ABSTRACT
Each cell produces its own responses even if it appears identical to other cells. To analyze these individual cell characteristics, we need to measure trace amounts of molecules in a single cell. Nucleic acids in a single cell can be easily amplified by polymerase chain reaction, but single-cell measurement of proteins and sugars will require de novo techniques. In the present study, we outline the techniques we have developed toward this end. For proteins, our ultrasensitive enzyme-linked immunosorbent assay (ELISA) coupled with thionicotinamide-adenine dinucleotide cycling can detect proteins at subattomoles per assay. For sugars, fluorescence correlation spectroscopy coupled with glucose oxidase-catalyzed reaction allows us to measure glucose at tens of nM. Our methods thus offer versatile techniques for single-cell-level analyses, and they are hoped to strongly promote single-cell biology as well as to develop noninvasive tests in clinical medicine.
KEYWORDS: adiponectin, fluorescence correlation spectroscopy, HIV-1 p24, insulin, thio-NAD cycling
Introduction
The exact detection and determination of proteins, nucleic acids, and sugars in a single cell are a romantic vision of modern biology and medical sciences.1,2 Even though a tissue consists of apparently similar cells, which belong to, for example, the same organ lobe, the same nerve nucleus, and even the same carcinomatous lesion, each of these cells has its own individuality and demonstrates various, complicated, and uneven responses. As we scientists secretly imagine, this individuality seems to create the complexity and magnificence of life. Realistically, however, we have so far been obliged to report data on the optical density of bands obtained by Southern, Northern, and Western blotting, in which a lot of cells are employed.3 Sometimes ‘single-cell-level amounts’ are calculated from many single cells after division by the number of cells. Needless to say, this practice occurs because it is too difficult to detect and determine the molecules in an actual single cell.
In this situation, trace amounts of nucleic acids have been made detectable by the invention of polymerase chain reaction (PCR) by Mullis.4 This success was based on the fact that nucleic acids can be amplified. Then, in the 1990s, the emergence of real-time PCR enabled us to determine the copy numbers of nucleic acids.5 Various derivative methods of real-time PCR are now applicable to cells and tissues,6 and in particular quantitative real-time PCR can detect the copy number, such as a few copies, of nucleic acids in a single cell.7–12 Although quantitative real-time PCR is not always suitable for a comprehensive analysis of gene expression, DNA microarray analysis can be used instead of PCR to examine nucleic acids for this purpose. For example, gene expression at the mRNA level can be easily examined if the sensitivity is allowed to be limited.8
More recently, digital PCR has been proposed as a new approach to the detection and quantification for nucleic acids.13–15 Real-time PCR carries out one reaction per single sample, whereas digital PCR also carries out a single reaction within a sample, however the sample is separated into a large number of partitions and the reaction is carried out in each partition individually. Some portions of these reactions contain the target molecule (positive) while others do not (negative). Following PCR analysis, the fraction of negative reactions is used to generate an absolute count of the number of target molecules in the sample, without reference to standards or endogenous controls. The digital PCR can be routinely used for clonal amplification of samples for next-generation sequencing.16 To sum up, almost all the analyses for nucleic acids can be performed at present using commercially available methods, ranging from a simultaneous and comprehensive analysis of multiple nucleic acids of cells and tissues to a precise analysis of a specific gene in a single cell.
On the other hand, a change in the amount of mRNAs in a cell does not always correspond to a change in the amount of translated proteins.9 That is, we have to accept that a quantitative analysis of nucleic acids offers important knowledge but does not conclude precisely the phenomenon of gene expression. Thus, we have to attempt to measure the amount of proteins, possibly at a single-cell level. In the present study, we introduce a de novo method we developed to measure trace amounts of proteins at a single-cell level.
Further, we describe a new way to measure sugars at very low concentrations. This highly sensitive detection method also helps to facilitate single-level studies in biological and medical fields. For example, single-cell metabolism has been vigorously examined, particularly for neurons and glia.17 Because metabolism is usually expressed as the sum of chemical processes that occur in living organisms, the data obtained from a single cell should be accumulated and analyzed to gain a better understanding of metabolism.
Ultrasensitive ELISA coupled with thio-NAD cycling for protein detection
Many attempts, including lab-on-a-chip studies, have been performed thus far to detect trace amounts of proteins in a single cell.18-21 Although some of these studies have achieved the ultimate sensitivity, they must employ specialized and expensive apparatuses for the experiments. Thus we decided to develop a system with special attention paid to ‘versatility’ and ‘user-friendliness’. Among the various techniques to quantify trace amounts of proteins, a ‘sandwich’ enzyme-linked immunosorbent assay (ELISA) may be the most versatile and user-friendly.22 ELISA is an easy, rapid, specific, and highly sensitive detection method and thus has been widely used in biology and clinical medicine. However, there is a limit to what conventional ELISA can do. For example, the limits of detection in the ELISA for insulin established in the 1980s and 1990s were on the order of μIU/mL, which corresponded to 10−14 moles/mL.23,24 So what should we do?
