Abstract
Williams-Beuren syndrome (WBS) manifests as supravalvular aortic stenosis, intellectual disability, developmental delay and characteristic facial features. The common WBS deletion region ranges from 1.55 to 1.84 Mb and primarily contains the ELN gene. We analyzed 10 patients diagnosed with 7q11.23 microdeletion syndrome by chromosomal microarray analysis. The clinical features of these patients varied from classic WBS to normal phenotype. All 10 patients exhibited different sizes and breakpoints of chromosome microdeletions ranging from 44 kb to 9.88 Mb. The hemizygosity of the ELN gene was detected in 7 patients, while a normal ELN gene was present in 3 other patients with small deletions. We observed that the phenotypic features of WBS varied in fetuses, children and adults, influenced by the genes, deletion size and breakpoint. Our findings provide more information on the genotype-phenotype correlations of WBS. However, further research is needed to explore the size and breakpoint effect and functions of the genes on chromosome 7q11.23.
Key Words: Atypical deletion, Chromosomal microarray analysis, ELN, Microdeletion 7q11.23
Williams-Beuren syndrome (WBS; OMIM 194050) is a complex multisystemic neurodevelopmental disorder caused by a hemizygous microdeletion in chromosome 7q11.23, with a prevalence of between 1/7,500 [Strømme et al., 2002] and 1/50,000 [Klein et al., 1990]. Patients with WBS demonstrate disorders including (1) cardiovascular manifestations, which are characterized by arterial stenosis, including supravalvular aortic stenosis (SVAS) and peripheral pulmonary stenosis, and hypertension; SVAS occurs in the majority (75%) of affected children [Burn, 1986]; (2) developmental delay or mild to moderate mental retardation; (3) behavioral and cognitive abnormalities, including overfriendliness, empathy for others, anxiety, hypersensitivity to sound, strengths in verbal short-term memory and communication skills, visuospatial construction impairments, and others; (4) characteristic facial features, which include bitemporal narrowing, periorbital fullness, short upturned nose, bulbous nasal tip, stellate irides, long philtrum, full lips, and dental abnormalities/malocclusion; (5) endocrine system problems, including infantile hypercalcemia, diabetes, subclinical hypothyroidism, and precocious puberty; (6) connective tissue abnormalities, and (7) renal anomalies and urinary tract defects, including hydronephrosis, kidney stones, renal agenesis, bladder diverticulum, voiding dysfunction, urinary tract infections, and undescended testis and hypospadias. Cardiovascular abnormalities occur in a large proportion (∼50-80%) of patients and account for most morbidity and mortality [Merla et al., 2010].
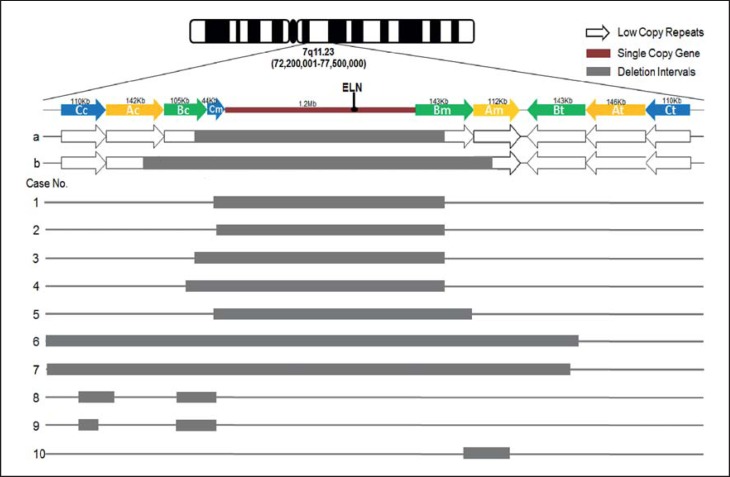
The WBS deletions are caused by intra- or interchromosomal NAHR in the 7q11.23 region. The common deletion interval ranges from 1.55 to 1.84 Mb and is composed of a single-copy gene region 1.2 Mb in size flanked by 3 large region-specific low-copy repeat (LCR) sequences, termed centromeric, medial and telomeric LCRs (fig. 1). LCRs are grouped into discrete blocks with a high degree of sequence identity known as A, B and C. Deletions arise as a consequence of misalignment of these repetitive sequences during meiosis due to the high similarity of the LCRs. Common deletions of ∼1.5 Mb in WBS patients display breakpoints within the centromeric and medial LCR block B, and the deletions of 1.8 Mb are caused by recombination between the centromeric and medial LCR block A.
