Abstract
For over half a century, tRNAs have been exclusively known as decoders of genomic information. However, recent reports evidenced that tRNA transcripts are also bearers of functional RNAs, which are able to execute various tasks through an array of mechanisms. Here, we succinctly review the diversity and functions of RNAs deriving from tRNA loci.
Keywords: 3′ETS, ITS, junk RNA, regulatory RNA, TLR, tRFs, tRNA, tRNA halves
Abbreviations
- 3′ETS
3′external transcribed spacer
- ITS
internal transcribed spacers
- TLR
tRNA-linked repeats
- tRFS
tRNA-derived fragments
- tRNA
transfert RNA
Transfer RNAs (tRNAs) are stable 70-90 nt long molecules found in all kingdoms of life.1,2 They are characterized by a universally conserved cloverleaf secondary structure composed of the D-loop, the anticodon loop and the T-loop, from 5′ to 3′CCA end (Fig. 1). tRNAs are commonly considered to be amino acid carriers that decode information contained in the nucleotide sequence of messenger RNAs (mRNAs) into specific polypeptide sequences and thus play a crucial role in protein synthesis. Besides this function, mature tRNAs are also involved in various biological processes including stress response, gene regulation and plasmid replication (for a review see refs.3,4). For example, in Escherichia coli, when cells are nutrient-starved, uncharged tRNAs act as signal molecules to activate the stringent response through induction of ppGpp alarmone and thereby promote cell survival.5 Under similar stress conditions, uncharged tRNAs play the role of ligands for T-box riboswitches, which are 5′untranslated region (5′ UTR) elements regulating the expression of downstream coding sequences.6
Figure 1.
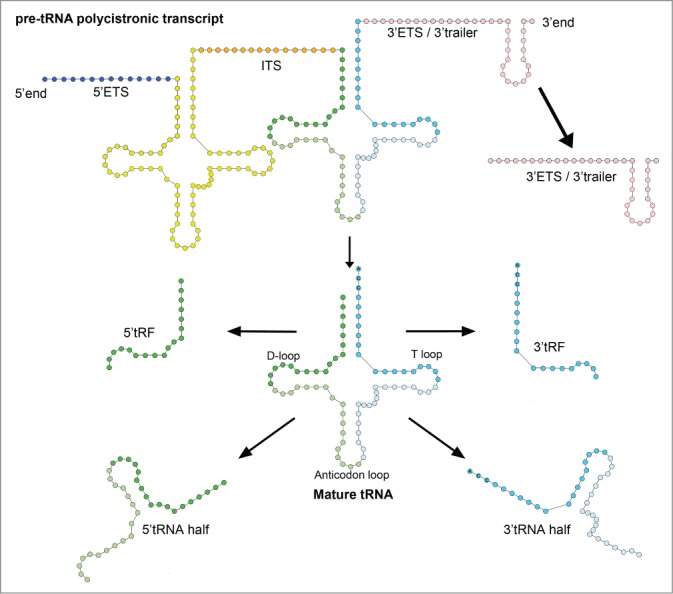
Overview of identified RNA fragments derived from tRNA transcripts. Various stable RNA molecules originate from mature tRNA [5’ and 3’ tRNA halves, 5’ and 3’ tRNA-derived fragments (tRFs)] and pre-tRNA transcript [3’external transcribed spacer (3’ETS) or 3’ trailer]. The 5’ and 3’ tRNA halves derive from both 5’ and 3’ sides of mature tRNA after cleavage at the close vicinity of the anticodon. The 5’ and 3’ tRFs originate from cleavage in the D-loop and T-loop, respectively. Recent findings describe internal transcribed spacers (ITS) as putative functional RNA molecules; however, there is still no evidence of free stable ITS in the cell.
