Abstract
IMPORTANCE
Probiotics have been hypothesized to affect immunologic responses to environmental exposures by supporting healthy gut microbiota and could therefore theoretically be used to prevent the development of type 1 diabetes mellitus (T1DM)–associated islet autoimmunity.
OBJECTIVE
To examine the association between supplemental probiotic use during the first year of life and islet autoimmunity among children at increased genetic risk of T1DM.
DESIGN, SETTING, AND PARTICIPANTS
In this ongoing prospective cohort study that started September 1, 2004, children from 6 clinical centers, 3 in the United States (Colorado, Georgia/Florida, and Washington) and 3 in Europe (Finland, Germany, and Sweden), were followed up for T1DM-related autoantibodies. Blood samples were collected every 3 months between 3 and 48 months of age and every 6 months thereafter to determine persistent islet autoimmunity. Details of infant feeding, including probiotic supplementation and infant formula use, were monitored from birth using questionnaires and diaries. We applied time-to-event analysis to study the association between probiotic use and islet autoimmunity, stratifying by country and adjusting for family history of type 1 diabetes, HLA-DR-DQ genotypes, sex, birth order, mode of delivery, exclusive breastfeeding, birth year, child’s antibiotic use, and diarrheal history, as well as maternal age, probiotic use, and smoking. Altogether 8676 infants with an eligible genotype were enrolled in the follow-up study before the age of 4 months. The final sample consisted of 7473 children with the age range of 4 to 10 years (as of October 31, 2014).
EXPOSURES
Early intake of probiotics.
MAIN OUTCOMES AND MEASURES
Islet autoimmunity revealed by specific islet autoantibodies.
RESULTS
Early probiotic supplementation (at the age of 0-27 days) was associated with a decreased risk of islet autoimmunity when compared with probiotic supplementation after 27 days or no probiotic supplementation (hazard ratio [HR], 0.66; 95% CI, 0.46-0.94). The association was accounted for by children with the DR3/4 genotype (HR, 0.40; 95% CI, 0.21-0.74) and was absent among other genotypes (HR, 0.97; 95% CI, 0.62-1.54).
CONCLUSIONS AND RELEVANCE
Early probiotic supplementation may reduce the risk of islet autoimmunity in children at the highest genetic risk of T1DM. The result needs to be confirmed in further studies before any recommendation of probiotics use is made.
A newborn infant’s immune system needs to quickly learn how to tolerate beneficial bacteria and defend against opportunistic pathogens. The intestinal microbiota can influence the balance between proinflammatory and regulatory immune resp onses.1 However, there are still unanswered questions as to how the immune system interacts with the microbiota.2,3
A healthy gut microbiota is believed to favorably regulate mucosal barrier function4 and reduce intestinal permeability.5,6 Abnormalities in gut permeability have been linked to the development of type 1 diabetes mellitus (T1DM).7 Healthy gut microbiota may also enhance the overall maturation of the infant immune system8,9 and exclude pathogens competitively.10 Imbalance in gut microbiota and a relative decrease in α-diversity are associated with T1DM according to a recent study.11 A larger proportion of the phylum Bacteroidetes has been observed in children with T1DM.12-14
Microbial colonization of the infant gut starts in utero,15 although frequent changes in gut microbiota, mainly in relative abundances of species, have been observed during the first 10 to 12 months of life.16-19 Early life events, such as mode of delivery, early environment, including hygiene measures, and early feeding, are thought to initially set the trajectory of colonization.20,21 Even though α-diversity may be large, strain composition within an individual typically remains constant throughout infancy.11
Probiotics have been defined as live organisms that, when administered in adequate amounts, confer a health benefit on the host.22 Administration of probiotics to healthy infants is considered safe.23,24 However, it is still unclear whether probiotics as an early dietary factor could modify the infant gut microbiota trajectory and disease susceptibility.
Studies25,26 on manipulation of gut microbiota by probiotics and consequent changes in the risk of developing T1DM-related autoimmunity have mainly used animal models. Probiotics induce favorable immunomodulation, and it has been suggested that probiotic treatment could prevent T1DM. The aim of this study is to examine the association between supplemental probiotic use during the first year of life and islet autoimmunity (IA) among children at increased risk of T1DM.
Methods
The Environmental Determinants of Diabetes in the Young (TEDDY) is a prospective cohort study with the primary goal to identify environmental causes of T1DM. It includes 6 clinical research centers (3 in the United States and 3 in Europe): University of Colorado Health Science Center, Georgia Regents University, Pacific Northwest Diabetes Research Institute, Turku University Hospital, Institute of Diabetes Research, and Lund University. Detailed study design and methods have been previously published.27,28 The study was approved by the local institutional review or ethics boards and is monitored by an external advisory board formed by the National Institutes of Health. Written informed consent was obtained for all study participants from a parent or primary caretaker for genetic screening and participation in prospective follow-up.
Study Population
From September 1, 2004, through February 28, 2010, a total of 424 788 newborns in the university hospitals affiliated with the clinical research centers were screened with parents’ consent for T1DM-associated HLA genotypes.29 The screening identified 21 589 eligible infants, of whom 8676 were enrolled in the follow-up study before the age of 4 months. Children who were followed up for less than 12 months (n = 1032) or whose islet autoantibody status was indeterminate (n = 55) were excluded. Because of HLA ineligibility, 116 additional study participants were removed from the analyses. The final sample for this study consisted of 7473 children with an age range of 4 to 10 years (as of October 31, 2014).
HLA Typing
Infants from the general population, with no first-degree relative (FDR) with T1DM, were eligible for the study if they had any one of the following HLA genotypes: (1) DR4-DQA1* 03-DQB1*03:02/DR3-DQA1*05:01-DQB1*02:01, (2) DR4-DQA1*03-DQB1*03:02/DR4-DQA1*03-DQB1*03:02, (3) DR4-DQA1*03-DQB1*03:02/DR8-DQA1*04:01-DQB1*04:02, and (4) DR3-DQA1*05:01-DQB1*02:01/DR3-DQA1*05:01-DQB1*02:01. Acceptable DQB1 alleles in any haplotype listed as DQB1*03:02 also include DQB1*03:04. For the above genotypes, any DR4 of the subtype DRB1*04:03 is ineligible.
Infants who have an FDR with T1DM were eligible for enrollment if they had any of the following HLA genotypes: (1) DR4-DQA1*03-DQB1*03:02/DR3-DQA1*05:01-DQB1*02: 01, (2) DR4-DQA1*03-DQB1*03:02/DR4-DQA1*03-DQB1*03: 02, (3) DR4-DQA1*03-DQB1*03:02/DR8-DQA1*04:01-DQB1*04: 02, (4) DR3-DQA1*05:01-DQB1*02:01/DR3-DQA1*05:01-DQB1*02:01, (5) DR4-DQA1*03-DQB1*03:02/DR4-DQA1*03-DQB1*02, (6) DR4-DQA1*03-DQB1*03:02/DR1-DQA1*01:01-DQB1*05:01, (7) DR4-DQA1*03-DQB1*03:02/DR13-DQA1*01: 02-DQB1*06:04, (8) DR4-DQA1*03-DQB1*03:02/DR9-DQA1*03-DQB1*03:03, and (9) DR3-DQA1*05:01-DQB1*02:01/DR9-DQA1*03-DQB1*03:03. Acceptable DQB1 alleles in any haplotype listed as DQB1*03:02 also include DQB1*03:04. All HLA genotypes are referred to in the text by their abbreviated names listing only DR alleles (i.e. DR3/4 for genotype [1] above).
