Abstract
Many proteins suffer from sub-optimal pharmacokinetics (PK) that limit their utility as drugs. The efficient synthesis of polymer conjugates of protein drugs with tunable PK to optimize their in vivo efficacy is hence critical. We report here the first study of the in vivo behavior of a site-specific conjugate of a zwitterionic polymer and a protein. To synthesize the conjugate, we first installed an initiator for atom transfer radical polymerization (ATRP) at the N-terminus of myoglobin (Mb-N-Br). Subsequently, in situ ATRP was carried out in aqueous buffer to grow an amine-functionalized polymer from Mb-N-Br. The cationic polymer was further derivatized to two zwitterionic polymers by reaction of the amine groups of the cationic polymer with iodoacetic acid to obtain poly(carboxybetaine methylacrylate) with a 1 carbon spacer (C1) (PCBMA), and sequentially with 3-iodopropionic acid and iodoacetic acid to obtain PCBMA(mix) with a mixture of 1 carbon (C1) and 2 carbon (C2) spacer. The Mb-N-PCBMA polymer conjugates had a longer in vivo plasma half-life than a PEG-like comb polymer conjugate of similar MW. The structure of the zwitterion plays a role in controlling the in vivo behavior of the conjugate, as the PCBMA conjugate with a C1 spacer had significantly longer plasma circulation than the conjugate with a mixture of C1 and C2 spacers.
Keywords: Zwitterionic, Site-specific modification, Protein-polymer conjugate, ATRP, Pharmacokinetics
Protein and peptide therapeutics show high biological activity, but their therapeutic efficacy can be limited by poor stability, short in vivo circulation, and immunogenicity.[1] Poly(ethylene glycol) (PEG) conjugates increase the half-life of proteins by retarding filtration through the kidneys, by increasing their apparent molecular weight (MW), and by preventing opsonization and premature clearance through the reticulo-endothelial system.[2] However, PEG conjugates can generate a antibody response that can neutralize their efficacy upon chronic administration.[3] In a search for alternatives to PEG, zwitterionic polymers have attracted interest, because of their resistance to nonspecific protein adsorption.[4] However, with the exception of a paper by Godwin et al. on the conjugation of a poly(2-methyacryloyloxyethyl phosphorylcholine) to interferon-α2a,[5] there have been no studies of the in vivo behavior of zwitterionic polymer conjugates, where the structure of the zwitterion is varied, and the site and stoichiometry of the conjugate are tightly controlled.
The goal of this paper is two-fold: first, we sought to examine the ability of a site-specific zwitterionic polymer conjugate to impart a long plasma half-life to a protein and second, we sought to understand the role of the charge pair on the in vivo properties of the conjugate. PCBMA, the zwitterionic polymer was chosen for this study, contains a cationic trimethyl ammonium and an anionic carboxylic acid that are linked by a methylene group in its side-chain. Because the distance between the cation and anion in the charge pair is important in modulating the physicochemical properties of zwitterions such as their pKa and dipole moment,[6] we hypothesized that it might