tRNAs are transcribed as monocistronic or polycistronic (with others tRNAs, rRNAs or mRNAs; only in prokaryotic organisms) pre-tRNA transcripts and are processed by ribonucleases and diverse modification enzymes to release mature tRNAs.1,2 In general, the 5′ external transcribed spacer (5′ETS) is removed by RNase P cleavage. Maturation of the 3′ETS, also named 3′ trailer, requires tRNase Z or RNase E depending on the organism (tRNase Z in Eukaryotes and Archaea; RNase E in Eubacteria). RNA fragments derived from tRNA transcripts were, for a long time, considered as non-functional “junk” RNA. However, the last few years have witnessed an abundance of reports revealing that tRNA loci also serve as a source of small RNA fragments in different organisms representing all domains of life. These RNA fragments, derived by cleavage of either mature tRNAs or pre-tRNAs, form an heterogeneous group, notably in size, processing pattern and function (for a review see refs.3,7,8). This emerging family of functional regulatory RNAs has been subdivided into 2 major classes based on tRNA fragment size and origin: tRNA halves and tRNA-derived fragments (tRFs) (Fig. 1).
tRNA halves
First described in E. coli, tRNA halves average 30-35 nt in length and can derive from both 5′ and 3′ parts of mature tRNAs after cleavage near the anticodon (for a review see refs.3,7-9). In bacteria, generation of tRNA halves is generally observed when cells are subjected to stress. For example, in E. coli, the tRNA anticodon nuclease PrrC specifically cleaves endogenous tRNALys in response to bacteriophage infection.9 Other tRNA halves can be generated by colicin D and colicin E5 cleavage from specific tRNAs.9 In Shigella, Salmonella and Leptospira, VapC toxin acts by cleaving tRNAfMet at a precise location in the anticodon loop.10,11 The specificity of tRNAs cleavage suggests that the processing of tRNA halves is not randomly performed. In a similar way, studies in eukaryotes unveiled the fact that the cleavage of tRNAs to produce half molecules also often occurs in stressful conditions. Indeed, the Rny1p protein, a member of the RNase T2 family in yeast, is induced in adverse conditions and is responsible for generating tRNA halves.12 Moreover, in mammalian cells, angiogenin, a member of the RNase A family, is responsible for the cleavage of tRNAs and accumulating evidence suggest a role of angiogenin in cell survival under harsh conditions.13
tRNA-derived fragments
tRNA-derived fragments (tRFs), which are usually shorter than tRNA halves, measure approximately 20 nt. They are subdivided into 3 major groups: 5′ tRFs (or tRFs-514), 3′ tRFs (also called tRFs-314 or 3′CCA tRFs15) deriving from mature tRNAs and 3′ trailer RNAs derived from pre-tRNAs (also called 3′ ETS,16 3′U tRFs15 or tRF-114). In Eukaryotes, the mechanisms adopted to generate 5′ and 3′ tRFs are not fully understood but often require Dicer. While 5′ tRFs originate from cleavage in the D-loop, 3′ tRFs originate from cleavage in the T-loop. In addition, 3′ trailers are sometimes longer (between 20 and 60 nt) and are released by RNase Z in Eukaryotes14 and by RNase E in E. coli.16
Upon close analysis of the RNAseq data reported by Ghosal et al.17, we noticed the presence of highly enriched tRNA fragments corresponding to the region between the anticodon loop and the T-loop of tRNAGlu, tRNAThr, tRNAGln, and tRNASer. It remains unclear whether these are merely by-products from the maturation process or yet another type of functional tRF. Further experiments are required to address this issue.
Functions of tRNA-derived RNA fragments
Overall, tRNA-derived RNA fragments seem to be involved in translation regulation and gene silencing.
In the archaeon Haloferax volcanii, a 26 nt-long 5′ tRF processed from mature tRNAVal targets the small ribosomal subunit, interferes with peptydil transferase activity and, as a consequence, reduces protein synthesis.18 In human cells, a similar mechanism has recently been reported.15
Accumulating evidence indicates that tRNA fragments can also act as miRNAs or siRNAs in post-transcriptional gene silencing as they have been shown to interact with Argonaute proteins. Indeed, a 22 nt-long 3′ tRF, generated by Dicer from tRNAGly and associated with Argonaute proteins, was shown to inhibit RPA1 protein synthesis, presumably by binding to its 3′ UTR.19 Additionally, Yeung et al.20 reported that an 18 nt-long 3′ tRF of tRNALys could target the primer binding sequence used to initiate reverse transcriptase in the HIV RNA genome, thereby inducing mRNA degradation. Recently, a 31 nt-long 5′ tRNA half, processed from tRNAGlu, has been reported to suppress apolipoprotein E receptor 2 (APOER2) expression by interacting with its 3′UTR.21 The repression of APOER2, which has been described as an antiviral protein, promotes respiratory syncytial virus (RSV) replication. Interestingly, RSV itself induces tRNAGlu half production and thus favors its own replication.
