Abstract
Duplications of the long arm of chromosome 1 are rare. Distal duplications are the most common and have been reported as either pure trisomy or unbalanced translocations. The paucity of cases with pure distal 1q duplications has made it difficult to delineate a partial distal trisomy 1q syndrome. Here, we report 2 patients with overlapping 1q duplications detected by G-banding. Array CGH and FISH were performed to characterize the duplicated segments, exclude the involvement of other chromosomes and determine the orientation of the duplication. Patient 1 presents with a mild phenotype and carries a 22.5-Mb 1q41q43 duplication. Patient 2 presents with a pure 1q42.13qter inverted duplication of 21.5 Mb, one of the smallest distal 1q duplications ever described and one of the few cases characterized by array CGH, thus contributing to a better characterization of distal 1q duplication syndrome.
Key Words: Array CGH, Distal partial duplication 1q, Duplication 1q41q43, Duplication 1q42.13, FISH
Established Facts
• There are few reports of patients with pure partial trisomy 1q syndrome.
Novel Insights
• The 2 patients described here help to better define the partial distal trisomy 1q syndrome.
• A possible role of RGS7, WNT9A and WNT3A in the etiology of cranial suture malformations is discussed.
Patients with pure partial distal trisomy 1q demonstrate a wide range of manifestations of variable severity making it difficult to define a ‘partial trisomy 1q syndrome’. In most cases, the duplication is the result of an unbalanced translocation with a possible imbalance of the other participating chromosome [Kimya et al., 1979; Concolino et al., 1998; Emberger et al., 2001; Percesepe et al., 2007; Utine et al., 2007]. It is difficult to evaluate the contribution of the 1q trisomy to the phenotype in cases involving another chromosome. Cases with pure partial distal trisomy 1q provide an opportunity to better define the partial distal trisomy 1q syndrome.
Here, we report 2 patients with overlapping 1q duplications detected by G-banding and characterized by array CGH and FISH.
Patients and Methods
Clinical Report
Patient 1 is the first child to healthy, nonconsanguineous parents (fig. 1A). She was born by normal delivery at term weighing 2,700 g (3rd-10th centile) and first seen by us at age 19 years. Clinical examination showed mild intellectual disability, moderate short stature (3rd-10th centile), low weight (41.4 kg, <3rd centile), normal head circumference (52,5 cm; 2nd-50th centile), prominent metopic suture, facial asymmetry, long eyelashes, high nasal root, hypoplastic alae nasi, high-arched palate, maxillary projection, narrow hands, a short fourth right metatarsus, scoliosis, and genu valgum.
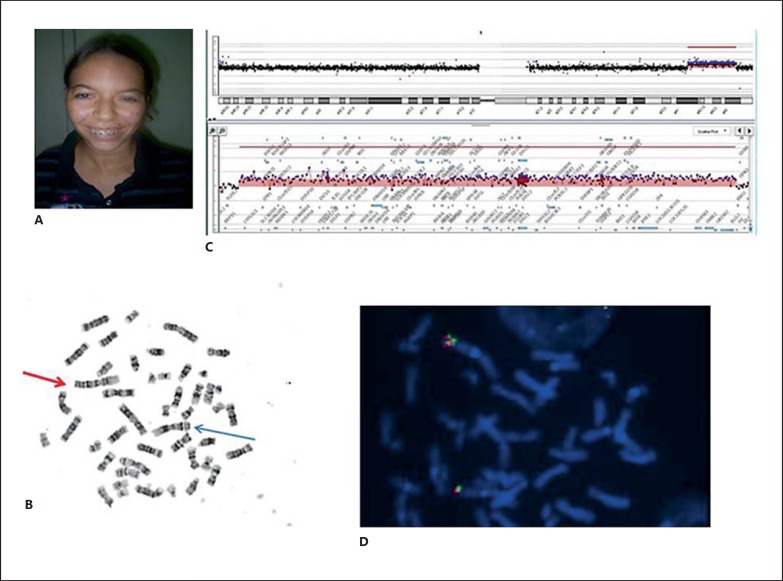
Fig. 1.