also play a role in mediating the in vivo response of PCBMA. We hence carried out a comparative study of the in vivo PK of two zwitterionic derivatives of a zwitterionic polymer —poly(carboxybetaine methylacrylate) (PCBMA)— wherein the distance between the ions in the charge pair in the repeat unit is varied. We benchmarked their in vivo performance against a protein conjugate of poly(oligo(ethylene glycol) methacrylate (POEGMA), a PEG-like comb polymer that we have previously shown has excellent protein resistance when grafted from a surface,[7] and long plasma half-life and good tumor accumulation when grafted from a protein.[8]
We synthesized site-specific (N-terminal) and stoichiometric (1:1 protein:polymer) Mb-N-PCBMA conjugates by a grafting from methodology, in which an ATRP initiator is attached to the N-terminus of Mb to synthesize Mb-N-Br followed by in situ ATRP from Mb-N-Br in aqueous buffer. We chose a post-polymerization strategy to synthesize Mb-N-PCBMA rather than growing PCBMA directly from Mb-N-Br, because initial scouting studies of ATRP of CBMA from the protein macroinitator failed due to the presence of -COOH groups that poison the ATRP catalyst.[9]
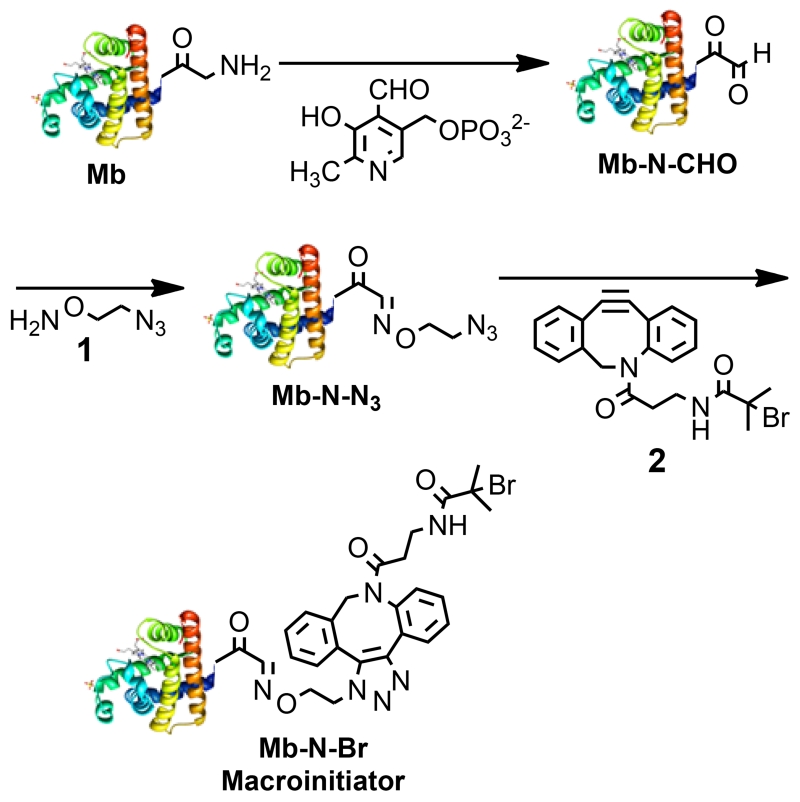
To synthesize a site-specific and stoichiometric PCBMA conjugate, first Mb-N-Br was synthesized in three steps from Mb, by a modified method from that previously reported (scheme 1).[8a] First, Mb was incubated with pyridoxal-5 phosphate (PLP) to yield an aldehyde (Mb-N-CHO).[8a, 10] The product was analyzed by high-performance liquid chromatography interfaced with a hybrid ion trap-orbitrap high resolution mass spectrometer (HPLC-IT-Orbitrap MS), which showed that Mb-N-CHO is in equilibrium with aldehyde hydrate, Mb-N-CH(OH)2 (peaks a and b, fig. S-1A). The measured m/z of peaks a and b are close to the theoretical MW of Mb-N-CHO and Mb-N-CH(OH)2 (16950.58 and 16968.60 Da, fig. S-1B and 1D) with only a −0.95 and −3.2 ppm mass error respectively (fig. S-1C and 1E). The product was next reacted with 1 to prepare Mb-N-N3. The product was analyzed by HPLC-IT-Orbitrap MS, and the major product (peak d, fig. S-3A) is close to the theoretical MW of Mb-N-N3 (17034.66 Da, fig. S-3B), with only a −7.7 ppm mass error (fig. S-3C).
Scheme 1.
Schematic illustration of N-terminal modification of Mb to Mb-N-Br.
Finally, Mb-N-Br was prepared by Cu-free click chemistry by reaction of Mb-N-N3 and 2.[11] The product was analyzed by HPLC-IT-Orbitrap MS and the measured MW of the major product (17459.99 Da, fig. S-5B) agrees with the theoretical MW, with only a −0.82 ppm mass error (fig. S-5C). We observed a peak that corresponds to unreacted Mb/Mb-N-CHO (1 Da mass difference, peak e, fig. S-5A) but no peak was observed that corresponds to Mb-N-N3, indicating quantitative conversion of Mb-N-N3 to Mb-N-Br. MALDI-TOF MS of Mb-N-Br showed a peak at 17460 (fig. 1A), which is close to the theoretical m/z of [Mb-N-Br+H]+ and thereby confirmed formation of Mb-N-Br. As a negative control, Mb was reacted with 1 and 2, skipping the reaction with PLP. MALDI and LC-MS showed that Mb-N-Br was not produced (fig. S-6A and 6B). The estimated conversion of the N-terminal amine of Mb to Mb-N-Br is ~70%.