Islet Autoimmunity
The primary outcome of this study was the development of persistent confirmed IA. Blood samples were drawn every 3 months between 3 and 48 months of age and every 6 months thereafter. Persistent IA was defined as confirmed positive antibodies to insulin, glutamic acid decarboxylase, or insulinoma antigen 2, which were analyzed by radiobinding assays,30,31 on at least 2 consecutive study visits. All positive islet autoantibodies and 5% of negative islet autoantibodies were confirmed in both central autoantibody laboratories, 1 located in the United States (Barbara Davis Center for Childhood Diabetes at the University of Colorado) and 1 in Europe (University of Bristol). Both laboratories have previously found high sensitivity and specificity32 and concordance. Positive results that were due to maternal IgG transmission when defining the child’s IA status led to omission from the IA-positive group. Date of persistent autoimmunity was defined as the draw date of the first of 2 consecutive samples that deemed the child’s IA status as persistent that were confirmed positive for a specific autoantibody (or any autoantibody). The mean (SD) age at first IA sampling was 33.4 (23.2) months among the seroconverters (n = 601), and the mean (SD) age at the last follow-up for children without IA was 65.6 (28.0) months.
Characteristics and Diet and Health Monitoring of the Study Population
Information about basic demographic characteristics and family history of diabetes was received from the infant screening form. A questionnaire on maternal medications, smoking habits, and probiotic dietary supplement use during pregnancy was mailed to the mothers of enrolled children and completed at 3 to 4 months post partum. After enrollment, parents also received a questionnaire on mode of delivery and child’s early diet, including the use of probiotics at 0 to 3 months of age. Parents were advised to consistently maintain a diary after the first clinic visit to collect information on child illnesses and diet. The start age of the probiotic supplement and each type of infant formula were recorded. Information about the mother’s educational level and birth order of the child was received from the primary caretaker questionnaire at the 9-month clinical visit. Probiotic exposure was defined as timing of first introduction of probiotics via dietary supplement or infant formula. Clinical center study nurses reviewed the questionnaires and diaries with the parent at clinic visits or over the telephone every 3 months to minimize missing and inaccurate information.
Statistical Analysis
The characteristics of probiotic users for the study children and their mothers were examined one by one using a Cochran-Mantel-Haenszel test and simultaneously using a logistic regression model adjusting for country. The association between the probiotic exposure age and IA was examined among those who were exposed to probiotics during the first year of life. For exploratory analyses, we categorized these individuals according to the probiotic exposure age into 3 equally sized groups (0-27 days, 28-90 days, and 91-365 days) and compared them with the individuals without probiotic exposure during the first year (>365 days) when studying the association with IA. A Cox proportional hazards regression model was applied to study the association between timing of probiotic exposure and occurrence of IA.
The Cox proportional hazards regression models were simultaneously adjusted for HLA-DR-DQ genotype (DR3/4 vs other), T1DM-related FDR status (yes/no), sex (female vs male), mode of delivery (cesarean delivery vs other), and exclusive breastfeeding duration (≥3 vs <3 months) and stratified for country using the STRATA statement within the model. The models were also adjusted for factors that were associated with probiotic use and could be associated with IA or T1DM: maternal age (≤24, 25-29 [reference], 30-34, and ≥35 years), maternal smoking during pregnancy (yes/no), maternal probiotic use during pregnancy (yes/no), birth year, birth order (first born child vs others), diarrheal history, and antibiotic use of the child (yes/no).33,34
Because early IA may have preceded the probiotic exposure, a sensitivity analysis of the association between timing of probiotic exposure and occurrence of IA was conducted by excluding the individuals who developed IA during the first year of life (n = 106) to eliminate the effect of the ordering between IA and probiotic exposure. The results from the sensitivity analysis indicated only minor changes in the estimated parameters (hazard ratios [HRs] and P values) and led to the same conclusions as when including all individuals in the model.
All tests for significance were 2-tailed with a significance level of .05. SAS statistical software, version 9.3 (SAS Institute Inc), was used for all statistical analyses.
Results
Probiotic supplementation, from dietary supplements or infant formula, varied by country (Table 1). It was most prevalent in Finland (869 [52.4%]) and Germany (237 [46.8%]) during the first year of life. Most of the Finnish children (827 [95.2%]) had received probiotics from dietary supplements, whereas in Germany, probiotic infant formulas (214 [90.3%]) were the most common sources of probiotics. The median age for first exposure to probiotics was 42 days. Overall, children born at the end of the recruitment period (2009-2010) were 4.9 times more likely (P < .001) to be given probiotics than those born in the beginning of the study period (2004-2005) (Table 2).
Table 1.
Distribution of Probiotic Exposure From Dietary Supplements and Infant Formulas During the First Year of Life by Age and Countrya
| Country |
|||||
|---|---|---|---|---|---|
| Variable | United States (n = 3046) |
Finland (n = 1658) |
Germany (n = 506) |
Sweden (n = 2263) |
All (N = 7473) |
| Probiotic use | |||||
|
| |||||
| During first 12 mo | 186 (6.1) | 869 (52.4) | 237 (46.8) | 345 (15.2) | 1637 (21.9) |
|
| |||||
| During first 3 mo | 70 (2.3) | 627 (37.8) | 123 (24.3) | 276 (12.2) | 1096 (14.7) |
|
| |||||
| Age at first exposure to probiotics, median (IQR), d | 137 (56-244) | 28 (14-105) | 84 (14-198) | 35 (21-70) | 42 (14-152) |
|
| |||||
| Source of first probiotic exposure among probiotic users | |||||
|
| |||||
| Dietary supplements | 124 (66.7) | 827 (95.2) | 11 (4.6) | 274 (79.4) | 1236 (75.5) |
|
| |||||
| Infant formula | 53 (28.5) | 25 (2.9) | 214 (90.3) | 40 (11.6) | 332 (20.3) |
|
| |||||
| Both | 9 (4.8) | 17 (1.9) | 12 (5.1) | 31 (9.0) | 69 (4.2) |
|
| |||||
| Timing of first probiotic exposure for users only, d | |||||
|
| |||||
| 0-27 | 20 (10.8) | 344 (39.5) | 74 (31.2) | 104 (30.0) | 542 (33.1) |
|
| |||||
| 28-90 | 53 (28.5) | 283 (32.6) | 49 (20.7) | 172 (50.0) | 557 (34.0) |
|
| |||||
| 91-365 | 113 (60.7) | 242 (27.9) | 114 (48.1) | 69 (20.0) | 538 (32.9) |
Abbreviation: IQR, interquartile range.
Data are presented as number (percentage) unless otherwise indicated.