A Patient 1 at 19 years of age showing mild facial dysmorphisms. B Metaphase after G-banding showing a 1q41q43 duplication. The red arrow points to the duplicated chromosome 1. The blue arrow points to the normal chromosome 1. C Mapping of the duplication on a 60k oligo array showing the duplicated region and genes. D Determination of orientation of the duplication by double-colored FISH. Hybridization with probes RP11-239E10 (green) and RP11-87P4 (red) shows the direct orientation of the 1q41q43 duplication in patient 1.
Patient 2 is the second child to healthy, nonconsanguineous parents (fig. 2A). Pregnancy was uneventful, and she was delivered by C-section at term with Apgar scores 3/10. She had neonatal asphyxia and jaundice. Her birth length was 52 cm (75th centile), birth weight 3,660 g (75th centile) and her head circumference was 36 cm (50th-98th centile). She was first seen by us at the age of 5 months with findings of mildly delayed neuropsychomotor development, large anterior fontanel, prominent metopic ridge, glabellar hemangioma, hypertelorism (inner canthal distance: 28 mm >98th centile; outer canthal distance: 90 mm >98th centile), upslanted palpebral fissures, epicanthal folds, large nasal bridge, bulbous nose, smooth philtrum, thin upper lip, large mouth, posteriorly rotated ears with elevated lobes, low posterior hairline, and short neck. She had pectus excavatum, leg edema and a hyperpigmented spot on the left leg. Her feet showed overlapping of the second and third toes. The echocardiogram revealed a patent foramen ovale. Follow-ups at 9, 15 and 18 months did not identify any other complications.
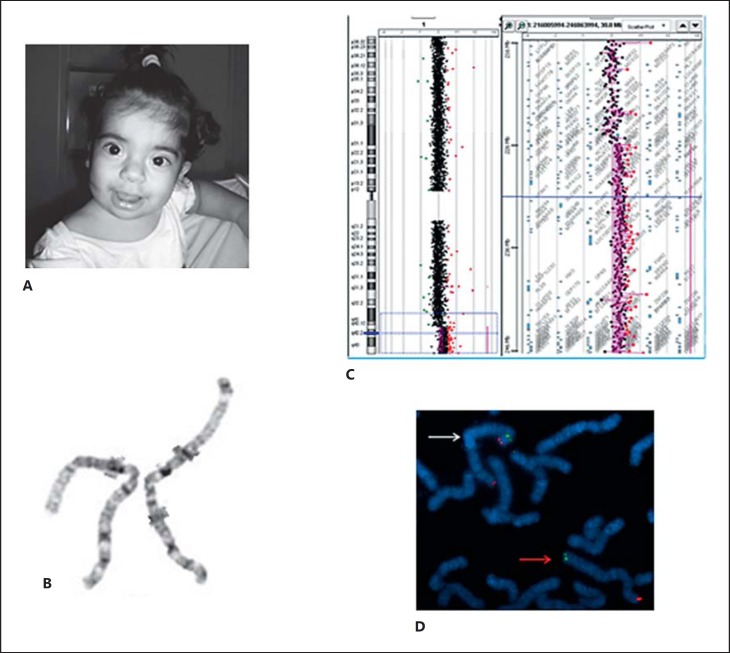
Fig. 2.
A Patient 2 at age 18 months showing facial dysmorphisms. B Metaphase after G-banding showing a 1q42q44 duplication: normal chromosome (left), duplicated chromosome (right). C Mapping of the duplication on a 60k oligo array, showing the duplicated region and genes. D Hybridization with TelVysion 1p SpectrumGreen and TelVysion 1q SpectrumOrange probes confirmed the inverted orientation of the 1q42q44 duplication in patient 2. The white arrow indicates the duplicated chromosome. The red arrow indicates the normal chromosome.