Fig. 1.
(A) Matrix-assisted laser desorption/ionization-time-of-flight (MALDI-TOF) MS of Mb-N-Br and unreacted Mb/Mb-N-CHO. (B) Measured isotope distribution with a characteristic isotopic pattern of Br in the b2 fragment ion of Mb-N-Br as determined by collisionally activated dissociation ion-ion proton transfer CAD/IIPT Orbitrap mass spectrometry. The values in parentheses are the difference between the experimentally measured mass and theoretical values.
Mb contains 19 lysine residues that could cross-react with the reagents used to modifiy the N-terminus. To investigate this possibility, we performed tandem MS (MS/MS) of the major product Mb-N-Br (peak f in fig. S-5A) by collisionally activated dissociation ion-ion proton transfer (CAD/IIPT) on a modified IT-Orbitrap mass spectrometer that allows direct sequencing of intact Mb-N-Br, a methodology reported by Hunt et al. to sequence intact proteins without prior protein digestion.[12] We observed a series of b and c ions corresponding to N-terminal fragment ions of Mb-N-Br, starting from initiator-GL, initiator-GLS, initiator-GLSD etc (fig. S-7C). These b ions and c ions (fig. S-8) exclusively have masses 507.0906 Da (monoisotopic mass of initiator) heavier than the theoretical b and c ions of unmodified Mb. The N-terminal fragment ions also showed the characteristic isotopic distribution pattern of Br (fig. 1b). In addition, no b or c ions corresponding to unmodified N-terminal fragment ions were detected, which suggest that modification occured on one of the first two amino acid residues (i.e. Gly or Ala) at the N-terminal side of Mb-N-Br. As Gly and Ala do not contain reactive side-chains, these results strongly support that modification occurred solely at the N-terminus of Mb.
Next, ATRP from Mb-N-Br was carried out in aqueous buffer for 20 h and 5 h at 37 °C to graft PDMAEMA and POEGMA respectively (scheme 2). We found that these reaction conditions provide conjugates with a MW that is much greater than the renal filtration cutoff, so that the effect of renal clearance on their PK could be eliminated. The Mb-N-polymer conjugates were purified by preparative size exclusion chromatography (SEC) and characterized by analytical SEC (fig. 2 and S-9), 1H-nuclear magnetic resonance (NMR) spectroscopy (fig. S-10 and S-11), and sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (fig. S-12).
Scheme 2.
Schematic illustration of synthesis of Mb-N-PDMAEMA by in situ ATRP from Mb-N-Br followed by post-polymerization derivatization of Mb-N-PDMAEMA to Mb-N-PCBMA and Mb-N-PCBMA (mix).
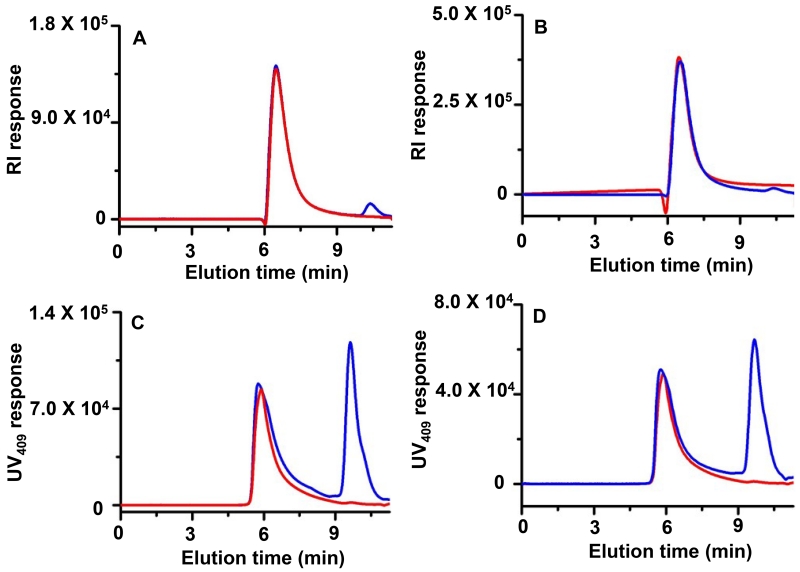
Fig. 2.
