Abstract
Purpose
We previously found that an epithelial-to-mesenchymal transition (EMT)-based gene expression signature was highly correlated to the first principal component (PC1) of 326 colorectal cancer (CRC) tumors and was prognostic. This study was designed to improve these signatures for better prediction of metastasis and outcome.
Experimental Design
468 CRC tumors including all stages (I–IV) and metastatic lesions were used to develop a new prognostic score (ΔPC1.EMT) by subtracting the EMT signature score from its correlated PC1 signature score. The score was validated on six other independent datasets with total 3697 tumors.
Results
ΔPC1.EMT was found to be far more predictive of metastasis and outcome than its parent scores. It performed well in Stages I–III, amongst MSI subtypes, and across multiple mutation-based subclasses, demonstrating a refined capacity to predict distant metastatic potential in tumors even with a “good” prognosis. For example, in the PETACC-3 clinical trial dataset it predicted worse overall survival in an adjusted multivariable model for Stage III patients (HR by IQR=1.50, 95%CI=1.25–1.81, P=0.000016, N=644). The improved performance of ΔPC1.EMT was related to its propensity of identifying epithelial-like subpopulations as well as mesenchymal-like subpopulations. Biologically, the signature was correlated positively with RAS signaling but negatively with mitochondrial metabolism. ΔPC1.EMT was a “best of assessed” prognostic score when compared to ten other known prognostic signatures.
Conclusion
The study developed a prognostic signature score with a propensity of detecting non-EMT features, including epithelial cancer stem cell-related properties, thereby improving its potential to predict metastasis and poorer outcome in Stages I-III patients.
Keywords: colorectal cancer, metastasis, prognosis, non-EMT, epithelial cancer stem cells
INTRODUCTION
The heterogeneity of colorectal cancer (CRC) makes it difficult to determine which patients will benefit from adjuvant therapy and which patients do not require further therapy beyond surgical resection. Thus, there is an urgent need for objective molecular classification to stratify adjuvant therapy for CRC patients (1–3). One major challenge is the identification of factors to evaluate the potential of distant metastasis that has contributed to most of CRC mortality. Metastasis is a complex series of steps including the tumor cell invasion and dissemination, survival in circulation, organ-specific targeting, tumor dormancy and reactivation for colonization at distant sites (4, 5). The epithelial-to-mesenchymal transition (EMT) has been intensely studied in various types of cancers (especially breast cancer) as a major mechanism promoting invasion and metastasis (5, 6). EMT has also been reported as one of the mechanisms contributing to the resistance to Cetuximab (anti-EGFR) therapy (7). However, the biology for most steps of metastasis, especially after the early steps of invasion and dissemination, is still poorly understood (4, 5). This has greatly restricted our ability to understand and predict metastatic potential in cancer patients and has led to generalized “one size fits all” approaches to the administration of adjuvant therapy.
We have previously shown that EMT gene expression signatures can predict poor outcome in CRC and breast cancer (3, 8, 9). In an unsupervised analysis, our past work yielded a list of top-ranked genes bearing positive and negative correlation with the first principal component (PC1) of CRC expression dataset of 326 tumors (8). Of many signatures tested, our “EMT signature”, derived from a gene expression analysis of 93 lung cancer cell lines sorted (based on their expression of CDH1 or VIM) into epithelial or mesenchymal groups, showed a very strong correlation (Pearson R=0.92, P<10−135) with PC1. This PC1 and EMT association was confirmed in 38 CRC cell lines, and was also verified by assessment of other known EMT-related genes and microRNAs in CRC tumors (8). Both PC1and EMT signatures were found to predict recurrence (indicating metastasis) (8).
To further assess the respective prognostic values of the PC1 and EMT signatures, we recently evaluated the outcomes on a new set of 468 CRC tumors (Moffitt468). The improvement in prognostic power noted in a bivariable survival model, when the two signatures were put in competition with each other, prompted us to generate a composite signature (ΔPC1.EMT) by subtracting EMT from PC1. Consequently, ΔPC1.EMT emerges as a new prognostic score for CRC prognosis, which could predominantly capture the non-EMT biological features to optimize prediction of metastasis and outcome.
MATERIALS AND METHODS
Tumor samples
The cohort of 468 colorectal adenocarcinoma patients (Moffitt468 dataset) from 468 distinct patients, including 367 primary lesions (306 Stage I–III and 61 Stage IV) and 101 metastatic lesions (49 from stage IV patients), with global gene expression analysis data from the surgical specimen, microsatellite instability (MSI) status, and targeted gene sequencing (Supplementary Table S1), with samples obtained between October 2006 and September 2010, was used to develop the “difference score” ΔPC1.EMT. Metastatic samples were included only for patients for whom primary samples were not sequenced. ΔPC1.EMT was then validated on 1544 independent primary and metastatic tumors). In all cases, tissue and clinical data were collected on patients under institutional review board approval as part of the Total Cancer Care® (TCC) project (10).
We assessed/selected five additional large, independent CRC datasets from public resources (GEO and ArrayExpress) for cohorts of colorectal cancer patients with more than 100 samples, gene expression profile as well as relevant clinical information (including stage and follow-up) to be used to validate prognostic value and to determine biological significance. These include PETACC3, ALMAC, LNCC, GEO41258 and GSE14333 (3, 11–14) (Supplementary Table S2). Notably, PETACC3 was selected because it is one of the largest gene expression profile set derived from Stage II & III patients recruited in a single clinical trial, while other datasets were retrospective collections of patients. Moreover, the TCGA adenocarcionoma dataset (15) was also used for biological interpretation.
ΔPC1.EMT score computation
Probe intensities were preprocessed using RMA. PC1 and EMT scores were calculated as previously described (8). Briefly, for each of the datasets, a score was computed for each of the 4 signatures (EMT.UP.score, EMT.DOWN.score, PC1.UP.score and PC1.DOWN.score) as the arithmetic mean of all probesets corresponding to gene symbols present in the corresponding gene signature (Supplementary Table S3). EMT and PC1 scores were then obtained as follows:
EMT.score = EMT.UP.score – EMT.DOWN.score
PC1.score = PC1.UP.score – PC1.DOWN.score
The ΔPC1.EMT score was computed as follows:
ΔPC1.EMT score = PC1.score – EMT.score
Scores were standardized by subtracting the score median and dividing by the score IQR. For Moffitt 468 dataset, the score median (interquartile range) for the PC1, EMT and ΔPC1.EMT scores are −0.29 (−0.42 to −0.18), −0.38 (−0.55 to 0.21) and −0.09 (−0.17 to 0.01), respectively.
Correlation analysis
Pearson’s product moment correlation coefficient was used to quantify the association between the scores, MSI status, and mutation status for various driver genes.
GO Process analysis
Pathway analysis of the non-overlapped genes of PC1 (i.e. PC1 genelist minus PC1 & EMT overlapped gene list) by GO Process was performed using the MetaCore package. A P-value cut-off of 0.05 unadjusted for multiplicity resulted in 35 significant dysregulated pathways.
Hierarchical cluster analysis
We performed hierarchical clustering of five datasets (PETACC3, ALMAC, LNCC, GEO41258 and GSE14333) in order to visualize how the genes included in the signature grouped across different cohorts and platforms. In order to do that across all the datasets we had to collapse the expression at the gene level selecting the probset which showed the higher variability (measured by median absolute deviation). We also tested the association of each gene to overall survival (OS) and Relapse Free Survival (RFS) using a meta-analytical approach.
