Abstract
Calorie restriction extends lifespan in a broad range of organisms, from yeasts to mammals. Numerous hypotheses have been proposed to explain this phenomenon, including decreased oxidative damage and altered energy metabolism. In Saccharomyces cerevisiae, lifespan extension by calorie restriction requires the NAD+-dependent histone deacetylase, Sir2 (ref. 1). We have recently shown that Sir2 and its closest human homologue SIRT1, a p53 deacetylase, are strongly inhibited by the vitamin B3 precursor nicotinamide2. Here we show that increased expression of PNC1 (pyrazinamidase/nicotinamidase 1), which encodes an enzyme that deaminates nicotinamide, is both necessary and sufficient for lifespan extension by calorie restriction and low-intensity stress. We also identify PNC1 as a longevity gene that is responsive to all stimuli that extend lifespan. We provide evidence that nicotinamide depletion is sufficient to activate Sir2 and that this is the mechanism by which PNC1 regulates longevity. We conclude that yeast lifespan extension by calorie restriction is the consequence of an active cellular response to a low-intensity stress and speculate that nicotinamide might regulate critical cellular processes in higher organisms.
Lifespan in the budding yeast S. cerevisiae is extended by a variety of stimuli such as heat stress, osmotic stress and the restriction of amino acids or glucose1,3–5. The latter two regimens are considered to be mimics of calorie restriction in higher organisms. In S. cerevisiae, replicative age is defined as the number of divisions that a cell undergoes before dying. The yeast SIR2 gene, which encodes the founding member of a conserved family of NAD+-dependent deacetylases6–9, is required for lifespan extension by glucose restriction1. Cells with an additional copy of SIR2 live 30% longer than the wild type, whereas sir2Δ strains age prematurely10 owing to increased recombination at the ribosomal DNA (rDNA) locus10,11. The importance of elucidating the yeast SIR2 pathway is underscored by increasing evidence that Sir2 proteins in higher organisms promote longevity and cell viability12–15.
Because Sir2 protein levels do not increase in response to calorie restriction16, lifespan extension must involve an increase in enzymatic activity of Sir2. One hypothesis is that Sir2 is activated by an increased availability of NAD+ (ref. 1). Nicotinamide, a product of the Sir2 reaction17, is a strong non-competitive inhibitor of Sir2-like enzymes in vitro2,17 and can accelerate yeast ageing by inhibiting Sir2 in vivo2. Thus an alternative explanation is that Sir2 is regulated by changes in nicotinamide levels.
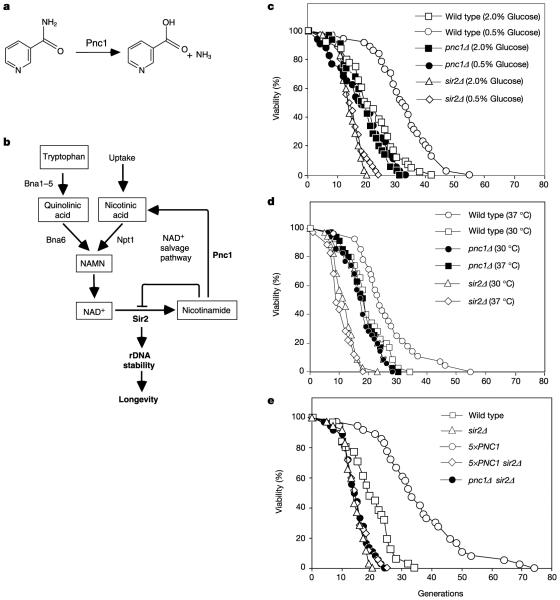
To explore the latter hypothesis, we focused on PNC1, a gene whose product converts nicotinamide to nicotinic acid in the NAD+ salvage pathway (Fig. 1a, b). Most wild-type yeast strains have an average lifespan of 21–23 divisions, with a maximum lifespan of about 40 divisions. A wild-type strain that was calorie restricted (0.5% glucose) or heat stressed (37 °C) exhibited a longer lifespan than an untreated control (2.0% glucose or 30 °C, respectively; Fig. 1c, d). The sir2Δ strain had a short lifespan, consistent with previous reports1,10, and neither calorie restriction (0.5% or 0.1% glucose) nor heat stress extended lifespan in this strain (Fig. 1c, d, and data not shown). The pnc1Δ strain did not exhibit a lifespan extension under either of these conditions, demonstrating that PNC1 is necessary for lifespan extension by calorie restriction and low-intensity stress.