We attempted to combine another assay to determine trace amounts of proteins with ELISA. This is an amplification technique that uses a continuous reaction of enzyme function. This is referred to as enzyme cycling.25,26 In our method, a cycling reaction is conducted by a dehydrogenase such as 3α-hydroxysteroid dehydrogenase (3α-HSD, EC. 1.1.1.50).27-33 3α-HSD catalyzes substrate cycling between 3α-hydroxysteroid and its corresponding 3-ketosteroid in the presence of an excess amount of NADH and thionicotinamide-adenine dinucleotide (thio-NAD), because 3α-HSD utilizes both NADH and thio-NAD as cofactors.34 In each turn of the cycle, one molecule of thio-NAD is reduced to thio-NADH. Thio-NADH has the absorbance at 400 nm (11900 M−1 cm−1). The other cofactors of 3α-HSD, such as thio-NAD, NAD, and NADH, have the absorbance maximums under 340 nm. Therefore, the amount of thio-NADH can be measured with a commercially available microplate reader with a 405 nm filter. These features make it possible to determine the amounts of 3α-hydroxysteroids with high sensitivity by measuring the cumulative quantity of thio-NADH. We call this enzyme cycling thio-NAD cycling.
The combination of a sandwich ELISA and a thio-NAD cycling method gives us an ultrasensitive detection of trace amounts of proteins (Figure 1).35 In our sandwich ELISA, we use 17β-methoxy-5β-androstan-3α-ol 3-phosphate as a synthetic substrate for alkaline phosphatase (ALP, EC. 3.1.3.1) linked to a secondary antibody, because 17β-methoxy-5β-androstan-3α-ol 3-phosphate is easily hydrolyzed by ALP to 17β-methoxy-5β-androstan-3α-ol, which shows highly efficient thio-NAD cycling. 17β-methoxy-5β-androstan-3α-ol is then oxidized to 17β-methoxy-5β-androstan-3-one under a catalytic reaction of 3α-HSD with the cofactor thio-NAD. By the opposite reaction, 17β-methoxy-5β-androstan-3-one is reduced to 17β-methoxy-5β-androstan-3α-ol with the cofactor NADH. In the ELISA, the detectable signal changes linearly against a measuring time; in thio-NAD cycling, the detectable signal also changes linearly against the measurement time. In the combination, however, the detectable signal, i.e., thio-NADH, accumulates in a quadratic function-like fashion during the cycling reaction within a short measurement time.
Figure 1.
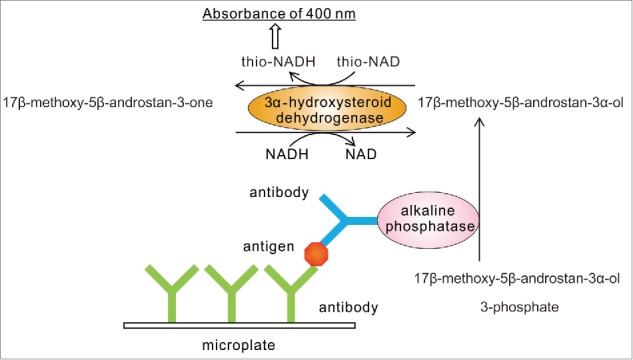
Scheme of ultrasensitive detection of proteins by ELISA coupled with thio-NAD cycling. Our ultrasensitive ELISA requires only the addition of a thio-NAD cycling solution, which includes androsterone derivatives, and 3α-hydroxysteroid dehydrogenase and its coenzymes, to the usual ELISA without any use of special instruments. Absorption (400 nm) of thio-NADH is measured with a commercially available microplate reader.
More specifically, because the thio-NADH signaling intensity depends on the number of thio-NADH molecules accumulated by the enzyme reactions, the thio-NADH signaling intensity
Here, a is the turnover ratio of ALP per min; b is the cycling ratio of 3α-HSD per min; and n = min of measurement time. The amount of insulin was calculated from the increase in absorbance at 405 nm. The claim for our novel method is that proteins themselves cannot be amplified, but a detectable signal for proteins can be amplified by means of a quadratic function-like response (i.e., triangular number). Our ultrasensitive ELISA coupled with thio-NAD cycling needs only a detection apparatus for absorbance at 405 nm, but does not need any specialized apparatus, specialized laboratory, or use of radio isotopes and fluorescent probes.
For an analysis at the single-cell level, we attempted to detect proteins at 10−18 moles/assay.36,37 We applied our ultrasensitive ELISA coupled with thio-NAD cycling to the detection of human proteins, such as insulin, HIV-1 (human immunodeficiency virus type 1) p24, and adiponectin. Insulin is a peptide hormone produced by β cells in the pancreas, and it regulates the metabolism of carbohydrates and fats by promoting the absorption of glucose from the blood to skeletal muscles and fat tissue.38,39 The limit of detection for insulin with our ultrasensitive ELISA was 8.0 × 10−19 moles/assay (0.0047 pg/assay) with the linear calibration curve of 8.9 × 10−19 – 8.9 × 10−18 moles/assay in Tris-buffered saline; the coefficient of variation was 4% for 8.9 × 10−19 moles/assay.35 We then applied our method to the measurement of immunoreactive insulin in blood serum.40 Because of this ultra-high sensitivity, only 5 μL of serum was needed. Further, a comparison between the data obtained from a commercially available immunoreactive insulin kit (i.e., a conventional ELISA) and the data obtained from our ultrasensitive ELISA using the same commercially available reference demonstrated that the correlation was very good, providing further evidence that our assay is also accurate in blood samples.