Weight contributions of individual signature genes
In order to characterize the three signatures (PC1, EMT and PC1.EMT), we estimated the average contribution of each gene to each of the signatures across the five datasets. Within each dataset, we first calculated a weight for each probe set in the PC1 and EMT signatures, respectively. The weight was defined as 1/P+ for probe sets with positive weight and −1/P− for probe sets with negative weight in the signature. Here, P+ and P- are the total number of probe sets with positive and negative weight, respectively. The contribution of a probe set to a signature in a given dataset was then defined as the product between the weight of the probe set and its average expression level across the dataset. By summing contributions for all probe sets corresponding to a given gene, we estimated gene-wise contributions to each signature. The contributions to the ΔPC1.EMT signature were obtained as the difference between the contributions to the PC1 and the EMT signatures. The final estimates of gene contributions to the three signatures were obtained as weighted averages of the gene contributions across all five datasets to obtain final estimates of the gene contributions to the three signatures. The weight for a data set in this sum was inversely proportional to the Euclidean norm of the vector of gene contributions to the PC1 and EMT signatures in the dataset. A linear contrast was used to test for a trend in gene expression score with increasing stage of primary disease to distant metastasis, using PROC GLM (SAS, version 9.2).
Association of gene expression with ΔPC1.EMT score
We tested the association of gene expression with the ΔPC1.EMT score within each of the five datasets plus TCGA CRC dataset (15) by a linear regression model with the score as the explanatory variable using the “limma” R package (version 3.16.3) (16), adjusting standard errors estimates by an empirical Bayes approach. P-values were combined across datasets using Fisher’s method (MADAM R package version 1.2.2). A Bonferroni correction was applied to control for false positive results introduced by multiple testing. Genes showing an adjusted P-value <0.00001 were split in two groups: those positively (N=2,983) and those negatively (N=2,221) correlated with the ΔPC1.EMT score.
Gene set enrichment analysis (GSEA) was performed to interpret the list of genes found to be correlating with ΔPC1.EMT score. The functional tool DAVID (http://david.abcc.ncifcrf.gov/) was employed to identify annotation terms enriched within each of the groups. We also performed GSEA using gene sets obtained from the MSig database (DB) (12) (MSigDB) which includes C2 (curated gene sets - Chemical and Genetic Perturbations, Biocarta and KEGG), C3 (transcription factors), C5 (GO biological process terms), C6 (oncogenic signature) and C7 (immunologic signatures). The analysis was done using the “Romer” algorithm (similar to GSEA (12)) and the same linear model used to identify genes correlating with ΔPC1.EMT score. The P-values obtained across the datasets were merged using Fisher’s method.
Survival analysis
We performed Kaplan Meier survival analysis on the Moffitt468 dataset, and used Cox proportional hazards regression models in the R package “survival” (version 2.37-7) to assess association of tumor scores with OS, RFS and/or Survival after Relapse (SAR) on the other five datasets.
Univariate analysis (OS and RFS) of other 10 known prognostic signatures
We selected a set of gene signatures known to be prognostic in CRC and could be computed from gene expression profiles. We computed the scores from 10 signatures (RAS Merck (17), RAS Astrazeneca (18), OncotypeDX colon (19), Veridex (20), MD Anderson (21) Decorin (9), MED12 (22), BRAF score (23) and ALM(12) on the five datasets as described in the original studies.
RESULTS
ΔPC1.EMT outperformed both PC1 and EMT in predicting metastasis and survival
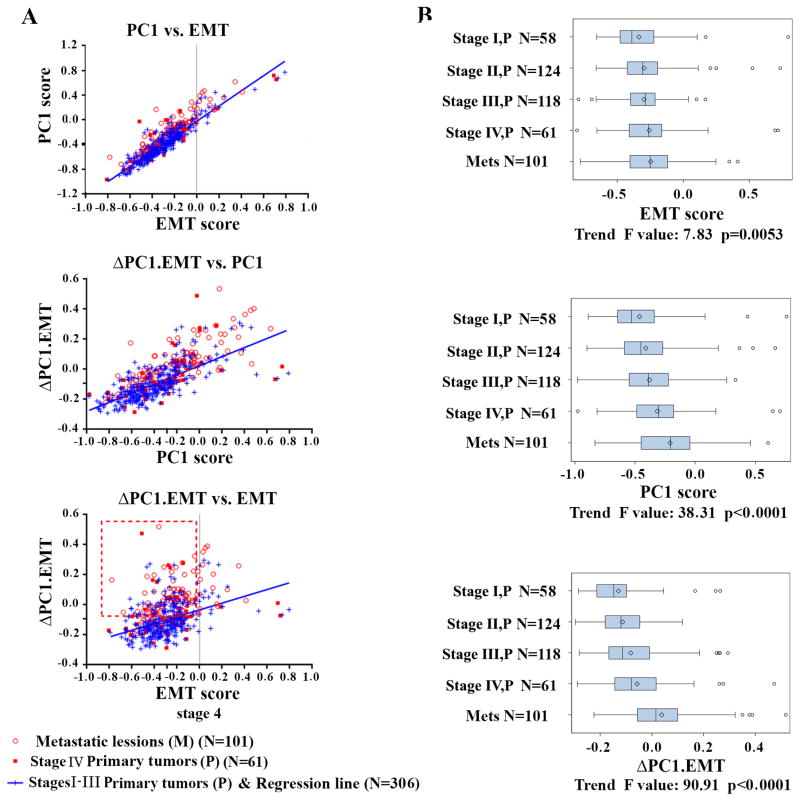
Our analysis of the Moffitt468 dataset showed that PC1 and EMT were highly correlated (Figure 1A, top panel, Pearson R=0.90, P<0.0001). The EMT score can be used to separate tumors with epithelial (<0) vs. mesenchymal (>0) features (8). The majority of metastatic tumors (who have poor overall survival) appear to be epithelial-like (EMT scores < 0, Figure 1A top panel). Notably, in the PC1 vs. EMT plot, tumors from metastatic patients or Stage IV primaries (with synchronous metastasis) (open and filled red cycles) appeared to cluster above the blue regression line (Stage I–III primary tumors), suggesting that metastatic tumors were more associated with PC1 than EMT.
Figure 1. Correlation of PC1, EMT and ΔPC1.EMT scores with each other and with stages, and metastasis on the Moffitt468 CRC dataset.
(A Top panel) PC1 vs. EMT shows strong correlation but metastatic tumor (open red circles) and Stage IV primary tumors with evidence of synchronous metastasis (filled red squares) displayed a slight propensity for higher PC1 scores than EMT scores compared to Stages I-III primary tumors (blue repression line). The gray line (EMT=0) is the dividing line as defxined (8) (EMT<0, non-EMT epithelial-like; EMT>0, EMT mesenchymal-like). (A, middle and bottom panels) ΔPC1.EMT outperformed EMT and PC1 in predicting metastasis. Red box highlights higher ΔPC1.EMT (above the median value)-captured non-EMT subpopulations (EMT<0). (B) Comparison between ΔPC1.EMT, PC1 and EMT scores in progressively deciphering metastatic potential of primary CRCs (stages I vs. II vs. III vs. IV) vs. metastatic lesions. Trend F and P values are given for the three scores. Six samples that lack stage information were removed.
Consistent with this figure, survival analysis using the univariate Cox proportional hazard regression model for overall survival (OS) on Moffitt468 indicates that the PC1 score was predictive of OS (HR=1.40, 95%CI: 1.18–1.66, P=0.0001), while the EMT score fell short of statistical significance (HR=1.13, 95%CI: 0.96–1.34, P=0.14). Interestingly, when the scores were used in a multivariable Cox survival model, the coefficients (logarithms of the HR) for PC1 and EMT were both highly significant – but of roughly equal magnitude and opposite numeric sign (i.e. for PC1, HR=3.75 (worse survival), 95%CI: 2.51–5.61, P<0.0001; for EMT, HR=0.36 (better survival), 95%CI: 0.24–0.53, P<0.0001, with log HRs = 1.32 and −1.02). The statistical interpretation of this result is that survival is best explained not by PC1 or EMT alone but by a score obtained by combining them into a new score to which the PC1 score contributes positively and the EMT score negatively, with roughly equal magnitudes. Thus, we elected to subtract the EMT score from the PC1 score to produce a “difference” score (ΔPC1.EMT). Subsequent univariate OS analysis indeed demonstrated that ΔPC1.EMT (HR=1.82, 95%CI: 1.51–2.18, P<0.0001) clearly outperformed not only EMT, but also PC1, which is supported by a significantly stronger association with metastatic tumors (Figure 1A middle and bottom panels). The ΔPC1.EMT score had a good association with EMT (Pearson R=0.38, P<0.0001), but displayed an even stronger correlation with PC1 (Pearson R=0.74, P<0.0001), suggesting that PC1 includes a non-EMT biological component (presuming that the EMT score captures the EMT component fairly completely). In support of this notion, higher ΔPC1.EMT scores better separate the metastatic and non-metastatic tumor tissues, most of which have EMT score < 0 indicating epithelial-like tumors (Figure 1A bottom panel, highlighted by red box). Moreover, it was clear that PC1, and especially ΔPC1.EMT, outperformed EMT in progressively deciphering the degree of tumor progression of primary CRCs (from increasing primary stage to metastatic lesions (Figure 1B).