Table 2.
Characteristics of Probiotic Supplement and/or Probiotic Formula Users and Nonusers
| Characteristic | Users (n = 1637) |
Nonusers (n = 5836) |
P Valuea | Adjusted ORb
(95% CI) |
|---|---|---|---|---|
| Maternal age, y | ||||
|
| ||||
| ≤24 | 151 (9.2) | 746 (12.8) | .003 | 0.68 (0.53-0.88) |
|
|
|
|||
| 25-29 | 520 (31.8) | 1664 (28.5) | 1 [Reference] | |
|
|
|
|||
| 30-34 | 598 (36.5) | 2062 (35.3) | 1.11 (0.94-1.30) | |
|
|
|
|||
| ≥35 | 368 (22.5) | 1364 (23.4) | 1.20 (0.99-1.45) | |
|
| ||||
| Maternal educational level of high school or more | 1407 (87.7) | 4534 (79.7) | .001 | 1.14 (0.93-1.39) |
|
| ||||
| Birth order, first child | 799 (50.2) | 2442 (43.1) | <.001 | 1.64 (1.42-1.90) |
|
| ||||
| Antibiotics use during pregnancy | 384 (23.7) | 1321 (22.9) | .45 | 1.04 (0.89-1.22) |
|
| ||||
| Probiotics use during pregnancy | 116 (7.1) | 169 (2.9) | <.001 | 2.69 (1.95-3.70) |
|
| ||||
| Smoking during pregnancy | 180 (11.1) | 704 (12.2) | <.001 | 0.74 (0.60-0.92) |
|
| ||||
| Cesarean delivery | 393 (24.0) | 1542 (26.4) | .008 | 1.11 (0.94-1.30) |
|
| ||||
| Birth year | ||||
|
| ||||
| 2004-2005 | 163 (10.0) | 1067 (18.3) | <.001 | 1 [Reference] |
|
|
|
|||
| 2006 | 228 (13.9) | 1084 (18.6) | 1.70 (1.32-2.18) | |
|
|
|
|||
| 2007 | 344 (21.0) | 1225 (21.0) | 2.79 (2.20-3.52) | |
|
|
|
|||
| 2008 | 379 (23.1) | 1149 (19.7) | 4.01 (3.17-5.08) | |
|
|
|
|||
| 2009-2010 | 523 (32.0) | 1311 (22.4) | 4.89 (3.89-6.14) | |
|
| ||||
| First-degree relative with T1DM | 203 (12.4) | 639 (11.0) | .38 | 0.97 (0.78-1.19) |
|
| ||||
| HLA genotype DR3/4 | 628 (38.4) | 2305 (39.5) | .13 | 1.08 (0.94-1.23) |
|
| ||||
| Female sex | 797 (48.7) | 2862 (49.0) | .72 | 1.04 (0.91-1.19) |
|
| ||||
| Exclusive breastfeeding at least 3 mo | 392 (24.0) | 1456 (25.0) | <.001 | 0.79 (0.68-0.92) |
|
| ||||
| Child’s antibiotics use during the first 12 mo | 910 (55.6) | 2435 (41.7) | <.001 | 1.68 (1.46-1.94) |
|
| ||||
| Diarrhea episode during the first 3 mo | 169 (10.3) | 494 (8.5) | <.001 | 1.50 (1.19-1.90) |
|
| ||||
| Common cold during the first 3 mo | 910 (55.6) | 3432 (58.8) | .04 | 1.14 (0.99-1.31) |
|
| ||||
| Gastroenteritis (infectious or noninfectious) during the first 12 mo |
582 (35.6) | 1723 (29.5) | <.001 | 1.38 (1.20-1.59) |
Abbreviations: OR, odds ratio; T1DM, type 1 diabetes mellitus.
P value from the Cochran-Mantel-Haenszel test for the association between probiotics and respondent characteristics; analyses adjusted for country.
Adjusted OR (95%CI) from multiple logistic regression; analyses adjusted for country.
Participant characteristics that were positively associated with probiotic use during the first year of life were probiotic use during pregnancy (P < .001), not smoking during pregnancy (P = .006), being first born (P < .001), later birth year (P < .001), shorter duration of exclusive breastfeeding (P = .003), use of antibiotics (P < .001), and having diarrhea (P < .001) or gastroenteritis (P < .001) (Table 2). Only 855 (11.4%) of the children used antibiotics before 3 months of age, whereas 2995 (40.1%) used antibiotics at 3 to 12 months of age. However, probiotic use, from dietary supplements or formula, was strongly associated with antibiotic use (P < .001) during the first year of life. Nevertheless, antibiotic use was not associated with IA. By the age of 3 months, 4342 (58.1%) of all TEDDY children had experienced at least one episode of common cold: 1670 (54.8%) in the United States, 757 (45.7%) in Finland, 274 (54.2%) in Germany, and 1641 (72.5%) in Sweden (P < .001, χ2 test). Gastroenteritis was strongly associated with probiotic use (P < .001) but not with IA.
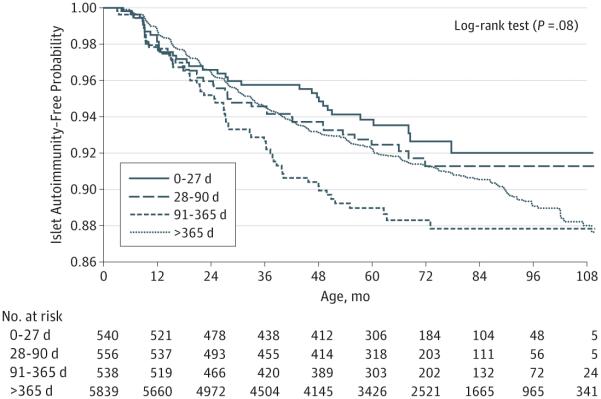
Kaplan-Meier curves of developing IA suggested that the earliest exposure of probiotics had the lowest risk of IA whereas the exposure of probiotics during 91 to 365 days had the highest risk of IA. However, the associations were not statistically significant (P = .08, log-rank test) (Figure). The estimated HRs and P values from the adjusted Cox proportional hazards regression models are listed in Table 3. Early exposure to probiotics during the first 27 days of life (n = 540) revealed decreased risk of IA (HR, 0.66; 95% CI, 0.45-0.96) in the TEDDY children when adjusting for FDR status, HLA-DR-DQ genotype (DR3/4 vs other), sex, birth order, mode of delivery, maternal age, maternal probiotic use, smoking during pregnancy, exclusive breastfeeding duration, birth year, child antibiotic use, and diarrhea. Results of the exploratory analysis suggested that very early exposure to probiotics may be important in relation to IA (Table 3). Therefore, we decided to focus on the early exposure (at 0-27 days) and the risk of IA in our further analyses. Early exposure of probiotics was associated with decreased risk of IA (HR, 0.66; 95% CI, 0.46-0.94) when compared with exposure after 27 days or no exposure and adjusted for FDR status (P < .001), HLA-DR-DQ genotype (P < .001), sex (P = .006), birth order (P = .15), mode of delivery (P = .46), maternal age (P = .98), maternal probiotic use (P = .82), smoking during pregnancy (P = .15), exclusive breastfeeding duration (P = .42), birth year (P = .70), child antibiotic use (P = .75), and diarrhea (P = .61).