Cytogenetic Analysis
Chromosome analysis of peripheral blood lymphocytes by GTG-banding was performed at 550 or greater banding level. Array CGH was performed using a whole-genome oligoarray platform consisting of 60,000 oligonucleotides (60K, Agilent Technologies, Santa Clara, Calif., USA). Labeling, hybridization and post-hybridization washing stages were performed according to the manufacturer's protocol. A scanned image of the microarray was processed using Feature Extraction software and analyzed with Genomic Workbench software (both from Agilent Technologies), with the statistical algorithm ADM-2, and a sensitivity threshold of 6.7. At least 3 consecutive oligonucleotides had to exhibit aberrant log2 ratios in order to be called by the software as a copy number alteration. FISH with probes mapped to the duplicated segments was performed to investigate the orientation of the duplications.
Results
Patient 1
Chromosome analysis revealed a duplication of the long arm of chromosome 1 (46,XX,dup(1)(q41q43); fig 1B). The mother has a normal karyotype, and the father was unavailable for study. Duplication was refined by array CGH to chromosomal region 1q41q43 to a size of 22.5 Mb (chr1: 217,201,257-239,696,544 Assembly NCBI36/hg18; 2006; fig. 1C). FISH experiments using probes RP11-239E10 (1q41) and RP11-87P4 (1q42.2) revealed the direct orientation of the duplication (fig. 1D). The patient's final karyotype was 46,XX,dup(1)(q41q43). arr 1q41q43(217,201,257-239,696,544)×3.
Patient 2
Chromosome analysis revealed a mosaic duplication of the long arm of chromosome 1 (46,XX,dup(1)(q43q44),inv(q43p36.3)[16]/46,XX[14]; fig. 2B). Both parents have a normal karyotype. Array-CGH analysis refined the duplication to chromosomal region 1q42.13q44 to a size of 21.5 Mb (chr1: 225,696,992-247,164,526 Assembly NCBI36/hg18; 2006; fig. 2C). No deletion at 1p36 was found. FISH experiments using TelVysion 1p SpectrumGreen and TelVysion 1q SpectrumOrange probes (Vysis Inc., Downers Grove, Ill., USA) confirmed the inverted orientation of the duplication (fig. 2D). The patient's final karyotype was 46,XX,dup(1)(q43q44),inv(q43p36.3)[16]/46,XX[14]. arr 1q42.13q44(225,696,992-247,164,526)×3.
Discussion
In this study, we report 2 cases with partially overlapping pure 1q duplications. Patient 1 has a previously unreported interstitial duplication involving only the segment 1q41q43. A partially overlapping duplication has been described in the DECIPHER database (251817) on a patient with facial abnormalities, proportionate short stature and intellectual disability, a phenotype concordant with our patient. Another reported case has a 19.4-Mb duplication in 1q41q43 [Bartnik et al., 2014] and clinical features including delayed psychomotor development, mild intellectual disability and dysmorphic features. Despite presenting with a large 22.5-Mb duplication, her overall phenotype is mild.
The duplication in patient 1 overlaps the critical region for 1q41q42 microdeletion syndrome [Shaffer at al., 2007; Mazzeu et al., 2010; Rosenfeld et al., 2011]. To the best of our knowledge, the reciprocal microduplication for the 1q41q42 segment has not been described to date. Since these patients present with duplications of much larger segments and have mild phenotypes, it is possible that the reciprocal 1q41q42 microduplication causes a mild effect or has no pathogenic effect.
The duplication in patient 2 is one of the few cases with pure distal 1q duplications characterized by array CGH involving only 1q43q44. Patients with pure partial distal trisomy 1q demonstrate a wide range of manifestations of variable severity. Distal 1q duplication syndrome is characterized by the signs present in almost all previously reported cases, namely intellectual disability/delayed neuropsychomotor development, a triangular face, slanted and narrow palpebral fissures, broad nasal bridge, pointed small nose, micrognathia, and low-set and posteriorly rotated ears with poorly formed helices (table 1). Heart defects have been found in all patients (including patient 2) except for those reported by Lungarotti et al. [1980] and Kimya et al. [1979]. Though not described in all patients, cardiac defects are a common finding and, therefore, should be evaluated in patients with distal duplications of 1q. Patient 2 presents with one of the smaller duplications reported (1q42.13qter), and it is mosaic. However, her clinical signs are consistent with the distal trisomy 1q phenotype, thus pointing to a more significant contribution of terminal duplicated segments to the distal 1q syndrome. The duplication is inverted and translocated to chromosome 1p without detectable distal deletion of 1pter as confirmed by array CGH and FISH. All previously described cases in which 1pter deletion has been investigated had 1p microdeletions.