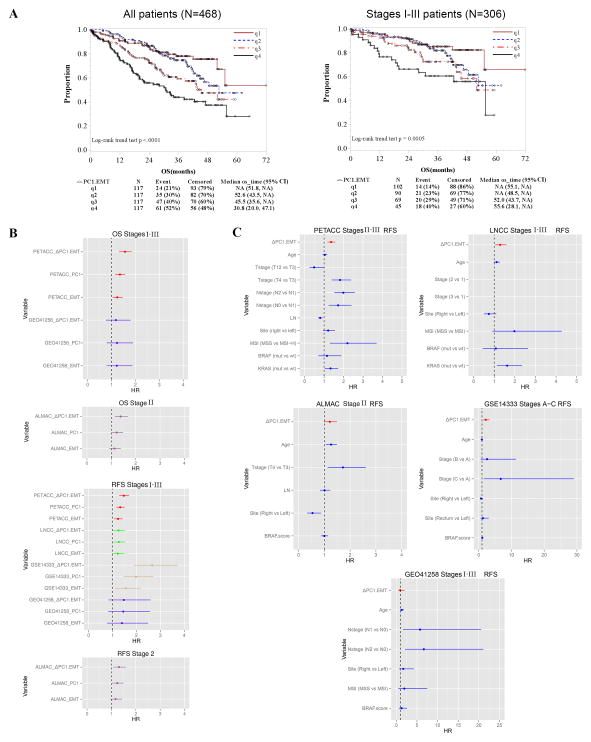
Furthermore, Kaplan Meier survival analysis shows that a higher ΔPC1.EMT score could better predict poorer OS for all patients (logrank trend, P<0.0001, Figure 2A left panel) than PC1 (logrank trend, P=0.0006) and EMT (logrank trend, P=0.1571) (Supplementary Figures S1A and S2A). Notably, ΔPC1.EMT predicted poorer OS for MSS (P<0.0001) and tended toward statistical significance for MSI (P=0.085) patients (Supplementary Figure S3A). Moreover, when limited to the 306 stage I-III primary tumors, ΔPC1.EMT clearly outperformed its parental scores (P=0.0005 for ΔPC1.EMT, Figure 2A right panel, as compared to P=0.1437 for PC1 and P=0.3313 for EMT, Supplementary Figures S1B and S2B). By contrast, for metastatic tumors, like its parental scores (Supplementary Figures S1C and S2C), ΔPC1.EMT did not predict poorer OS (Supplementary Figure S3B).
Figure 2. ΔPC1.EMT is associated with poor overall survival on the Moffitt468 CRC dataset and on independent CRC datasets.
(A) Kaplan Meier (KM) survival analysis of quartile scores on Moffitt468 shows that a higher ΔPC1.EMT predicted poorer overall survival (OS) for all 468 patients (left panel), and also when limited to the 306 stage I–III primary tumors (right panel). Also see Supplementary Figures S1–S3 for additional analyses on MSS, MSI, and metastatic tumors as well as for PC1 and EMT scores. (B) Forest plot summary of OS and RFS univariate analyses of EMT, PC1 and ΔPC1.EMT scores on PETACC3, LNCC, GSE14333, GEO41258 and ALMAC datasets. See Supplementary Table S4 for detailed information with other clinico-pathological and molecular variables. Note: signature scores were standardized by IQR. (C) Forest plot summary of RFS multivariable analyses of the ΔPC1.EMT score on PETACC3, LNCC, GSE14333, GEO41258 and ALMAC datasets. Note: the solid lines represent 95%CI and signature scores were standardized by IQR. See Supplementary Figure S4 for OS multivariable analyses. Also see Supplementary Tables S5–S9 for detailed information for PC1 and EMT scores as well as other clinico-pathological and molecular variables.
Validation of ΔPC1.EMT’s prognostic value
The prognostic value of ΔPC1.EMT was also tested and confirmed by a univariate Cox regression analysis in a Moffitt dataset with 1544 independent cases, showing that ΔPC1.EMT robustly predicted worse OS (HR=1.49, 95% CI: 1.36-1.64, P=2.2Δ 10−16), or when restricted to 981 stage I-III primary tumors (HR=1.43, 95% CI:1.26–1.63, P=3.6× 10−8).
These findings were validated when ΔPC1.EMT was further tested for OS, relapse free survival (RFS), and survival after relapse (SAR) on the PETACC3 dataset (n=752) (3) (Table 1a). As observed on Moffitt468, ΔPC1.EMT outperformed both PC1 and EMT scores. For instance, in a univariate model for OS with the Stage III patients (n=644, Table 1b), ΔPC1.EMT had the most significant P-value of the three signatures, with an HR of 1.69 (P=8.22×10−09) compared to that of 1.41 (P=5.13×10−05) and 1.28 (P=8.21×10−03) for PC1 and EMT, respectively. In the multivariable modeling including PC1 and EMT on the same dataset (Table 1c), the HR for PC1 was 3.22 while the HR for EMT was 0.37 (coefficients 1.17 and −0.99). The statistical meaning is that also in this cohort a contrast of these two scores is significantly better than either of them alone, and quantitatively similarly as in the Moffitt data the simple difference (coefficients +1 and −1) is close to the optimally fitting combination.
Table 1.
Cox Proportional Hazard Regression models for survival on PETACC3 dataset.