tRFs could also compete with miRNAs/siRNAs for RISC loading. Lee et al.14 noticed that tRF-1001, a tRF released from the 3′ ETS of a tRNASer precursor by the tRNA endonuclease ELAC2 (homolog of RNase Z), seems to be necessary for cell viability. According to Haussecker et al.22, this tRF could be involved in gene silencing modulation.
tRFs can also use uncommon mechanisms such as protein sequestration23 or buffering of other regulatory RNAs.16
In breast cancer cells, tRFs can sequester YBX1, a versatile RNA binding protein that stabilizes oncogenic transcripts and enhances their translation. By displacing YBX1 from the 3′ untranslated regions (3′ UTR) of these transcripts, the tRFs antagonize YBX1 activity and suppress metastatic progression.23 This post-transcriptional control is reminiscent of motif mimicry used by some sRNAs in bacteria (e.g., CsrB/C chelating CsrA protein24).
Buffering of regulatory RNAs via tRFs can occur in bacterial cells, as demonstrated by our group in early 2015.16 The use of a novel technique to identify RNA-RNA interactions led us to discover that 2 small regulatory RNAs (i.e. RyhB and RybB) of E. coli interact with the 3′ ETS of the glyW-cysT-leuZ pre-tRNA transcript. This ∼50 nt-long 3′ ETS is released from the pre-tRNA by RNase E cleavage close to the 3′ CCA of leuZ and is detectable in vivo in various normal and stressful growth conditions. The interaction of this tRF with both RyhB and RybB sRNAs was first thought to be a demonstration of tRNA levels being regulated by sRNAs. However, this hypothesis promptly shifted when it was revealed that mature levels of tRNA GlyW, CysT, or LeuZ were unaffected by either sRNAs. Through different methods, we were able to unveil the legitimate role of this tRF, which is to suppress RyhB and RybB transcriptional noise in non-stressful conditions. Because sRNA promoters are remarkably strong to properly respond to environmental stress, they can hardly be fully repressed. We demonstrated that cells developed a sponge-based system to nullify sRNA transcriptional noise. Indeed, the sequestration of sRNAs by 3′ ETSleuZ sets a concentration threshold, which sRNAs need to overcome in order to regulate their targetome. This tRF-based mechanism prevents regulation of targets in absence of stress, thus improving bacterial fitness. Despite being detected in lower cellular concentration than mature tRNAs, the 3′ ETSleuZ molecule fulfills versatile roles. This molecule directly buffers regulatory RNAs and indirectly plays an important role in post-transcriptional regulation of a broad pool of mRNAs. Moreover, this unique bacterial tRF acting as an RNA sponge is even able to modulate cellular sensitivity to antibiotic and adaptation to carbon sources. Coupling this information with the high sequence conservation of the 3′ ETSleuZ in organisms expressing RyhB or RybB–like sRNAs suggests the strong significance of such tRFs.
Recently brought to light by Ghosal et al.17, bacterial tRFs and tRNA halves can be found in outer membrane vesicles (OMV) released by E. coli and, consequently, in the extracellular medium. Although it is not known whether these RNAs serve any particular function, their presence outside the bacterial cell is reminiscent of the work by Dhahbi et al.25, whose report demonstrated that eukaryotic 5′ tRNA halves circulate in blood. The authors proposed that circulating 5′ tRNA halves might act as signaling molecules. Along the same lines, it is tempting to speculate that extracellular tRFs and tRNA halves could play a role in exchanges between bacteria.
Other putative functional elements encoded in tRNA transcripts
Pre-tRNA transcripts can conceal other elements with the potential to be functional such as internal transcribed spacers (ITS) and tRNA-linked repeats (TLR).
ITS are short sequences (between 1 and 61 nt in E. coli) found between 2 tRNAs of the same polyscistronic transcript. In the light of our work,16 ITS appear to have a functional role beyond tRNA maturation. Indeed, our group demonstrated that both ITSmetZ-metW and ITSmetW-metV, from the metZ-metW-metV pre-tRNA transcript, are able to interact with 2 sRNAs: RybB and MicF.16 Since both ITS cannot be detected in vivo as separate entities, we suggest that different forms of the pre-tRNA transcript act as functional RNAs. However, their exact roles have yet to be described. Our preliminary data already suggest a distinct mechanism of action from 3′ETSleuZ.
