Abstract
Neutral lipids are packed into dedicated intracellular compartments termed lipid droplets (LDs). LDs are spherical structures delineated by an unusual lipid monolayer and they harbor a specific set of proteins, many of which function in lipid synthesis and lipid turnover. In mammals, LDs are covered by abundant scaffolding proteins, the perilipins (PLIN1–5). LDs in yeast are functionally similar to that of mammalian cells, but they lack the perilipins. We have previously shown that perilipins (PLIN1–3) are properly targeted to LDs when expressed in yeast and that they promote LD formation from the ER membrane enriched in neutral lipids. Here we address the question whether PLIN5 (OXPAT) has a similar function. Both human and murine PLIN5 were properly targeted to yeast LDs, but the protein localized to the cytosol and its steady-state level was reduced when expressed in yeast mutants lacking the capacity to synthesize storage lipids. When expressed in cells containing high levels of neutral lipids within the membrane of the endoplasmatic reticulum, PLIN5 promoted the formation of LDs. Interestingly, PLIN5 was properly targeted to LDs, irrespective of whether these LDs were filled with triacylglycerol or steryl esters, indicating that PLIN5 did not exhibit targeting specificity for a particular subtypes of LDs as was reported for mammalian cells.
Keywords: endoplasmatic reticulum, lipid droplets, organelle biogenesis, perilipins, PLIN5 (OXPAT), steryl esters, Saccharomyces cerevisiae, triacylglycerol
Present in many prokaryotic and most eukaryotic cells, lipid droplets (LDs) serve to store metabolic energy as well as precursors for membrane proliferation in the form of neutral lipids, particularly triacylglycerol (TAG) and steryl esters (STE).1 LDs are metabolically, functionally and possibly also structurally connected to the membrane of the ER with which they exchange lipids and proteins.2,3 In plant seeds and animal cells LDs are covered by abundant scaffolding proteins, the oleosins and perilipins (PLINs), respectively. The yeast genome, however, lacks recognizable homologues of these scaffolding factors; nevertheless, yeast cells make LDs that are functionally equivalent to those of plants and animal cells. Perilipin-1 (PLIN1), Adipophilin (PLIN2), and Tip47 (PLIN3), are the founding member of the PAT/PERILIPIN superfamily, which was then expanded by the addition of PLIN4 (S3–12) and PLIN5 (OXPAT/LSDP5).4-7 Perilipins play important functions in regulating lipolysis and they display tissue specific expression patterns.5,8 PLIN4 (S3–12) is induced during adipocyte differentiation and coats nascent LDs.6,9,10 PLIN5 (OXPAT/LSDP5) is expressed in highly oxidative tissues such as liver or cardiomyocytes and the expression of both PLIN4 (S3–12) and PLIN5 (OXPAT/LSDP5) in adipose tissue is regulated by PPARγ (peroxisome proliferator-activated receptor).7,11-13
We have previously shown that PLIN1–3 are properly targeted to LDs when expressed in yeast cells and that these proteins became cytosolic and lost their membrane association when expressed in cells lacking LDs.14 In addition, expression of these LD scaffolding proteins is sufficient to induce LD formation in cells having high levels of neutral lipids in the ER membrane, such as mutants lacking the phosphatidate phosphatase Pah1/Lipin.14 Here we extend these studies to one additional member of the perilipin family, PLIN5 (OXPAT).
