ABSTRACT
Protein secretion systems that mediate interbacterial competition secret a wide repertoire of antibacterial toxins. A major player in these competitions is the newly discovered bacterial type VI secretion system (T6SS). We recently found that a subset of polymorphic MIX-effectors, which are a widespread class of effectors secreted by T6SSs, are horizontally shared between marine bacteria and are used to diversify their T6SS effector repertoires, thus enhancing their environmental fitness. In this commentary, I expand on the ideas that were introduced in the previous report, and further speculate on the possible mobility of other MIX-effectors. In addition, I discuss the possible role of horizontal gene transfer in the dissemination of MIX-effectors through bacterial genomes, as well as its possible role in diversifying the T6SS effector repertoire.
Keywords: environmental fitness, interbacterial competition, MIX-effectors, toxin, T6SS
Introduction
The type VI secretion system (T6SS) is a contact-dependent protein delivery apparatus that is found in many Gram-negative bacteria.1 The data that have accumulated over the past few years point to interbacterial competition as being a major role of this secretion system.2,3 The interbacterial toxicity is mediated by proteins called effectors which are delivered by the T6SS into the competing bacterium, where they exert their toxic effects.4 Notably, bacteria are immune against self-intoxication because they encode cognate immunity proteins that inactivate their T6SS effectors. These effector/immunity (E/I) pairs are encoded together as bicistronic units.3,5
Several families of T6SS effectors with antibacterial activities have been described, and can be divided into groups based on their activities.4 We previously identified a widespread class of polymorphic T6SS effectors that share an N-terminal domain we named MIX (Marker for type sIX effectors) and variable C-terminal toxin domains.6 MIX domains are grouped into 5 distinct clans based on their sequences and named MIX I-V. In a recent report, we characterized the 2 T6SSs found in the marine pathogen Vibrio alginolyticus 12G01.7 We determined that both T6SSs mediate bacterial-killing and are differentially regulated by environmental conditions. T6SS1 was active in low salt concentrations at 30°C, whereas T6SS2 was active in high salt concentrations at both 30°C and 37°C. In addition, we used comparative proteomics and genetics to identify the E/I repertoires of both T6SSs. We found that T6SS1 delivers at least 4 effectors with antibacterial toxic activities into competing bacteria, of which 3 belong to the MIX-effector class. Whereas one of the MIX-effectors was encoded as part of the T6SS1 gene cluster, the other 2, Va16152 and Va02265, were “orphan” effectors encoded in bicistronic E/I cassettes outside of the T6SS1 gene cluster.
Previously, we8 and others9 observed that T6SSs can not only mediate inter-species competition, but also intra-species competition, suggesting that the E/I repertoire differs between isolates of the same bacterial species. This was later elegantly demonstrated by Unterweger et al. who sequenced several V. cholerae isolates and found variable T6SS E/I repertoires.10 These observations prompted us to ask whether the E/I repertoire of the MIX-secreting T6SS1 in V. alginolyticus 12G01 is the same in other isolates. We found that the bicistronic cassettes encoding 3 of the 4 E/I pairs, including the MIX-effector encoded within the T6SS gene cluster and the “orphan” MIX-effector Va16152, were found in all available genomic sequences of V. alginolyticus isolates, suggesting that these 3 effectors are the “core repertoire” of the V. alginolyticus T6SS1. However, the “orphan” cassette encoding the MIX-effector Va02265 was not found in any of the other V. alginolyticus isolates, suggesting that this bicistronic cassette was a recent acquisition by the 12G01 isolate.