Also deriving from pre-tRNAs transcripts, another type of tRFs found in bacterial cells are tRNA-linked repeats (TLR). At least 20 TLRs are present in E. coli. These RNA molecules, located immediately downstream of tRNA genes, share the last 18-19nt of the 3′ end of tRNA sequences. According to Rudd,26 “the TLR repeats may be remnants of repeated utilization of tRNA genes for phage or plasmid integrations and excisions, which can leave small regions of duplicated tRNA gene ends behind as debris from horizontal gene transmissions.” Contradicting the hypothesis of debris is the tyrTV operon, where 3 178-long repeats can be found. Each of those repeats, rtV1, rtV2, and rtV3, contains 19 nt of the 3′end of tRNATyr.27 Whereas the most studied TLR is rtV1, which encodes a small protamine-like protein called Tpr, rtV2, and rtV3 act as Rho-dependent transcriptional terminators. Given that no other TLR has been studied so far, we cannot exclude an additional functional role for these elements.
Conclusion
Long considered as “junk” RNAs, tRFs and tRNA halves turn out to be important functional elements of eukaryotic and prokaryotic cells. With the help of high-throughput sequencing and genome-wide analysis, recent reports have described surprising functions for these molecules. Although roles of most identified tRNA fragments are still elusive, latest discoveries demonstrate that they could used similar mechanisms than well-characterized regulatory RNAs (gene silencing, protein sequestration, sRNA buffering). Taking into consideration the latest findings in this field, it appears clear that tRNAs represent an impressive source of various functional RNAs.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work has been supported by an operating grant from the Canadian Institutes of Health Research (CIHR) to EM. MCC holds a MSc studentship from the Fonds de Recherche du Québec – Nature et Technologies (FRQNT). EM is a senior scholar from the Fonds de la Recherche en santé du Québec (FRSQ).
References
- 1.Shepherd J, Ibba M. Bacterial transfer RNAs. FEMS Microbiol Rev 2015; 39:280-300; PMID:25796611; http://dx.doi.org/ 10.1093/femsre/fuv004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev 2010; 24:1832-60; PMID:20810645; http://dx.doi.org/ 10.1101/gad.1956510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raina M, Ibba M. tRNAs as regulators of biological processes. Front Genet 2014; 5:171; PMID:24966867; http://dx.doi.org/ 10.3389/fgene.2014.00171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wegrzyn G, Wegrzyn A. Is tRNA only a translation factor or also a regulator of other processes? J Appl Genet 2008; 49:115-22; PMID:18263978; http://dx.doi.org/ 10.1007/BF03195257 [DOI] [PubMed] [Google Scholar]
- 5.Goldman E, Jakubowski H. Uncharged tRNA, protein synthesis, and the bacterial stringent response. Mol Microbiol 1990; 4:2035-40; PMID:1708437; http://dx.doi.org/ 10.1111/j.1365-2958.1990.tb00563.x [DOI] [PubMed] [Google Scholar]
- 6.Green NJ, Grundy FJ, Henkin TM. The T box mechanism: tRNA as a regulatory molecule. FEBS Lett 2010; 584:318-24; PMID:19932103; http://dx.doi.org/ 10.1016/j.febslet.2009.11.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson P, Ivanov P. tRNA fragments in human health and disease. FEBS Lett 2014; 588(23):4297-304; PMID: 25220675; http://dx.doi.org/ 10.1016/j.febslet.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol 2013; 10:1798-806; PMID:24351723; http://dx.doi.org/ 10.4161/rna.27177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masaki H, Ogawa T. The modes of action of colicins E5 and D, and related cytotoxic tRNases. Biochimie 2002; 84:433-8; PMID:12423786; http://dx.doi.org/ 10.1016/S0300-9084(02)01425-6 [DOI] [PubMed] [Google Scholar]
