Abstract
Transcription repression plays a central role in gene regulation. Transcription repressors utilize diverse strategies to mediate transcriptional repression. We have recently demonstrated that BEND3 (BANP, E5R and Nac1 domain) protein represses rDNA transcription by stabilizing a NoRC component. We discuss the role of BEND3 as a global regulator of gene expression and propose a model whereby BEND3 associates with chromatin remodeling complexes to modulate gene expression and heterochromatin organization.
Keywords: BEND3, NuRD, NoRC, rDNA, transcription repression, telomeres
Abbreviations
- BEND3
BANP, E5R and Nac1 domain
- NoRC
nucleolar remodeling complex
- NuRD
Nucleosome Remodeling Deacetylase
- rDNA
rDNA
- USP21
ubiquitin specific peptidase 21
- Tip5
TTF-1-interacting protein-5
- PRC2
Polycomb Repressive Complex 2
Introduction
Transcriptional repression is a multifaceted process, involving a huge repertoire of proteins with varied functionalities. Multiple modes can ensure gene repression; one such mode involves the creation of an epigenetically repressive chromatin environment. This is achieved by changing chromatin architecture through recruitment of nucleosome remodeling and histone modifying enzymes. Histone modifying enzymes, such as histone deacetylases (HDACs) and histone methyltransferase including H3 lysine 9 methyltransferase (Suv39h), H4 lysine 20 methyltransferase (Suv420h) and H3 lysine 27 methyltransferase (EZH2), give rise to a local heterochromatin-like state.1-3 Heterochromatin maintenance is critical for proper chromosome segregation, mitotic progression, genome stability and aging.4,5 In this article, we present a comprehensive analysis of the role of BEND3, a quadruple BEN-domain-containing protein in transcriptional repression and heterochromatin organization.
BEND3 Represses rDNA Transcription
We have previously shown that a BEN domain-containing protein, BEND3, is a transcriptional repressor.6 BEND3 localizes to heterochromatic structures and causes transcriptional repression when targeted to an artificial in vivo locus. Furthermore, overexpression of BEND3 causes hyper-heterochromatinization. In a recent proteomic screen employed to identify proteins associated with promoters and enhancers, BEND3 was found to associate exclusively with gene promoters, indicative of its role in transcriptional regulation.7 However, information regarding direct endogenous targets of BEND3 as well as its physiological role was lacking. We have recently demonstrated that BEND3 mediates rRNA gene repression.8 BEND3 localizes to nucleolus and associates with rDNA and directly binds to the rDNA promoter in a sequence-specific manner. Loss-of-function analyses showed that BEND3 efficiently represses rDNA transcription. Further, overexpression of BEND3 causes changes in histone modifications leading to a general repressive chromatin environment at the rDNA locus. In addition, we demonstrate that BEND3 interacts with the nucleolar-remodeling complex (NoRC). NoRC is a well-characterized complex and is known to establish as well as maintain gene silencing at rDNA loci.9-11 In an attempt to study the biochemical role of BEND3 as part of NoRC and its role in rDNA silencing, we found that the SUMOylation status of BEND3 is critical for NoRC stability. We show that Tip5, a bona fide member of the NoRC, undergoes ubiquitin-mediated degradation, and that SUMOylated BEND3 stabilizes Tip5 by preventing its ubiquitination. Finally we show that BEND3 stabilizes Tip5 via its interaction with a deubiquitinase, ubiquitin specific peptidase 21 (USP21).12 Based on these results, we propose that BEND3-mediated rDNA silencing occurs by stabilization of the NoRC. This leads to epigenetic changes at the rDNA locus thereby causing transcriptional repression.
Role of BEND3 in Global Gene Repression
BEND3-NuRD connection
In our previous report, we had shown that BEND3 interacts with Sall4 and HDAC1, members of the NuRD complex.6 NuRD complex contains chromatin remodeling and histone deacetylase activity and is known to mediate transcriptional repression.13,14 The NuRD complex was recently shown to establish a specific chromatin environment at the rRNA genes that are transcriptionally inactive but are poised for activation.15 In addition to the rRNA genes, the NuRD complex has been implicated in regulating various physiological processes including maintenance of pluripotency,16-18 reprogramming of neural stem cell into iPSC,19 S phase progression and pericentric heterochromatin formation.20 In a recent report, mass spectrometry analyses of BEND3-associated proteins revealed that BEND3 associates strongly with most of the NuRD complex members.21 In light of these findings, it is highly probable that BEND3, in association with NuRD, plays an important role in global gene regulation. Since NuRD plays such critical roles in ESCs, it would be very interesting to map the genome wide occupancy of NuRD and BEND3 in ESCs. Comparative analyses of genome wide occupancies would give us insights into whether BEND3 colocalizes with NuRD at most loci and/or if BEND3 binds to a set of targets distinct from that of the NuRD complex. In either case, it would provide sufficient impetus to probe deeper into a BEND3-NuRD connection in gene regulation.