Figure.
Islet Autoimmunity Risk by First Probiotic Exposure Age of the Child
Table 3.
First Probiotic Exposure of the Child via Infant Formula and/or Dietary Supplement During the First Year of Life and Risk of IA
| No. (%) of Infants |
|||
|---|---|---|---|
| Variable | Developed IA (n = 601) |
Did Not Develop IA (n = 6872) |
HR (95% CI)a |
| Country | |||
|
| |||
| United States | 201 (33.4) | 2845 (41.4) | … b |
|
| |||
| Finland | 151 (25.1) | 1507 (21.9) | … |
|
| |||
| Germany | 46 (7.7) | 460 (6.7) | … |
|
| |||
| Sweden | 203 (33.8) | 2060 (30.0) | … |
|
| |||
| Timing of first probiotic exposure, d | |||
|
| |||
| 0-27 | 34 (5.7) | 506 (7.4) | 0.66 (0.45-0.96) |
|
| |||
| 28-90 | 41 (6.8) | 515 (7.5) | 0.85(0.61-1.19) |
|
| |||
| 91-365 | 57 (9.5) | 481 (7.0) | 1.16 (0.86-1.57) |
|
| |||
| After 1 year or no exposure | 469 (78.0) | 5370 (78.1) | 1 [Reference] |
|
| |||
| FDR with T1DM | 126 (21.0) | 716 (10.4) | 2.30 (1.87-2.84) |
|
| |||
| High-risk HLA-DR- DR3/4 | 304 (50.6) | 2629 (38.3) | 1.76 (1.50-2.07) |
|
| |||
| Female sex | 262 (43.6) | 3397 (49.4) | 0.79 (0.67-0.94) |
Abbreviations: FDR, first-degree relative; HR, hazard ratio; IA, islet autoimmunity; T1DM, type 1 diabetes mellitus.
The HRs were adjusted for FDR status, HLA-DR genotype, sex, and the following nonsignificant covariates: birth order, mode of delivery, exclusive breastfeeding duration, birth year, child antibiotic use, diarrhea, maternal age, maternal probiotic use, and maternal smoking during pregnancy and stratified for country.
Ellipses indicate data not applicable.
Our analyses also revealed an interaction (P = .02) between early probiotic exposure (at 0-27 days) and HLA genotype in relation to IA. Separate analyses by HLA-DR-DQ genotype revealed a strong inverse association between early probiotic exposure and IA among those with an HLA genotype of DR3/4 (HR, 0.40; 95% CI, 0.21-0.74) but not among other genotypes (HR, 0.97; 95% CI, 0.62-1.54).
We did not find a statistically significant interaction between early probiotic exposure (0-27 days) and country (P = .34). The country-specific HRs were not heterogeneous (United States: HR, 0.98; 95% CI, 0.14-7.02; Finland: HR, 0.60; 95% CI, 0.38-0.97; Germany: HR, 0.98; 95% CI, 0.38-2.50; Sweden: HR, 0.73; 95% CI, 0.32-1.67), reflecting a possible protective association between early probiotic exposure and IA.
Discussion
In this multinational cohort study of children at increased genetic risk of T1DM, we observed a reduction in the risk of IA in the children who had received probiotics via dietary supplements and/or via fortified infant formula before or at the age of 27 days compared with those who had first received probiotics after 27 days or not at all. Early probiotic exposure was associated with 60% decrease in the risk of IA among children with the DR3/4 genotype but not among other genotypes.
The strengths of the study included a large international sample with consistent recording of the information on child diet, including probiotics, and health conditions covering the whole first year of life. A limitation of the study was that the species and amounts of microbes from probiotics were not studied. Most of the supplements used by TEDDY children contained mixtures of various Lactobacillus and Bifidobacterium species along with other commonly used probiotics. Therefore, the effect of specific species could not be evaluated as has been done in co ntrolled clinical trials.35 In addition, the stability of probiotic bacteria is dependent on many factors (eg, storage conditions).36,37 Therefore, it would not have been possible to compare doses of probiotics.
Probiotic use varied between TEDDY countries. Giving probiotic supplements to neonates is a fairly new trend, particularly in the United States.38 Even today probiotics are not recommended as a routine supplementation in the United States even though adverse events associated with probiotic use are extremely rare.23,39,40 Smaller population-based interventions on probiotic use and health outcomes among infants in Finland40 have received considerable attention by the media, which may have boosted the consumption of probiotics in that country.
Antibiotic medication use was common at the ages of 3 to 12 months, and probiotic use was often associated with administration of antibiotics. There is also evidence that antibiotic use may increase the risk of T1DM.34 This could contribute to the fact that we did not find a protective association between later introduction of probiotics and the risk of IA. We also have to consider that the probiotics may be able to modify gut microbiota only in early life because after introduction of solid foods, diet may have an overly dominant effect on gut microbiota composition, making probiotic supplementation less successful.41 Young infants are particularly susceptible to infectious diseases.42 Lönnrot et al43 also noticed that respiratory infections during early life in TEDDY were often accompanied by gastrointestinal symptoms, such as diarrhea, which we found to be strongly associated with probiotic use. The very early weeks of life may open a window for probiotics to modify gut microbiota favorably, thus helping the immune system of an infant’s gut to gain the maturity needed for correctly processing new environmental exposures, such as pathogens.
Approximately 70% of all gastroenteritis cases are caused by a virus,44 making an antibiotic treatment ineffective. Probiotics are often prescribed instead to shorten diarrheal episodes.44,45 Probiotic use was strongly associated with gastroenteritis in TEDDY. Shortened duration of gastroenteritis by probiotic treatment may protect infant gut from extended inflammation and adverse immunity-suppressing consequences.45 This may partly explain why we did not find an association between gastroenteritis and IA even though such an association has been suggested earlier.46,47
Both T1DM and T1DM-related autoimmunity have multifactorial origins.33,48,49 Contributing factors and their interplay with probiotics could also vary among countries. The recent observation of a plateau in the incidence of T1DM in Finland50 suggests changes in environmental exposures, for example, in serum 25-hydroxyvitamin D concentrations.51 The increasing trend of probiotic use40,52 also preceded the plateauing rates of T1DM. Respiratory tract infections at an early age have been found to be associated with IA.53 Early use of probiotics and relatively lower rates of the common cold before 3 months of age in Finland also warrant further study. However, the changes in the exposures and their putative connection to the incidence of IA and T1DM in Finland still lack conclusive evidence.
Finding the larger protective association between early probiotic exposure and IA among children with the DR3/4 genotype, when compared with other genotypes, suggests a gene-environment interaction. The genotype may influence the interaction of the host immune system with the bacteria present in the probiotic supplement. An earlier study54 has also suggested that the HLA genotype may modify the association between the timing of dietary exposure and IA.