Figure 1.
Calorie restriction and heat stress extend lifespan in a PNC1-dependent manner. a, Pnc1 converts nicotinamide to nicotinic acid. b, In S. cerevisiae, NAD+ is synthesized de novo from tryptophan via Bna1–6 or recycled from nicotinamide. c, Average lifespan on 2.0% (w/v) glucose: wild type, 21.6 generations; pnc1Δ, 19.1; sir2Δ, 14.2. Average lifespan on 0.5% glucose: wild type, 32.7 generations; pnc1D, 18.1; sir2Δ, 14.7. d, At 30 °C, average lifespans: wild type, 19.4 generations; pnc1Δ, 18.5; sir2Δ, 12.0. At 37 °C, average lifespans: wild type, 23.4 generations; pnc1Δ, 17.5; sir2Δ, 10.6. e, Average lifespans on 2.0% glucose at 30 °C: wild type, 19.7 generations; 5×PNC1, 36.1; sir2Δ, 14.2; 5×PNC1 sir2Δ, 15.1; pnc1Δ sir2Δ, 14.4.
Strikingly, under non-stressing conditions (2% glucose, 30 °C), a strain with additional copies of PNC1 (5×PNC1) lived 70% longer than the wild type and some cells lived for more than 70 divisions, which is the longest reported lifespan extension in this organism (Fig. 1e). Neither calorie restriction nor heat stress further increased the lifespan of the 5×PNC1 strain (not shown). Deletion of SIR2 in the 5×PNC1 background shortened lifespan to that of the sir2Δ mutant (Fig. 1e). Furthermore, the pnc1Δ sir2Δ double mutant had a lifespan similar to that of the sir2Δ mutant (Fig. 1e) and its lifespan was unaffected by glucose restriction (not shown). These findings indicate that PNC1 and SIR2 function in the same pathway and that PNC1 increases lifespan through SIR2. Together, these results show that PNC1 is necessary for lifespan extension by both calorie restriction and heat stress, and that additional PNC1 is sufficient to mimic these stimuli.
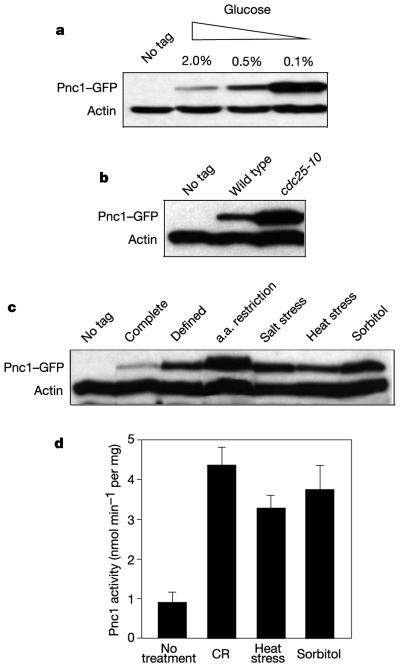
Given that additional PNC1 is sufficient to extend lifespan, we examined whether PNC1 expression is upregulated in response to stimuli that extend lifespan. We found that Pnc1 levels were greatly induced in a dose-dependent manner by glucose restriction (Fig. 2a) and in cells carrying a cdc25-10 allele, which mimics calorie restriction1 (Fig. 2b). MSN2 and MSN4, which encode transcription factors that coordinate the response to carbon source starvation and intense stress, were not required for Pnc1 induction (not shown). This is consistent with the previous observation that these two genes are not required for lifespan extension by glucose restriction1.
Figure 2.