Characterization of Mb-N-PDMAEMA and Mb-N-POEGMA conjugates by SEC with RI (A and B) and UV detection at 409 nm (C and D) respectively. Conjugates before and after purification are shown in blue and red respectively.
Refractive index (RI) and UV-vis absorbance at 409 nm (HEME) and 280 nm (protein) enabled quantification of the unmodified Mb and Mb-N-polymer conjugates, as conjugates appeared at lower elution time than Mb in SEC (fig. 2 and S-9). Quantification of the peak area indicated that the yield of the Mb-N-PDMAEMA and Mb-N-POEGMA conjugate was ~65% and 70 % respectively; which are similar to those reported previously by in situ ATRP of OEGMA from the N-terminus of Mb.[8a] Fig. 2 and S-9 show that the conjugates could be completely purified by preparative SEC. The weight-average molecular weight (MW) of conjugates were determined by size exclusion chromatography multi-angle light scattering (SEC-MALS) (table 1). The MWs of Mb-N-POEGMA and Mb-N-PDMAEMA measured by SEC-MALS were 612 and, 425 kDa, with a polydspersity index (PDI) of 1.33 and 1.14 respectively.
Table 1.
MW and light scattering characterization of protein-polymer conjugates.
| Conjugates | MW (kDa) |
MW (kDa) |
Rg (nm) |
Rh (nm) |
Rg/Rh (ρ) |
|---|---|---|---|---|---|
| Mb-N-PDMAEMA | 425a | 421c | 43.0 | 24.5 | 1.7 |
| Mb-N-PCBMA | 575b | 643c | 44.3 | 25.9 | 1.7 |
| Mb-N-PCBMA (mix) | 600b | 653c | 44.9 | 26.2 | 1.7 |
| Mb-N-POEGMA | 612a | 626c | 37.1 | 20.1 | 1.8 |
Obtained from SEC-MALS.
Obtained from NMR.
Obtained from SLS.
Next, the side-chain amine groups of PDMAEMA were derivatized with Iodoacetic acid to yield Mb-N-PCBMA (C1 spacer) with ~ 97 % yield as determined by 1H-NMR (fig. S-10). Mb-N-PCBMA (mix) was obtained by sequential reaction of PDMAEMA with 3-iodopropionic and iodoacetic acid. We synthesized Mb-N-PCBMA (mix) because reaction with 3-Iodopropionic acid only led to ~65% conversion of the amine groups in PDMAEMA, while the sequential reaction succeeded in converting ~ 97 % of the amine groups, with a mixture of C1 and C2 spacers in a ~ 1:1.8 ratio as determined by 1H-NMR (fig. S-10).
The calculated MW of Mb-N-PCBMA and Mb-N-PCBMA (mix) by side chain analysis of their 1H-NMR spectra were 585 and 600 kDa respectively. SDS-PAGE showed bands of higher MW for conjugates than Mb-N-Br (fig. S-12). The hydrodynamic radius (Rh) and radius of gyration (Rg) of the conjugates were measured by dynamic light scattering (DLS) and static light scattering (SLS) respectively, and are shown in table 1. The Rh of Mb-N-PDMAEMA, Mb-N-PCBMA, Mb-N-PCBMA (mix) and Mb-N-POEGMA were 25, 26, 26 and 20 nm respectively, consistent with their similar MWs. The form factor (ρ) of Mb-N-PDMAEMA, Mb-N-POEGMA, Mb-N-PCBMA and Mb-N-PCBMA (mix) were 1.7-1.8, suggesting that the polymers are in a random coil conformation.[13]
To address a concern that the post-polymerization derivatization of PDMAEMA could also react with the lysine residues in Mb, Mb-N-PCBMA conjugates were subjected to tryptic digestion followed by analysis by MS/MS. MS/MS showed that 14 lys residues in the Mb-N-PCBMA conjuate were unmodified (fig S-13 IV), while 5 lys residues were not detected. Of these 5 residues, peptide FDKFK could be digested into FDK and FK, which are too short to be retained on a C18 column and are also outside the detection range. The IPIK peptide is not detected for similar reasons. The successful detection of the HPGDFGADAQGAMTK peptide suggests that K119 was unmodified, as proteins are digested at the C-terminal end of Lys and Arg unless they are followed by Pro or their side chains are modified.[14] These results suggest that none of Lys residues were affected by post-polymerization dervatization of the conjugate.