| a. Univariate models for overall survival (OS), relapse free survival (RFS) and/or survival after relapse (SAR) by ΔPC1.EMT score on PETACC3 Stage II&III and Stage III patients | |||
|---|---|---|---|
| Covariates | HR (95% CI) | p | n |
| OS for Stage II and III | 1.56 (1.32–1.84) | 1.16e-07 | 752 |
|
| |||
| OS for Stage III | 1.69 (1.42–2.03) | 8.22e-09 | 644 |
|
| |||
| RFS for Stage II and III | 1.47 (1.28–1.69) | 8.98e-08 | 752 |
|
| |||
| RFS for Stage III | 1.55 (1.33–1.81) | 3.99e-08 | 644 |
|
| |||
| SAR for Stage II and III | 1.20 (1.02–1.42) | 3.11e-02 | 291 |
|
| |||
| SAR for Stage III | 1.26 (1.04–1.51) | 1.54e-02 | 241 |
| b. Univariate models for OS by ΔPC1.EMT, PC1 and EMT scores as well as other clinic-pathological and molecular variables on PETACC3 Stage III patients (n=644) | ||
|---|---|---|
| Covariates | HR (95% CI) | p |
| ΔPC1.EMT | 1.69 (1.42–2.03) | 8.22e-09 |
| PC1 | 1.41 (1.20–1.67) | 5.13e-05 |
| EMT | 1.28 (1.07–1.53) | 8.21e-03 |
| MSI (MSS vs. MSI-H) | 1.49 (0.81–2.75) | 2.00e-01 |
| BRAF (wt vs. mut) | 0.57 (0.34–0.95) | 3.04e-02 |
| site (right vs. left) | 1.42 (1.06–1.91) | 1.84e-02 |
| T stage (T1,2 vs. T3) | 0.36 (0.16–0.82) | 1.47e-02 |
| T stage (T4 vs. T3) | 2.03 (1.45–2.86) | 4.09e-05 |
| grade (G-3,4 vs. G-1,2) | 1.89 (1.26–2.82) | 2.07e-03 |
| SMAD4 (Any Loss vs. No Loss) (45) | 1.57 (1.13–2.16) | 6.75e-03 |
| KRAS (mut vs. wt) | 1.49 (1.10–2.01) | 9.93e-03 |
| BRAF.score (23) | 1.34 (1.20–1.50) | 1.84e-07 |
| Age | 1.09 (0.94–1.25) | 2.49e-01 |
| LN (No. of lymph nodes) | 0.81 (0.68–0.97) | 1.95e-02 |
| c. Multivariate models for OS including PC1 and EMT scores as well as other clinic-pathological and molecular variables on PETACC3_Stage III patients (n=644) | ||
|---|---|---|
| Covariates | HR (95% CI) | p |
| PC1 | 3.22 (1.82–5.7) | 5.62e-005 |
| EMT | 0.37 (0.2–0.67) | 1.14e-003 |
| Age | 1.08 (0.93–1.25) | 3.14e-001 |
| T stage (T1,2 vs T3) | 0.48 (0.21–1.11) | 8.65e-002 |
| T stage (T4 vs T3) | 1.96 (1.39–2.77) | 1.32e-004 |
| N stage (N2 vs N1) | 2.11 (1.57–2.85) | 9.50e-007 |
| LN (No. of lymph nodes) | 0.73 (0.6–0.88) | 9.13e-004 |
| site (right vs left) | 1.75 (1.27–2.4) | 5.75e-004 |
| MSI (MSS vs MSI-H) | 1.92 (1–3.67) | 4.96e-002 |
| BRAF (wt vs mut) | 0.87 (0.5–1.52) | 6.34e-001 |
| d. Multivariate models for OS including ΔPC1.EMT or PC1or EMT scores as well as other clinic-pathological and molecular variables on PETACC3_Stage III patients (n=644) | ||||||
|---|---|---|---|---|---|---|
| ΔPC1.EMT | PC1 | EMT | ||||
| Covariates | HR (95% CI) | p | HR (95% CI) | p | HR (95% CI) | p |
| Scores | 1.50 (1.25–1.81) | 1.61e-05 | 1.32 (1.11–1.58) | 2.12e-03 | 1.21 (1.00–1.46) | 4.97e-02 |
| Age | 1.04 (0.89–1.20) | 6.37e-01 | 1.04 (0.90–1.21) | 5.95e-01 | 1.03 (0.89–1.20) | 6.64e-01 |
| Tstage (T12 vs T3) | 0.51 (0.22–1.16) | 1.09e-01 | 0.51 (0.22–1.19) | 1.20e-01 | 0.48 (0.21–1.11) | 8.50e-02 |
| Tstage (T4 vs T3) | 2.07 (1.46–2.92) | 3.92e-05 | 2.09 (1.48–2.96) | 3.03e-05 | 2.12 (1.50–3.00) | 2.36e-05 |
| Nstage (N2 vs N1) | 2.18 (1.61–2.94) | 3.87e-07 | 2.23 (1.65–3.00) | 1.46e-07 | 2.25 (1.67–3.03) | 9.43e-08 |
| LN | 0.73 (0.60–0.88) | 8.57e-04 | 0.73 (0.61–0.88) | 1.03e-03 | 0.73 (0.61–0.88) | 1.01e-03 |
| Site (right vs left) | 1.70 (1.24–2.34) | 1.03e-03 | 1.61 (1.17–2.21) | 3.51e-03 | 1.60 (1.16–2.20) | 3.86e-03 |
| MSI (MSS vs MSI-H) | 1.90 (0.99–3.65) | 5.22e-02 | 2.19 (1.14–4.21) | 1.80e-02 | 2.22 (1.15–4.27) | 1.69e-02 |
| BRAF (mut vs wt) | 1.48 (0.82–2.65) | 1.93e-01 | 1.75 (0.98–3.11) | 5.65e-02 | 1.83 (1.03–3.26) | 3.86e-02 |
| KRAS (mut vs wt) | 1.59 (1.14–2.21) | 5.73e-03 | 1.64 (1.18–2.28) | 3.32e-03 | 1.64 (1.18–2.28) | 3.54e-03 |
Note: the scores were standardized by IQR.
The validation was then expanded to include additional independent datasets (n=1401 CRC tumors from the other 4 datasets) (Supplementary Table S2) along with various clinico-pathological and molecular variables including age, T and N stages, number of examined lymph nodes, tumor site (left and right), MSI status, BRAF mutation, BRAF score, and/or KRAS mutation. Generally, in univariate models, ΔPC1.EMT outperformed PC1, which performed better than EMT (this ordering held in 11 of 15 models) (Figure 2B and Supplementary Table S4).
Furthermore, the independent prognostic value of ΔPC1.EMT was confirmed in 3 out of 5 datasets when analyzed in multivariate models including other clinico-pathological and molecular parameters such as MSI, BRAF and/or KRAS mutations (Figure 2C, Supplementary Figure S4 and Supplementary Table S5). The signature performance was further verified by additional analyses, as shown by the survival vs. score curves (Supplementary Figures S5 and S6) as well as the observed vs. predicted survival probability curves (Supplementary Figures S7 and S8).
In addition, in agreement with the univariate results, overall, ΔPC1.EMT significantly outperformed both EMT and PC1 scores in multivariate OS and RFS analyses when they were compared with each other by individually (Table 1d, Supplementary Tables S5 (ΔPC1.EMT) vs. S6 (PC1) vs. S7 (EMT), or in combinations (Supplementary Tables S8 (ΔPC1.EMT vs.PC1) and S9 (ΔPC1.EMT vs. EMT)). For example, when compared individually for OS on PETACC Stage III, n=642), HR (95%CI)=1.50 (1.25–1.81), P=1.61×10−05 (ΔPC1.EMT) vs. 1.32 (1.11–1.58), P=2.12×10−03 (PC1) vs. 1.21 (1.00–1.46), P=4.97×10−02 (EMT), while for RFS on the same dataset, HR (95%CI) =1.41 (1.20–1.65), P=2.84×10−05 (ΔPC1.EMT) vs. 1.28 (1.09–1.49), P=1.92×10−03 (PC1) vs. 1.19 (1.01–1.40), P=4.04×10−02 (EMT).
It is noteworthy that currently only few CRC datasets exist and are accessible where survival and expression profiles having >100 patients. For example, GEO41258 is our smallest dataset in which we could not also find correlation with survival, neither in univariate nor in multivariate models, for well-known variables such as MSI status, tumor side, T-stage and stage. This may suggest that this population is not representative of CRC patients. However, we decided to include it for an unbiased report of our results.