HIV is a lentivirus that causes HIV infection and acquired immune deficiency syndrome (AIDS). HIV infection occurs by the transfer of blood, semen, vaginal fluid, breast milk, and so on. To reduce the window period between HIV-1 infection and the ability to diagnose it, a fourth-generation immunoassay including the detection of a viral protein, HIV-1 p24, has been developed.41,42 Our ultrasensitive ELISA enabled us to detect p24 at 2.3 × 10−18 moles/assay (0.0055 IU/assay) as the limit of detection.43,44 The linear calibration curves were obtained in the range of 2.1 × 10−18 – 2.1 × 10−17 moles/assay. The coefficient of variation was 8% for 2.1 × 10−17 moles/assay. The spike-and-recovery tests were performed in which the HIV-1 p24 antigen was added into the control serum. The results demonstrated that the ratio was about 100% for 1.0 × 10−17 moles/assay (0.5 IU/mL) of HIV-1 p24, which was less than the value (2 IU/mL) required for a CE-marked HIV antigen/antibody assay. Thus, our ultrasensitive method is able to detect HIV-1 p24 antigen in human blood obtained from patients in the very early period after infection.
Briefly, we explain the spike-and-recovery test. The spike-and-recovery test is a technique for analyzing and accessing the accuracy of ELISA for particular sample types, such as serum, plasma, saliva, urine, etc. It is used to determine whether analyte detection can be affected by the difference between the diluent used for preparation and the experimental sample matrix. To perform a spike-and-recovery test, a known amount of analyte (i.e., target protein in our case) is added to a matrix (i.e., blood or urine in our case). This ‘addition’ is called ‘spike’. The concentration of the added analyte in the matrix is determined from standard curves prepared. The concentrations denote the spike recovered in the matrix.
Adiponectin is a protein hormone that modulates a number of metabolic processes, including glucose regulation and fatty acid oxidation.45 Adiponectin is exclusively secreted from adipose tissue into the bloodstream and is very abundant in plasma relative to many hormones. Levels of the hormone are inversely correlated with body fat percentage in adults.46 However, the patterns of change in urinary adiponectin levels in various diseases remain unknown, because the urinary adiponectin levels are thought to be extremely low.47,48 We thus examined whether our ultrasensitive ELISA was applicable to measure urinary adiponectin.49 Spike-and-recovery tests using urine confirmed the reliability of our ultrasensitive ELISA. The limit of detection for adiponectin was 2.3 × 10−19 moles/assay (0.069 pg/assay) with a linear calibration curve of 4.2 × 10−18 – 1.7 × 10−16 moles/assay. The coefficient of variation was 4% for 1.7 × 10−17 moles/assay of adiponectin. The pilot study showed that the urinary adiponectin levels, which were corrected by the creatinine concentration, were higher in diabetes mellitus patients than in healthy subjects. Further, these urinary adiponectin levels tended to increase with the progression of diabetes mellitus accompanied by nephropathy.
Fluorescence correlation spectroscopy for detection of sugars
Glucose is the primary source of energy for cells.3 Glucose is transported from the intestines or liver to bodily cells via the bloodstream, and is made available for cell absorption via the hormone insulin, produced primarily in the pancreas. Recently, the examination of single-cell metabolism has begun, particularly in the brain,17 and thus a highly sensitive measurement of sugars is also needed for this purpose.
Glucose oxidase (GOD) reduces oxygen O2 to hydrogen peroxide H2O2 in the presence of glucose.50 This reaction occurs in a glucose dose-dependent manner, and thus the concentration of H2O2 corresponds to that of glucose. Thus, we focused on the development of a system to detect trace amounts of H2O2. If H2O2 is detected with high sensitivity, various substrates, such as cholesterol, choline, glutamate, lactate, NAD(P)H, urate, and xanthine, can be also detected with high sensiti-vity with the help of their corresponding oxidases, because H2O2 is generated by these oxidase-catalyzed reactions.51-54 That is, the use of oxidase-catalyzed reactions offers us a chance to develop a versatile system.
For this purpose, we selected a special method, fluorescence correlation spectroscopy (FCS), which, along with its derived methods, has been applied to characterize and determine fluorescent components in aqueous solution.55–57 FCS is a technique in which spontaneous fluorescence intensity fluctuations are measured in a microscopic detection volume of about 10−15 L (fL) defined by a tightly focused laser beam. Analysis of fluorescence fluctuation offers the information on mobility and the concentrations of fluorescent components in sample solution.55 Fluorescence intensity fluctuations measured by FCS represent changes in either the number or the fluorescence quantum yield of molecules resident in the detection volume. When a fluorescent probe of low molecular weight binds to a protein in a sample solution, the slow-diffusing component (protein labeled with fluorescent probes) increases with the decrease in the fast-diffusing component (fluorescent probe), which affects the fluorescence autocorrelation curve in FCS. The fluorescence autocorrelation curve is obtained after the fluctuations are recorded as a function of time and is statistically analyzed by autocorrelation analysis. The average residence time and the absolute numbers of slow- and fast-diffusing components in a small volume can be deduced by the fluorescence autocorrelation function calculated from the fluorescence autocorrelation curve.