BEND3-NoRC connection
In our recent work, we have shown that BEND3 is a novel interactor of the NoRC.8 The role of NoRC in rDNA silencing is well documented.9,10,22 Overexpression of BEND3 phenocopies most of the Tip5 gain-of-function phenotypes observed with regard to epigenetic changes at the rDNA locus. In addition to silencing rRNA genes, NoRC has been shown to regulate heterochromatin formation at centromeres and telomeres, in part through its interaction with Suv4–20h2, an enzyme responsible for H4K20 trimethylation.11,23 Similar to Tip5, we have shown that YFP-BEND3 localizes to telomeric ends of mitotic chromosomes (Fig. 1A).8 Note the bright BEND3 foci (green) represent the BEND3 localizing to the rDNA (Fig. 1A) A recent study identified BEND3 to be associated with telomeres and showed that BEND3 colocalizes with telomeric repeat binding factor 2 (TRF2), a marker for telomeres. Furthermore, we show here that BEND3 can interact strongly with Suv4–20h2 (Fig. 1B). Immunoprecipitation (IP) carried out on lysates from cells expressing YFP-Suv4–20h2 and HA-BEND3 shows that BEND3 associates strongly with Suv4–20h2 (Fig 1Ba). Reciprocal IP using GFP antibody confirmed the interaction (Fig 1Bb). Taken together, these findings indicate that BEND3 and NoRC play a concerted role not only in rRNA gene repression but also in global heterochromatin organization.
Figure 1.
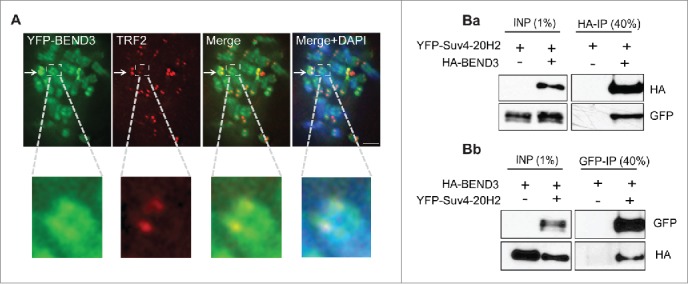
(A) Immunostaining of telomere marker TRF2 (red) on mitotic chromosomes of U2OS cells expressing YFP-BEND3 (green). Magnified inset with higher exposure in all channels shows YFP-BEND3 colocalizing with TRF2. Arrow indicates a representative YFP-BEND3 focus localized to rDNA locus. Scale bar represents 5 μm. (B) BEND3 associates with Suv4–20h2. (Ba) Immunoprecipitation was performed using HA antibody in lysates containing YFP-Suv4–20h2 with or without HA-BEND3. (Bb) Reciprocal immunoprecipitation performed using GFP antibody in lysates containing HA-BEND3 with or without YFP-Suv4–20h2.
BEND3 in a megacomplex
An elegant study from the Dejardin laboratory showed that in the absence of Suv39h or DNMTs, the pericentromeric heterochromatin undergoes a switch from constitutive to facultative heterochromatin, as a safeguard mechanism to maintain genomic integrity.21 Facultative heterochromatin is marked with H3K27 trimethylation that is catalyzed by EZH2 methyltransferase of the PRC2 complex. The authors demonstrate that in the absence of DNA methylation or H3K9me3 marks, BEND3 enables the recruitment of EZH2, and hence H3K27me3, at the pericentromeric heterochromatin. Their data strongly suggest that BEND3 is critical for this switch from constitutive to facultative heterochromatin. In Suv39h KO or DNMT KO mESCs, there is a dramatic enrichment of BEND3 at the pericentromeric heterochromatin. In addition to BEND3, members of the PRC2, NuRD and NoRC complexes are also highly enriched. It is interesting to note that the BEND3 overexpression-induced heterochromatinized regions within the chromatin show enrichment of H3K27me3 marks. These data strongly argue that BEND3 shares a functional interaction with the PRC2 complex. Given the role of PRC2 in differentiation and development,24,25 it is tempting to propose that BEND3 plays a concerted role with PRC2 in differentiation and development. These are indeed very fascinating findings, especially in light of our work, which implicates BEND3 as a part of both NuRD and NoRC complexes. Perhaps, association of BEND3 with NoRC, NuRD or PRC2 complex is highly context-specific or these complexes are not mutually exclusive; they exhibit a high degree of crosstalk and therefore function as a megacomplex at heterochromatic regions within the genome.