The HEME absorbance at 409 nm of Mb, Mb-N-Br and conjugates was identical (fig. S-14) which suggested that the tertiary structure of Mb was unaffected by synthesis of the conjugate. The peroxidase-like activity of Mb was similar for the conjugates and the unmodified protein[15] suggesting that polymer conjugation had mininmal effect on the functional activity of Mb (fig. S-15).
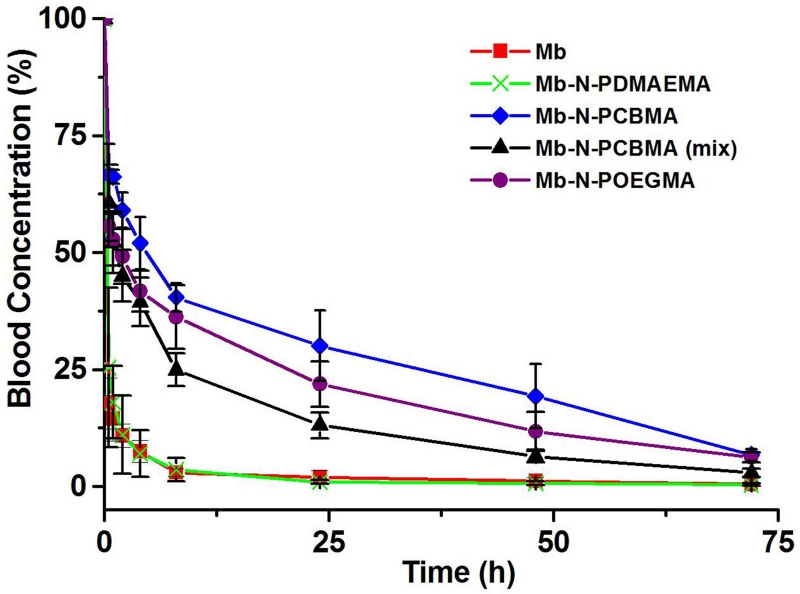
Next, the PK of the conjugates were determined by intravenously administering 125I-labeled Mb and conjugates to nude mice, followed by collecting blood samples at various time intervals and quantifying their radioactivity (fig. 3). A two-compartment model was used to fit the data and to obtain the area under the curve (AUC), total body clearance (CL), and the half-life for the distribution and elimination phase (T1/2α and T1/2β respectively) (table 2). Mb-N-PCBMA, Mb-N-PCBMA (mix), Mb-N-POEGMA and Mb-N-PDMAEMA showed ~ 15.1, 8.2, 11.9, and 0.8 -fold higher AUC and CL respectively than Mb. Mb and Mb-N-PDMAEMA have a relatively short T1/2β of 3.1 and 3.2 h respectively. In contrast, Mb-N-PCBMA, Mb-N-PCBMA (mix), and Mb-N-POEGMA have a significantly longer T1/2β of 17, 10 and 13 h respectively. These results suggest that N-terminal conjugation significantly enhances the cumulative exposure of Mb in the blood in the order Mb-N-PCBMA > Mb-N-POEGMA > Mb-N-PCBMA (mix) > Mb (one way ANOVA, Scheffe post-hoc tests, p < 0.05). Although the MW of the Mb-N-PCBMA and Mb-N-POEGMA are similar (and are well above the renal cut-off), if we discount the increased half-life of the Mb-N-PCBMA compared to the Mb-N-POEGMA because Rg and Rh of Mb-N-POEGMA slightly smaller than Mb-N-PCBMA, the most conservative conclusion that can be drawn is that the Mb-N-PCBMA have at least as long a plasma half-life and exposure as the Mb-N-POEGMA. Furthermore, these data also suggest that the structure of the charge pair has an impact on its in vivo behavior, as the Mb-N-PCBMA (C1 spacer) has a longer systemic circulation and greater plasma exposure than the Mb-N-PCBMA (mix) (mixture of C1 and C2 spacer). In contrast, the precursor conjugate, Mb-N-PDMAEMA showed no improvement in circulation half-life compared to Mb, despite its significantly larger size, which agrees with reports that showed positively charged nanoparticles are rapidly cleared from the body.[16]
Fig. 3.