Pnc1 levels and activity are elevated in response to calorie restriction and low-intensity stress. a, Western analysis of Pnc1–GFP under conditions of glucose restriction. b, Pnc1–GFP in wild-type or cdc25-10 cells. c, Detection of Pnc1–GFP in cells subjected to mild stress as indicated (a.a., amino acid). d, Measurement of nicotinamide deamination by Pnc1. Activity (nmol ammonia min−1 per mg protein) from three experiments (means ± s.d): no treatment (2% glucose), 0.9 ± 0.26; calorie restriction (CR; 0.1% glucose), 4.38 ± 0.43; heat stress (37 °C), 3.28 ± 0.32; sorbitol (1 M), 3.75 ± 0.65.
Pnc1 levels were elevated under every other condition known to extend yeast lifespan, including amino acid restriction, salt stress and heat stress (Fig. 2c), in agreement with whole-genome mRNA analyses of stressed yeast cells18. Pnc1 activity in extracts from treated cells was correlated with Pnc1 concentrations in western blots (Fig. 2d), showing that these cells have increased rates of nicotinamide hydrolysis.
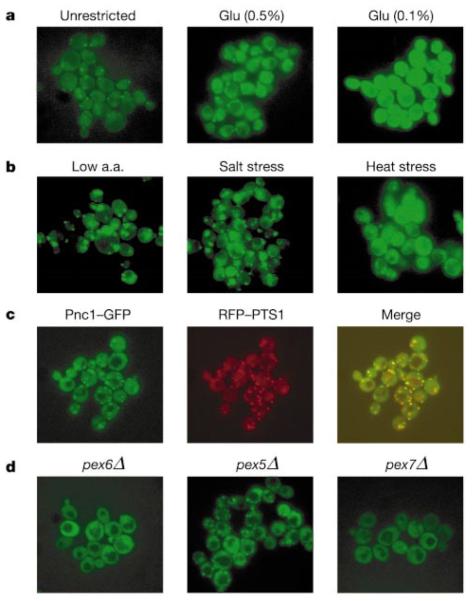
We and others have previously shown that two other enzymes in the NAD+ salvage pathway, Npt1 (nicotinic acid phosphoribosyltransferase) and Nma2 (nicotinic acid mononucleotide adenylyltransferase), are concentrated in the nucleus16,19. Surprisingly, a fusion protein of Pnc1 with green fluorescent protein (Pnc1–GFP) was not only localized in the nucleus and the cytoplasm but was also concentrated in three to six discrete cytoplasmic foci per cell (Fig. 3a–d). Calorie-restricted (Fig. 3a) or stressed (Fig. 3b) cells showed a marked increase in the intensity of fluorescence, consistent with the western data. Interestingly, under conditions of amino acid restriction or salt stress, the fluorescence was predominantly localized to the foci (Fig. 3b), suggesting that Pnc1 localization is regulated.
Figure 3.
Pnc1–GFP is localized in the nucleus and cytoplasm, and concentrated in peroxisomes. a, Pnc1–GFP fluorescence in glucose-restricted cells (Glu 0.5% and 0.1%). b, Pnc1–GFP fluorescence under conditions of mild stress (a.a., amino acid). c, Co-localization of Pnc1–GFP (green) and RFP–PTS1 (red). Yellow indicates overlap. d, Localization of Pnc1–GFP in cells from peroxisomal mutant strains, pex6Δ, pex5Δ and pex7Δ.
To determine the identity of the foci, we searched for cellular markers that co-localized with Pnc1–GFP and observed significant overlap with a peroxisomally targeted red fluorescent protein (RFP) (Fig. 3c). Pnc1–GFP foci were no longer observed in a peroxisome-deficient pex6Δ mutant, confirming that Pnc1–GFP was peroxisomal (Fig. 3d). Because our studies indicated that the localization of Pnc1 to peroxisomes might be regulated, we sought to identify the transporter responsible for its import into this organelle. Although Pex5 imports the vast majority of peroxisomal proteins, the localization of Pnc1 to peroxisomes required the less-used transporter Pex7 (Fig. 3d). The localization of Pnc1 to sites outside the nucleus implies that this enzyme could regulate proteins other than Sir2 (such as the homologues of Sir2, Hst1–Hst4). The peroxisomal localization of Pnc1 is of particular interest because these organelles are a major source of reactive oxygen species and have been implicated in mammalian ageing20.