Role of BEND3 SUMOylation in Gene Regulation
Our laboratory has previously shown that BEND3 undergoes SUMOylation at K20 and K512 and that SUMOylation is important for its repressive ability in a reporter based assay.6 However, a detailed understanding of the molecular and biochemical role of SUMOylation in BEND3-mediated repression was lacking. In our recent work, we show that SUMOylation of BEND3 is important for the stability of NoRC.8 We find that Tip5 undergoes ubiquitin-mediated degradation in the presence of a BEND3 sumo double mutant (BEND3-SDM) but not in the presence of the wild type protein (BEND3-WT). Tip5 is deubiquitinated in the presence of BEND3-WT, thereby stabilizing and preventing its degradation. We show that the deubiquitinase USP21, interacts with BEND3 and Tip5, and can efficiently deubiquitinate exogenous Tip5. However, the stability of USP21 also depends on SUMOylation status of BEND3. In the presence of BEND3-SDM, USP21 undergoes dramatic degradation. Therefore, we propose that BEND3 influences the stability of Tip5 indirectly by stabilizing USP21, which then deubiquitinates Tip5.
Some key questions arise from these findings: How does SUMOylation of BEND3 prevent ubiquitination of its interactors? How does SUMOylated BEND3 stabilize USP21? We hypothesize that SUMOylation of BEND3 prevents ubiquitination of its interactors by limiting E3 ligase access to its substrates. The SUMO moiety could possibly mask the ubiquitination sites on the BEND3-interacting proteins or cause steric hindrance, thereby preventing E3 ligase to transfer ubiquitin. Alternatively, it is possible that BEND3-SDM interacts with an E3 ligase whereas BEND3-WT does not. Although there are no reports suggesting potential interaction of BEND3 with any known E3 ligase, it would be critical to perform quantitative mass spectrometry analysis of proteins associated with BEND3-WT and BEND3-SDM. This would provide key insights into the role of SUMOylation in BEND3-mediated protein stabilization. The use of Chromatin Immunoprecipitation—quantitative Mass Spectrometry (ChIP-qMS) 7,26 to observe quantitative changes with regard to global protein stability on chromatinand correlate them with gene expression analysis would enable us to monitor changes in chromatin landscape in the presence of BEND3.WT or BEND3-SDM.
Conclusion
BEN domain-containing proteins are emerging rapidly as an important class of proteins involved in gene regulation.27 BEN domain has been shown to posses sequence specific DNA binding activity28,29 and, indeed, we have also shown that BEND3 can directly bind to DNA as well.8 Apart from BEND3, the only other BEN domain-containing protein in mammals studied so far is BEND6 which is a BEN-solo domain protein involved in Notch signaling.29 We have shown that BEND3, a quadruple BEN domain-containing protein, plays an important role in rDNA silencing. BEND3 associates with members of NuRD, NoRC and PRC2 complexes. BEND3 regulates gene transcription as well as heterochromatin organization in a chromatin context-dependent manner (Fig. 2). Dissecting the genome-wide association of BEND3 would yield important insights into its role in gene regulation. Others and we provided ample evidence that BEND3 plays diverse roles in gene regulation and chromatin organization and therefore prompt a deeper study for a comprehensive understanding of its physiological functions.
Figure 2.
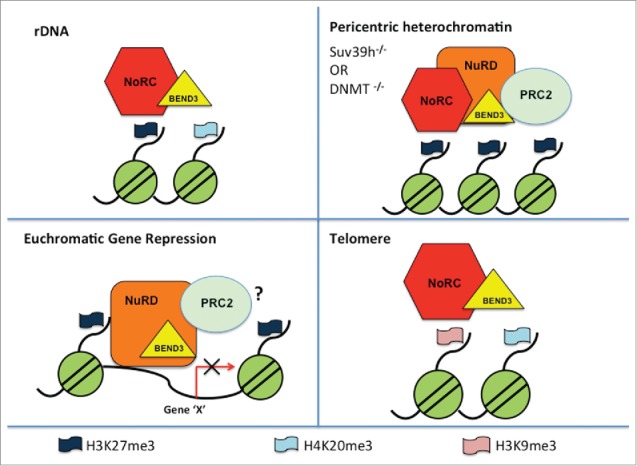
A model for BEND3 mediated gene repression and heterochromatin organization based on current understanding.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank members of the Prasanth laboratory for discussions and suggestions. We thank Drs. M. Dundr, D. Spector and B. Stillman for providing reagents and suggestions. We thank Dr. K. Prasanth for critical reading of the manuscript.