This situation is in marked contrast to conventional fluorescence photometry, which is carried out in sample volumes of around 0.1 - 1.0 mL (about 1012 times larger than FCS measurement volumes) and provides only the macroscopic average of diffusion-dependent intensity fluctuations. In a typical FCS measurement, the fluorescence intensity is recorded for a small number of molecules in the detection volume (e.g., a few molecules/fL, equivalent to an approximately nanomolar concentration) over a time range from about 1 μs to 1 s. That is, even though FCS does not require physical separation between free and bound fluorescent probes, it enables us to detect the molecules at a nanomolar level.
Then, to measure FCS, we considered a small volume of a fluorescent probe and a large volume of a protein that should be labeled with the fluorescent probe. We used tyramide labeled with tetramethyl rhodamine (tyramide-TMR) as a fluorescent probe and bovine serum albumin (BSA) as a protein. When tyramide was applied at lower concentrations to BSA in the horseradish peroxidase (HRP)-catalyzed oxidation with H2O2, tyramide radical bound to a tyrosine residue of BSA, resulting in TMR-labeled BSA (Figure 2).58,59 Under the optimized conditions, TMR-labeled BSA (the fraction of the slow-diffusing component) measured by FCS was found to be proportional to the H2O2 concentration of the sample solution.58,59
Figure 2.
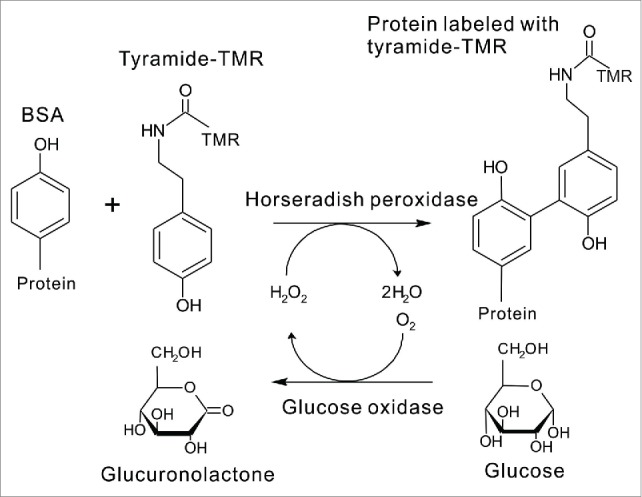
Scheme of ultrasensitive detection of sugars by FCS.The concentrations of H2O2 correspond to those of the proteins labeled with tyramide-tetramethyl rhodamine (TMR). H2O2 is produced by the reaction between glucose and glucose oxidase, and thus we can determine the concentrations of H2O2 and deduce the concentrations of glucose by FCS.
Further, by the coupling with the GOD-mediated reaction, our H2O2 detection method can be applied to the selective assay of glucose (Figure 2). Although various methods for the determination of glucose have been reported, enzymatic ones using GOD have been widely used due to their simplicity and selectivity.50 Therefore, we attempted to develop a highly sensitive method for determining glucose by coupling our FCS method with GOD-catalyzed oxidation of glucose (Figure 2).
Concerning H2O2 and β-D-glucose, we obtained the following results. The limit of detection for H2O2 was 8 nM with a linear calibration curve of 28 nM to 300 nM; the detection limit for β-D-glucose was 24 nM with a linear calibration curve of 80 nM to 1500 nM.58 The latter enabled us to assay glucose in human plasma with only a very small amount of plasma (20 nL). A recent study demonstrated that an amperometric needle-type electrochemical glucose sensor can detect the μM-order glucose in the tear of μL fluid.60 Our FCS method also has a sufficient sensitivity for a tear.
Conclusion
When we have access to 2 suitable antibodies of a sandwich ELISA for a target protein, our ultrasensitive system described in the present study can be widely applied to detect trace amounts of various proteins in blood or urine only by application of the thio-NAD cycling reagents to the conventional sandwich ELISA system. Further, when we combine the peroxidase-catalyzed and the specific oxidase-catalyzed reactions, we can detect trace amounts of sugars and other molecules with the FCS. We really hope that our new methods for the detection of proteins and sugars at the single-cell level can contribute to the further evolution of single-cell studies as well as pave the way for further noninvasive clinical tests.
Abbreviations
- 3α-HSD
3α-hydroxysteroid dehydrogenase
- AIDS
acquired immune deficiency syndrome
- ALP
alkaline phosphatase
- BSA
bovine serum albumin
- ELISA
enzyme-linked immunosorbent assay
- FCS
fluorescence correlation spectroscopy
- GOD
glucose oxidase
- HRP
horseradish peroxidase
- PCR
polymerase chain reaction
- thio-NAD
thionicotinamide-adenine dinucleotide
- tyramide-TMR
tyramide labeled with tetramethyl rhodamine
Disclosure of potential conflicts of interest
SW is an employee of BL Co., Ltd., and MM and KN are employees of TAUNS Laboratories, Inc.. MK, AN, MN, TY, TM, and EI declare that they have no conflicts of interest.