Because PNC1 converts nicotinamide to nicotinic acid as part of the NAD+ salvage pathway, it could theoretically activate Sir2 either by increasing the availability of its co-substrate, NAD+, or by depleting levels of the inhibitor nicotinamide. Although these mechanisms are not mutually exclusive, and mutations that alter NAD+ levels can affect silencing1,3,19,21, current evidence favours the nicotinamide model. We and others have been unable to detect increases in NAD+ levels or the NAD+/NADH ratio in calorie-restricted cells22 or in genetic mimics of calorie restriction16, even when unbound NAD+ was measured (R.M.A., A. R. Neves, H. Santos and D.A.S., unpublished observations). In addition, we have previously shown that Sir2 is inhibited in vitro by physiological concentrations of nicotinamide and that exogenous nicotinamide can abolish silencing in vivo2. Perhaps most persuasive is the observation that cells lacking PNC1 have a silencing defect, yet they show no change in NAD+ levels19. Although these observations are supportive of the nicotinamide model, we sought more conclusive evidence.
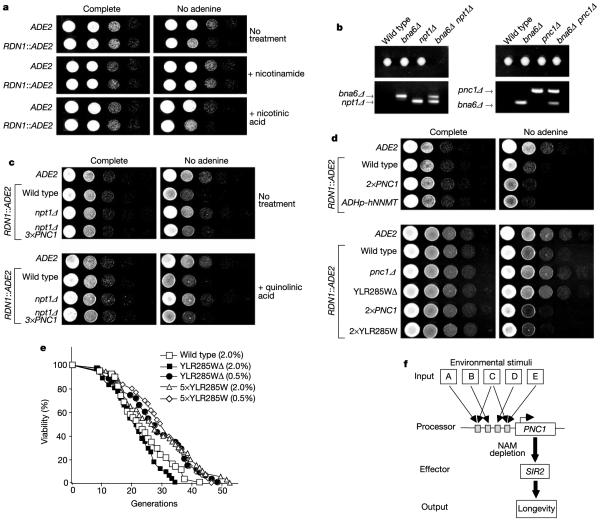
First, we reasoned that if Pnc1 activates Sir2 by stimulating the NAD+ salvage pathway (by converting nicotinamide to nicotinic acid), then an increase in the intracellular nicotinic acid pool should have the same effect as increasing Pnc1 levels (see Fig. 1b). Exogenous nicotinic acid is readily taken up by yeast cells and can significantly increase the intracellular pool (R.M.A., A. R. Neves, H. Santos and D.A.S., unpublished observations)23. A common indicator of Sir2 activity is the extent to which a reporter gene inserted at the rDNA locus (RDN1) is silenced. As shown in Fig. 4a, exogenous nicotinic acid did not increase rDNA silencing, indicating that nicotinic acid is not limiting for the salvage pathway. Furthermore, genetic analysis demonstrates that the contribution of PNC1 to NAD+ synthesis is minimal, even in the absence of NAD+ synthesis de novo (Fig. 4b). Taken together, these data argue against a model in which Pnc1 stimulates Sir2 by providing additional nicotinic acid for NAD+ synthesis.
Figure 4.
Manipulation of nicotinamide metabolism alters silencing and lifespan. a, To monitor silencing, ADE2 was integrated at the rDNA locus. Increased growth on medium lacking adenine indicates decreased silencing. Serial dilutions of cells spotted on plates containing nicotinic acid or nicotinamide (5 mM). b, PNC1 does not have a critical role in NAD+ biosynthesis, even in the absence of the de novo NAD+ synthesis pathway. BNA6 encodes an enzyme in the NAD+ de novo synthesis pathway (see Fig. 1b). NPT1 encodes a phosphoribosyltransferase that converts nicotinic acid to nicotinic acid mononucleotide in the NAD+ salvage pathway. Spore colonies from heterozygous bna6Δ npt1Δ or bna6Δ pnc1Δ diploids. Genotypes determined by PCR with a colony/ microcolony genomic template. c, Partial rescue of silencing by additional PNC1 in the absence of the NAD+ salvage pathway and complete rescue in the presence of quinolinic acid (5 mM). d, Manipulation of genes involved in nicotinamide metabolism alters rDNA silencing. e, Manipulation of YLR285W affects lifespan. f, Model for the regulation of Sir2 activity and lifespan by PNC1 and nicotinamide (NAM).