Funding
This work was supported by NSF career (1243372) and NIH (1RO1GM099669) awards to SGP. The authors declare no competing financial interests.
References
- 1.Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 1996; 272:408-11; PMID:8602529; http://dx.doi.org/ 10.1126/science.272.5260.408 [DOI] [PubMed] [Google Scholar]
- 2.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell 2007; 128:707-19; PMID:17320508; http://dx.doi.org/ 10.1016/j.cell.2007.01.015 [DOI] [PubMed] [Google Scholar]
- 3.Fodor BD, Shukeir N, Reuter G, Jenuwein T. Mammalian Su(var) genes in chromatin control. Annu Rev Cell Dev Biol 2010; 26:471-501; PMID:19575672; http://dx.doi.org/ 10.1146/annurev.cellbio.042308.113225 [DOI] [PubMed] [Google Scholar]
- 4.Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al.. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 2001; 107:323-37; PMID:11701123; http://dx.doi.org/ 10.1016/S0092-8674(01)00542-6 [DOI] [PubMed] [Google Scholar]
- 5.Larson K, Yan SJ, Tsurumi A, Liu J, Zhou J, Gaur K, Guo D, Eickbush TH, Li WX. Heterochromatin formation promotes longevity and represses ribosomal RNA synthesis. PLoS Genet 2012; 8:e1002473; PMID:22291607; http://dx.doi.org/ 10.1371/journal.pgen.1002473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sathyan KM, Shen Z, Tripathi V, Prasanth KV, Prasanth SG. A BEN-domain-containing protein associates with heterochromatin and represses transcription. J Cell Sci 2011; 124:3149-63; PMID:21914818; http://dx.doi.org/ 10.1242/jcs.086603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ji X, Dadon DB, Abraham BJ, Lee TI, Jaenisch R, Bradner JE, Young RA. Chromatin proteomic profiling reveals novel proteins associated with histone-marked genomic regions. Proc Natl Acad Sci U S A 2015; 112:3841-6; PMID:25755260; http://dx.doi.org/ 10.1073/pnas.1422916112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khan A, Giri S, Wang Y, Chakraborty A, Ghosh AK, Anantharaman A, Aggarwal V, Sathyan KM, Ha T, Prasanth KV, et al.. BEND3 represses rDNA transcription by stabilizing a NoRC component via USP21 deubiquitinase. Proc Natl Acad Sci U S A 2015; 112:8338-43; PMID:26100909; http://dx.doi.org/ 10.1073/pnas.1424705112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strohner R, Nemeth A, Jansa P, Hofmann-Rohrer U, Santoro R, Langst G, Grummt I. NoRC–a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J 2001; 20:4892-900; PMID:11532953; http://dx.doi.org/ 10.1093/emboj/20.17.4892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santoro R, Li J, Grummt I. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat Genet 2002; 32:393-6; PMID:12368916; http://dx.doi.org/ 10.1038/ng1010 [DOI] [PubMed] [Google Scholar]
- 11.Guetg C, Lienemann P, Sirri V, Grummt I, Hernandez-Verdun D, Hottiger MO, Fussenegger M, Santoro R. The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J 2010; 29:2135-46; PMID:20168299; http://dx.doi.org/ 10.1038/emboj.2010.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Chen C, Hou X, Gao Y, Lin F, Yang J, Gao Z, Pan L, Tao L, Wen C, et al.. Identification of the E3 deubiquitinase ubiquitin-specific peptidase 21 (USP21) as a positive regulator of the transcription factor GATA3. J Biol Chem 2013; 288:9373-82; PMID:23395819; http://dx.doi.org/ 10.1074/jbc.M112.374744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene 2007; 26:5433-8; PMID:17694084; http://dx.doi.org/ 10.1038/sj.onc.1210611 [DOI] [PubMed] [Google Scholar]
- 14.Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell 1998; 2:851-61; PMID:9885572; http://dx.doi.org/ 10.1016/S1097-2765(00)80299-3 [DOI] [PubMed] [Google Scholar]