Funding
This study was supported by a grant for the Development of Systems and Technology for Advanced Measurement and Analysis from JST, by a grant for the Regional Innovation Strategy Support Program 2009 from MEXT, by a grant for the New Regional Consortium Research and Development Project from METI, and by the Hokkaido Bureau of Economy, Trade, and Industry to E.I. and the others.
References
- [1].Spiller DG, Wood CD, Rand DA, White MR. Measurement of single-cell dynamics. Nature 2010; 465:736-45; PMID:20535203; http://dx.doi.org/ 10.1038/nature09232 [DOI] [PubMed] [Google Scholar]
- [2].Zenobi R. Single-cell metabolomics: analytical and biological perspectives. Science 2013; 342:1243259; PMID:24311695; http://dx.doi.org/ 10.1126/science.1243259 [DOI] [PubMed] [Google Scholar]
- [3].Alberts B, Johnson A, Lewis J, Morgan D, Raff M, Roberts K, Walter P. Molecular Biology of the Cell, 6th edition Garland Science, New York, USA: 2014 [Google Scholar]
- [4].Mullis KB, Faloona FA. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol 1987; 155:335-50; PMID:3431465; http://dx.doi.org/ 10.1016/0076-6879(87)55023-6 [DOI] [PubMed] [Google Scholar]
- [5].Higuchi R, Fockler C, Dollinger G, Watson R. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnol (NY) 1993; 11:1026-30; http://dx.doi.org/ 10.1038/nbt0993-1026 [DOI] [PubMed] [Google Scholar]
- [6].Hatakeyama D, Okuta A, Otsuka E, Lukowiak K, Ito E. Consolidation of long-term memory by insulin in Lymnaea is not brought about by changing the number of insulin receptors. Commun Integr Biol 2013; 6:e23955; PMID:23710281; http://dx.doi.org/ 10.4161/cib.23955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wagatsuma A, Sadamoto H, Kitahashi T, Lukowiak K, Urano A, Ito E. Determination of the exact copy numbers of particular mRNAs in a single cell by quantitative real-time RT-PCR. J Exp Biol 2005; 208:2389-98; PMID:15939778; http://dx.doi.org/ 10.1242/jeb.01625 [DOI] [PubMed] [Google Scholar]
- [8].Azami S, Wagatsuma A, Sadamoto H, Hatakeyama D, Usami T, Fujie M, Koyanagi R, Azumi K, Fujito Y, Lukowiak K, et al.. Altered gene activity correlated with long-term memory formation of conditioned taste aversion in Lymnaea. J Neurosci Res 2006; 84:1610-20; PMID:16941636; http://dx.doi.org/ 10.1002/jnr.21045 [DOI] [PubMed] [Google Scholar]
- [9].Hatakeyama D, Sadamoto H, Watanabe T, Wagatsuma A, Kobayashi S, Fujito Y, Yamashita M, Sakakibara M, Kemenes G, Ito E. Requirement of new protein synthesis of a transcription factor for memory consolidation: Paradoxical changes in mRNA and protein levels of C/EBP. J Mol Biol 2006; 356:569-77; PMID:16403525; http://dx.doi.org/ 10.1016/j.jmb.2005.12.009 [DOI] [PubMed] [Google Scholar]
- [10].Wagatsuma A, Azami S, Sakura M, Hatakeyama D, Aonuma H, Ito E. De novo synthesis of CREB in a presynaptic neuron is required for synaptic enhancement involved in memory consolidation. J Neurosci Res 2006; 84:954-60; PMID:16886187; http://dx.doi.org/ 10.1002/jnr.21012 [DOI] [PubMed] [Google Scholar]
- [11].Sadamoto H, Kitahashi T, Fujito Y, Ito E. Learning-dependent gene expression of CREB1 isoforms in the molluscan brain. Front Behav Neurosci 2010; 4:25; PMID:20631825; http://dx.doi.org/ 10.3389/fnbeh.2010.00025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Murakami J, Okada R, Sadamoto H, Kobayashi S, Mita K, Sakamoto Y, Yamagishi M, Hatakeyama D, Otsuka E, Okuta A, et al.. Involvement of insulin-like peptide in long-term synaptic plasticity and long-term memory of the pond snail Lymnaea stagnalis. J Neurosci 2013; 33:371-83; PMID:23283349; http://dx.doi.org/ 10.1523/JNEUROSCI.0679-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sykes PJ, Neoh SH, Brisco MJ, Hughes E, Condon J, Morley AA. Quantitation of targets for PCR by use of limiting dilution. Biotechniques 1992; 13:444-9; PMID:1389177 [PubMed] [Google Scholar]