So where did this “orphan” E/I cassette come from? The answer to this question became apparent once we focused on the identity of the MIX-domain found in this effector. Va02265 has a MIX domain that belongs to the MIX V clan.6,7 When we examined the identities of all other members of the MIX V clan, we realized that all of them (except for one) were encoded as “orphan” effectors, that they were only found in the genomes of bacteria that occupy marine habitats, and that many of them neighbor transposable elements such as transposases or integrases. Moreover, homologous MIX V E/I cassettes are found in various bacterial species, but in a different synteny in each bacterium. We thus concluded that effectors that belong to the MIX V clan are mobile toxins that can be shared between marine bacteria via horizontal gene transfer and used to diversify their T6SS effector repertoires. We hypothesized that if that were the case, bacterial cells that acquire a new mobile MIX V E/I cassette can utilize the new effector and deliver it via the T6SS. These ‘evolved’ cells would have a competitive advantage as they can now intoxicate their parental kin cells using the new MIX V effector, because the parental kin cells did not acquire the cognate immunity gene and, therefore, cannot protect themselves in such a self-intoxication event. Indeed, we showed that a bacterium expressing a MIX-secreting T6SS, that acquired a new MIX V E/I cassette can use it to enhance its own fitness by gaining T6SS-mediated advantage over its parental kin during competition.
Possible mechanisms for horizontal gene transfer of MIX-effectors
How can these mobile MIX V E/I cassettes move to new bacterial genomes? One possibility is by being transferred into a new bacterium via a conjugative plasmid. This seems to be the case for the MIX V E/I cassette found in Aliivibrio salmonicida.7 This MIX V E/I cassette resides in a conjugative plasmid flanked by transposases, and indeed it appears to have been duplicated into the bacterial chromosome.11
Another possibility is a direct result of the bactericidal activity of the T6SS itself (Fig. 1). This possibility is based on a recent observation by Borgeaud et al., who found that the T6SS in V. cholerae is co-regulated with the DNA uptake machinery. They further demonstrated that after the prey cell dies and releases its DNA to the environment, the V. cholerae attacker can take up the released DNA and incorporate it into its own genome.12 Therefore, we propose that other marine bacteria can also take up DNA released by dead prey cells after they launched a T6SS-mediated attack. If the released DNA harbored a mobile MIX V E/I cassette, this cassette can incorporate itself into the attacker's chromosome, thereby diversifying its T6SS effector repertoire and enhancing its fitness. Acquisition of a new MIX V E/I pair with antibacterial toxic activities would thus provide competitive advantage to the cell against its neighboring parental kin who do not possess immunity against the newly acquired effector. If this is the case, the cell that acquired the new MIX V E/I pair can take over the population. Thus, it is plausible that acquisition of new MIX V E/I pairs drives bacterial evolution.
Figure 1.
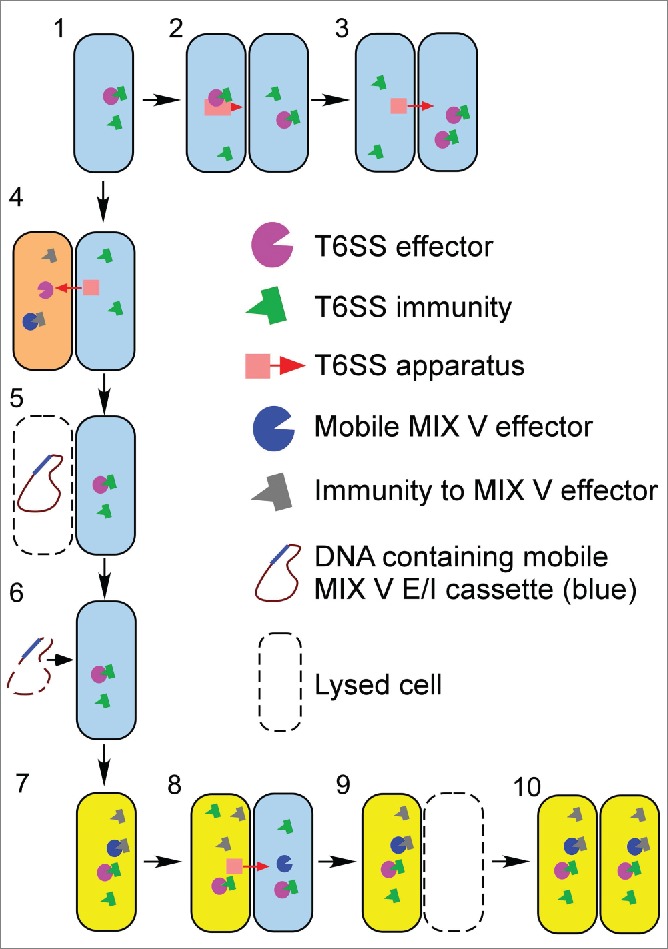