Table 1.
Clinical signs reported in patients with distal trisomy 1q including the patients described here
| Utine et al., 2007 patient 1 | Flatz and Fonatsch, 1979 | Steffen sen et al., 1977 | Balasubramanian et al., 2009 | Clark et al., 1994 | Nowaczyk et al., 2003 | Polity ko et al., 2005 patient B | Duba et al., 1997 | Kimya et al., 1979 | Bartsch et al., 2001 | Present case 1 | Present case 2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cytogenetic studies | FISH (MCB) | oaCGH, G-banding, FISH, MLPA | G-banding, FISH (WCP), aCGH | FISH (MCB) | G-banding, FISH | G-banding, FISH | aCGH, G-banding, FISH | aCGH, G-banding, FISH | ||||
| Cytogenetic aberration | dup(1) (q31q43~44) | dup(1) (q32q44) | dup(1) (q32q44) | inv dup(l) (q31q43) | dup(1) (q32qter) | dup(l) (q32q44) | dup(l) (q41q44) | dup(l) (q32qter) | dup(l) (q32q44) | dup(l) (q32q44) | inv dup(l) (q41q43) | dup(1) (q42.13q44) |
| Features Triangular face | + | + | + | + | + | + | ? | + | + | + | + | − |
| Long face | + | + | + | + | ? | ? | ? | ? | ? | − | − | − |
| Long and smooth philtrum | ? | ? | − | ? | ? | ? | + | ? | ? | − | − | + |
| Micrognathia | + | ? | − | ? | ? | ? | ? | ? | + | + | − | + |
| Narrow palpebral fissures | + | + | + | + | + | + | ? | + | ? | − | − | + |
| Microphthalmia | + | ? | − | ? | ? | ? | ? | ? | ? | − | − | − |
| Wide, flat nasal bridge | + | + | + | + | ? | + | + | + | + | + | − | + |
| Cleft lip/palate | + | − | − | − | − | − | ? | + | − | − | − | − |
| High-arched palate | ? | ? | ? | ? | ? | + | ? | ? | + | − | + | + |
| Low-set ears | + | + | + | + | + | + | + | + | + | − | − | + |
| Posteriorly rotated ears | + | + | + | + | + | + | + | + | ? | + | − | + |
| Poorly formed helices | + | + | + | + | + | − | ? | + | ? | − | − | + |
| Patent ductus arteriosus | − | ? | + | − | − | + | ? | − | − | − | − | − |
| Patent foramen ovale | − | ? | + | − | − | ? | ? | − | − | − | − | + |
| Other heart defects | + | ? | + | + | + | + | + | + | − | + | − | − |
| Double right renal artery | + | − | − | − | − | ? | ? | − | − | − | − | − |
| Meckel diverticulum | + | − | − | − | − | ? | ? | − | − | − | − | − |
| Long fingers and toes | + | − | + | − | − | − | ? | − | − | − | − | + |
| Camptodactyly | + | − | − | − | − | − | ? | − | − | + | − | − |
| Preaxial polydactyly | + | − | − | − | − | − | ? | + | + | − | − | − |
aCGH = Array CGH; MCB = multicolor banding; oaCGH = oligonucleotide array CGH; WCP = whole chromosome paint; + = present; − = absent; ? = not mentioned. Adapted from Utine et al. [2007].
Both patients described here have a prominent metopic suture suggesting the presence of a candidate gene for time regulation of cranial sutures closing in the overlapping segment of the 2 duplications. In previous studies, other genetic disorders have been described in patients with metopic craniosynostosis such as a point mutation in FGFR1 [Kress et al., 2000] and microdeletion in PTPRD [Choucair et al., 2015]. In several types of cranial bone formation, factors such as bone morphogenetic proteins, fibroblast growth factors and WNTs play an important role in controlling this process, confirming that the pattern of gene expression in this pathology is heterogeneous and complex [Zhou et al., 2009].