ΔPC1.EMT identified metastatic tumors with non-EMT features
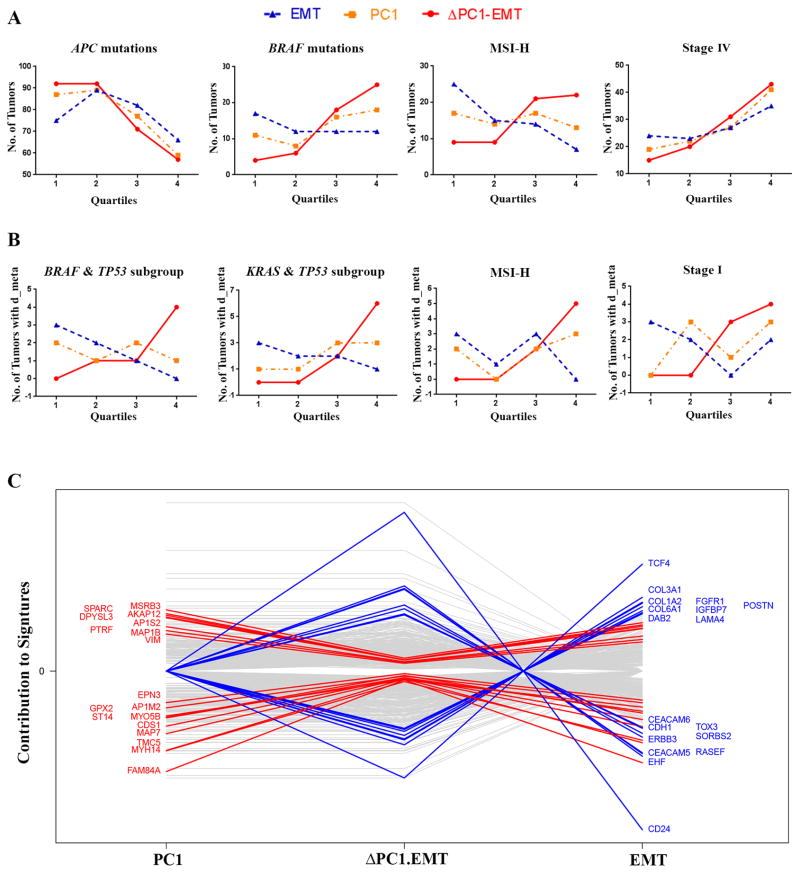
To explore the molecular basis for the observed prognostic improvement of ΔPC1.EMT from its parent PC1 and EMT scores, we examined quartile trends of these three scores versus the number of tumors harboring observed mutations of several known “driver” genes on Moffitt468, as this may provide insights into the mechanisms underlying the signature. The ΔPC1.EMT score had stronger trends (relative to PC1 and EMT) with tumors harboring APC truncated mutations (negative) and BRAF (V600E) mutations (positive), as well as tumors identified as MSI-H (positive) or Stage IV (positive) (Figure 3A; for Stage I–III patients, see Supplementary Figure S9). Notably, while percentage of distant metastatic tumors overall increased across the quartiles for all three scores, for some subgroups of combined mutations (KRAS & TP53, or BRAF & TP53), as well as in MSI-H and Stage I cases, the positive trend was more pronounced for ΔPC1.EMT in contrast to the negative trend for EMT (Figure 3B), further supporting the notion that ΔPC1.EMT might be measuring non-EMT components of metastasis.
Figure 3. ΔPC1.EMT identified several subgroups of CRC and appeared to have propensity to measure non-EMT components of CRC.
(A and B) Analysis of quartile trends (from low 1st to high 4th quartiles) of ΔPC1.EMT, PC1 and EMT scores on the Moffitt468 dataset (for all patients). (A) The ΔPC1.EMT score trended well (relative to PC1 and EMT) with APC truncated mutation (downward) and BRAF V600E (upward), and with tumors identified as MSI-H (upward) and Stage IV (upward). See Supplementary Figure S9 for Stage I-III patients. (B) In the subgroups of combined mutations (KRAS&TP53 or BRAF&TP53) as well as in MSI-H and Stage I cases, ΔPC1.EMT and EMT trended in opposite directions with respect to the number of patients with distant metastases (d_meta). (C) Weighted analysis of individual genes contributing to PC1 and EMT vs. ΔPC1.EMT signatures on five datasets (PETACC3, ALMAC, LNCC, GEO41258, and GSE14333) datasets suggests that ΔPC1.EMT was represented by more non-EMT components when compared with the other two scores. The genes that were the most changed from EMT or PC1 to ΔPC1.EMT are highlighted.
The ΔPC1.EMT score was found to be associated with several clinico-pathological and molecular variables using the The Cancer Genome Atlas (TCGA) dataset (15) (Supplementary Figure S10), with BRAF mutation, MSI status, and mucinous tumors showing the strongest positive associations (P<0.001). It is noteworthy that for the Moffitt468 data, MSI was positively correlated with ΔPC1.EMT, but uncorrelated with PC1 and negatively correlated with EMT (Supplementary Table S10).
Hierarchical clustering and contribution analyses of the signature genes
To better understand the molecular underpinnings of ΔPC1.EMT, gene expression clustering analysis was performed on the five datasets. Data show areas of strong overlap of PC1 and EMT genes, especially in the middle of the OS and RFS heatmaps, accounting for their high correlation, but also show isolated, non-overlapping genes (Supplementary Figure S11), providing the potential for ΔPC1.EMT to improve outcome. Notably, the high correlation between these two signatures was shown in Supplementary Figure S12. Since the contributions of VIM (a mesenchymal gene used to create the EMT signature) and other overlapped genes were effectively diminished in ΔPC1.EMT, we suspected that ΔPC1.EMT might better measure non-EMT features of CRC. An analysis of the GO Process of those non-overlapping genes indicates that a number of the pathways were related to cell adhesion and cellular remodeling, which are frequently associated with metastasis (Supplementary Table S11). To further address this issue, we analyzed respective weighted contributions of individual signature genes on the five datasets to identify the genes whose contributions changed the most from PC1 or EMT to ΔPC1.EMT (Figure 3C). ΔPC1.EMT was represented by more epithelial and less mesenchymal gene contributions as evidenced by the increased contribution of the epithelial marker CDH1, whereas the mesenchymal marker VIM and other EMT-related genes including SPARC, TCF4, COL1A2 and COL3A1 decreased.
Identification of ΔPC1.EMT-correlated genes and pathways
To further explore the biological implication of ΔPC1.EMT, we performed another association analysis and identified a list of top-ranked genes whose expression was either positively or negatively correlated with ΔPC1.EMT (Table 2) in a linear model on the five datasets plus the TCGA CRC dataset (15). Many of the identified genes have been reported to have biological functions related to metastasis and cancer stem cell-like properties, as discussed below. Notably, 13/20 of them belong to PC1 and/or EMT signature genes. To interpret the biological meaning of identified ΔPC1.EMT-correlated genes, we also carried out extensive gene set enrichment analysis (GSEA) and identified a variety of biological processes correlated with ΔPC1.EMT, including negatively correlated mitochondrial metabolism (Supplementary Tables S12 and S13).
Table 2.
Genes most correlated with ΔPC1.EMT score*
| a.Top ten genes positively correlated with ΔPC1.EMT | ||||||
|---|---|---|---|---|---|---|
| Gene.Symbol | EntrezID | S | num.p | p.adj | Sum t statistics | Signature genes** |
| CD109 | 135228 | 869.79 | 5 | <0.0001 | 75.05 | |
| AHNAK2 | 113146 | 837.41 | 6 | <0.0001 | 79.32 | |
| GAS1 | 2619 | 806.50 | 6 | <0.0001 | 76.57 | |
| PRKCDBP | 112464 | 806.43 | 6 | <0.0001 | 77.90 | |
| MEIS2 | 4212 | 779.02 | 6 | <0.0001 | 77.16 | |
| NXN | 64359 | 772.64 | 5 | <0.0001 | 70.33 | |
| GFPT2 | 9945 | 727.95 | 6 | <0.0001 | 72.26 | PC1&EMT Up |
| OPMP22 | 5376 | 711.36 | 6 | <0.0001 | 73.46 | EMT Up |
| WWTR1 | 25937 | 692.29 | 6 | <0.0001 | 72.07 | PC1 Down |
| PTRF | 284119 | 688.52 | 6 | <0.0001 | 71.22 | PC1&EMT Up |
| b.Top ten genes negatively correlated with ΔPC1.EMT | ||||||
|---|---|---|---|---|---|---|
| Gene.Symbol | EntrezID | S | num.p | p.adj | Sum t statistics | Signature genes** |
| CDX1 | 1044 | 860.61 | 6 | <0.0001 | −80.16 | PC1 Down |
| CDX2 | 1045 | 845.27 | 6 | <0.0001 | −79.41 | |
| C10orf99 | 387695 | 767.33 | 5 | <0.0001 | −67.82 | PC1 Down |
| DDC | 1644 | 752.19 | 6 | <0.0001 | −73.57 | |
| GPA33 | 10223 | 726.29 | 6 | <0.0001 | −72.98 | PC1 Down |
| FAM84A | 151354 | 720.55 | 5 | <0.0001 | −67.43 | |
| NR1I2 | 8856 | 697.98 | 6 | <0.0001 | −70.24 | PC1 Down |
| MYB | 4602 | 630.56 | 6 | <0.0001 | −68.13 | PC1 Down |
| C2orf89 | 129293 | 616.89 | 5 | <0.0001 | −60.62 | PC1&EMT Down |
| EPHB2 | 2048 | 597.82 | 6 | <0.0001 | −66.42 | PC1 Down |
Note:
identified in a linear model on the six datasets
The genes correlated with ΔPC1.EMT that are overlapped with the PC1 and EMT signature genes (also see Supplementary Table S3).