Fig. 1.
Map of chromosome 7q11.23 WBS critical region and the deletions in 10 WBS cases. a = 1.5-Mb typical deletion in WBS. b = 1.8-Mb deletion in WBS. Cases 1-5 consisted of typical deletions (1.40-1.55 Mb); cases 6 and 7 consisted of larger atypical deletions (4.16 and 9.88 Mb); cases 8 and 9 carried 2 separated deletions, and case 10 carried a smaller atypical deletion (145 kb).
Because the WBS microdeletion is caused by NAHR between LCRs, the existence of the reciprocal microduplication syndrome is not unexpected [Dixit et al., 2013]. Microduplication of the 7q11.23 region, known as Sommerville-van der AA syndrome (OMIM 609757), was first reported by Somerville et al. [2005]. The prevalence of microduplications is only half that of microdeletions [Dutra et al., 2012]. Phenotypes found in patients with 7q11.23 microduplications are milder, more variable, and even affect the patient in different ways, such as by producing a broad nose, a short philtrum, speech delay, and deficits in social interaction ability. The features of WBS microduplication are so indistinct that the disease is often mis- or undiagnosed.
Here, we present 10 patients diagnosed with 7q11.23 microdeletion syndrome by chromosomal microarray analysis (CMA). CMA, which includes SNPs and aCGH, can precisely detect genomic imbalances by high-resolution genome-wide analysis.
Patients and Methods
We retrospectively reviewed 10 patients diagnosed with 7q11.23 microdeletion using CMA from May 2013 to December 2015, including 2 fetuses, 7 children (aged 3 weeks-7 years) and 1 adult. Among the 10 patients, 5 patients presented with cardiovascular abnormalities, including SVAS, pulmonary stenosis, patent foramen ovale, ventricular septal defect, and mitral valve insufficiency. Five patients showed developmental delay and intellectual disability ranging from mild to moderate. Two patients had distinctive facial features. Other abnormalities included a urinary fistula, cleft palate, patellar dislocation, and upper eyelid edema. One fetus showed duodenal atresia and a single umbilical artery on the left side, and the other showed a right pelvic ectopic kidney in the ultrasound examination. Patients 3 and 6 had typical cardiovascular abnormalities and were diagnosed with WBS by an ultrasonic cardiogram before the disease was detected by CMA. Patient 10 is a 32-year-old woman without any abnormalities in intelligence and development. She delivered a baby diagnosed with atelencephalia according to manifestations of epilepsy, spasm and muscle tremor. The baby died at 4 months of age, and neither an autopsy nor a molecular diagnosis was performed. The clinical features of all patients are described in table 1.
Table 1.