Fifty-one OMIM genes map to the region of intersection between the duplications on chromosome 1 (chr1: 225,696,992-239,696,544g19/hg18), including WNT9A and WNT3A. From hydra to humans, WNT genes play indisputably important roles in patterning [Cadigan and Nusse, 1997], including facial [Brugmann et al., 2007] and cranial morphogenesis [Behr et al., 2010]. Wnt signaling can stimulate differentiation of early osteoblasts and inhibit the mineralization of mature osteoblasts [Eijken et al., 2008].
WNT9A is associated with chondrogenic development of craniofacial structures. Previous studies show that wnt9a expression in zebrafish is essential for extension of the median palatine processes during the palatogenesis [Dougherty et al., 2013] and the lower jaw formation [Curtin et al., 2011]. WNT3A may suppress the chondrogenesis and gene expression in chondrocytes [Reinhold et al., 2006]. Although the metopic suture is closed through intramembranous ossification, it is supported by cartilage, which should disappear before calcification [Holmbeck et al., 2003; Sahar et al., 2005]. Previous studies in embryo mice showed Wnt9a mRNA in calvarian osteoblasts, and the supracalvarial injection of Wnt3a resulted in complete cartilage degradation [Zhou et al., 2009].
In addition to the WNT genes, Mefford et al. [2010] described a patient with coronal synostosis and a 2.5-Mb microduplication at 1q43 (chr1: 23,934-24,099/hg18) and proposed the RGS7 gene as a candidate for craniosynostosis. This gene is the last gene duplicated in patient 1, close to the distal breakpoint and in the overlapping segment of the 2 duplications. No other genes in the overlapping segment have been proposed as candidates for craniosynostosis and, therefore, our data reinforces a possible role of RGS7, WNT9A and WNT3A in the etiology of cranial suture malformations.
Statement of Ethics
This study was approved by the institutional Ethics Committee with the written informed consent of the families.
Disclosure Statement
The authors have no conflict of interest to declare.
Acknowledgements
We thank the Rede Sarah de Hospitais de Reabilitação for conducting part of the G-banding and FISH experiments. This study was supported by grants from the Decanato de Pesquisa e Pós-graduação, Universidade de Brasília and the Fundação de Amparo à Pesquisa do Distrito Federal e Ministério da Saúde, PPSUS 2010 e Fundação de Amparo à Pesquisa do Estado de São Paulo.
References
- 1.Balasubramanian M, Barber JC, Collinson MN, Huang S, Maloney VK, et al. Inverted duplication of 1q32.1 to 1q44 characterized by array CGH and review of distal 1q partial trisomy. Am J Med Genet A. 2009;149A:793–797. doi: 10.1002/ajmg.a.32463. [DOI] [PubMed] [Google Scholar]
- 2.Bartnik M, Nowakowska B, Derwińska K, Wiśniowiecka-Kowalnik B, Kedzior M, et al. Application of array comparative genomic hybridization in 256 patients with developmental delay or intellectual disability. J Appl Genetics. 2014;55:125–144. doi: 10.1007/s13353-013-0181-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartsch C, Aslan M, Köhler J, Miny P, Horst J, et al. Duplication dup(1)(q32q44) detected by comparative genomic hybridization (CGH): further delineation of trisomies 1q. Fetal Diagn Ther. 2001;16:265–273. doi: 10.1159/000053926. [DOI] [PubMed] [Google Scholar]