cDNAs encoding both murine and human PLIN5 (OXPAT) were placed under transcriptional control of a constitutively active alcohol dehydrogenase 1 (ADH1) promoter and N-terminally fused in frame with GFP and expressed from a centromeric plasmid. When expressed in wild-type cells, PLIN5 stained circular intracellular structures that were co-stained with the lipophilic fluorescent dye Nile Red, indicating LD localization of PLIN5 (Fig. 1A). LDs formed in the presence of PLIN5, however, frequently appear to be slightly bigger (average diameter 0.85 µm) and clustered compared to those formed in wild-type cells expressing the LD marker Erg6-GFP (average diameter 0.6 µm) (Fig. 1B) When expressed in cells lacking the capacity to synthesize neutral lipids due to the deletion of 3 of the genes required for neutral lipid synthesis (are1Δ are2Δ dga1Δ) and placement of the fourth gene under transcriptional control of the glucose repressible GAL1 promoter (GAL-LRO1), PLIN5 showed cytosolic staining and Nile Red stained endomembranes. Western blot analysis of PLIN5 indicated higher steady-state levels of the protein when expressed in cells containing LDs, suggesting that the proteins is unstable when expressed in cells lacking LDs (Fig. 1C). Analysis of protein turnover after treatment of cells with cycloheximide to block translation, however, indicates that PLIN5 turnover is not dramatically accelerated in cell lacking LDs, but that its steady-state levels are decreased (Fig. 1D). PLIN5 thus behaves similar to ADRP/PLIN2 and TIP47/PLIN3 in that the protein is stable even in the absence of LDs. This is in contrast to Oleosin and PLIN1, which are both rapidly degraded in cells lacking LDs. 14
Figure 1.
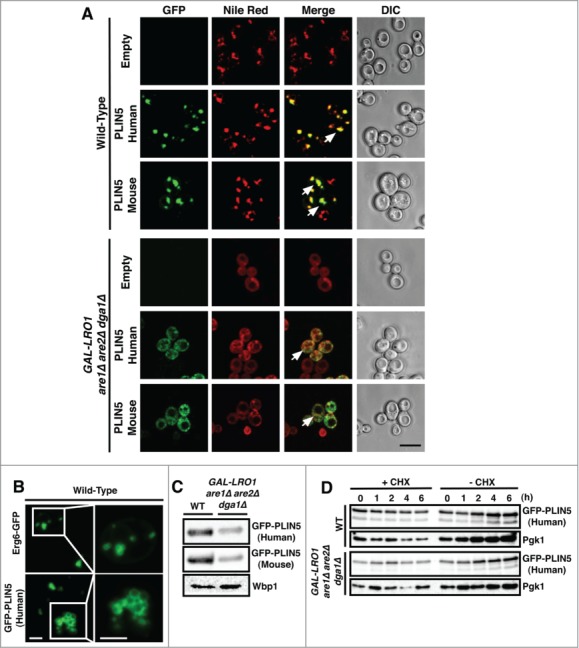
PLIN5 localizes to LDs when expressed in yeast cells. (A) Wild-type cells and cells lacking LDs (GAL-LRO1 are1Δ are2Δ dga1Δ) expressing either human or murine GFP-tagged PLIN5 were cultivated overnight in synthetic media (SC) media at 24°C. Cells were collected and stained with Nile Red and living cells were analyzed by confocal microscopy. Arrows in the merge indicate colocalization. Bar, 5 μm. (B) LD clustering in cells expressing PLIN5. LD morphology was analyzed by confocal microscopy in wild-type cells expressing either Erg6-GFP or GFP-PLIN5. The LDs in the boxed region are enlarged 2.5-fold in the panel to the right. Bar, 7 µm and 3 µm, respectively. (C) Total cell proteins from wild-type or cells lacking LDs were TCA precipitated and expression of GFP-tagged PLIN5 was analyzed by western blotting. Wbp1, a subunit of the oligosaccharyl transferase complex served as a loading control. (D) Cells were treated with cycloheximide (CHX, 50 µg/ml) and samples were removed at the indicated time points. Proteins were TCA precipitated, resolved by SDS-PAGE and probed with antibodies against GFP and against Pgk1, phosphoglycerate kinase, which served as a loading control.