Mobile MIX V-effectors can drive population dynamics and evolution. 1) A parental bacterial cell (blue) harbors a T6SS and antibacterial E/I pairs. 2) When encountering a kin cell, it can activate its T6SS and deliver antibacterial effectors. 3) The recipient kin cell survives the attack as it encodes and expresses the immunity proteins against its own (and its kin's) effectors. 4) A foreign bacterium (orange) that encodes a mobile MIX V E/I pair comes in contact with the parental cell, which attacks it using its T6SS. Antibacterial effectors are delivered into the "foreign intruder" cell. 5) The "foreign intruder" cell dies and lyses as a result of the antibacterial activities of the delivered T6SS effectors, as it does not encode the cognate immunity proteins. 6) The lysed cell releases its DNA containing a mobile MIX V E/I cassette, which can be taken up by the attacker. 7) The mobile MIX V E/I cassette from the DNA of the lysed cell is incorporated into the attacker's genome, and this “evolved” cell (yellow) now expresses the new MIX V E/I pair. 8) The “evolved” cell attacks its neighboring parental kin using the T6SS and delivers the newly acquired antibacterial MIX V effector into the recipient parental kin. 9) The recipient parental kin cell dies as a result of this attack as it does not encode an immunity protein against the new MIX V effector delivered by its “evolved” neighbor. 10) The “evolved” cell containing the new MIX V E/I cassette, which now has competitive advantage over its parental kin, propagates and takes over the bacterial population.
MIX V-effectors only found in marine bacteria
It is puzzling why these mobile MIX V E/I cassettes are only found in the genomes of marine bacteria. One possibility is that marine bacteria that harbor these mobile cassettes only interact with each other, as they share the same aquatic habitat, thus non-marine bacteria are not often exposed to them and are not likely to acquire these cassettes. However, this seems unlikely as MIX V E/I cassettes are common in Vibrios, many of which are pathogens of terrestrial animals. Therefore, these Vibrios often encounter non-marine bacteria during animal infections and could have passed them their mobile MIX V E/I cassettes. Another, more likely possibility is that mobile MIX V E/I cassettes are being transferred into the genomes of non-marine bacteria, yet there is no evolutionary pressure that drives their persistence in the population as they do not confer any selective advantage, perhaps because T6SSs of non-marine bacteria lack a feature (that maybe is unique to T6SSs found in MIX-secreting marine bacteria) that is required to utilize these MIX V effectors in bacterial competition.
Mobility of other MIX-effector clans
Whereas genes encoding MIX V effectors are mostly "orphan" and neighbor transposable elements, they are not the only clan of MIX-effectors that present these features. Previously, we found that many members of the MIX III clan neighbor transposable elements.6 Moreover, examples of "orphan" MIX E/I cassettes can be found in all other MIX clans, such as the V. alginolyticus MIX IV E/I cassette encoding Va16152/Va16147.7 Hence, it is possible that members of other MIX-effector clans, which are "orphan" (and thus not genetically linked to a specific T6SS), can be transferred between bacterial genomes. As we showed previously that a given T6SS can deliver members of various MIX clans,6,7 acquisition of new MIX E/I cassettes that do not belong to the MIX V clan should provide enhanced fitness as well.
In addition, we previously observed another type of MIX-effector mobility, as we reported that MIX-effectors of clans II, III, and IV which are part of T6SS gene cluster can be duplicated into other regions of the genome as "orphan" E/I cassettes.6 Presumably, these duplication events can give rise to new toxins as they accumulate mutations and evolve. MIX effectors that are part of a T6SS gene cluster may also be transferred between bacterial genomes together with the T6SS itself, as many T6SS clusters are flanked by transposable elements.