- 4.Behr B, Longaker MT, Quarto N. 2010. Differential activation of canonical Wnt Signaling determines cranial sutures fate: a novel mechanism for sagittal suture craniosynostosis. Dev Biol. 2010;344:922–940. doi: 10.1016/j.ydbio.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 5.Brugmann SA, Goodnough LH, Gregorieff A, Leucht P, ten Berge D, et al. Wnt signaling mediates regional specification in the vertebrate face. Development. 2007;134:3283–3295. doi: 10.1242/dev.005132. [DOI] [PubMed] [Google Scholar]
- 6.Cadigan KM, Nusse R. Wnt signaling: a common theme in animal development. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 7.Choucair N, Micnon-Ravix C, Cacciagli P, Abou-Ghoch J, Fawaz A, et al. Evidence that homozygous PTPRD gene microdeletion causes trigonocephaly, hearing loss, and intellectual disability. Mol Cytogenet. 2015;8:39. doi: 10.1186/s13039-015-0149-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark BK, Lowther GW, Lee WR. Congenital ocular defects associated with an abnormality of the human chromosome 1: trisomy 1q32-qter. J Pediatr Ophthalmol Strabismus. 1994;31:41–45. doi: 10.3928/0191-3913-19940101-09. [DOI] [PubMed] [Google Scholar]
- 9.Concolino D, Cinti R, Ferraro L, Moricca MT, Strisciuglio P. Partial trisomy 1(q42→qter): a new case with a mild phenotype. J Med Genet. 1998;35:75–77. doi: 10.1136/jmg.35.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curtin E, Hickey G, Kamel G, Davidson AJ, Liao EC. Zebrafish wnt9a is expressed in pharyngeal ectoderm and is required for palate and lower jaw development. Mech Dev. 2011;128:104–115. doi: 10.1016/j.mod.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Doughert M, Kamel G, Grimaldi M, Gfrerer L, Shubinets V, et al. Distinct requirements for wnt9a and irf6 in extension and integration mechanisms during zebrafish palate morphogenesis. Development. 2013;140:76–81. doi: 10.1242/dev.080473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duba HC, Erdel M, Löffler J, Bereuther L, Fisher H, et al. Detection of a de novo duplication of 1q32-qter by fluorescence in situ hibridisation in a boy with multiple malformations: further delineation of the trisomy 1q syndrome. J Med Genet. 1997;34:309–313. doi: 10.1136/jmg.34.4.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eijken M, Meijer IM, Westbroek I, Koedam M, Chiba H, et al. Wnt signaling acts and is regulated in a human osteoblast differentiation dependent manner. J Cell Biochem. 2008;104:568–579. doi: 10.1002/jcb.21651. [DOI] [PubMed] [Google Scholar]
- 14.Emberger W, Petek E, Kroisel PM, Zierler H, Wagner K. Clinical and molecular cytogenetic characterization of two patients with partial trisomy 1q41-qter: further delineation of partial trisomy 1q syndrome. Am J Med Genet. 2001;104:312–318. doi: 10.1002/ajmg.10096. [DOI] [PubMed] [Google Scholar]
- 15.Flatz S, Fonatsch C. Partial trisomy 1q due to tandem duplication. Clin Genet. 1979;15:541–542. doi: 10.1111/j.1399-0004.1979.tb00839.x. [DOI] [PubMed] [Google Scholar]
- 16.Holmbeck K, Bianco P, Chrysovergis K, Yamada S, Birkedal-Hansen H. MT1-MMP-dependent, apoptotic remodeling of unmineralized cartilage: a critical process in skeletal growth. J Cell Biol. 2003;163:661–671. doi: 10.1083/jcb.200307061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimya Y, Yakut T, Egeli U, Ozerkan K. Prenatal diagnosis of a fetus with pure partial trisomy 1q32-44 due to a familial balanced rearrangement. Prenat Diagn. 1979;22:957–961. doi: 10.1002/pd.403. [DOI] [PubMed] [Google Scholar]
- 18.Kress W, Petersen B, Collmann H, Grimm T. An unusual FGFR1 mutation (fibroblast growth factor receptor 1 mutation) in a girl with non-syndromic trigonocephaly. Cytogenet Cell Genet. 2000;91:138–140. doi: 10.1159/000056834. [DOI] [PubMed] [Google Scholar]