Clinical features and CMA results for 10 cases
| Case | Sex, age | CMA results |
ELN gene deletion | Other CNVs | Karyotype | Results of parental tests | Clinical features | |
|---|---|---|---|---|---|---|---|---|
| deletion | size | |||||||
| 1 | F, 27 gw | arr7q11.2 (72,701,018–74,141,746)×1 | 1.44 Mb | yes | 46,XX | maternal FISH: 7q11.23 deletion | right pelvic ectopic kidney | |
| 2 | M, 3 w | arr7q11.23 (72,718,277–74,142,190)×1 | 1.42 Mb | yes | – | – | SVAS, PFO, PS, distinctive facial features | |
| 3 | F, 3 mo | arr7q11.23 (72,643,631–74,142,190)×1 | 1.50 Mb | yes | – | – | PS, PFO | |
| 4 | M, 4.5 ys | arr7q11.23 (72,611,954–74,143,060)×1 | 1.53 Mb | yes | 15q11.2 microduplication | 46,XY | parental CMA: normal | GDD |
| 5 | M, 5 ys | arr7q11.23 (72,701,018–74,253,352)×1 | 1.55 Mb | yes | 46,XY | – | SVAS, PFO GDD | |
| 6 | F, 1 y | arr7q11.22q11.23 (70,463,755–74,629,034)×1 | 4.16 Mb | yes | SVAS, GDD, mitral valve insufficiency, urinary fistula, distinctive facial features | |||
| 7 | F, 7 ys | arr7q11.21q11.23 (64,721,473–74,610,673)×1 | 9.88 Mb | yes | 11q14.3 microduplication | 46,XX | parental FISH: normal | mental retardation, feeding difficulties, impaired social interaction, upper eye lid edema |
| 8 | M, 13+4 gw | arr7q11.23 (72,404,175–72,481,934)×1 (72,621,798–72,718,124)×1 | 78 kb 96 kb | no | – | – | duodenal atresia, single umbilical artery | |
| 9 | M, 5 ys | arr7q11.23 (72,414,624–72,458,643)×1 (72,621,722–72,710,124)×1 | 44 kb 96 kb | no | 46,XY | parental FISH: normal | AS, VSD, GDD, cleft palate, patellar dislocation | |
| 10 | F, 32 ys | arr7q11.23 (74,242,166–74,384,749)×1 | 145 kb | no | 46,XX | phenotypically normal, her baby had epilepsy, spasm, muscle tremor, and died at 4 months. | ||
AS = Aortic stenosis; F = female; GDD = global developmental delay; gw = gestational weeks; M = male; mo = months; PFO = patent foramen ovale; PS = pulmonary stenosis; SVAS = supravalvular aortic stenosis; VSD = ventricular septal defect; w = weeks; ys = years.
Peripheral blood (2 ml) was obtained from 7 children, 1 adult and 4 pairs of parents, a cordocentesis was performed on fetus 1, and an amniocentesis was performed on fetus 8 to obtain samples. Genomic DNA was extracted from the samples using a QIAamp DNA Mini Kit (Qiagen, Valencia, Calif., USA). The DNA was then amplified, labeled and hybridized to the Affymetrix CytoScan HD array chip following the manufacturer's protocol. The CytoScan HD array is a high-density microarray including more than 2,700,000 functional markers across the entire genome. It contains about 750,000 SNP probes and 2,000,000 probes for chromosomal CNVs. Original data were obtained by the CytoScan array scanning platform and were analyzed with the Chromosome Analysis Suite software (Affymetrix). The results were annotated with genome version GRCH37 (hg19). To analyze the clinical significance of each result, several online genetic databases were referred to, including the Database of Genomic Variants (DGVS), Database of Chromosomal Imbalance and Phenotype in Humans using Ensemble Resources (DECIPHER), Online Mendelian Inheritance in Man (OMIM), and the International Standards for Cytogenomic Arrays (ISCA) Consortium.
Karyotyping analysis using conventional G-banded techniques (500-band resolution) was performed on 6 patients. FISH was performed following the standard technique using ELN probes on 3 pairs of parents and CMA on 1 pair. Six pairs of parents did not undergo any test due to lack of collaboration.
Results
7q11.23 Microdeletion
All 10 patients had different sizes and loci of chromosome microdeletions in the 7q11.23 region, ranging from 44 kb to 9.88 Mb. ELN gene deletions were found by CMA in 7 patients, and among them, 5 patients presented with typical WBS deletions (1.40-1.55 Mb), and the other 2 patients presented with larger atypical deletions (4.16 Mb and 9.88 Mb). The ELN gene was not contained in the deletions of the remaining 3 patients in which 2 children carried 2 separated deletions and 1 adult carried a small atypical deletion (145 kb). The deletions in chromosome 7q11.23 are depicted in figure 1.