Blood concentration as a function of time post intravenous injection.
Table 2.
PK parameters calculated from a two-compartment model.
| Protein and conjugates |
AUC (h % of blood concentration/mL) |
CL (mL/h) |
T1/2α (h) |
T1/2β (h) |
|---|---|---|---|---|
| Mb | 1.21 × 102 | 1.46 | 0.05 | 3.1 |
| Mb-N-PDMAEMA | 0.91 × 102 | 1.94 | 0.14 | 3.2 |
| Mb-N-PCBMA | 1.82 × 103 | 0.097 | 0.15 | 17.0 |
| Mb-N-PCBMA (mix) | 9.89 × 102 | 0.18 | 0.10 | 10.0 |
| Mb-N-POEGMA | 1.44 × 103 | 0.12 | 0.13 | 13.0 |
While we successfully used a post-polymerization derivatization method to prepare Mb-N-PCBMA without detectable modification of lysine residues in Mb, this method runs a risk of side-reaction of the lysine residues that are ubiquitous of most proteins. Future work will attempt to avoid this limitation by exploring a larger set of ATRP reaction conditions (beyond the limited set we explored in this paper) and alternative living polymerization methods such as reversible addition fragmentation chain transfer polymerization.
In conclusion, this study is the first demonstration of a site-specific (N-terminal) and stoichiometric (1:1) conjugate of a zwitterionic protein-polymer conjugate by in situ ATRP. There are two notable findings from this study: first, this study showed that a zwitterionic polymer conjugate significantly prolongs the circulation half-life of a short-lived protein upon systemic administration to mice. Second, the distance between the cation and anion appears to be a critical factor in modulating the in vivo behavior of the zwitterionic protein-polymer conjugate. This new class of zwitterionic protein-polymer conjugates provides a new tool to control the in vivo behavior of protein therapeutics for a range of biomedical applications.
Supplementary Material
Acknowledgments
We thank Dr. Erik Soderblom and Meredith Mayer-Salman of the Proteomics and Metabolomics Core Facility at Duke University for MS analysis of tryptic digests. This work was supported by the NIH by a grant (R01-DK092665) to A.C. IW thanks the NSF for a graduate research fellowship (NSF-DGE-1106401).
Footnotes
Supporting information for this article is given via a link at the end of the document.
Contributor Information
Dr. Wei-Han Wang, Department of Chemistry, University of Virginia, Charlottesville, VA 22904
Dr. Donald F. Hunt, Department of Chemistry, University of Virginia, Charlottesville, VA 22904; Department of Pathology, Health Sciences Center, University of Virginia, Charlottesville, VA 22908
References
- [1].a) Nischan N, Hackenberger CPR. J. Org. Chem. 2014;79:10727–10733. doi: 10.1021/jo502136n. [DOI] [PubMed] [Google Scholar]; b) Pelegri-O’Day EM, Lin E-W, Maynard HD. J. Am. Chem. Soc. 2014;136:14323–14332. doi: 10.1021/ja504390x. [DOI] [PubMed] [Google Scholar]
- [2].a) Caliceti P, Veronese FM. Adv. Drug Delivery Rev. 2003;55:1261–1277. doi: 10.1016/s0169-409x(03)00108-x. [DOI] [PubMed] [Google Scholar]; b) Liu M, Leroux J-C, Johansen P, Zabel F, Gauthier MA. Nat. Commun. 2014;5:5526. doi: 10.1038/ncomms6526. [DOI] [PubMed] [Google Scholar]
- [3].Mima Y, Hashimoto Y, Shimizu T, Kiwada H, Ishida T. Mol. Pharmaceutics. 2015:2429–2435. doi: 10.1021/acs.molpharmaceut.5b00144. [DOI] [PubMed] [Google Scholar]