Comparison of ΔPC1.EMT with other known prognostic signatures
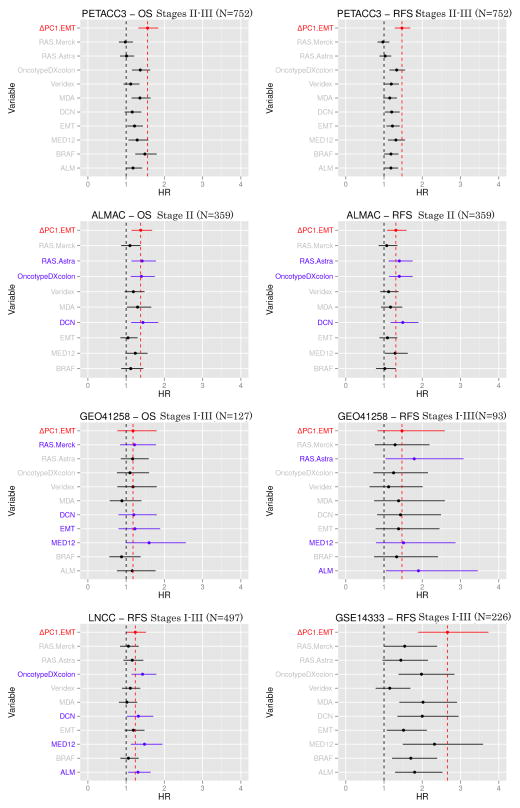
Finally, we compared the ΔPC1.EMT score with an expanded set of other known prognostic signatures on the five datasets in a univariate analysis. Results show that overall, ΔPC1.EMT was among the best prognostic signature scores for OS and RFS analyses when compared to ten other known prognostic signatures across eight comparisons, with a higher HR more often than all other scores except DCN, with which it was tied (Figure 4). It is of interest to mention that ΔPC1.EMT showed a partial correlation with the OncotypeDX colon signature (GH) which had exploited cell proliferation as a potential prognostic marker (19) (Supplementary Table S14).
Figure 4. Univariate analysis (OS and RFS) of ΔPC1.EMT and other 10 known prognostic signatures.
on five datasets (PETACC3, ALMAC, LNCC, GEO41258 and GSE14333). We computed the scores from 10 signatures (RAS Merck, RAS Astrazeneca, OncotypeDX colon, Veridex, MD Anderson (MDA), Decorin (DCN), EMT, MED12, BRAF score and ALM on the five datasets as described in the original studies. ΔPC1.EMT is colored in red, while signatures showing relative higher HR are colored in blue. Note: the solid lines represent 95%CI and prognostic signature scores were standardized by IQR.
DISCUSSION
Here we present the first evidence using human tissues revealing that although EMT is a dominant molecular program of colorectal cancer (8), the non-EMT features captured by ΔPC1.EMT appear to be necessary to optimally predict distant metastasis. In support of this notion, the ΔPC1.EMT score demonstrated a strong non-EMT signature propensity in predicting distant metastasis (Figure 1). It also displayed a refined capacity to detect non-EMT-related metastatic potential in tumors harboring subgroups of combined mutations (KRAS & TP53 or BRAF & TP53) with abnormal RAS activation as well as in MSI-H and Stage I cases generally classified with a “good” prognosis (Figure 3). Our findings are in agreement with the recent notion that the epithelial phenotype may be critical for the successful seeding and propagation of cancer cells at distant sites (4, 24–29). For instance, from a clinico-pathological point of view, cohesive epithelial migration was often observed as the predominant pattern in CRC (30), although the related biology is not clear yet.
The result of analyzing contributions of the signature genes sheds light on the molecular underpinnings of ΔPC1.EMT. Compared to EMT, the gene with the greatest contribution increase in ΔPC1.EMT was CD24 (Figure 3C), previously reported as a metastasis-associated gene (31), and a marker of colon cancer stem cells (CSC) whose properties are thought to contribute to “metastatic traits” and therapeutic resistance (5, 32). Thus, ΔPC1.EMT captures both epithelial and CSC features, which are supported by a recent report demonstrating that in breast cancer, CDH1 and CD24 were highly enriched in the epithelial CSCs (ALDH1-positive), while their expression was down-regulated in the mesenchymal CSCs (CD44+CD24-)(33). ERBB3, a member of the EGFR family (34), was also identified as one of the genes whose contribution was increased in ΔPC1.EMT (Figure 3C). In agreement with this, we observed that ΔPC1.EMT, but not EMT, was associated with activation of the RAS/MAPK pathway, evidenced by its positive correlation with various RAS signature scores (Supplementary Table S10). Thus, we speculated that ΔPC1.EMT-associated poor prognosis might, in part, result from RAS/MAPK activation-mediated drug resistance (34) in epithelial-like CRC.
Moreover, many of identified most strongly ΔPC1.EMT-correlated genes (Table 2) have been reported to relate to metastasis and/or CSCs. For instance, CD109 (the top positively-correlated gene) has recently identified by proteomic analyses as a metastasis-associated protein marker (35) and was highly expressed in ALDH1-characterized epithelioid sarcoma CSCs (36). Meanwhile, CDX1 and CDX2 (the two most negatively correlated genes) were reported as putative tumor suppressor genes whose expression was epigenetically repressed in CRC, and reduced expression of CDX1 inhibited CSC stem cell differentiation and thus promoted CSC renewal (35). In support of this, HCT116, an epithelial, MSI CRC cell line that lacks expression of CDX1 was recently classified as a colon CSC cell line (2). In addition, reduced expression of EPHB2 was associated with metastasis (37) while its overexpression induced EMT (38), whereas the cell cycle gene MYB, when ectopically expressed, contributed to cell migration and invasion but to also prevent metastasis (39). Thus, identification of EPHB2 and MYB as strong negatively-correlated genes of ΔPC1.EMT further supports the notion of non-EMT contributions to metastasis. In agreement with this, the ΔPC1.EMT-correlated GH prognostic signature was negatively correlated with cell cycle genes such as MYBL2 (19), although how GH may be potentially related to metastasis is unknown.
In further support of the negative association of ΔPC1.EMT with cell proliferation, pathway analyses show that ΔPC1.EMT was clearly correlated with negative regulation of mitochondrial metabolism (Supplementary Tables S12 and S13). It is noteworthy that the metastasis suppressor gene KISS1 was recently reported to promote normal mitochondrial metabolism, an anti-metastasis mechanism (40). Moreover, it has recently been reported that the mitochondrial pyruvate carrier (MPC) played a repressor role in the Warburg effect in CRC and results indicated that inhibition of mitochondrial metabolism was connected to the maintenance and fate of cancer stem cells (41).
