman MS, Ferrari G, et al. Vaccine protection against acquisition of neutralization-resistant SIV challenges in rhesus monkeys. Nature. 2012;482:89–93. doi: 10.1038/nature10766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley JM, Paiardini M, Knox KS, Asher AI, Cervasi B, Asher TE, Scheinberg P, Price DA, Hage CA, Kholi LM, et al. Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood. 2008;112:2826–2835. doi: 10.1182/blood-2008-05-159301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, Kazzaz Z, Bornstein E, Lambotte O, Altmann D, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchinato V, Trindade CJ, Laurence A, Heraud JM, Brenchley JM, Ferrari MG, Zaffiri L, Tryniszewska E, Tsai WP, Vaccari M, et al. Altered balance between Th17 and Th1 cells at mucosal sites predicts AIDS progression in simian immunodeficiency virus-infected macaques. Mucosal Immunology. 2008;1:279–288. doi: 10.1038/mi.2008.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks SG, Kitchen CM, Liu L, Guo H, Gascon R, Narvaez AB, Hunt P, Martin JN, Kahn JO, Levy J, et al. Immune activation set point during early HIV infection predicts subsequent CD4+ T-cell changes independent of viral load. Blood. 2004;104:942–947. doi: 10.1182/blood-2003-09-3333. [DOI] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Estes JD, Gordon SN, Zeng M, Chahroudi AM, Dunham RM, Staprans SI, Reilly CS, Silvestri G, Haase AT. Early resolution of acute immune activation and induction of PD-1 in SIV-infected sooty mangabeys distinguishes nonpathogenic from pathogenic infection in rhesus macaques. Journal of Immunology. 2008;180:6798–6807. doi: 10.4049/jimmunol.180.10.6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner SR, Holtz LR, Jiang Y, Rajendran P, Franz CJ, Zhao G, Kang G, Wang D. Human stool contains a previously unrecognized diversity of novel astroviruses. Virol J. 2009;6:161. doi: 10.1186/1743-422X-6-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgi JV, Hultin LE, McKeating JA, Johnson TD, Owens B, Jacobson LP, Shih R, Lewis J, Wiley DJ, Phair JP, et al. Shorter survival in advanced human immunodeficiency virus type 1 infection is more closely associated with T lymphocyte activation than with plasma virus burden or virus chemokine coreceptor usage. J Infect Dis. 1999;179:859–870. doi: 10.1086/314660. [DOI] [PubMed] [Google Scholar]
- Gori A, Tincati C, Rizzardini G, Torti C, Quirino T, Haarman M, Ben Amor K, van Schaik J, Vriesema A, Knol J, et al. Early impairment of gut function and gut flora supporting a role for alteration of gastrointestinal mucosa in human immunodeficiency virus pathogenesis. Journal of Clinical Microbiology. 2008;46:757–758. doi: 10.1128/JCM.01729-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley SA, Thackray LB, Zhao G, Presti R, Miller AD, Droit L, Abbink P, Maxfield LF, Kambal A, Duan E, et al. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell. 2012;151:253–266. doi: 10.1016/j.cell.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt PW, Brenchley J, Sinclair E, McCune JM, Roland M, Page-Shafer K, Hsue P, Emu B, Krone M, Lampiris H, et al. Relationship between T cell activation and CD4+ T cell count in HIV-seropositive individuals with undetectable plasma HIV RNA levels in the absence of therapy. J Infect Dis. 2008;197:126–133. doi: 10.1086/524143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Research. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klase Z, Ortiz A, Deleage C, Mudd JC, Quiñones M, Schwartzman E, Klatt NR, Canary L, Estes JD, Brenchley JM. Dysbiotic bacteria translocate in progressive SIV infection. Mucosal immunology. 2015;8:1009–20. doi: 10.1038/mi.2014.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatt NR, Estes JD, Sun X, Ortiz AM, Barber JS, Harris LD, Cervasi B, Yokomizo LK, Pan L, Vinton CL, et al. Loss of mucosal CD103+ DCs and IL-17+ and IL-22+ lymphocytes is associated with mucosal damage in SIV infection. Mucosal immunology. 2012;5:646–657. doi: 10.1038/mi.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde R. pheatmap: Pretty Heatmaps. R package version 0.7.7 2013 [Google Scholar]
- Liu Z, Cumberland WG, Hultin LE, Prince HE, Detels R, Giorgi JV. Elevated CD38 antigen expression on CD8+ T cells is a stronger marker for the risk of chronic HIV disease progression to AIDS and death in the Multicenter AIDS Cohort Study than CD4+ cell count, soluble immune activation markers, or combinations of HLA-DR and CD38 expression. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;16:83–92. doi: 10.1097/00042560-199710010-00003. [DOI] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Applied and environmental microbiology. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra U, Holte S, Zhu T, Delpit E, Huntsberry C, Sette A, Shankarappa R, Maenza J, Corey L, McElrath MJ. Early induction and maintenance of Env-specific T-helper cells following human immunodeficiency virus type 1 infection. Journal of Virology. 2003;77:2663–2674. doi: 10.1128/JVI.77.4.2663-2674.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. The ISME Journal. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna P, Hoffmann C, Minkah N, Aye PP, Lackner A, Liu Z, Lozupone CA, Hamady M, Knight R, Bushman FD. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog. 2008;4:e20. doi: 10.1371/journal.ppat.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S. Phyloseq: a bioconductor package for handling and analysis of high-throughput phylogenetic sequence data. Pacific Symposium on Biocomputing Pacific Symposium on Biocomputing. 2012:235–246. [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Computational Biology. 2014;10:e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutlu EA, Keshavarzian A, Losurdo J, Swanson G, Siewe B, Forsyth C, French A, Demarais P, Sun Y, Koenig L, et al. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathogens. 2014;10:e1003829. doi: 10.1371/journal.ppat.1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson EC, Storm RJ, Bauer J, Johansson SM, Lookene A, Angstrom J, Hedenstrom M, Eriksson TL, Frangsmyr L, Rinaldi S, et al. The GD1a glycan is a cellular receptor for adenoviruses causing epidemic keratoconjunctivitis. Nature medicine. 17:105–109. doi: 10.1038/nm.2267. [DOI] [PubMed] [Google Scholar]
- Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC, Kambal A, Monaco CL, Zhao G, Fleshner P, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160:447–460. doi: 10.1016/j.cell.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H. vegan: Community Ecology Pacakge. ( http://cran.r-project.org/web/packages/vegan/index.html), pp. GNU Public License (GPL)
- Pérez-Brocal V, García-López R, Vázquez-Castellanos JF, Nos P, Beltrán B, Latorre A, Moya A. Study of the viral and microbial communities associated with Crohn’s disease: a metagenomic approach. Clinical and Translational Gastroenterology. 2013;4:e36. doi: 10.1038/ctg.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffatellu M, Santos RL, Verhoeven DE, George MD, Wilson RP, Winter SE, Godinez I, Sankaran S, Paixao TA, Gordon MA, et al. Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nature Medicine. 2008;14:421–428. doi: 10.1038/nm1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda K, Oh K, Ren B, Tickle TL, Franzosa EA, Wachtman LM, Miller AD, Westmoreland SV, Mansfield KG, Vallender EJ, et al. Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell Host & Microbe. 2015;17:385–391. doi: 10.1016/j.chom.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