Do transposable elements play a role in the polymorphism of the MIX-effectors toxin domains?
Polymorphic antibacterial toxins are found as secreted protein substrates of various protein secretion systems.13 Two recently characterized examples are the polymorphic toxins of the contact-dependent growth inhibition system (CDI)14 and members of the Rearrangement hotspot (Rhs) toxin class.15 CDI and Rhs toxins are modular proteins with polymorphic C-terminal toxin domains. MIX-effectors are also modular proteins containing an N-terminal MIX domain and polymorphic C-terminal toxin domains.6 We have noticed that homologous toxin domains are found in multiple classes of toxins. For example, a MIX V effector from V. proteolyticus contains a C-terminal CNF1 domain which is homologous to both the C-terminal CNF1 domain found in Rhs effectors and the C-terminus of VopC, an effector of the type III secretion system from V. parahaemolyticus.16 Strikingly, the same V. proteolyticus strain encodes a homologous MIX V effector which harbors a different C-terminal domain (Fig. 2). Thus, it appears that the C-terminal toxin domains are swappable between different classes of toxins, and it is therefore compelling to consider a role for toxin mobility in the evolution of these modular proteins.
Figure 2.
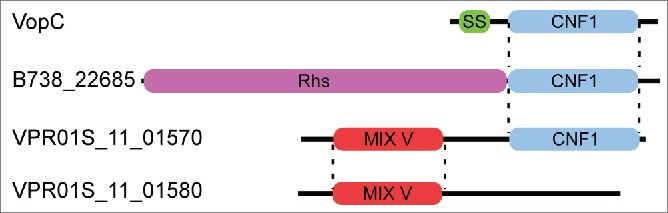
Modular MIX-effectors contain swappable toxin domains. The V. proteolyticus MIX-effector VPR01S_11_01570 (GI: 543441460) contains an N-terminal MIX V domain that is homologous to the MIX V domain of the V. proteolyticus MIX-effector VPR01S_11_01580 (GI: 543441461). The C-terminal CNF1 domain of the VPR01S_11_01570 is homologous to the CNF1 domain found in the Photorhabdus temperata Rhs-containing toxin B738_22685 (GI: 530707815) and the V. parahaemolyticus type III secretion system effector VopC (GI: 28901176). SS = type III secretion signal. Dashed lines mark borders of homologous regions.
Conclusion
In our recent study, we found a subset of MIX-effectors that belong to the MIX V clan, which are mobile and shared between marine bacteria. These mobile MIX-effectors diversify T6SS toxin repertoires and enhance fitness. Thus, this phenomenon could be a driving force for the evolution of these bacteria and the rise of pathogenic isolates, as the population dynamics would change once a cell acquired a new MIX E/I pair that provided it with a competitive advantage against its parental kin. It remains to be determined whether horizontal gene transfer is used as a spreading mechanism for members of other MIX-effector clans or other T6SS effector classes as well.
Abbreviations
- CDI
contact-dependent growth inhibition system.
- E/I
effector/immunity
- MIX
marker for type VI effectors
- T6SS
type VI secretion system
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
I would like to thank members of the Kim Orth lab for helpful discussions and suggestions.
Funding
DS is supported by the National Institute of Allergy And Infectious Diseases of the NIH under Award Number K99AI116948.