To examine whether PLIN5 could also induce formation of LDs, as we previously observed for both oleosin and PLINs,14 we expressed PLIN5 in mutants lacking the phosphatidate phosphatase Pah1, the yeast homolog of human lipin.15,16 Pah1p catalyzes the dephosphorylation of phosphatidic acid to diacyl-glycerol (DAG), which is then further converted to triacylglycerol. Cells lacking Pah1 display a defect in LD formation and show elevated levels of neutral lipids in the ER membrane.17 When stained with Nile Red, pah1Δ mutant cells did not show any detectable LDs and the dye instead stained the endomembranes, particularly the perinuclear ER (Fig. 2A). However, when these cells expressed PLIN5, punctuate structures that stained with both Nile Red and GFP-tagged PLIN5 became apparent, indicating that PLIN5 was able to induce formation of LDs in a pah1Δ mutant background. The LDs formed under these conditions were not only enriched in neutral lipids as indicated by the Nile Red staining, but they also contained a bona fide LD marker protein, Erg6, as indicated by the colocalization of Erg6-RFP with GFP-tagged human PLIN5 (Fig. 2B).
Figure 2.
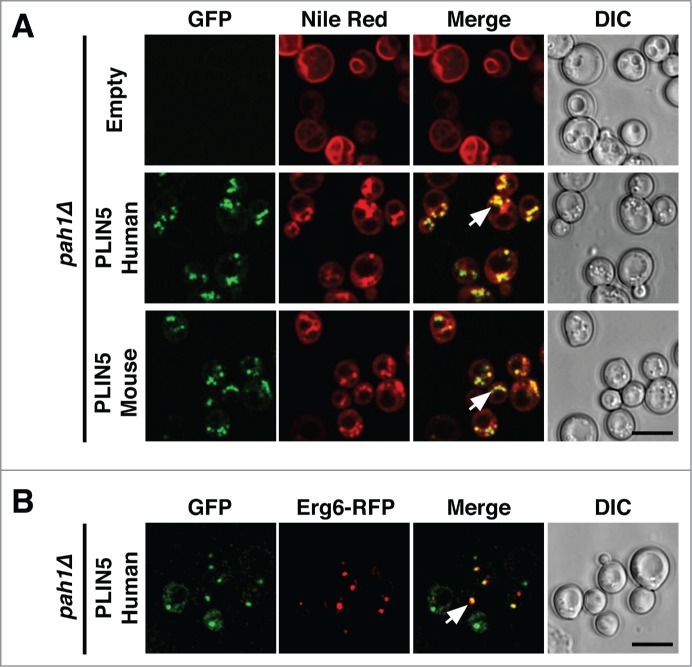
PLIN5 expression promotes formation of LDs in a sensitized strain. (A) PLIN5 expression results in induction of LDs in cells lacking Pah1. Cells of the indicated genotype were cultivated, stained with Nile Red and analyzed by confocal microscopy, as described in Figure 1. (B) PLIN5 colocalizes with Erg6. Pah1 mutant cells expressing human GFP-PLIN5 and Erg6-RFP were cultivated as described above and colocalization was assessed by imaging of living cells. Arrows in the merge indicate colocalization. Bar, 5 μm.
In mammalian adrenocortical cells some perilipins are preferentially stabilized in the presence of exogenous lipids, either oleic acid or cholesterol, indicating that they specifically target to LDs enriched with either STE or TAG.18 PLIN5, for example, preferentially targets to TAG containing LDs at the cell periphery.18 To examine whether PLIN5 would exhibit target-specificity also when expressed in yeast, we analyzed its localization in cells lacking the 2 ACAT (acyl-CoA cholesterol acyltransferase) enzymes Are1 and Are2, as well as in cells lacking the TAG biosynthetic enzymes Dga1 and Lro1.19 Both human and mouse PLIN5 co-localized with Nile Red positive punctuate structures in both double mutants strains, indicating that LD-targeting of PLIN5 did not exhibit a strong preference for LDs subtypes enriched with either STE or TAG (Fig. 3). In addition, the Nile Red stained punctuate structures to which PLIN5 localized in cells synthesizing either STE or TAG also contained the LD marker Erg6, indicating the formation of functional LDs (Fig. 4). Taken together, the results presented here indicate that PLIN5 is targeted to LDs when expressed in yeast, that the protein is expressed at increased steady-state levels in the presence of LDs and that it promotes LD formation from the ER membrane of a sensitized strain which accumulates high levels of neutral lipids within its ER bilayer.