This is the first time, to our knowledge, that the association between probiotic use and T1DM-related IA has been studied in a longitudinal, observational setting among genetically high-risk children. Of importance, a protective association between early probiotic use and T1DM-related IA has been observed. Previous studies11,12,14 have provided evidence that imbalance in gut microbiota may be connected with autoimmune disorders, such as T1DM, and that changes in the microbiota precede the pathogenic condition. However, studies reporting a successful manipulation of gut microbiota by probiotics in humans are scarce. In any case, influencing the gut microbiota with ingested probiotics would be expected to be more effective very early in life, as we observed.
Conclusions
Early exposure to supplemental probiotics may decrease the risk of IA among children at elevated risk of T1DM. However, a randomized clinical trial should confirm the association, and mechanistic analyses are needed to identify potential environmental factors (eg, infections that could mediate the association). These results have to be confirmed before making recommendations on the use of probiotic supplementation.
At a Glance.
Probiotics are live organisms that may confer health benefits on the host.
The aim of this study was to examine the association between early probiotic exposure and islet autoimmunity among children in The Environmental Determinants of Diabetes in the Young study.
Early administration of probiotics, during the first 27 days of life, may be associated with reduced risk of islet autoimmunity (hazard ratio [HR], 0.66; 95% CI, 0.45-0.96) among children who were genetically at increased risk for type 1 diabetes mellitus.
This reduced risk of islet autoimmunity was primarily observed in children with the highest-risk HLA genotype of DR3/4 (HR, 0.40; 95% CI, 0.21-0.74) but not in children with the other, moderately higher-risk genotypes (HR, 0.97; 95% CI, 0.62-1.54).
Acknowledgments
Funding/Support: This study was supported by grants U01 DK63829, U01 DK63861, U01 DK63821, U01 DK63865, U01 DK63863, U01 DK63836, U01 DK63790, UC4 DK63829, UC4 DK63861, UC4 DK63821, UC4 DK63865, UC4 DK63863, UC4 DK63836, UC4 DK95300, and UC4 DK100238 and contract HHSN267200700014C from the National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Allergy and Infectious Diseases, National Institute of Child Health and Human Development, National Institute of Environmental Health Sciences, Juvenile Diabetes Research Foundation, and Centers for Disease Control and Prevention. This work is supported in part by the National Institutes of Health/National Center for Advancing Translational Sciences Clinical and Translational Science Awards UL1 TR000064 (University of Florida) and UL1 TR001082 (University of Colorado).
Role of the Funder/Sponsor: The sponsors of this study were represented on the Steering Committee and played a role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication. The corresponding author had the final decision to submit the manuscript for publication.
Author Affiliations.
Health Informatics Institute, Morsani College of Medicine, University of South Florida, Tampa (Uusitalo, Liu, Yang, Butterworth, Krischer); Department of Clinical Sciences, Lund University, Malmö, Sweden (Aronsson, Lernmark); Institute of Diabetes Research, Helmholtz Zentrum München and Forschergruppe Diabetes, Klinikum rechts der Isar, Technische Universität München and Forschergruppe Diabetes e.V., Munich, Germany (Hummel, Ziegler); Barbara Davis Center for Childhood Diabetes, University of Colorado School of Medicine, Aurora (Rewers); Pacific Northwest Diabetes Research Institute, Seattle, Washington (Hagopian); Medical College of Georgia, Georgia Regents University, Augusta (She); Department of Pediatrics, University of Turku and Turku University Hospital, Turku, Finland (Simell, Toppari); Department of Physiology, Institute of Biomedicine, University of Turku, Turku, Finland (Toppari); National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, Maryland (Akolkar); Department of Epidemiology, Colorado School of Public Health, University of Colorado Denver, Aurora (Norris); National Institute for Health and Welfare, Nutrition Unit, Helsinki, Finland (Virtanen); School of Health Sciences and Center for Child Health Research, University of Tampere and Tampere University Hospital, Tampere, Finland (Virtanen); The Science Center of Pirkanmaa Hospital District, Tampere, Finland (Virtanen).
Author Contributions: Drs Norris and Virtanen contributed equally to this study. Drs Uusitalo and Liu had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Uusitalo, Lernmark, Rewers, Hagopian, She, Simell, Toppari, Ziegler, Akolkar, Krischer, Virtanen.
Acquisition, analysis, or interpretation of data: Uusitalo, Liu, Yang, Aronsson, Hummel, Butterworth, Rewers, Hagopian, She, Simell, Toppari, Ziegler, Krischer, Norris, Virtanen.
Drafting of the manuscript: Uusitalo, Lernmark.
Critical revision of the manuscript for important intellectual content: Liu, Yang, Aronsson, Hummel, Butterworth, Lernmark, Rewers, Hagopian, She, Simell, Toppari, Ziegler, Akolkar, Krischer, Norris, Virtanen.
Statistical analysis: Liu, Krischer.
Obtained funding: Akolkar, Lernmark, Rewers, Hagopian, She, Simell, Toppari, Ziegler, Krischer.
Administrative, technical, or material support: Hummel, Rewers, Hagopian, Simell, Toppari, Ziegler, Krischer, Virtanen.
Study supervision: Uusitalo, Lernmark, Hagopian, She, Toppari, Ziegler, Krischer, Norris, Virtanen.
Conflict of Interest Disclosures: None reported.
Group Information: TEDDY Study Group: Colorado Clinical Center: Marian Rewers, MD, PhD, (principal investigator), Kimberly Bautista, Judith Baxter, Ruth Bedoy, Daniel Felipe-Morales, Brigitte Frohnert, MD, Patricia Gesualdo, Michelle Hoffman, Rachel Karban, Edwin Liu, MD, Jill Norris, PhD, Adela Samper-Imaz, Andrea Steck, MD, Kathleen Waugh, Hali Wright; University of Colorado, Anschutz Medical Campus, Barbara Davis Center for Childhood Diabetes.
Georgia/Florida Clinical Center: Jin-Xiong She, PhD (principal investigator), Desmond Schatz, MD, Diane Hopkins, Leigh Steed, Jamie Thomas, Katherine Silvis, Michael Haller, MD, Meena Shankar, Eleni Sheehan, Melissa Gardiner, Richard McIndoe, PhD, Ashok Sharma, Joshua Williams, Gabriela Foghis, Stephen W. Anderson, MD, Richard Robinson; Center for Biotechnology and Genomic Medicine, Georgia Regents University, University of Florida, and Pediatric Endocrine Associates, Atlanta.
Germany Clinical Center: Anette G. Ziegler, MD (principal investigator), Andreas Beyerlein, PhD, Ezio Bonifacio, PhD, Michael Hummel, MD, Sandra Hummel, PhD, Kristina Foterek, Mathilde Kersting, PhD, Annette Knopff, Sibylle Koletzko, MD, Claudia Peplow, Roswith Roth, PhD, Joanna Stock, Elisabeth Strauss, Katharina Warncke, MD, Christiane Winkler, PhD; Forschergruppe Diabetes e.V. and Institute of Diabetes Research, Helmholtz Zentrum München, and Klinikum rechts der Isar, Technische Universität München; Center for Regenerative Therapies, TU Dresden; Dr. von Hauner Children’s Hospital, Department of Gastroenterology, Ludwig Maximillians University Munich; and Research Institute for Child Nutrition, Dortmund.