Second, we tested whether the manipulation of PNC1 could increase silencing even when its contribution to NAD+ synthesis was blocked. In S. cerevisiae, the only other NAD+ salvage pathway gene that can be deleted without a loss of viability is NPT1 (see Fig. 1b). We have previously shown that additional copies of PNC1 increase rDNA silencing in wild-type cells16. Additional copies of PNC1 led to a partial rescue of the silencing defect in the npt1Δ strain (Fig. 4c). Because cells lacking NPT1 have NAD+ levels one-half of those in wild-type strains6, we included an NAD+ precursor, quinolinic acid, in the medium, which in mammals has been shown to compensate for a low NAD+ concentration24. In the presence of this compound, additional PNC1 restored rDNA silencing in the npt1Δ strain to near wild-type levels (Fig. 4c), showing that Pnc1 can increase Sir2 activity even in the absence of the NAD+ salvage pathway.
Last, if PNC1 regulates Sir2 activity by modulating nicotinamide levels, we reasoned that manipulation of nicotinamide using a gene outside the NAD+ salvage pathway should have the same effect. In humans, nicotinamide is converted to N-methylnicotinamide by nicotinamide N-methyltransferase (NNMT)25 and then excreted. As predicted by the nicotinamide model, overexpression of human NNMT in yeast increased rDNA silencing (Fig. 4d). By homology we also identified a putative S. cerevisiae NNMT gene, YLR285W. The predicted protein contains the four signature motifs of S-adenosylmethionine-dependent methyltranferases26 and its core domain is 23% identical to that of human NNMT25. Additional copies of YLR285W increased silencing, whereas deletion of this gene led to a loss of silencing, similar to the effect of manipulating PNC1 (ref. 16) (Fig. 4d). Additional copies of YLR285W increased yeast lifespan and this effect was not enhanced by glucose restriction (Fig. 4e). Unlike PNC1, YLR285W is not a true longevity regulator because its expression is not apparently modulated by stimuli that extend lifespan18, and its deletion does not abolish lifespan extension by glucose restriction (Fig. 4e).
Our results show that lifespan extension by either calorie restriction or mild stress is the result of an active cellular response that requires the upregulation of a specific longevity gene, PNC1 (Fig. 4f). This system of longevity regulation explains how multiple, disparate stimuli can lead to the same longevity response and how species can rapidly evolve strategies to suit a changing environment. We also provide multiple lines of evidence that PNC1 regulates Sir2 by modulating intracellular nicotinamide. It has been proposed that Sir2 is regulated by passive means, by changes in either NAD+ availability1,3,6,21 or the NAD+/NADH ratio1,3,21. We do not exclude the possibility that these mechanisms can function in tandem with nicotinamide depletion. However, an attractive feature of nicotinamide-based regulation is that it does not require the modulation of NAD+, an essential cofactor involved in cellular homeostasis.
Nicotinamide has been shown to promote apoptosis in mammalian cells by inhibiting the Sir2 homologue SIRT1 (refs 2, 15), a regulator of p53 (refs 14, 15). Moreover, the poly(ADP-ribose) polymerase family of proteins, which are involved in many processes including DNA repair, telomere-length regulation and the opening of chromatin associated with stress-activated genes, are also inhibited by nicotinamide27. Interestingly, an increased expression of NNMT is correlated with tumorigenesis28 and a decreased expression is correlated with radiosensitivity29. These findings raise the possibility that nicotinamide regulates critical cellular processes in higher organisms.