Other Microdeletions or/and Microduplications
Patient 4 exhibited 15q11.2 microduplications, and patient 7 showed 11q14.3 microduplications.
Karyotype Analysis
No abnormal karyotype was found in the 6 patients who underwent G-banded karyotype analysis.
Parental Results
The results for the 3 pairs of parents who underwent FISH showed that 1 mother had a 7q11.23 microdeletion. All results are described in table 1.
Discussion
Among the 10 cases diagnosed with 7q11.23 microdeletion syndrome in our report, 5 had rather typical WBS deletions and 2 had much larger deletions overlapping the typical deletion. Seven of 10 affected individuals demonstrated typical WBS features including SVAS, distinctive facies and developmental delay.
To date, 28 genes have been associated with WBS on chromosome 7q11.23 [Merla et al., 2010]. Genotype-phenotype correlations have been established for a few genes such as GTF2IRD2, CLIP2, LIMK1, FKBP36, LAT2, and RFC2 [Schubert, 2009; Antonell et al., 2010; Ferrero et al., 2010; Honjo et al., 2012; Vandeweyer et al., 2012], although some correlations are unclear. The identification of affected patients with small atypical deletions that only involve few genes has provided valuable information regarding genotype-phenotype correlations. Using the Chromosome Analysis Suite software (Affymetrix), we were able to detect all genes involved in the deletion region of each patient (table 2).
Table 2.
Single genes involved in the WBS region
| LCR block | Gene | Type | Case |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |||
| Cc | POM121 | coding | − | − | + | + | ||||||
| NSUN5C | noncoding | − | − | − | − | |||||||
| TRIM74 | coding | − | − | − | − | |||||||
| FKBP6-like | noncoding | − | − | − | ||||||||
| Ac | STAG3L3 | noncoding | − | − | − | |||||||
| PMS2L | noncoding | − | − | + | ||||||||
| WBSCR19 | noncoding | − | − | |||||||||
| Bc | GTF2IP | noncoding | + | − | − | |||||||
| NCF1B | noncoding | + | − | − | − | + | − | |||||
| GTF2IRD2P | noncoding | − | − | − | − | − | − | |||||
| Cm | POM121B | noncoding | − | − | − | − | + | − | ||||
| NUSN5 | coding | − | + | − | − | − | − | − | ||||
| TRIM50 | coding | − | − | − | − | − | − | − | ||||
| FKBP6 | coding | − | − | − | − | − | − | − | ||||
| Single-copy gene | FZD9 | coding | − | − | − | − | − | − | − | |||
| BAZ1B | coding | − | − | − | − | − | − | − | ||||
| BCL7B | coding | − | − | − | − | − | − | − | ||||
| TBL2 | coding | − | − | − | − | − | − | − | ||||
| MLXIPL | coding | − | − | − | − | − | − | − | ||||
| VPS37D | coding | − | − | − | − | − | − | − | ||||
| DNAJC30 | coding | − | − | − | − | − | − | − | ||||
| WBSCR22 | coding | − | − | − | − | − | − | − | ||||
| STX1A | coding | − | − | − | − | − | − | − | ||||
| ABHD11 | coding | − | − | − | − | − | − | − | ||||
| CLDN3 | coding | − | − | − | − | − | − | − | ||||
| CLDN4 | coding | − | − | − | − | − | − | − | ||||
| WBSCR27 | coding | − | − | − | − | − | − | − | ||||
| WBSCR28 | coding | − | − | − | − | − | − | − | ||||
| ELN | coding | − | − | − | − | − | − | − | ||||
| LIMK1 | coding | − | − | − | − | − | − | − | ||||
| EIF4H | coding | − | − | − | − | − | − | − | ||||
| LAT2 | coding | − | − | − | − | − | − | − | ||||
| RFC2 | coding | − | − | − | − | − | − | − | ||||
| CLIP2 | coding | − | − | − | − | − | − | − | ||||
| GTF2IRD1 | coding | − | − | − | − | − | − | − | ||||
| WBSCR23 | unknown | − | − | − | − | − | − | − | ||||
| Bm | GTF2I | coding | + | + | + | + | − | − | − | |||
| NCF1 | coding | − | − | − | ||||||||
| GTF2IRD2 | coding | + | − | − | + | |||||||
| Am | STAG3L2 | noncoding | − | − | − | |||||||
| PMS2L5 | noncoding | − | − | − | ||||||||
| GATSL | noncoding | − | − | + | ||||||||
| WBSCR16 | coding | − | − | |||||||||
| Bt | GTF2IED2B | coding | − | − | ||||||||
| NCF1C | noncoding | − | − | |||||||||
| GTF2IP1 | noncoding | + | + | |||||||||
| At | GATSL | noncoding | ||||||||||
| WBSCR19 | noncoding | |||||||||||
| PMS2L | noncoding | |||||||||||
| STAG3L1 | noncoding | |||||||||||
| Ct | FKBP6-like | noncoding | ||||||||||
| TRIM73 | coding | |||||||||||
| NSUN5B | coding | |||||||||||
| POM121 | coding | |||||||||||
− = Deleted genes within the microdeletions of each patient; + = genes disrupted by the breakpoints.