- [14].Kalinina O, Lebedeva I, Brown J, Silver J. Nanoliter scale PCR with TaqMan detection. Nucleic Acids Res 1997; 25:1999-2004; PMID:9115368; http://dx.doi.org/ 10.1093/nar/25.10.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci USA 1999; 96:9236-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Pettersson E, Lundeberg J, Ahmadian A. Generations of sequencing technologies. Genomics 2009; 93:105-11; PMID:18992322; http://dx.doi.org/ 10.1016/j.ygeno.2008.10.003 [DOI] [PubMed] [Google Scholar]
- [17].San Martín A, Sotelo-Hitschfeld T, Lerchundi R, Fernández-Moncada I, Ceballo S, Valdebenito R, Baeza-Lehnert F, Alegría K, Contreras-Baeza Y, Garrido-Gerter P, et al.. Single-cell imaging tools for brain energy metabolism: a review. Neurophotonics 2014; 1:011004; http://dx.doi.org/ 10.1117/1.NPh.1.1.011004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Huang B, Wu H, Bhaya D, Grossman A, Granier S, Kobilka BK, Zare RN. Counting low-copy number proteins in a single cell. Science 2007; 315:81-4; PMID:17204646; http://dx.doi.org/ 10.1126/science.1133992 [DOI] [PubMed] [Google Scholar]
- [19].Polat AN, Özlü N. Towards single-cell LC-MS phosphoproteomics. Analyst 2014; 139:4733-49; PMID:25068147; http://dx.doi.org/ 10.1039/C4AN00463A [DOI] [PubMed] [Google Scholar]
- [20].Devi RV, Doble M, Verma RS. Nanomaterials for early detection of cancer biomarker with special emphasis on gold nanoparticles in immunoassays/sensors. Biosens Bioelectron 2015; 68:688-98; PMID:25660660; http://dx.doi.org/ 10.1016/j.bios.2015.01.066 [DOI] [PubMed] [Google Scholar]
- [21].Klepárník K. Recent advances in combination of capillary electrophoresis with mass spectrometry: methodology and theory. Electrophoresis 2015; 36:159-78; http://dx.doi.org/ 10.1002/elps.201400392 [DOI] [PubMed] [Google Scholar]
- [22].Crowther JR. The ELISA Guidebook, 2nd edition Springer, New York, USA: 2009 [Google Scholar]
- [23].Bürgi W, Briner M, Franken N, Kessler AC. One-step sandwich enzyme immunoassay for insulin using monoclonal antibodies. Clin Biochem 1988; 21:311-4; PMID:3069245; http://dx.doi.org/ 10.1016/S0009-9120(88)80087-0 [DOI] [PubMed] [Google Scholar]
- [24].Storch MJ, Alexopoulos A, Kerp L. Evaluation of a solid-phase monoclonal antibody-based enzyme immunoassay for insulin in human serum. J Immunol Methods 1989; 119:53-7; PMID:2651525; http://dx.doi.org/ 10.1016/0022-1759(89)90380-3 [DOI] [PubMed] [Google Scholar]
- [25].Lowry OH, Passonneau JV, Schulz DW, Rock MK. The measurement of pyridine nucleotides by enzymatic cycling. J Biol Chem 1961; 236:2746-55; PMID:144669814351123 [PubMed] [Google Scholar]
- [26].Kato T, Berger SJ, Carter JA, Lowry OH. An enzymatic cycling method for nicotinamide-adenine dinucleotide with malic and alcohol dehydrogenases. Anal Biochem 1973; 53:86-97; PMID:4351123; http://dx.doi.org/ 10.1016/0003-2697(73)90409-0 [DOI] [PubMed] [Google Scholar]
- [27].Mashige F, Imai K, Osuga T. A simple and sensitive assay of total serum bile acids. Clin Chim Acta 1976; 70:79-86; PMID:947625; http://dx.doi.org/ 10.1016/0009-8981(76)90007-3 [DOI] [PubMed] [Google Scholar]
- [28].Mashige F, Tanaka N, Maki A, Kamei S, Yamanaka M. Direct spectrophotometry of total bile acids in serum. Clin Chem 1981; 27:1352-6; PMID:6895053 [PubMed] [Google Scholar]
- [29].Yoshimura T, Kurosawa T, Ikegami S. Tohma M. Substrate specificity of 3a-hydroxysteroid dehydrogenase for the oxidation of fetal bile acids. Bunseki Kagaku 1995; 44:865-9; http://dx.doi.org/ 10.2116/bunsekikagaku.44.865 [DOI] [Google Scholar]
- [30].Ueda S, Oda M, Imamura S, Ohnishi M. Kinetic study of the enzymatic cycling reaction conducted with 3a-hydroxysteroid dehydrogenase in the presence of excessive thio-NAD+ and NADH. Anal Biochem 2004; 332:84-89; PMID:15301952; http://dx.doi.org/ 10.1016/j.ab.2004.04.035 [DOI] [PubMed] [Google Scholar]
- [31].Zhang GH, Cong AR, Xu GB, Li CB, Yang RF, Xia TA. An enzymatic cycling method for the determination of serum total bile acids with recombinant 3α-hydroxysteroid dehydrogenase. Biochem Biophys Res Commun 2005; 326:87-92; PMID:15567156; http://dx.doi.org/ 10.1016/j.bbrc.2004.11.005 [DOI] [PubMed] [Google Scholar]