Finland Clinical Center: Jorma Toppari, MD, PhD (principal investigator), Olli G. Simell, MD, PhD, Annika Adamsson, PhD, Heikki Hyöty, MD, PhD, Jorma Ilonen, MD, PhD, Sanna Jokipuu, Tiina Kallio, Miia Kähönen, Mikael Knip, MD, PhD, Annika Koivu, Mirva Koreasalo, Kalle Kurppa, MD, PhD, Maria Lönnrot, MD, PhD, Elina Mäntymäki, Katja Multasuo, Juha Mykkänen, PhD, Tiina Niininen, Mia Nyblom, Petra Rajala, Jenna Rautanen, Anne Riikonen, Minna Romo, Satu Simell, MD, PhD, Tuula Simell, PhD, Ville Simell, Maija Sjöberg, Aino Stenius, Maria Särmä, Sini Vainionpää, Eeva Varjonen, Riitta Veijola, MD, PhD, Suvi M. Virtanen, MD, PhD, Mari Vähä-Mäkilä, Mari Åkerlund; University of Turku, University of Tampere, University of Oulu, Turku University Hospital, Hospital District of Southwest Finland, Tampere University Hospital, Oulu University Hospital, National Institute for Health and Welfare, Finland, and University of Kuopio.
Sweden Clinical Center: Åke Lernmark, PhD (principal investigator), Daniel Agardh, MD, PhD, Carin Andrén Aronsson, Maria Ask, Jenny Bremer, Ulla-Marie Carlsson, Corrado Cilio, PhD, MD, Emelie Ericson-Hallström, Lina Fransson, Thomas Gard, Joanna Gerardsson, Rasmus Bennet, Monica Hansen, Gertie Hansson, Susanne Hyberg, Fredrik Johansen, Berglind Jonasdottir, MD, Linda Jonsson, Helena Elding Larsson, MD, PhD, Sigrid Lenrick Forss, Maria Månsson-Martinez, Maria Markan, Jessica Melin, Zeliha Mestan, Kobra Rahmati, Anita Ramelius, Anna Rosenquist, Falastin Salami, Sara Sibthorpe, Birgitta Sjöberg, Ulrica Swartling, PhD, Evelyn Tekum Amboh, Erika Trulsson, Carina Törn, PhD, Anne Wallin, Åsa Wimar, Sofie Åberg; Lund University.
Washington Clinical Center: William A. Hagopian, MD, PhD (principal investigator), Xiang Yan, MD, Michael Killian, Claire Cowen Crouch, Jennifer Skidmore, Stephen Ayres, Kayleen Dunson, Diana Heaney, Rachel Hervey, Corbin Johnson, Rachel Lyons, Arlene Meyer, Denise Mulenga, Emma Schulte, Elizabeth Scott, Joshua Stabbert, John Willis; Pacific Northwest Diabetes Research Institute.
Pennsylvania Satellite Center: Dorothy Becker, MD, Margaret Franciscus, MaryEllen Dalmagro-Elias Smith, Ashi Daftary, MD, Mary Beth Klein, Chrystal Yates; Children’s Hospital of Pittsburgh of UPMC.
Data Coordinating Center: Jeffrey P. Krischer, PhD (principal investigator), Michael Abbondondolo, Sarah Austin-Gonzalez, Sandra Baethke, Rasheedah Brown, Brant Burkhardt, PhD, Martha Butterworth, Joanna Clasen, David Cuthbertson, Christopher Eberhard, Steven Fiske, Dena Garcia, Jennifer Garmeson, Veena Gowda, David Hadley, PhD, Kathleen Heyman, Hye-Seung Lee, PhD, Shu Liu, Xiang Liu, PhD, Kristian Lynch, PhD, Jamie Malloy, Cristina McCarthy, Wendy McLeod, Steven Meulemans, Chris Shaffer, Laura Smith, PhD, Susan Smith, Roy Tamura, PhD, Ulla Uusitalo, PhD, Kendra Vehik, PhD, Ponni Vijayakandipan, Keith Wood, Jimin Yang, PhD, RD; University of South Florida.
Project scientist: Beena Akolkar, PhD.; National Institutes of Diabetes and Digestive and Kidney Diseases.
Other contributors: Kasia Bourcier, PhD, National Institute of Allergy and Infectious Diseases; Thomas Briese, PhD, Columbia University; Suzanne Bennett Johnson, PhD, Florida State University; Steve Oberste, PhD, Centers for Disease Control and Prevention; Eric Triplett, PhD; University of Florida.
Autoantibody Reference Laboratories: Liping Yu, MD, Dongmei Miao, MD, Polly Bingley, MD, FRCP, Alistair Williams, Kyla Chandler, Saba Rokni, Claire Caygill, Nicholas Lovis, Claire Williams, Rebecca Wyatt. Barbara Davis Center for Childhood Diabetes, University of Colorado, Denver, and School of Clinical Sciences, University of Bristol, Bristol, England.
Cortisol Laboratory: Elisabeth Aardal Eriksson, MD, PhD, Ing-Marie Lundgren, Ewa Lönn Karlsson, Dzeneta Nezirevic Dernroth, PhD; Department of Clinical Chemistry, Linköping University Hospital, Linköping, Sweden.
Dietary Biomarkers Laboratory: Iris Erlund, PhD, Irma Salminen, Jouko Sundvall, Jaana Leiviskä, Nina Kangas; National Institute for Health and Welfare, Helsinki, Finland.
HbA1c Laboratory: Randie R. Little, PhD, Alethea L. Tennill; Diabetes Diagnostic Laboratory, Department of Pathology, University of Missouri School of Medicine.
HLA Reference Laboratory: Henry Erlich, PhD, Steven J. Mack, PhD, Anna Lisa Fear; Center for Genetics, Children’s Hospital Oakland Research Institute.
Metabolomics Laboratory: Oliver Fiehn, PhD, Bill Wikoff, PhD, Brian Defelice, Dmitry Grapov, PhD, Tobias Kind, PhD, Mine Palazoglu, Luis Valdiviez, Benjamin Wancewicz, Gert Wohlgemuth, Joyce Wong; UC Davis Metabolomics Center.
Microbiome and Viral Metagenomics Laboratory: Joseph F. Petrosino, PhD; Alkek Center for Metagenomics and Microbiome Research, Department of Molecular Virology and Microbiology, Baylor College of Medicine.
OGTT Laboratory: Santica M. Marcovina, PhD, ScD, Vinod P. Gaur, PhD; Northwest Lipid Metabolism and Diabetes Research Laboratories, University of Washington.
Proteomics Laboratory: Richard D. Smith, PhD, Thomas O. Metz, PhD, Charles Ansong, PhD, Bobbie-Jo Webb-Robertson, PhD, Hugh D. Mitchell, PhD; Pacific Northwest National Laboratory.
Repository: Heather Higgins, Sandra Ke; National Institute of Diabetes and Digestive and Kidney Diseases Biosample Repository at Fisher BioServices.