The ELN gene, a gene coding for elastic fibers of the extracellular matrix throughout the body, is the most critical and extensively explored gene in WBS. Hemizygosity of the ELN gene in WBS contributes to cardiovascular and connective tissue abnormalities, such as SVAS, hernias, bladder or bowel diverticulum, and lax skin. In our study, a deletion of the ELN gene was detected in 7 patients, only 4 of whom manifested cardiovascular and connective tissue abnormalities. Patient 9 showed cardiovascular and connective tissue abnormalities even though his ELN gene was normal, which suggested that some unknown genes or factors produce these cardiovascular abnormalities. Recent studies have suggested that, in addition to the genes missing because of the deleted segment, multiple factors such as position effects related to CNV size, variations in the nondeleted alleles, epigenetic mechanisms, and regulatory sequences may be important in determining the variable expressivity of 7q11.23 CNV phenotypes [Merla al., 2010; Euteneuer et al., 2014].
Besides the ELN gene, there could be other genes and factors associated with cardiovascular anomalies and developmental delay. Patient 9 deserves particular attention. He showed aortic stenosis and ventricular septal defect, global developmental delay, cleft palate, and patellar dislocation, but only 2 slightly separated deletions of 44 kb and 96 kb were detected in the uncommon region. Two structural genes located in LCR Bm are involved in the deletion region, POM121 and TRIM74, which are not hemizygotes for 1.5 Mb or 1.8 Mb WBS patients. POM121 (OMIM 615753) encodes a membrane-bound nuclear pore complex (NPC) protein that interacts with other NPC subunits. Disruption of NPC caused cytoplasmic distribution of NPC subunits and inner nuclear membrane proteins [Mitchell et al., 2010]. TRIM74 (OMIM 612550) belongs to the tripartite motif gen family; the other 2 members of this family are TRIM50 in LCR Cm and TRIM73 in LCR Ct. TRIM74 is 94% identical to TRIM50, and 99.6% to TRIM73. Typical WBS patients are hemizygous for TRIM50, but not for the paralogous TRIM73 and TRIM74 copies. The function of TRIM74 is still unclear, while TRIM50 encodes an E3-ubiquitin-ligase in ubiquitin-mediated proteasome pathway that may be involved in the WBS phenotype [Micale et al., 2008]. Based on the present knowledge, none of the 2 genes seems to be related to the phenotypic features presented in the patient. Although the microdeletions do not overlap with the typical 7q11.23 deleted regions, it is not sufficient to exclude a causative role of the deleted genes in determining the clinical phenotype of the patient.