- 19.Lungarotti MS, Falomi A, Calabro A, Passalacqua F, Dallapiccola B. De novo duplication 1q32-q42: variability of phenotypic features in partial 1q trisomics. J Med Genet. 1979;17:398–402. doi: 10.1136/jmg.17.5.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mazzeu JF, Vianna-Morgante AM, Krepischi AC, Oudakker A, Rosenberg C, et al. Deletions encompassing 1q41q42.1 and clinical features of autosomal dominant Robinow syndrome. Clin Genet. 2010;77:404–407. doi: 10.1111/j.1399-0004.2009.01355.x. [DOI] [PubMed] [Google Scholar]
- 21.Mefford HC, Shafer N, Antonacci F, Tsai JM, Park SS, et al. Copy number variation analysis in single-suture craniosynostosis: multiple rare variants including RUNX2 duplication in two cousins with metopic craniosynostosis. Am J Med Genet A. 2010;152:2203–2210. doi: 10.1002/ajmg.a.33557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nowaczyk MJM, Bayani J, Freeman V, Watts J, Squire J, Xu J. De novo 1q32q44 duplication and distal 1q trisomy syndrome. Am J Med Genet A. 2003;120A:229–233. doi: 10.1002/ajmg.a.20028. [DOI] [PubMed] [Google Scholar]
- 23.Percesepe A, Lugli L, Pierluigi M, Cavani S, Malacarne M, et al. Pure segmental trisomy 1q42-qter in a boy with a severe phenotype. Am J Med Genet A. 2007;143A:2339–2342. doi: 10.1002/ajmg.a.31890. [DOI] [PubMed] [Google Scholar]
- 24.Polityko A, Starke H, Rumyantseva N, Claussen U, Liehr T, Raskin S. Three cases with rare interstitial rearrangements of chromosome 1 characterized by multicolor banding. Cytogenet Genome Res. 2005;111:171–174. doi: 10.1159/000086388. [DOI] [PubMed] [Google Scholar]
- 25.Reinhold MI, Kapadia RM, Liao Z, Naski MC. The Wnt-inducible transcription fator Twist1 inhibits chondrogenesis. J Biol Chem. 2006;281:1381–1388. doi: 10.1074/jbc.M504875200. [DOI] [PubMed] [Google Scholar]
- 26.Rosenfeld JA, Lacassie Y, El-Khechen D, Escobar LF, Reggin J, et al. New cases and refinement of the critical region in the 1q41q42 microdeletion syndrome. Eur J Med Genet. 2011;54:42–49. doi: 10.1016/j.ejmg.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Sahar DE, Longaker MT, Quarto N. Sox9 neural crest determinant gene controls patterning and closure of the posterior frontal cranial suture. Dev Biol. 2005;280:344–361. doi: 10.1016/j.ydbio.2005.01.022. [DOI] [PubMed] [Google Scholar]
- 28.Shaffer LG, Theisen A, Bejjani BA, Ballif BC, Aylsworth AS, et al. The discovery of microdeletion syndromes in the post-genomic era: review of the methodology and characterization of a new 1q41q42 microdeletion syndrome. Genet Med. 2007;9:607–616. doi: 10.1097/gim.0b013e3181484b49. [DOI] [PubMed] [Google Scholar]
- 29.Steffensen DM, Chu EHY, Speert DP, Wall PM, Meilinger K, Kelch RP. Partial trisomy of the long arm of human chromosome 1 as demonstrated by in situ hybridization with 5S ribosomal RNA. Hum Genet. 1977;36:25–33. doi: 10.1007/BF00390432. [DOI] [PubMed] [Google Scholar]
- 30.Utine GE, Aktas D, Alanay Y, Gücer S, Tuncbilek E, et al. 2007. Distal partial trisomy 1q: report of two cases and review of the literature. Prenat Diagn. 2007;27:865–871. doi: 10.1002/pd.1788. [DOI] [PubMed] [Google Scholar]
- 31.Zhou H, Mak W, Kalak R, Street J, Fong-Yee C, et al. Glucocorticoid-dependent Wnt signaling by mature osteoblasts is a key regulator of cranial skeletal development in mice. Development. 2009;136:427–436. doi: 10.1242/dev.027706. [DOI] [PubMed] [Google Scholar]