RNA Laboratory and Gene Expression Laboratory: Jin-Xiong She, PhD (principal investigator), Richard McIndoe, PhD, Haitao Liu, MD, John Nechtman, Yansheng Zhao, Na Jiang, MD, Yanna Tian, MS, Guangkuo Dong, MS; Jinfiniti Biosciences, LLC.
SNP Laboratory: Stephen S. Rich, PhD, Wei-Min Chen, PhD, Suna Onengut-Gumuscu, PhD, Emily Farber, Rebecca Roche Pickin, PhD, Jordan Davis, Dan Gallo, Jessica Bonnie, Paul Campolieto; Center for Public Health Genomics, University of Virginia.
REFERENCES
- 1.Hara N, Alkanani AK, Ir D, et al. The role of the intestinal microbiota in type 1 diabetes. Clin Immunol. 2013;146(2):112–119. doi: 10.1016/j.clim.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Kaplan JL, Shi HN, Walker WA. The role of microbes in developmental immunologic programming. Pediatr Res. 2011;69(6):465–472. doi: 10.1203/PDR.0b013e318217638a. [DOI] [PubMed] [Google Scholar]
- 3.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336(6086):1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tlaskalová-Hogenová H, Stěpánková R, Kozáková H, et al. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cell Mol Immunol. 2011;8(2):110–120. doi: 10.1038/cmi.2010.67. doi:10.1038/cmi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaarala O, Atkinson MA, Neu J. The “perfect storm” for type 1 diabetes: the complex interplay between intestinal microbiota, gut permeability, and mucosal immunity. Diabetes. 2008;57(10):2555–2562. doi: 10.2337/db08-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cani PD, Possemiers S, Van de Wiele T, et al. Changes in gut microbiota c ontrol inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut. 2009;58(8):1091–1103. doi: 10.1136/gut.2008.165886. doi:10.1136/gut.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bosi E, Molteni L, Radaelli MG, et al. Increased intestinal permeability precedes clinical onset of type 1 diabetes. Diabetologia. 2006;49(12):2824–2827. doi: 10.1007/s00125-006-0465-3. [DOI] [PubMed] [Google Scholar]
- 8.Siggers RH, Siggers J, Thymann T, Boye M, Sangild PT. Nutritional modulation of the gut microbiota and immune system in preterm neonates susceptible to necrotizing enterocolitis. J Nutr Biochem. 2011;22(6):511–521. doi: 10.1016/j.jnutbio.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 9.Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489(7415):231–241. doi: 10.1038/nature11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flint HJ, Scott KP, Louis P, Duncan SH. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol. 2012;9(10):577–589. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- 11.Kostic AD, Gevers D, Siljander H, et al. DIABIMMUNE Study Group The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe. 2015;17(2):260–273. doi: 10.1016/j.chom.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giongo A, Gano KA, Crabb DB, et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 2011;5(1):82–91. doi: 10.1038/ismej.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murri M, Leiva I, Gomez-Zumaquero JM, et al. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. BMC Med. 2013;11:46. doi: 10.1186/1741-7015-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mejía-León ME, Petrosino JF, Ajami NJ, Domínguez-Bello MG, de la Barca AMC. Fecal microbiota imbalance in Mexican children with type 1 diabetes. Sci Rep. 2014;4:3814. doi: 10.1038/srep03814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6(237):237ra65. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahrné S, Lönnermark E, Wold AE, et al. Lactobacilli in the intestinal microbiota of Swedish infants. Microbes Infect. 2005;7(11-12):1256–1262. doi: 10.1016/j.micinf.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 17.Sekirov I, Russell SL, Antunes LCM, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90(3):859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 18.Salami F, Abels M, Hyöty H, et al. TEDDY study group Detection of Lactobacilli in monthly mail-in stool samples from 3-18 months old infants at genetic risk for type 1 diabetes. Int J Probiotics Prebiotics. 2012;7(3-4):135–144. [PMC free article] [PubMed] [Google Scholar]
- 19.Walker WA. Initial intestinal colonization in the human infant and immune homeostasis. Ann Nutr Metab. 2013;63(suppl 2):8–15. doi: 10.1159/000354907. [DOI] [PubMed] [Google Scholar]
- 20.Grönlund MM, Nuutila J, Pelto L, et al. Mode of delivery directs the phagocyte functions of infants for the first 6 months of life. Clin Exp Immunol. 1999;116(3):521–526. doi: 10.1046/j.1365-2249.1999.00902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Penders J, Thijs C, Vink C, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118(2):511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 22.Food and Agriculture Organization of the United Nations and World Health Organization . Health and Nutritional Properties of Probiotics in Food Including Powder Milk With Live Lactic Acid Bacteria: Report of a Joint FAO/WHO Expert Consultation on the Health and Nutritional Properties of Probiotics in Food Including Powder Milk With Live Lactic Acid Bacteria. Food and Agriculture Organization of the United Nations; Rome, Italy: 2001. [Google Scholar]
- 23.Bergmann H, Rodríguez JM, Salminen S, Szajewska H. Probiotics in human milk and probiotic supplementation in infant nutrition: a workshop report. Br J Nutr. 2014;112(7):1119–1128. doi: 10.1017/S0007114514001949. [DOI] [PubMed] [Google Scholar]
- 24.Martín R, Langa S, Reviriego C, et al. Human milk is a source of lactic acid bacteria for the infant gut. J Pediatr. 2003;143(6):754–758. doi: 10.1016/j.jpeds.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 25.Calcinaro F, Dionisi S, Marinaro M, et al. Oral probiotic administration induces interleukin-10 production and prevents spontaneous autoimmune diabetes in the non-obese diabetic mouse. Diabetologia. 2005;48(8):1565–1575. doi: 10.1007/s00125-005-1831-2. [DOI] [PubMed] [Google Scholar]
- 26.Lau K, Benitez P, Ardissone A, et al. I nhibition of type 1 diabetes correlated to a Lactobacillus johnsonii N6.2-mediated Th17 bias. J Immunol. 2011;186(6):3538–3546. doi: 10.4049/jimmunol.1001864. [DOI] [PubMed] [Google Scholar]
- 27.TEDDY Study Group The Environmental Determinants of Diabetes in the Young (TEDDY) study: study design. Pediatr Diabetes. 2007;8(5):286–298. doi: 10.1111/j.1399-5448.2007.00269.x. [DOI] [PubMed] [Google Scholar]