In the “cell cooperativity” model (27), Tsuji et al. demonstrated that primary tumors were heterogeneous and contained both mesenchymal and epithelial cell types (with mesenchymal cells populating the invasive front), but metastatic tumors contained only the cells originating from the epithelial type. This model postulates that the canonical epithelial-to-mesenchymal transition (EMT) does not fully explain metastatic potential, and should have strong epithelial features. Accordingly, the ΔPC1.EMT score reported here captures predominantly non-EMT features. Although ΔPC1.EMT is certainly distinct from EMT, it still retains some correlation with it (Supplementary Table S10 and Supplementary Figure S12). Indeed, ΔPC1.EMT was positively correlated with the EMT-related pathways associated with response to wounding, cell motility, extracellular matrix remodeling, activation of TGFbeta signaling, and angiogenesis (Supplementary Tables S12 and S13), as well as three important EMT-related pathways centered around SLUG1 (Supplementary Table S11). It is noteworthy that SLUG1 was reported to cooperate with SOX9 to convert differentiated mammary epithelial cells to stem cells (42) and stromal gene expression (EMT-related) was also recently reported to define poor-prognosis subtypes in colorectal cancer (43, 44). The reason for a significant EMT-correlation for ΔPC1.EMT is not yet clear. According to the recent notion of epithelial plasticity (25), EMT is not a “black and white” program in human cancers, and there likely exist a variety of “gray” EMT states in most tumors especially CRC, which may be a part of the intrinsic heterogeneous nature of the disease. PC1 and EMT scores, which have quite different lists of signature genes, may differ in their abilities to measure the degree of “gray”. Thus, the “EMT” components might be only partially canceled out by subtracting the EMT score from the PC1 score, resulting in the significant “residual” correlation between ΔPC1.EMT and EMT.
The results of this study are compelling and suggest that ΔPC1.EMT may be strongly predictive of adverse outcomes (metastasis, and diminished survival), which should help determine which patients may need adjuvant chemotherapy in Stage II/III disease. However, few high quality datasets exist where molecular data have been collected with clinical data in patients with identified adjuvant chemotherapy history. Probably for this reason, we observed that the ΔPC1.EMT score was found significantly correlated with survival in the majority but not all the test sets. This also happened for ten other known prognostic signatures when tested on the same datasets (Figure 4); but overall, ΔPC1.EMT appeared to be a “best of assessed” prognostic score with an “optimized” capacity to predict metastasis. However, excluding the PETACC dataset, the analyzed cohorts were derived from retrospective collections of patients, hampering the generalization of our findings. Therefore, there is a clear need for further investigation of ΔPC1.EMT in a prospective clinical trial to determine its prognostic value in predicting which patients will metastasize and thus possibly benefit from adjuvant chemotherapy.
In conclusion, our findings suggest that poor RFS and OS can be predicted by a robust gene expression signature, ΔPC1.EMT, preferentially based on non-EMT stem cell biology. ΔPC1.EMT had a refined capacity to detect poorer overall, and in various subgroups of CRC using preferentially non-EMT features (including epithelial cancer stem cell-related properties), thereby potentially providing new targets for therapy of distant disease. The score may have utility in identifying Stages II and III patients with high risk of metastasis. Thus, we believe that there should be considerable enthusiasm about further examination of this signature in a prospective clinical trial.
Supplementary Material
Statement of Translational Relevance.
The heterogeneity of colorectal cancer (CRC) results in an urgent need for objective molecular classification to stratify adjuvant therapy for CRC patients. One major challenge is to evaluate the potential of distant metastasis that has contributed to most to CRC mortality. The new prognostic signature score developed and validated in this study was shown to be a “best of assessed” prognostic signature score demonstrating a refined capacity to predict metastasis and outcomes in CRC, with a non-EMT propensity including epithelial cancer stem cell-related properties. The finding has clinical utility to determine which patients will metastasize and which will not, in otherwise good and poor prognosis lesions (all stages, MSI, and within mutational subgroups). The signature may have potential to identify which patients may or may not benefit from adjuvant chemotherapy—a problem for which there is no current solution.
Acknowledgments
This work was supported by National Institutes of Health grant U01CA157960 (to TJY). Total Cancer Care® is enabled, in part, by the support of the DeBartolo Family, and we thank the patients who provided data and tissue to the Total Cancer Care Consortium. Our study also received assistance from the Biostatistics and Cancer Informatics Core Facilities at the H. Lee Moffitt Cancer Center & Research Institute, an NCI designated Comprehensive Cancer Center, supported under NIH grant P30-CA76292. This work was also supported by a grant of the Swiss State Secretariat for Education, Research and Innovation to the SIB Swiss Institute of Bioinformatics (group M.D.) for Service and infrastructure resources. We also thank the Fondation Medic for financial support. Author Gregory Bloom was not available to confirm coauthorship, but the corresponding author, Timothy J. Yeatman, affirms that Gregory Bloom contributed to the manuscript and thus confirms author Gregory Bloom’s coauthorship status.
Footnotes
Competing financial interests: M.J.S and T.J.Y report involvement as part of a provisional patent submitted to The United States Patent and Trademark Office, during the conduct of the study. All other authors declare no competing financial interests.
References
- 1.De Sousa EMF, Wang X, Jansen M, Fessler E, Trinh A, de Rooij LP, et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med. 2013;19(5):614–8. doi: 10.1038/nm.3174. [DOI] [PubMed] [Google Scholar]
- 2.Sadanandam A, Lyssiotis CA, Homicsko K, Collisson EA, Gibb WJ, Wullschleger S, et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19(5):619–25. doi: 10.1038/nm.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Budinska E, Popovici V, Tejpar S, D’Ario G, Lapique N, Sikora KO, et al. Gene expression patterns unveil a new level of molecular heterogeneity in colorectal cancer. J Pathol. 2013;231(1):63–76. doi: 10.1002/path.4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giancotti FG. Mechanisms governing metastatic dormancy and reactivation. Cell. 2013;155(4):750–64. doi: 10.1016/j.cell.2013.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vanharanta S, Massague J. Origins of metastatic traits. Cancer Cell. 2013;24(4):410–21. doi: 10.1016/j.ccr.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331(6024):1559–64. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 7.Brand TM, Iida M, Wheeler DL. Molecular mechanisms of resistance to the EGFR monoclonal antibody cetuximab. Cancer biology & therapy. 2011;11(9):777–92. doi: 10.4161/cbt.11.9.15050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loboda A, Nebozhyn MV, Watters JW, Buser CA, Shaw PM, Huang PS, et al. EMT is the dominant program in human colon cancer. BMC Med Genomics. 2011;4:9. doi: 10.1186/1755-8794-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farmer P, Bonnefoi H, Anderle P, Cameron D, Wirapati P, Becette V, et al. A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat Med. 2009;15(1):68–74. doi: 10.1038/nm.1908. [DOI] [PubMed] [Google Scholar]
- 10.Fenstermacher DA, Wenham RM, Rollison DE, Dalton WS. Implementing personalized medicine in a cancer center. Cancer journal. 2011;17(6):528–36. doi: 10.1097/PPO.0b013e318238216e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jorissen RN, Gibbs P, Christie M, Prakash S, Lipton L, Desai J, et al. Metastasis-Associated Gene Expression Changes Predict Poor Outcomes in Patients with Dukes Stage B and C Colorectal Cancer. Clin Cancer Res. 2009;15(24):7642–51. doi: 10.1158/1078-0432.CCR-09-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kennedy RD, Bylesjo M, Kerr P, Davison T, Black JM, Kay EW, et al. Development and independent validation of a prognostic assay for stage II colon cancer using formalin-fixed paraffin-embedded tissue. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29(35):4620–6. doi: 10.1200/JCO.2011.35.4498. [DOI] [PubMed] [Google Scholar]