Figure 3.
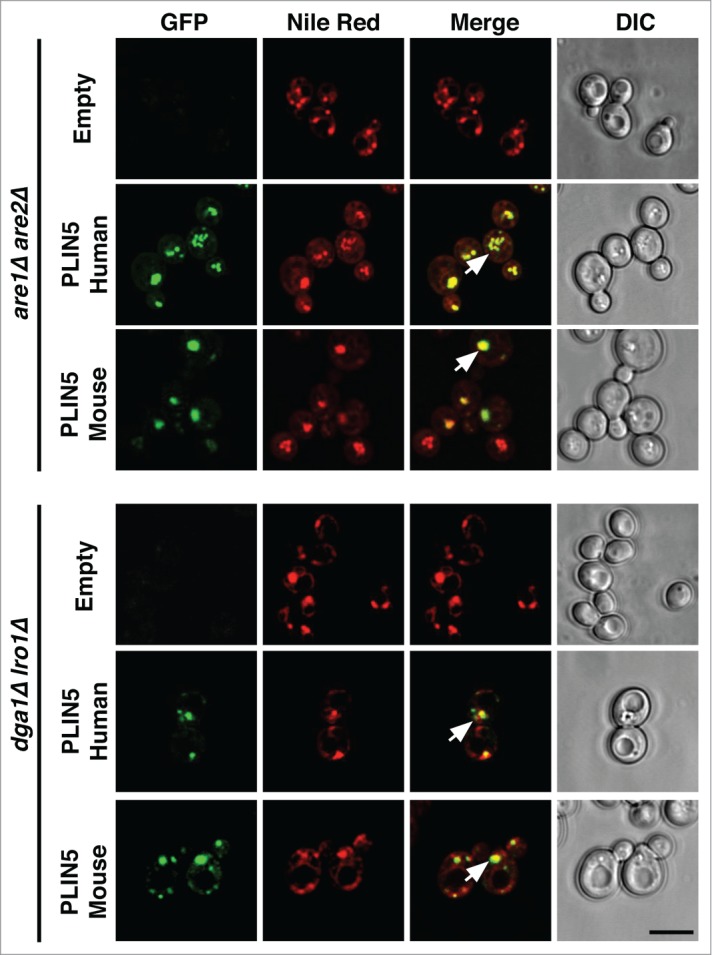
PLIN5 localizes to LDs irrespective of whether they are filled with either TAG or STE. GFP-PLIN5 was expressed in are1Δ are2Δ or in dga1Δ lro1Δ double mutant strains, cells were stained with Nile Red and analyzed by confocal microscopy as described for Figure 1. Arrows in the merge indicate colocalization. Bar, 5 μm.
Figure 4.