- [32].Matsuoka T, Ueda S, Matsumoto H, Kawakami M. An ultrasensitive enzymatic method for measuring mevalonic acid in serum. J Lipid Res 2012; 53:1987-92; PMID:22715156; http://dx.doi.org/ 10.1194/jlr.D028621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Iwai A, Yoshimura T, Wada K, Watabe S, Sakamoto Y, Ito E, Miura T. Spectrophotometric method for the assay of steroid 5α-reductase activity of rat liver and prostate microsomes. Anal Sci 2013; 29:455-9; PMID:23574674; http://dx.doi.org/ 10.2116/analsci.29.455 [DOI] [PubMed] [Google Scholar]
- [34].Skålhegg BA. 3a-hydroxysteroid dehydrogenase from Pseudomonas testosteroni: kinetic properties with NAD and its thionicotinamide analogue. Eur J Biochem 1975; 50:603-9; http://dx.doi.org/ 10.1111/j.1432-1033.1975.tb09901.x [DOI] [PubMed] [Google Scholar]
- [35].Watabe S, Kodama H, Kaneda M, Morikawa M, Nakaishi K, Yoshimura T, Iwai A, Miura T, Ito E. Ultrasensitive enzyme-linked immunosorbent assay (ELISA) of proteins by combination with the thio-NAD cycling method. Biophysics 2014; 10:49-54; http://dx.doi.org/ 10.2142/biophysics.10.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Niemeyer CM, Adler M, Wacker R. Immuno-PCR: high sensitivity detection of proteins by nucleic acid amplification. Trends Biotechnol 2005; 23:208-16; PMID:15780713; http://dx.doi.org/ 10.1016/j.tibtech.2005.02.006 [DOI] [PubMed] [Google Scholar]
- [37].Rissin DM, Kan CW, Campbell TG, Howes SC, Fournier DR, Song L, Piech T, Patel PP, Chang L, Rivnak AJ, et al.. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat Biotechnol 2010; 28:595-599; PMID:20495550; http://dx.doi.org/ 10.1038/nbt.1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Dwarakanathan A. Diabetes update. J Insur Med 2006; 38:20-30; PMID:16642640 [PubMed] [Google Scholar]
- [39].Cersosimo E, Solis-Herrera C, Trautmann ME, Malloy J, Triplitt CL. Assessment of pancreatic β-cell function: review of methods and clinical applications. Curr Diabetes Rev 2014; 10:2-42; PMID:24524730; http://dx.doi.org/ 10.2174/1573399810666140214093600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ito E, Kaneda M, Kodama H, Morikawa M, Tai M, Aoki K, Watabe S, Nakaishi K, Hashida S, Tada S, et al.. Immunoreactive insulin in DM-patient sera detected by ultrasensitive ELISA with thio-NAD cycling. BioTechniques 2015; 59:359-67; PMID:26651515; http://dx.doi.org/19535523 10.2144/000114355 [DOI] [PubMed] [Google Scholar]
- [41].Pandori MW, Hackett J Jr, Louie B, Vallari A, Dowling T, Liska S, Klausner JD. Assessment of the ability of a fourth-generation immunoassay for human immunodeficiency virus (HIV) antibody and p24 antigen to detect both acute and recent HIV infections in a high-risk setting. J Clin Microbiol 2009; 47:2639-42; PMID:19535523; http://dx.doi.org/ 10.1128/JCM.00119-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].George CRR, Robertson PW, Lusk MJ, Whybin R, Rawlinson W. Prolonged second diagnostic window for human immunodeficiency virus type 1 in a fourth-generation immunoassay: Are alternative testing strategies required? J Clin Microbiol 2014; 52:4105-8; PMID:25210068; http://dx.doi.org/ 10.1128/JCM.01573-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Nakatsuma A, Kaneda M, Kodama H, Morikawa M, Watabe S, Nakaishi K, Yamashita M, Yoshimura T, Miura T, Ninomiya M, et al.. Detection of HIV-1 p24 at attomole level by ultrasensitive ELISA with thio-NAD cycling. Plos One 2015; 10:e0131319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Nakatsuma A, Kaneda M, Kodama H, Morikawa M, Watabe S, Nakaishi K, Yamashita M, Yoshimura T, Miura T, Ninomiya M, et al.. Ultrasensitive colorimetric detection of HIV-1 p24. Clin Lab Int 2015; 10(6):20-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Matsuzawa Y, Funahashi T, Nakamura T. Molecular mechanism of metabolic syndrome X: contribution of adipocytokines adipocyte-derived bioactive substances. Ann N Y Acad Sci 1999; 892:146-54; PMID:10842660; http://dx.doi.org/ 10.1111/j.1749-6632.1999.tb07793.x [DOI] [PubMed] [Google Scholar]