Patient 10 had a 145-kb deletion between Bm and Am which resulted in the loss of 4 genes, the transcription factor GTF2IRD2 and noncoding genes STAG3L2, PMS2L5 and GATSL. Compared to WBS patients with a typical 1.5-Mb deletion, only 2 additional genes, NCF1 and GTF2IRD2, are deleted in those with a larger 1.8-Mb size deletion. After analyzing cognitive, behavioral and psychological functioning in 55 WBS patients, Porter et al. [2012] showed that those with 1.8-Mb deletions encompassing GTF2IRD2 had significantly poorer cognitive functions including spatial functioning, social reasoning, and executive function. The GTF2IRD1 and GTF2I are the transcription factor TFII-I family members encoding BEN and TFII-I proteins, respectively. Haploinsufficiency of TFII-I proteins causes multiple developmental anomalies including microcephaly, retarded growth, and skeletal and craniofacial defects which are associated with WBS [Enkhmandakh et al., 2009]. Those authors also provided the evidence that homozygous loss of either GTF2IRD1 or GTF2I results in embryonic lethality, brain hemorrhage, vasculogenic defects, and craniofacial and neural tube defects in mice. However, there is no abnormality of clinical manifestations, development and cognitive impairment in patient 10 based on a detailed examination. We suppose that haploinsufficiency of the products encoded by GTF2IRD1 is not sufficient to cause the phenotypic features, the different size and the dose of deletions determining the variable expressivity and effect of the genes involved. Still, further research is needed to explore the causative role of genes in the deletion region that is not overlapped.
The condition of patient 4 was complicated with a 15q11.2 microduplication (312 kb). Reports of copy number alteration on chromosome 15q11.2 suggest an association with autism, developmental delay, motor and language delay, and behavioral problems [Burnside et al., 2011]. Therefore, we presumed that her manifestations were due to the superposition of the 7q11.23 microdeletion and the 15q11.2 microduplication. Patient 7, who had an 11q14.3 microduplication (1.77 Mb), showed developmental delay, feeding difficulties, upper eyelid edema, weakness of the limbs, and poor language skills. Although there is little evidence for the relationship of CNVs on 11q14.3 and his phenotypic features, we cannot exclude the contribution of the 11q14.3 microduplication in patient 7. However, no additional pathological CNVs were identified in patients 8, 9 and 10 who carried atypical deletions. So it can be considered that their phenotypes are consequences of the 7q11.23 microdeletion only.
Fetuses 1 and 8 showed atypical phenotypes including duodenal atresia, a single umbilical artery and ectopic kidney. To our knowledge, only 8 WBS patients detected by prenatal diagnosis were reported and the main (62.5%) referral reason was intrauterine growth retardation [Marcato et al., 2014]. Since the limitation of prenatal ultrasound and less pronounced phenotypes, it is not easy to diagnose WBS in fetus.
Another abnormality in WBS critical region is the inversion between the centromeric and medial LCRs, which is accompanied by interstitial deletions [Osborne et al., 2001]. It is possible that the deletions in patients 8 and 9 were caused by this inversion because they each carried 2 interrupted deletions between the centromeric and medial LCRs. In the population of WBS patients, 33% of the chromosome-transmitting parents carried a paracentric inversion of the WBS locus on chromosome 7 [Osborne et al., 2001]. Carriers of inversions are often phenotypically normal because breakpoints are external to the WBS single-copy gene region and actively expressed genes are not disrupted, but carriers are at risk for generating gametes harboring the WBS deletion due to an increase in mispairings in chromosome 7 during meiosis [Scherer et al., 2005; Tam et al., 2008]. The inversions cannot be detected by CMA. Inversions may exist in the asymptomatic parents of patient 9 in whom standard FISH results were normal.
Fetus 1 had an ectopic kidney, which was the only indication for prenatal diagnosis. Patient 6 had a urinary fistula and repeated urinary tract infections. To our knowledge, urological abnormalities are not listed as a diagnostic criterion in most scoring systems for the clinical evaluation of WBS. Pankau et al. [1996] reported that, compared with the normal population, the risk of structural abnormalities of the kidneys and urinary tract is increased by 12 to 36-fold in WBS patients. Sammour et al. [2014] found that in 79 patients, 47 (59.5%) showed abnormalities on physical examination, whereas 36 (45.6%) had at least one uroradiological abnormality. There is no association between urological abnormalities and cardiovascular defects. Therefore, the possibility of WBS may be taken into account when a child or fetus shows abnormalities of the kidneys or urinary tract.