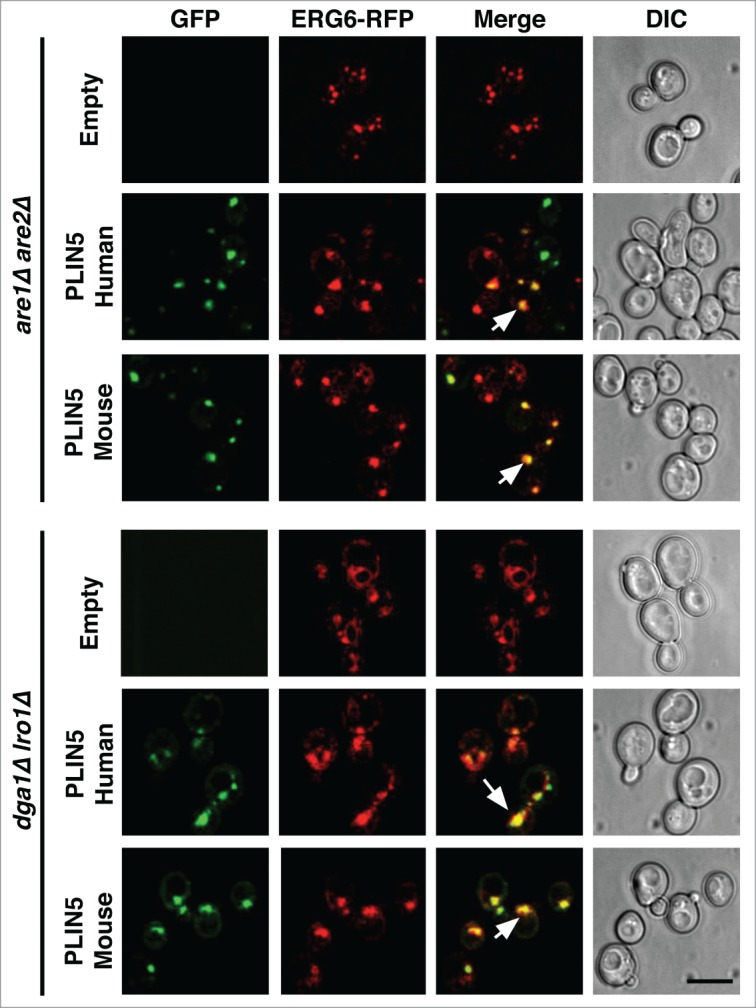
PLIN5 co-localizes with the LD marker protein Erg6 on LDs filled with either TAG or STE. GFP-PLIN5 and Erg6-RFP were co-expressed in are1Δ are2Δ or in dga1Δ lro1Δ double mutant strains and cells were analyzed by confocal microscopy. Arrows in the merge indicate colocalization. Bar, 5 μm.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the Canton of Fribourg and the Swiss National Science Foundation (31003A_134742).
References
- 1.Walther TC, Farese RVJ. Lipid droplets and cellular lipid metabolism. Annu Rev Biochem 2012; 81:687-714; PMID:22524315; http://dx.doi.org/ 10.1146/annurev-biochem-061009-102430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jacquier N, Choudhary V, Mari M, Toulmay A, Reggiori F, Schneiter R. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Sci 2011; 124:2424-37; PMID:21693588; http://dx.doi.org/ 10.1242/jcs.076836 [DOI] [PubMed] [Google Scholar]
- 3.Wilfling F, Wang H, Haas JT, Krahmer N, Gould TJ, Uchida A, Cheng JX, Graham M, Christiano R, Fröhlich F, et al.. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev Cell 2013; 24:384-99; PMID:23415954; http://dx.doi.org/ 10.1016/j.devcel.2013.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kimmel AR, Brasaemle DL, McAndrews-Hill M, Sztalryd C, Londos C. Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-f amily of intracellular lipid storage droplet proteins. J Lipid Res 2010; 51:468-71; PMID:19638644; http://dx.doi.org/ 10.1194/jlr.R000034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brasaemle DL. Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J Lipid Res 2007; 48:2547-59; PMID:17878492; http://dx.doi.org/ 10.1194/jlr.R700014-JLR200 [DOI] [PubMed] [Google Scholar]
- 6.Wolins NE, Skinner JR, Schoenfish MJ, Tzekov A, Bensch KG, Bickel PE. Adipocyte protein S3–12 coats nascent lipid droplets. J Biol Chem 2003; 278:37713-21; PMID:12840023; http://dx.doi.org/ 10.1074/jbc.M304025200 [DOI] [PubMed] [Google Scholar]
- 7.Wolins NE, Quaynor BK, Skinner JR, Tzekov A, Croce MA, Gropler MC, Varma V, Yao-Borengasser A, Rasouli N, Kern PA, et al.. OXPAT/PAT-1 is a PPAR-induced lipid droplet protein that promotes fatty acid utilization. Diabetes 2006; 55:3418-28; PMID:17130488; http://dx.doi.org/ 10.2337/db06-0399 [DOI] [PubMed] [Google Scholar]
- 8.Bickel PE, Tansey JT, Welte MA. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim Biophys Acta 2009; 1791:419-40; PMID:19375517; http://dx.doi.org/ 10.1016/j.bbalip.2009.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scherer PE, Bickel PE, Kotler M, Lodish HF. Cloning of cell-specific secreted and surface proteins by subtractive antibody screening. Nat Biotechnol 1998; 16:581-6; PMID:9624692; http://dx.doi.org/ 10.1038/nbt0698-581 [DOI] [PubMed] [Google Scholar]