References
- [1]. Ho BT, Dong TG, Mekalanos JJ. A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 2014; 15:9–21; PMID:24332978; http://dx.doi.org/ 10.1016/j.chom.2013.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2]. Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, et al. . A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 2010; 7:25–37; PMID:20114026; http://dx.doi.org/ 10.1016/j.chom.2009.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3]. Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 2011; 475:343–7; PMID:21776080; http://dx.doi.org/ 10.1038/nature10244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4]. Russell AB, Peterson SB, Mougous JD. Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol 2014; 12:137–48; PMID:24384601; http://dx.doi.org/ 10.1038/nrmicro3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5]. Russell AB, LeRoux M, Hathazi K, Agnello DM, Ishikawa T, Wiggins PA, Wai SN, Mougous JD. Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature 2013; 496:508–12; PMID:23552891; http://dx.doi.org/ 10.1038/nature12074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6]. Salomon D, Kinch LN, Trudgian DC, Guo X, Klimko JA, Grishin NV, Mirzaei H, Orth K. Marker for type VI secretion system effectors. Proc Natl Acad Sci U S A 2014; 111:9271–6; PMID:24927539; http://dx.doi.org/ 10.1073/pnas.1406110111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7]. Salomon D, Klimko JA, Trudgian DC, Kinch LN, Grishin NV, Mirzaei H, Orth K. Type VI Secretion System Toxins Horizontally Shared between Marine Bacteria. PLoS Pathog 2015; 11:e1005128; PMID:26305100; http://dx.doi.org/ 10.1371/journal.ppat.1005128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Salomon D, Gonzalez H, Updegraff BL, Orth K. Vibrio parahaemolyticus type VI secretion system 1 is activated in marine conditions to target bacteria, and is differentially regulated from system 2. PLoS One 2013; 8:e61086; PMID:23613791; http://dx.doi.org/ 10.1371/journal.pone.0061086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9]. Unterweger D, Kitaoka M, Miyata ST, Bachmann V, Brooks TM, Moloney J, Sosa O, Silva D, Duran-Gonzalez J, Provenzano D, et al. . Constitutive type VI secretion system expression gives Vibrio cholerae intra- and interspecific competitive advantages. PLoS One 2012; 7:e48320; PMID:23110230; http://dx.doi.org/ 10.1371/journal.pone.0048320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10]. Unterweger D, Miyata ST, Bachmann V, Brooks TM, Mullins T, Kostiuk B, Provenzano D, Pukatzki S. The Vibrio cholerae type VI secretion system employs diverse effector modules for intraspecific competition. Nat Commun 2014; 5:3549; PMID:24686479; http://dx.doi.org/ 10.1038/ncomms4549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11]. Hjerde E, Lorentzen MS, Holden MT, Seeger K, Paulsen S, Bason N, Churcher C, Harris D, Norbertczak H, Quail MA, et al. . The genome sequence of the fish pathogen Aliivibrio salmonicida strain LFI1238 shows extensive evidence of gene decay. BMC Genomics 2008; 9:616; PMID:19099551; http://dx.doi.org/ 10.1186/1471-2164-9-616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12]. Borgeaud S, Metzger LC, Scrignari T, Blokesch M. The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science 2015; 347:63–7; PMID:25554784; http://dx.doi.org/ 10.1126/science.1260064 [DOI] [PubMed] [Google Scholar]
- [13].Jamet A, Nassif X. New players in the toxin field: polymorphic toxin systems in bacteria. MBio 2015; 6:e00285-15; PMID:25944858; http://dx.doi.org/ 10.1128/mBio.00285-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14]. Aoki SK, Diner EJ, de Roodenbeke CT, Burgess BR, Poole SJ, Braaten BA, Jones AM, Webb JS, Hayes CS, Cotter PA, et al. . A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature 2010; 468:439–42; PMID:21085179; http://dx.doi.org/ 10.1038/nature09490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Koskiniemi S, Lamoureux JG, Nikolakakis KC, t'Kint de Roodenbeke C, Kaplan MD, Low DA, Hayes CS. Rhs proteins from diverse bacteria mediate intercellular competition. Proc Natl Acad Sci U S A 2013; 110:7032–7; PMID:23572593; http://dx.doi.org/ 10.1073/pnas.1300627110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhang L, Krachler AM, Broberg CA, Li Y, Mirzaei H, Gilpin CJ, Orth K. Type III effector VopC mediates invasion for Vibrio species. Cell reports 2012; 1:453–60; PMID:22787576; http://dx.doi.org/ 10.1016/j.celrep.2012.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]