Generally, patients with WBS are identified through recognizable clinical phenotypes during childhood. However, laboratory tests using molecular biology techniques are necessary due to the wide heterogeneity of phenotypes. Although FISH on metaphase chromosomes using a probe for the ELN or LIMIK gene is widely regarded as the gold standard for the diagnosis of WBS, it cannot estimate the specific size of the microdeletion/microduplication. It is difficult to choose which FISH probe to order for patients who are clinically suspected of having WBS. In addition, the commercial FISH assay with a probe for ELN is not capable of detecting 7q11.23 microdeletion or microduplication in which the ELN gene was not contained, such as patients 8, 9 and 10. However, CMA can cover the whole genome with a high resolution, allowing the detection of the exact size and locus of the microdeletion/microduplication, and the result can be obtained within 2-4 days, which is beneficial to prenatal diagnoses.
Statement of Ethics
All patients and their parents have given written consent for the research, and the study has been approved by our ethics committee.
Disclosure Statement
The authors have no conflict of interest to declare.
Acknowledgment
We are grateful to the patients and their families for their kind cooperation.
References
- 1.Antonell A, Del Campo M, Magano LF, Kaufmann L, de la Iglesia JM, et al. Partial 7q11.23 deletions further implicate GTF2I and GTF2IRD1 as the main genes responsible for the Williams-Beuren syndrome neurocognitive profile. J Med Genet. 2010;47:312–320. doi: 10.1136/jmg.2009.071712. [DOI] [PubMed] [Google Scholar]
- 2.Burn J. Williams syndrome. J Med Genet. 1986;23:389–395. doi: 10.1136/jmg.23.5.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burnside RD, Pasion R, Mikhail FM, Carroll AJ, Robin NH, et al. Microdeletion/microduplication of proximal 15q11.2 between BP1 and BP2: a susceptibility region for neurological dysfunction including developmental and language delay. Hum Genet. 2011;130:517–528. doi: 10.1007/s00439-011-0970-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dixit A, McKee S, Mansour S, Mehta SG, Tanteles GA, et al. 7q11.23 Microduplication: a recognizable phenotype. Clin Genet. 2013;83:155–161. doi: 10.1111/j.1399-0004.2012.01862.x. [DOI] [PubMed] [Google Scholar]
- 5.Dutra RL, Honjo RS, Kulikowski LD, Fonseca FM, Pieri PC, et al. Copy number variation in Williams-Beuren syndrome: suitable diagnostic strategy for developing countries. BMC Res Notes. 2012;5:13. doi: 10.1186/1756-0500-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enkhmandakh B, Makeyev AV, Erdenechimeg L, Ruddle FH, Chimge NO, et al. Essential functions of the Williams-Beuren syndrome-associated TFII-I genes in embryonic development. Proc Natl Acad Sci USA. 2009;106:181–186. doi: 10.1073/pnas.0811531106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Euteneuer J, Carvalho CM, Kulkarni S, Vineyard M, Mark GR, et al. Molecular and phenotypic characterization of atypical Williams-Beuren syndrome. Clin Genet. 2014;86:487–491. doi: 10.1111/cge.12305. [DOI] [PubMed] [Google Scholar]
- 8.Ferrero GB, Howald C, Micale L, Biamino E, Augello B, et al. An atypical 7q11.23 deletion in a normal IQ Williams-Beuren syndrome patient. Eur J Hum Genet. 2010;18:33–38. doi: 10.1038/ejhg.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Honjo RS, Dutra RL, Nunes MM, Gomy I, Kulikowski LD, et al. Atypical deletion in Williams-Beuren syndrome critical region detected by MLPA in a patient with supravalvular aortic stenosis and learning difficulty. J Genet Genomics. 2012;39:571–574. doi: 10.1016/j.jgg.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Klein AJ, Armstrong BL, Greer MK, Brown FR., 3rd Hyperacusis and otitis media in individuals with Williams syndrome. J Speech Hear Disord. 1990;55:339–444. doi: 10.1044/jshd.5502.339. [DOI] [PubMed] [Google Scholar]