- 10.Wolins NE, Quaynor BK, Skinner JR, Schoenfish MJ, Tzekov A, Bickel PE. S3–12, Adipophilin, and TIP47 package lipid in adipocytes. J Biol Chem 2005; 280:19146-55; PMID:15731108; http://dx.doi.org/ 10.1074/jbc.M500978200 [DOI] [PubMed] [Google Scholar]
- 11.Dalen KT, Schoonjans K, Ulven SM, Weedon-Fekjaer MS, Bentzen TG, Koutnikova H, Auwerx J, Nebb HI. Adipose tissue expression of the lipid droplet-associating proteins S3–12 and perilipin is controlled by peroxisome proliferator-activated receptor-gamma. Diabetes 2004; 53:1243-52; PMID:15111493; http://dx.doi.org/ 10.2337/diabetes.53.5.1243 [DOI] [PubMed] [Google Scholar]
- 12.Yamaguchi T, Matsushita S, Motojima K, Hirose F, Osumi T. MLDP, a novel PAT family protein localized to lipid droplets and enriched in the heart, is regulated by peroxisome proliferator-activated receptor alpha. J Biol Chem 2006; 281:14232-40; PMID:16571721; http://dx.doi.org/ 10.1074/jbc.M601682200 [DOI] [PubMed] [Google Scholar]
- 13.Dalen KT, Dahl T, Holter E, Arntsen B, Londos C, Sztalryd C, Nebb HI. LSDP5 is a PAT protein specifically expressed in fatty acid oxidizing tissues. Biochim Biophys Acta 2007; 1771:210-27; PMID:17234449; http://dx.doi.org/ 10.1016/j.bbalip.2006.11.011 [DOI] [PubMed] [Google Scholar]
- 14.Jacquier N, Mishra S, Choudhary V, Schneiter R. Expression of oleosin and perilipins in yeast promotes formation of lipid droplets from the endoplasmic reticulum. J Cell Sci 2013; 126:5198-209; PMID:24006263; http://dx.doi.org/ 10.1242/jcs.131896 [DOI] [PubMed] [Google Scholar]
- 15.Carman GM, Han GS. Roles of phosphatidate phosphatase enzymes in lipid metabolism. Trends Biochem Sci 2006; 31:694-9; PMID:17079146; http://dx.doi.org/ 10.1016/j.tibs.2006.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Csaki LS, Dwyer JR, Fong LG, Tontonoz P, Young SG, Reue K. Lipins, lipinopathies, and the modulation of cellular lipid storage and signaling. Prog Lipid Res 2013; 52:305-16; PMID:23603613; http://dx.doi.org/ 10.1016/j.plipres.2013.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adeyo O, Horn PJ, Lee S, Binns DD, Chandrahas A, Chapman KD, Goodman JM. The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J Cell Biol 2011; 192:1043-55; PMID:21422231; http://dx.doi.org/ 10.1083/jcb.201010111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsieh K, Lee YK, Londos C, Raaka BM, Dalen KT, Kimmel AR. Perilipin family members preferentially sequester to either triacylglycerol-specific or cholesteryl-ester-specific intracellular lipid storage droplets. J Cell Sci 2012; 125:4067-76; PMID:22685330; http://dx.doi.org/ 10.1242/jcs.104943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Czabany T, Athenstaedt K, Daum G. Synthesis, storage and degradation of neutral lipids in yeast. Biochim Biophys Acta 2007; 1771:299-309; PMID:16916618; http://dx.doi.org/ 10.1016/j.bbalip.2006.07.001 [DOI] [PubMed] [Google Scholar]