- [46].Cnop M, Havel PJ, Utzschneider KM, Carr DB, Sinha MK, Boyko EJ, Retzlaff BM, Knopp RH, Brunzell JD, Kahn SE. Relationship of adiponectin to body fat distribution, insulin sensitivity and plasma lipoproteins: evidence for independent roles of age and sex. Diabetologia 2003; 46:459-69; PMID:12687327 [DOI] [PubMed] [Google Scholar]
- [47].Koshimura J, Fujita H, Narita T, Shimotomai T, Hosoba M, Yoshioka N, Kakei M, Fujishima H, Ito S. Urinary adiponectin excretion is increased in patients with overt diabetic nephropathy. Biochem Biophys Res Commun 2004; 316:165-9; PMID:15003525; http://dx.doi.org/ 10.1016/j.bbrc.2004.02.032 [DOI] [PubMed] [Google Scholar]
- [48].Shimotomai T, Kakei M, Narita T, Koshimura J, Hosoba M, Kato M, Komatsuda A, Ito S. Enhanced urinary adiponectin excretion in IgA-nephropathy patients with proteinuria. Ren. Fail 2005; 27:323-8; http://dx.doi.org/ 10.1081/JDI-200056597 [DOI] [PubMed] [Google Scholar]
- [49].Morikawa M, Naito R, Mita K, Watabe S, Nakaishi K, Yoshimura T, Miura T, Hashida S, Ito E. Subattomole detection of adiponectin in urine by ultrasensitive ELISA coupled with thio-NAD cycling. Biophys Physicobiol 2015; 12:79-86; http://dx.doi.org/ 10.2142/biophysico.12.0_79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Free AH. Enzymatic determination of glucose. Adv Clin Chem. 1963; 6:67-96; PMID:14281819; http://dx.doi.org/ 10.1016/S0065-2423(08)60238-3 [DOI] [PubMed] [Google Scholar]
- [51].Zhou M, Diwu Z, Panchuk-Voloshina N, Haugland RP. A stable nonfluorescent derivative of resorfin for the fluorometric de termination of trace hydrogen peroxide: Applications in detecting the activity of phagocyte NADPH oxidase and other oxidases. Anal Biochem 1997; 253:162-8; PMID:9367498; http://dx.doi.org/ 10.1006/abio.1997.2391 [DOI] [PubMed] [Google Scholar]
- [52].Nagaraja P, Shivakumar A, Shrestha AK. Quantification of hydrogen peroxide and glucose using 3-methyl-2-benzothiazolinonehydrazone hydrochloride with 10,11-dihydro-5H-benz(b,f)azepine as chromogenic probe. Anal Biochem 2009; 395:231-6; PMID:19686697; http://dx.doi.org/ 10.1016/j.ab.2009.07.053 [DOI] [PubMed] [Google Scholar]
- [53].Luo W, Li YS, Yuan J, Zhu L, Liu Z, Tamg H, Liu S. Ultrasensitive fluorometric determination of hydrogen peroxide and glucose by using multiferroic BiFeO3 nanoparticles as a catalyst. Talanta 2010; 81:901-7; PMID:20298871; http://dx.doi.org/ 10.1016/j.talanta.2010.01.035 [DOI] [PubMed] [Google Scholar]
- [54].Hu Y, Zhang Z. Determi nation of free cholesterol based on a novel flow-injection chemiluminescence method by immobilizing enzyme. Luminescence 2008; 23:338-43; PMID:18500700; http://dx.doi.org/ 10.1002/bio.1042 [DOI] [PubMed] [Google Scholar]
- [55].Kim SA, Heinze KG, Schwille P. Fluorescence correlation spectroscopy in living cells. Nature Methods 2007; 4:963-73; PMID:17971781; http://dx.doi.org/ 10.1038/nmeth1104 [DOI] [PubMed] [Google Scholar]
- [56].Nomura Y, Nakamura T, Feng Z, Kinjo M. Direct quantification of gene expression using fluorescence correlation spectroscopy. Curr Pharm Biotechnol 2007; 8:286-90; PMID:17979726; http://dx.doi.org/ 10.2174/138920107782109958 [DOI] [PubMed] [Google Scholar]
- [57].Sadamoto H, Saito K, Muto H, Kinjo M, Ito E. Direct observation of dimerization between different CREB1 isoforms in a living cell. PLoS ONE 2011; 6:e20285; PMID:21673803; http://dx.doi.org/ 10.1371/journal.pone.0020285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Watabe S, Sakamoto Y, Morikawa M, Okada R, Miura T, Ito E. Highly sensitive determination of hydrogen peroxide and glucose by fluorescence correlation spectroscopy. PLoS One 2011; 6:e22955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Ito E, Watabe S, Morikawa M, Kodama H, Okada R, Miura T. Detection of H2O2 by fluorescence correlation spectroscopy. Methods Enzymol 2013; 526:135-43; PMID:23791098; http://dx.doi.org/ 10.1016/B978-0-12-405883-5.00008-9 [DOI] [PubMed] [Google Scholar]
- [60].Yan Q, Peng B, Su G, Cohan BE, Major TC, Meyerhoff ME. Measurement of tear glucose levels with amperometric glucose biosensor/capillary tube configuration. Anal Chem 2011; 83:8341-6; PMID:21961809; http://dx.doi.org/ 10.1021/ac201700c [DOI] [PubMed] [Google Scholar]