Methods
Media and strains
All strains were grown at 30 °C in complete 2.0% (w/v) glucose (YPD) medium except where stated otherwise. Glucose restriction medium contained 0.5% or 0.1% glucose. Mild stress conditions were one of the following: defined medium (SD); amino acid restriction (SD lacking non-essential amino acids); salt stress (NaCl, 300 mM); heat stress (37 °C); sorbitol (1 M). In all experiments, auxotrophic markers were matched between strains by integrating empty vector. The copy number of integrated genes was determined by Southern blotting. Deletions were generated with a kan-MX6 PCR-based technique16 and confirmed by PCR. Additional copies of PNC1 were integrated as described previously16. The entire open reading frame and 700 bases of the upstream sequence of YLR285W were amplified from genomic DNA and cloned into pSP400, then sequenced and integrated as described previously16. A GFP cassette was integrated in frame at the 3 ′ end of the native PNC1 gene as described previously16. The RFP–PTS1 (for peroxisomal targeting signal 1) plasmid (pSG421) was a gift from S. J. Gould (Johns Hopkins University). The coding region of human NNMT was subcloned from p91023(B), a gift from R. Weinshilboum (Mayo Clinic), into pSP400 downstream of the ADH1 promoter. Strains derived from PSY316AT16 were used for lifespan analysis. Strains derived from W303AR11 were used for western blots, microscopy and silencing assays. W303AR cdc25-10 was a gift from L. Guarente (MIT).
Yeast assays
Lifespan measurements were performed as described previously16 except for the heat-stress experiments, in which strains were incubated after each dissection at 37 °C. Silencing was assayed as described previously16.
Protein expression analysis
Strains were pretreated under the indicated conditions and grown to mid-exponential phase. Western blots were performed as described16 with whole-cell extracts (75 mg). Proteins were detected with anti-GFP antibodies (Santa Cruz) or anti-actin antibodies (Chemicon). Fluorescent microscopy images were captured at the same exposure (1 s) at ×100 magnification with a Hamamatsu Orca100 CCD camera and processed with Openlab software. Cultures were grown in complete medium containing 0.5% glucose to enhance the detection of fluorescence.
Nicotinamidase activity assay
The activity of Pnc1 in extracts obtained from pretreated mid-exponential-phase cultures was determined as described previously30. In brief, 0.16 mg of protein was incubated with either 0 or 8 mM nicotinamide for 45 min at 30 °C in a final volume of 400 μl of 10 mM Tris-HCl pH 7.5, 150 mM NaCl and 1 mM MgCl2. Pnc1 activity was determined by measuring the final concentration of the reaction product, ammonia, with an ammonia diagnostic kit (Sigma). Baseline ammonia was accounted for by subtracting a no-nicotinamide control. Nicotinamidase activity was expressed as nmol ammonia min−1 per mg total protein. Pnc1 activity was obtained by subtracting the background value for the pnc1Δ strain (0.06 ± 0.004 nmol min−1 per mg).
Acknowledgements
We thank members of the Sinclair laboratory, R. Veech, C. Wolberger, W. Forrester, S. Luikenhuis and D. Finkelstein, for reagents and discussions. This work was supported by the NIA and the Harvard–Armenise Foundation. D.S. is an Ellison Medical Research Foundation Special Fellow. R.A. is supported by a John Taplan Postdoctoral Fellowship, J.W. by a National Science Foundation Scholarship, and K.B. and O.M. by the American Federation of Aging Research.
Footnotes
Competing interests statement The authors declare that they have no competing financial interests.