- [4].Kane RS, Deschatelets P, Whitesides GM. Langmuir. 2003;19:2388–2391. [Google Scholar]
- [5].Lewis A, Tang Y, Brocchini S, Choi J.-w., Godwin A. Bioconjugate Chem. 2008;19:2144–2155. doi: 10.1021/bc800242t. [DOI] [PubMed] [Google Scholar]
- [6].a) Shao Q, Jiang S. Adv. Mater. 2015;27:15–26. doi: 10.1002/adma.201404059. [DOI] [PubMed] [Google Scholar]; b) Weers JG, Rathman JF, Axe FU, Crichlow CA, Foland LD, Scheuing DR, Wiersema RJ, Zielske AG. Langmuir. 1991;7:854–867. [Google Scholar]
- [7].a) Hucknall A, Rangarajan S, Chilkoti A. Adv. Mater. 2009;21:2441–2446. doi: 10.1002/adma.200803125. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hucknall A, Kim D-H, Rangarajan S, Hill RT, Reichert WM, Chilkoti A. Adv. Mater. 2009;21:1968–1971. doi: 10.1002/adma.200803125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].a) Gao W, Liu W, Mackay JA, Zalutsky MR, Toone EJ, Chilkoti A. Proc. Natl. Acad. Sci. U. S. A. 2009;106:15231–15236. doi: 10.1073/pnas.0904378106. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Gao W, Liu W, Christensen T, Zalutsky MR, Chilkoti A. Proc. Natl. Acad. Sci. U. S. A. 2010;107:16432–16437. doi: 10.1073/pnas.1006044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Matyjaszewski K, Xia J. Chem. Rev. 2001;101:2921–2990. doi: 10.1021/cr940534g. [DOI] [PubMed] [Google Scholar]
- [10].Gilmore JM, Scheck RA, Esser-Kahn AP, Joshi NS, Francis MB. Angew. Chem., Int. Ed. 2006;45:5307–5311. doi: 10.1002/anie.200600368. [DOI] [PubMed] [Google Scholar]
- [11].Jewett JC, Bertozzi CR. Chem. Soc. Rev. 2010;39:1272–1279. doi: 10.1039/b901970g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].a) Coon JJ, Ueberheide B, Syka JEP, Dryhurst DD, Ausio J, Shabanowitz J, Hunt DF. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9463–9468. doi: 10.1073/pnas.0503189102. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Anderson LC, English AM, Wang W-H, Bai DL, Shabanowitz J, Hunt DF. Int. J. Mass Spectrom. 2014:617–624. doi: 10.1016/j.ijms.2014.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].a) Burchard W, Schmidt M, Stockmayer WH. Macromolecules. 1980;13:1265–1272. [Google Scholar]; b) Niu A, Liaw D-J, Sang H-C, Wu C. Macromolecules. 2000;33:3492–3494. [Google Scholar]
- [14].a) Garcia BA, Mollah S, Ueberheide BM, Busby SA, Muratore TL, Shabanowitz J, Hunt DF. Nat. Protoc. 2007;2:933–938. doi: 10.1038/nprot.2007.106. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Switzar L, Giera M, Niessen WMA. J. Proteome Res. 2013;12:1067–1077. doi: 10.1021/pr301201x. [DOI] [PubMed] [Google Scholar]; c) Polgar L. Cell. Mol. Life Sci. 2005;62:2161–2172. doi: 10.1007/s00018-005-5160-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hayashi T, Hitomi Y, Ando T, Mizutani T, Hisaeda Y, Kitagawa S, Ogoshi H. J. Am. Chem. Soc. 1999;121:7747–7750. [Google Scholar]
- [16].a) Arvizo RR, Miranda OR, Moyano DF, Walden CA, Giri K, Bhattacharya R, Robertson JD, Rotello VM, Reid JM, Mukherjee P. PLoS One. 2011;6:e24374. doi: 10.1371/journal.pone.0024374. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Alexis F, Pridgen E, Molnar LK, Farokhzad OC. Mol Pharm. 2008;5:505–515. doi: 10.1021/mp800051m. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.