- 28.TEDDY Study Group The Environmental Determinants of Diabetes in the Young (TEDDY) Study. AnnNY Acad Sci. 2008;1150:1–13. doi: 10.1196/annals.1447.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hagopian WA, Erlich H, Lernmark A, et al. TEDDY Study Group The Environmental Determinants of Diabetes in the Young (TEDDY): genetic criteria and international diabetes risk screening of 421 000 infants. Pediatr Diabetes. 2011;12(8):733–743. doi: 10.1111/j.1399-5448.2011.00774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bonifacio E, Yu L, Williams AK, et al. Harmonization of glutamic acid decarboxylase and islet antigen-2 autoantibody assays for national institute of diabetes and digestive and kidney diseases consortia. J Clin Endocrinol Metab. 2010;95(7):3360–3367. doi: 10.1210/jc.2010-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Babaya N, Yu L, Miao D, et al. Comparison of insulin autoantibody: polyethylene glycol and micro-IAA 1-day and 7-day assays. Diabetes Metab Res Rev. 2009;25(7):665–670. doi: 10.1002/dmrr.1014. [DOI] [PubMed] [Google Scholar]
- 32.Törn C, Mueller PW, Schlosser M, Bonifacio E, Bingley PJ, Participating Laboratories Diabetes Antibody Standardization Program: evaluation of assays for autoantibodies to glutamic acid decarboxylase and islet antigen-2. Diabetologia. 2008;51(5):846–852. doi: 10.1007/s00125-008-0967-2. [DOI] [PubMed] [Google Scholar]
- 33.Vehik K, Dabelea D. The changing epidemiology of type 1 diabetes: why is it going through the roof? Diabetes Metab Res Rev. 2011;27(1):3–13. doi: 10.1002/dmrr.1141. [DOI] [PubMed] [Google Scholar]
- 34.Kilkkinen A, Virtanen SM, Klaukka T, et al. Use of antimicrobials and risk of type 1 diabetes in a population-based mother-child cohort. Diabetologia. 2006;49(1):66–70. doi: 10.1007/s00125-005-0078-2. [DOI] [PubMed] [Google Scholar]
- 35.Gomes AC, Bueno AA, de Souza RGM, Mota JF. Gut microbiota, probiotics and diabetes. Nutr J. 2014;13:60. doi: 10.1186/1475-2891-13-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Achour M, Mtimet N, Cornelius C, et al. Application of the accelerated shelf life testing method ASLT to study the survival rates of freeze-dried Lactococcus starter cultures. J Chem Technol Biotechnol. 2001;76(6):624–628. [Google Scholar]
- 37.Senz M, van Lengerich B, Bader J, Stahl U. Control of cell morphology of probiotic Lactobacillus acidophilus for enhanced cell stability during industrial processing. Int J Food Microbiol. 2015;192:34–42. doi: 10.1016/j.ijfoodmicro.2014.09.015. [DOI] [PubMed] [Google Scholar]
- 38.Armstrong C. AAP Reports on use of probiotics and prebiotics in children. Am Fam Physician. 2011;83(7):849–852. [Google Scholar]
- 39.Sung V, Collett S, de Gooyer T, Hiscock H, Tang M, Wake M. Probiotics to prevent or treat excessive infant crying: systematic review and meta-analysis. JAMA Pediatr. 2013;167(12):1150–1157. doi: 10.1001/jamapediatrics.2013.2572. [DOI] [PubMed] [Google Scholar]
- 40.Salminen MK, Tynkkynen S, Rautelin H, et al. Lactobacillus bacteremia during a rapid increase in probiotic use of Lactobacillus rhamnosus GG in Finland. Clin Infect Dis. 2002;35(10):1155–1160. doi: 10.1086/342912. [DOI] [PubMed] [Google Scholar]
- 41.Fallani M, Amarri S, Uusijarvi A, et al. INFABIO team Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiology. 2011;157:1385–1392. doi: 10.1099/mic.0.042143-0. pt 5. [DOI] [PubMed] [Google Scholar]
- 42.McIntyre AF, Gonzalez-Feliciano AG, Bryan LN, Santibanez TA, Williams WW, Singleton JA, Centers for Disease Control and Prevention (CDC) Seasonal influenza vaccination coverage - United States, 2009-10 and 2010-11. MMWR Surveill Summ. 2013;62(suppl 3):65–68. [PubMed] [Google Scholar]
- 43.Lönnrot M, Lynch K, Larsson HE, et al. TEDDY Study Group A method for reporting and classifying acute infectious diseases in a prospective study of young children: TEDDY. BMC Pediatr. 2015;15:24. doi: 10.1186/s12887-015-0333-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ciccarelli S, Stolfi I, Caramia G. Management strategies in the treatment of neonatal and pediatric gastroenteritis. Infect Drug Resist. 2013;6:133–161. doi: 10.2147/IDR.S12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hsieh MH, Versalovic J. The human microbiome and probiotics: implications for pediatrics. Curr Probl Pediatr Adolesc Health Care. 2008;38(10):309–327. doi: 10.1016/j.cppeds.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Honeyman MC, Coulson BS, Stone NL, et al. Association between rotavirus infection and pancreatic islet autoimmunity in children at risk of developing type 1 diabetes. Diabetes. 2000;49(8):1319–1324. doi: 10.2337/diabetes.49.8.1319. [DOI] [PubMed] [Google Scholar]
- 47.Galleri L, Sebastiani G, Vendrame F, Grieco FA, Spagnuolo I, Dotta F. Viral infections and diabetes. Adv Exp Med Biol. 2012;771:252–271. doi: 10.1007/978-1-4614-5441-0_20. [DOI] [PubMed] [Google Scholar]
- 48.Virtanen SM, Knip M. Nutritional risk predictors of beta cell autoimmunity and type 1 diabetes at a young age. Am J Clin Nutr. 2003;78(6):1053–1067. doi: 10.1093/ajcn/78.6.1053. [DOI] [PubMed] [Google Scholar]
- 49.Knip M, Virtanen SM, Åkerblom HK. Infant feeding and the risk of type 1 diabetes. Am J Clin Nutr. 2010;91(5):1506S–1513S. doi: 10.3945/ajcn.2010.28701C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harjutsalo V, Sund R, Knip M, Groop PH. Incidence of type 1 diabetes in Finland. JAMA. 2013;310(4):427–428. doi: 10.1001/jama.2013.8399. [DOI] [PubMed] [Google Scholar]
- 51.Mäkinen M, Simell V, Mykkänen J, et al. An increase in serum 25-hydroxyvitamin D concentrations preceded a plateau in type 1 diabetes incidence in Finnish children. J Clin Endocrinol Metab. 2014;99(11):E2353–E2356. doi: 10.1210/jc.2014-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ylinen S, Hämeen-Anttila K, Sepponen K, Lindblad AK, Ahonen R. The use of prescription medicines and self-medication among children--a population-based study in Finland. Pharmacoepidemiol Drug Saf. 2010;19(10):1000–1008. doi: 10.1002/pds.1963. [DOI] [PubMed] [Google Scholar]
- 53.Beyerlein A, Wehweck F, Ziegler AG, Pflueger M. Respiratory infections in early life and the development of islet autoimmunity in children at increased type 1 diabetes risk: evidence from the BABYDIET study. JAMA Pediatr. 2013;167(9):800–807. doi: 10.1001/jamapediatrics.2013.158. [DOI] [PubMed] [Google Scholar]
- 54.Norris JM, Barriga K, Klingensmith G, et al. Timing of initial cereal exposure in infancy and risk of islet autoimmunity. JAMA. 2003;290(13):1713–1720. doi: 10.1001/jama.290.13.1713. [DOI] [PubMed] [Google Scholar]