References
- 1.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 2.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast Sir2 and human SIRT1. J. Biol. Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 3.Kaeberlein M, Andalis AA, Fink GR, Guarente L. High osmolarity extends life span in Saccharomyces cerevisiae by a mechanism related to calorie restriction. Mol. Cell. Biol. 2002;22:8056–8066. doi: 10.1128/MCB.22.22.8056-8066.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang JC, Jaruga E, Repnevskaya MV, Jazwinski SM. An intervention resembling caloric restriction prolongs life span and retards aging in yeast. FASEB J. 2000;14:2135–2137. doi: 10.1096/fj.00-0242fje. [DOI] [PubMed] [Google Scholar]
- 5.Swiecilo A, Krawiec Z, Wawryn J, Bartosz G, Bilinski T. Effect of stress on the life span of the yeast Saccharomyces cerevisiae. Acta Biochim. Pol. 2000;47:355–364. [PubMed] [Google Scholar]
- 6.Smith JS, et al. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl Acad. Sci. USA. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 8.Tanny JC, Moazed D. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: Evidence for acetyl transfer from substrate to an NAD breakdown product. Proc. Natl Acad. Sci. USA. 2001;98:415–420. doi: 10.1073/pnas.031563798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landry J, et al. The silencing protein Sir2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl Acad. Sci. USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaeberlein M, McVey M, Guarente L. The Sir2/3/4 complex and Sir2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sinclair DA, Guarente L. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 12.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 13.Rogina B, Helfand SL, Frankel S. Longevity regulation by Drosophila Rpd3 deacetylase and caloric restriction. Science. 2002;298:1745. doi: 10.1126/science.1078986. [DOI] [PubMed] [Google Scholar]
- 14.Vaziri H, et al. hSIR2(SIRT1) Functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 15.Luo J, et al. Negative control of p53 by Sir2α promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 16.Anderson RM, et al. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J. Biol. Chem. 2002;277:18881–18890. doi: 10.1074/jbc.M111773200. [DOI] [PubMed] [Google Scholar]
- 17.Landry J, Slama JT, Sternglanz R. Role of NAD+ in the deacetylase activity of the SIR2-like proteins. Biochem. Biophys. Res. Commun. 2000;278:685–690. doi: 10.1006/bbrc.2000.3854. [DOI] [PubMed] [Google Scholar]
- 18.Gasch AP, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sandmeier JJ, Celic I, Boeke JD, Smith JS. Telomeric and rDNA silencing in Saccharomyces cerevisiae are dependent on a nuclear NAD+ salvage pathway. Genetics. 2002;160:877–889. doi: 10.1093/genetics/160.3.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perichon R, Bourre JM, Kelly JF, Roth GS. The role of peroxisomes in aging. Cell Mol. Life Sci. 1998;54:641–652. doi: 10.1007/s000180050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin SJ, et al. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- 22.Lin SS, Manchester JK, Gordon JI. Enhanced gluconeogenesis and increased energy storage as hallmarks of aging in Saccharomyces cerevisiae. J. Biol. Chem. 2001;276:36000–36007. doi: 10.1074/jbc.M103509200. [DOI] [PubMed] [Google Scholar]
- 23.Unkefer CJ, London RE. In vivo studies of pyridine nucleotide metabolism in Escherichia coli and Saccharomyces cerevisiae by carbon-13 NMR spectroscopy. J. Biol. Chem. 1984;259:2311–2320. [PubMed] [Google Scholar]
- 24.Grant RS, Kapoor V. Murine glial cells regenerate NAD, after peroxide-induced depletion, using either nicotinic acid, nicotinamide, or quinolinic acid as substrates. J. Neurochem. 1998;70:1759–1763. doi: 10.1046/j.1471-4159.1998.70041759.x. [DOI] [PubMed] [Google Scholar]
- 25.Aksoy S, Szumlanski CL, Weinshilboum RM. Human liver nicotinamide N-methyltransferase. cDNA cloning, expression, and biochemical characterization. J. Biol. Chem. 1994;269:14835–14840. [PubMed] [Google Scholar]
- 26.Niewmierzycka A, Clarke S. S-Adenosylmethionine-dependent methylation in Saccharomyces cerevisiae. Identification of a novel protein arginine methyltransferase. J. Biol. Chem. 1999;274:814–824. doi: 10.1074/jbc.274.2.814. [DOI] [PubMed] [Google Scholar]
- 27.Virag L, Szabo C. The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol. Rev. 2002;54:375–429. doi: 10.1124/pr.54.3.375. [DOI] [PubMed] [Google Scholar]
- 28.Lal A, et al. A public database for gene expression in human cancers. Cancer Res. 1999;59:5403–5407. [PubMed] [Google Scholar]
- 29.Kassem H, Sangar V, Cowan R, Clarke N, Margison GP. A potential role of heat shock proteins and nicotinamide N-methyl transferase in predicting response to radiation in bladder cancer. Int. J. Cancer. 2002;101:454–460. doi: 10.1002/ijc.10631. [DOI] [PubMed] [Google Scholar]
- 30.Ghislain M, Talla E, Francois JM. Identification and functional analysis of the Saccharomyces cerevisiae nicotinamidase gene, PNC1. Yeast. 2002;19:215–224. doi: 10.1002/yea.810. [DOI] [PubMed] [Google Scholar]