- 10.Winther KS, Gerdes K. Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc Natl Acad Sci U S A 2011; 108:7403-7; PMID:21502523; http://dx.doi.org/ 10.1073/pnas.1019587108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopes AP, Lopes LM, Fraga TR, Chura-Chambi RM, Sanson AL, Cheng E, Nakajima E, Morganti L, Martins EA. VapC from the leptospiral VapBC toxin-antitoxin module displays ribonuclease activity on the initiator tRNA. PLoS One 2014; 9:e101678; PMID:25047537; http://dx.doi.org/ 10.1371/journal.pone.0101678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thompson DM, Parker R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J Cell Biol 2009; 185:43-50; PMID:19332891; http://dx.doi.org/ 10.1083/jcb.200811119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li S, Hu GF. Emerging role of angiogenin in stress response and cell survival under adverse conditions. J Cell Physiol 2012; 227:2822-6; PMID:22021078; http://dx.doi.org/ 10.1002/jcp.23051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev 2009; 23:2639-49; PMID:19933153; http://dx.doi.org/ 10.1101/gad.1837609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sobala A, Hutvagner G. Small RNAs derived from the 5′ end of tRNA can inhibit protein translation in human cells. RNA Biol 2013; 10:553-63; PMID:23563448; http://dx.doi.org/ 10.4161/rna.24285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lalaouna D, Carrier MC, Semsey S, Brouard JS, Wang J, Wade JT, Masse E. A 3′ External Transcribed Spacer in a tRNA Transcript Acts as a Sponge for Small RNAs to Prevent Transcriptional Noise. Mol Cell 2015; 58:393-405; PMID:25891076; http://dx.doi.org/ 10.1016/j.molcel.2015.03.013 [DOI] [PubMed] [Google Scholar]
- 17.Ghosal A, Upadhyaya BB, Fritz JV, Heintz-Buschart A, Desai MS, Yusuf D, Huang D, Baumuratov A, Wang K, Galas D, et al.. The extracellular RNA complement of Escherichia coli. Microbiologyopen 2015; PMID:25611733; http://dx.doi.org/ 10.1002/mbo3.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gebetsberger J, Zywicki M, Kunzi A, Polacek N. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea 2012; 2012:260909; PMID:23326205; http://dx.doi.org/ 10.1155/2012/260909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K, Dalla-Favera R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci U S A 2013; 110:1404-9; PMID:23297232; http://dx.doi.org/ 10.1073/pnas.1206761110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeung ML, Bennasser Y, Watashi K, Le SY, Houzet L, Jeang KT. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res 2009; 37:6575-86; PMID:19729508; http://dx.doi.org/ 10.1093/nar/gkp707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng J, Ptashkin RN, Chen Y, Cheng Z, Liu G, Phan T, Deng X, Zhou J, Lee I, Lee YS, et al.. Respiratory syncytial virus utilizes a tRNA fragment to suppress antiviral responses through a novel targeting mechanism. Mol Ther 2015; 23(10):1622-9; PMID:26156244; http://dx.doi.org/ 10.1038/mt.2015.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. Rna 2010; 16:673-95; PMID:20181738; http://dx.doi.org/ 10.1261/rna.2000810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SF. Endogenous tRNA-Derived Fragments Suppress Breast Cancer Progression via YBX1 Displacement. Cell 2015; 161:790-802; PMID:25957686; http://dx.doi.org/ 10.1016/j.cell.2015.02.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Camacho MI, Alvarez AF, Chavez RG, Romeo T, Merino E, Georgellis D. Effects of the global regulator CsrA on the BarA/UvrY two-component signaling system. J Bacteriol 2015; 197:983-91; PMID:25535275; http://dx.doi.org/ 10.1128/JB.02325-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dhahbi JM. 5′ tRNA Halves: The Next Generation of Immune Signaling Molecules. Front Immunol 2015; 6:74; PMID:25745425; http://dx.doi.org/ 10.3389/fimmu.2015.00074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rudd KE. Novel intergenic repeats of Escherichia coli K-12. Res Microbiol 1999; 150:653-64; PMID:10673004; http://dx.doi.org/ 10.1016/S0923-2508(99)00126-6 [DOI] [PubMed] [Google Scholar]
- 27.Timms AR, Bridges BA. The tyrT locus of Escherichia coli B. J Bacteriol 1996; 178:2469-70; PMID:8636060 [DOI] [PMC free article] [PubMed] [Google Scholar]