- 11.Marcato L, Turolla L, Pompilii E, Dupont C, Gruchy N, et al. Prenatal phenotype of Williams-Beuren syndrome and of the reciprocal duplication syndrome. Clin Case Rep. 2014;2:25–32. doi: 10.1002/ccr3.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merla G, Brunetti-Pierri N, Micale L, Fusco C. Copy number variants at Williams-Beuren syndrome 7q11.23 region. Hum Genet. 2010;128:3–26. doi: 10.1007/s00439-010-0827-2. [DOI] [PubMed] [Google Scholar]
- 13.Micale L, Fusco C, Augello B, Napolitano LM, Dermitzakis ET, et al. Williams-Beuren syndrome TRIM50 encodes an E3 ubiquitin ligase. Eur J Hum Genet. 2008;16:1038–1049. doi: 10.1038/ejhg.2008.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitchell JM, Mansfeld J, Capitanio J, Kutay U, Wozniak RW. Pom121 links two essential subcomplexes of the nuclear pore complex core to the membrane. J Cell Biol. 2010;191:505–521. doi: 10.1083/jcb.201007098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osborne LR, Li M, Pober B, Chitayat D, Bodurtha J, et al. A 1.5 million-base pair inversion polymorphism in families with Williams-Beuren syndrome. Nat Genet. 2001;29:321–325. doi: 10.1038/ng753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pankau R, Partsch CJ, Winter M, Gosch A, Wessel A. Incidence and spectrum of renal abnormalities in Williams-Beuren syndrome. Am J Med Genet. 1996;63:301–304. doi: 10.1002/(SICI)1096-8628(19960503)63:1<301::AID-AJMG49>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 17.Porter MA, Dobson-Stone C, Kwok JB, Schofield PR, Beckett W, Tassabehji M. A role for transcription factor GTF2IRD2 in executive function in Williams-Beuren syndrome. PLoS One. 2012;7:e47457. doi: 10.1371/journal.pone.0047457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sammour ZM, Gomes CM, de Bessa JJ, Pinheiro MS, Kim CA, et al. Congenital genitourinary abnormalities in children with Williams-Beuren syndrome. J Pediatr Urol. 2014;10:804–809. doi: 10.1016/j.jpurol.2014.01.013. [DOI] [PubMed] [Google Scholar]
- 19.Scherer SW, Gripp KW, Lucena J, Nicholson L, Bonnefont JP, et al. Observation of a parental inversion variant in a rare Williams-Beuren syndrome family with two affected children. Hum Genet. 2005;117:383–388. doi: 10.1007/s00439-005-1325-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schubert C. The genomic basis of the Williams-Beuren syndrome. Cell Mol Life Sci. 2009;66:1178–1197. doi: 10.1007/s00018-008-8401-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Somerville MJ, Mervis CB, Young EJ, Seo EJ, Del Campo M, et al. Severe expressive-language delay related to duplication of the Williams-Beuren locus. N Engl J Med. 2005;353:1694–1701. doi: 10.1056/NEJMoa051962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strømme P, Bjørnstad PG, Ramstad K. Prevalence estimation of Williams syndrome. J Child Neurol. 2002;17:269–271. doi: 10.1177/088307380201700406. [DOI] [PubMed] [Google Scholar]
- 23.Tam E, Young EJ, Morris CA, Marshall CR, Loo W, et al. The common inversion of the Williams-Beuren syndrome region at 7q11.23 does not cause clinical symptoms. Am J Med Genet A. 2008;146A:1797–1806. doi: 10.1002/ajmg.a.32360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vandeweyer G, Van der Aa N, Reyniers E, Kooy RF. The contribution of CLIP2 haploinsufficiency to the clinical manifestations of the Williams-Beuren syndrome. Am J Hum Genet. 2012;90:1071–1078. doi: 10.1016/j.ajhg.2012.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]