- 15.Xie W, Ling T, Zhou Y, Feng W, Zhu Q, Stunnenberg HG, Grummt I, Tao W. The chromatin remodeling complex NuRD establishes the poised state of rRNA genes characterized by bivalent histone modifications and altered nucleosome positions. Proc Natl Acad Sci U S A 2012; 109:8161-6; PMID:22570494; http://dx.doi.org/ 10.1073/pnas.1201262109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaji K, Caballero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol 2006; 8:285-92; PMID:16462733; http://dx.doi.org/ 10.1038/ncb1372 [DOI] [PubMed] [Google Scholar]
- 17.Zhu D, Fang J, Li Y, Zhang J. Mbd3, a component of NuRD/Mi-2 complex, helps maintain pluripotency of mouse embryonic stem cells by repressing trophectoderm differentiation. PLoS One 2009; 4:e7684; PMID:19888462; http://dx.doi.org/ 10.1371/journal.pone.0007684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van den Berg DL, Snoek T, Mullin NP, Yates A, Bezstarosti K, Demmers J, Chambers I, Poot RA. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell 2010; 6:369-81; PMID:20362541; http://dx.doi.org/ 10.1016/j.stem.2010.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.dos Santos RL, Tosti L, Radzisheuskaya A, Caballero IM, Kaji K, Hendrich B, Silva JC. MBD3/NuRD facilitates induction of pluripotency in a context-dependent manner. Cell Stem Cell 2014; 15:102-10; PMID:24835571; http://dx.doi.org/ 10.1016/j.stem.2014.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sims JK, Wade PA. Mi-2/NuRD complex function is required for normal S phase progression and assembly of pericentric heterochromatin. Mol Biol Cell 2011; 22:3094-102; PMID:21737684; http://dx.doi.org/ 10.1091/mbc.E11-03-0258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saksouk N, Barth TK, Ziegler-Birling C, Olova N, Nowak A, Rey E, Mateos-Langerak J, Urbach S, Reik W, Torres-Padilla ME, et al.. Redundant mechanisms to form silent chromatin at pericentromeric regions rely on BEND3 and DNA methylation. Mol Cell 2014; 56:580-94; PMID:25457167; http://dx.doi.org/ 10.1016/j.molcel.2014.10.001 [DOI] [PubMed] [Google Scholar]
- 22.Zhou Y, Santoro R, Grummt I. The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J 2002; 21:4632-40; PMID:12198165; http://dx.doi.org/ 10.1093/emboj/cdf460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Postepska-Igielska A, Krunic D, Schmitt N, Greulich-Bode KM, Boukamp P, Grummt I. The chromatin remodelling complex NoRC safeguards genome stability by heterochromatin formation at telomeres and centromeres. EMBO Rep 2013; 14:704-10; PMID:23797874; http://dx.doi.org/ 10.1038/embor.2013.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature 2011; 469:343-9; PMID:21248841; http://dx.doi.org/ 10.1038/nature09784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aldiri I, Vetter ML. PRC2 during vertebrate organogenesis: a complex in transition. Dev Biol 2012; 367:91-9; PMID:22565092; http://dx.doi.org/ 10.1016/j.ydbio.2012.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engelen E, Brandsma JH, Moen MJ, Signorile L, Dekkers DH, Demmers J, Kockx CE, Ozgür Z, van IJcken WF, van den Berg DL, et al.. Proteins that bind regulatory regions identified by histone modification chromatin immunoprecipitations and mass spectrometry. Nat Commun 2015; 6:7155; PMID:25990348; http://dx.doi.org/ 10.1038/ncomms8155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abhiman S, Iyer LM, Aravind L. BEN: a novel domain in chromatin factors and DNA viral proteins. Bioinformatics 2008; 24:458-61; PMID:18203771; http://dx.doi.org/ 10.1093/bioinformatics/btn007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dai Q, Ren A, Westholm JO, Serganov AA, Patel DJ, Lai EC. The BEN domain is a novel sequence-specific DNA-binding domain conserved in neural transcriptional repressors. Genes Dev 2013; 27:602-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai Q, Andreu-Agullo C, Insolera R, Wong LC, Shi SH, Lai EC. BEND6 is a nuclear antagonist of Notch signaling during self-renewal of neural stem cells. Development 2013; 140:1892-902; PMID:23468431; http://dx.doi.org/ 10.1242/dev.087502 [DOI] [PMC free article] [PubMed] [Google Scholar]