- 13.Marisa L, de Reynies A, Duval A, Selves J, Gaub MP, Vescovo L, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS medicine. 2013;10(5):e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheffer M, Bacolod MD, Zuk O, Giardina SF, Pincas H, Barany F, et al. Association of survival and disease progression with chromosomal instability: a genomic exploration of colorectal cancer. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(17):7131–6. doi: 10.1073/pnas.0902232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.GKS . Limma: linear models for microarray data. In: Gentleman RCV, Huber W, et al., editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 17.Loboda A, Nebozhyn M, Klinghoffer R, Frazier J, Chastain M, Arthur W, et al. A gene expression signature of RAS pathway dependence predicts response to PI3K and RAS pathway inhibitors and expands the population of RAS pathway activated tumors. BMC medical genomics. 2010;3:26. doi: 10.1186/1755-8794-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dry JR, Pavey S, Pratilas CA, Harbron C, Runswick S, Hodgson D, et al. Transcriptional pathway signatures predict MEK addiction and response to selumetinib (AZD6244) Cancer research. 2010;70(6):2264–73. doi: 10.1158/0008-5472.CAN-09-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Connell MJ, Lavery I, Yothers G, Paik S, Clark-Langone KM, Lopatin M, et al. Relationship between tumor gene expression and recurrence in four independent studies of patients with stage II/III colon cancer treated with surgery alone or surgery plus adjuvant fluorouracil plus leucovorin. J Clin Oncol. 2010;28(25):3937–44. doi: 10.1200/JCO.2010.28.9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang Y, Casey G, Lavery IC, Zhang Y, Talantov D, Martin-McGreevy M, et al. Development of a clinically feasible molecular assay to predict recurrence of stage II colon cancer. J Mol Diagn. 2008;10(4):346–54. doi: 10.2353/jmoldx.2008.080011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oh SC, Park YY, Park ES, Lim JY, Kim SM, Kim SB, et al. Prognostic gene expression signature associated with two molecularly distinct subtypes of colorectal cancer. Gut. 2012;61(9):1291–8. doi: 10.1136/gutjnl-2011-300812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang S, Holzel M, Knijnenburg T, Schlicker A, Roepman P, McDermott U, et al. MED12 controls the response to multiple cancer drugs through regulation of TGF-beta receptor signaling. Cell. 2012;151(5):937–50. doi: 10.1016/j.cell.2012.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Popovici V, Budinska E, Tejpar S, Weinrich S, Estrella H, Hodgson G, et al. Identification of a poor-prognosis BRAF-mutant-like population of patients with colon cancer. J Clin Oncol. 2012;30(12):1288–95. doi: 10.1200/JCO.2011.39.5814. [DOI] [PubMed] [Google Scholar]
- 24.Tsuji T, Ibaragi S, Shima K, Hu MG, Katsurano M, Sasaki A, et al. Epithelial-mesenchymal transition induced by growth suppressor p12CDK2-AP1 promotes tumor cell local invasion but suppresses distant colony growth. Cancer research. 2008;68(24):10377–86. doi: 10.1158/0008-5472.CAN-08-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nieto MA. Epithelial plasticity: a common theme in embryonic and cancer cells. Science. 2013;342(6159):1234850. doi: 10.1126/science.1234850. [DOI] [PubMed] [Google Scholar]
- 26.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481(7381):306–13. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsuji T, Ibaragi S, Hu GF. Epithelial-mesenchymal transition and cell cooperativity in metastasis. Cancer research. 2009;69(18):7135–9. doi: 10.1158/0008-5472.CAN-09-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiang X, Deng Z, Zhuang X, Ju S, Mu J, Jiang H, et al. Grhl2 determines the epithelial phenotype of breast cancers and promotes tumor progression. PloS one. 2012;7(12):e50781. doi: 10.1371/journal.pone.0050781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yae T, Tsuchihashi K, Ishimoto T, Motohara T, Yoshikawa M, Yoshida GJ, et al. Alternative splicing of CD44 mRNA by ESRP1 enhances lung colonization of metastatic cancer cell. Nature communications. 2012;3:883. doi: 10.1038/ncomms1892. [DOI] [PubMed] [Google Scholar]
- 30.Chui MH. Insights into cancer metastasis from a clinicopathologic perspective: Epithelial-Mesenchymal Transition is not a necessary step. Int J Cancer. 2013;132(7):1487–95. doi: 10.1002/ijc.27745. [DOI] [PubMed] [Google Scholar]
- 31.Smith SC, Oxford G, Wu Z, Nitz MD, Conaway M, Frierson HF, et al. The metastasis-associated gene CD24 is regulated by Ral GTPase and is a mediator of cell proliferation and survival in human cancer. Cancer research. 2006;66(4):1917–22. doi: 10.1158/0008-5472.CAN-05-3855. [DOI] [PubMed] [Google Scholar]
- 32.Ashley N, Yeung TM, Bodmer WF. Stem cell differentiation and lumen formation in colorectal cancer cell lines and primary tumors. Cancer research. 2013;73(18):5798–809. doi: 10.1158/0008-5472.CAN-13-0454. [DOI] [PubMed] [Google Scholar]
- 33.Liu S, Cong Y, Wang D, Sun Y, Deng L, Liu Y, et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem cell reports. 2014;2(1):78–91. doi: 10.1016/j.stemcr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guinney J, Ferte C, Dry J, McEwen R, Manceau G, Kao KJ, et al. Modeling RAS phenotype in colorectal cancer uncovers novel molecular traits of RAS dependency and improves prediction of response to targeted agents in patients. Clin Cancer Res. 2014;20(1):265–72. doi: 10.1158/1078-0432.CCR-13-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karhemo PR, Ravela S, Laakso M, Ritamo I, Tatti O, Makinen S, et al. An optimized isolation of biotinylated cell surface proteins reveals novel players in cancer metastasis. Journal of proteomics. 2012;77:87–100. doi: 10.1016/j.jprot.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emori M, Tsukahara T, Murase M, Kano M, Murata K, Takahashi A, et al. High expression of CD109 antigen regulates the phenotype of cancer stem-like cells/cancer-initiating cells in the novel epithelioid sarcoma cell line ESX and is related to poor prognosis of soft tissue sarcoma. PloS one. 2013;8(12):e84187. doi: 10.1371/journal.pone.0084187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu G, Gao Y, Ni C, Chen Y, Pan J, Wang X, et al. Reduced expression of EphB2 is significantly associated with nodal metastasis in Chinese patients with gastric cancer. Journal of cancer research and clinical oncology. 2011;137(1):73–80. doi: 10.1007/s00432-010-0861-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao Q, Liu W, Cai J, Li M, Gao Y, Lin W, et al. EphB2 promotes cervical cancer progression by inducing epithelial-mesenchymal transition. Human pathology. 2014;45(2):372–81. doi: 10.1016/j.humpath.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 39.Knopfova L, Benes P, Pekarcikova L, Hermanova M, Masarik M, Pernicova Z, et al. c-Myb regulates matrix metalloproteinases 1/9, and cathepsin D: implications for matrix-dependent breast cancer cell invasion and metastasis. Molecular cancer. 2012;11:15. doi: 10.1186/1476-4598-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Favre C, Zhdanov A, Leahy M, Papkovsky D, O’Connor R. Mitochondrial pyrimidine nucleotide carrier (PNC1) regulates mitochondrial biogenesis and the invasive phenotype of cancer cells. Oncogene. 2010;29(27):3964–76. doi: 10.1038/onc.2010.146. [DOI] [PubMed] [Google Scholar]
- 41.Schell JC, Olson KA, Jiang L, Hawkins AJ, Van Vranken JG, Xie J, et al. A role for the mitochondrial pyruvate carrier as a repressor of the warburg effect and colon cancer cell growth. Molecular cell. 2014;56(3):400–13. doi: 10.1016/j.molcel.2014.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148(5):1015–28. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calon A, Lonardo E, Berenguer-Llergo A, Espinet E, Hernando-Momblona X, Iglesias M, et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nature genetics. 2015;47(4):320–9. doi: 10.1038/ng.3225. [DOI] [PubMed] [Google Scholar]
- 44.Isella C, Terrasi A, Bellomo SE, Petti C, Galatola G, Muratore A, et al. Stromal contribution to the colorectal cancer transcriptome. Nature genetics. 2015;47(4):312–9. doi: 10.1038/ng.3224. [DOI] [PubMed] [Google Scholar]
- 45.Roth AD, Tejpar S, Delorenzi M, Yan P, Fiocca R, Klingbiel D, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28(3):466–74. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.