A unique feature of preimplantation development is the drastic, genome-wide changes occurring to nuclear architecture. In this review, Borsos and Torres-Padilla discuss the peculiarities of nuclear compartments and chromatin organization during mammalian preimplantation development in the context of the transition from totipotency to pluripotency.
Keywords: mouse embryo, nuclear architecture, totipotency
Abstract
In mammals, epigenetic reprogramming, the acquisition and loss of totipotency, and the first cell fate decision all occur within a 3-d window after fertilization from the one-cell zygote to the formation of the blastocyst. These processes are poorly understood in molecular detail, yet this is an essential prerequisite to uncover principles of stem cells, chromatin biology, and thus regenerative medicine. A unique feature of preimplantation development is the drastic genome-wide changes occurring to nuclear architecture. From studying somatic and in vitro cultured embryonic stem cells (ESCs) it is becoming increasingly established that the three-dimensional (3D) positions of genomic loci relative to each other and to specific compartments of the nucleus can act on the regulation of gene expression, potentially driving cell fate. However, the functionality, mechanisms, and molecular characteristics of the changes in nuclear organization during preimplantation development are only now beginning to be unraveled. Here, we discuss the peculiarities of nuclear compartments and chromatin organization during mammalian preimplantation development in the context of the transition from totipotency to pluripotency.
Totipotency is the capacity to form all cells in a new organism, including embryonic and extraembryonic tissues. A totipotent zygote is created with the fusion of two highly differentiated cells: the oocyte and the sperm that undergoes epigenetic reprogramming. Upon fertilization, the oocyte completes the second meiotic division, and the maternal chromosomes and the sperm chromatin decondense to form separate pronuclei. The paternal chromatin is quickly assembled into nucleosomes upon removal of the sperm-derived protamines. In contrast, the maternal chromatin inherits a nucleosomal configuration from oogenesis. The subsequent cell divisions are termed cleavages because the size of the daughter cells remains approximately half the size of their mother cells (Aiken et al. 2004). After five divisions, the mouse embryo reaches the blastocyst stage, which is initially composed of the inner cell mass (ICM) and the trophectoderm. While the ICM is pluripotent, the trophectoderm is considered a differentiated, epithelia-like tissue. Thus, the transition from a totipotent state in the zygote to a pluripotent state in the blastocyst occurs in only ∼3 d. Although nuclear size progressively shrinks from the zygote to the blastocyst stage, the nucleocytoplasmic ratio increases due to the cleavage nature of divisions. Another peculiarity of preimplantation development is the inheritance of a large protein pool from the oocyte, which enables the earliest steps of development until the embryo undergoes the maternal-to-zygotic transition and activates its own genome.
Nuclear organization refers to the spatial position of nuclear compartments as well as the position that specific regions of the genome adopt with respect to each other and the nuclear compartments. Functionally, increasing evidence suggests that the three-dimensional (3D) position of genomic regions during interphase may be causally related to the regulation of gene expression and chromatin architecture (Meister et al. 2011; Talamas and Capelson 2015). The physical proximity of genomic loci can promote an environment that is permissive or repressive for gene expression locally but can also favor translocations resulting in genome instability (Roix et al. 2003; Eskeland et al. 2010; Zhang et al. 2012). The organization of the nucleus can change over time in both the composition of nuclear compartments and the 3D positioning of the genome. Such changes have been documented to occur during development and upon reprogramming of cellular identity but also during disease progression (Noordermeer et al. 2011; Andrey et al. 2013; Lupianez et al. 2015). Nuclear organization has therefore emerged as a key player underlying the architectural definition of the epigenome.
Concerning the organization of the genome itself, chromosomes are the most well-defined structural entities. They contain clearly distinctive blocks of telomeric and pericentromeric domains, which are typically assembled into constitutive heterochromatin (Fig. 1A). Between them, the chromosomal arms harbor genes and also the repetitive units of rDNA, which cluster in the nucleolar organizer regions (NORs) distributed along six chromosomes in mice and five in humans (Henderson et al. 1972; Gibbons et al. 2015). Chromosome territories, the physical space occupied by individual chromosomes, were originally suggested as one of the main organizational and functional landmarks of the nucleus (Cremer et al. 2006). In general, gene-rich chromosomes localize toward the interior of the nucleus (Mayer et al. 2005). Additionally, gene-dense regions of a chromosome tend to loop out of its territory, while gene-poor domains cluster in the territory's core. These observations have had an enormous impact on our initial conception of nuclear organization functionally and have been reached by both microscopic approaches (e.g., DNA fluorescent in situ hybridization [FISH]) and using recent high-coverage techniques to uncover genomic interactions (e.g., HiC) (Boyle et al. 2011; Kalhor et al. 2012). Topologically associated domains (TADs) are large self-associating domains of chromosomes ranging several hundreds of kilobases. Their identification has led to a model for genome organization in which most genomic interactions are stable across cell types, and more specific gene regulation occurs within TADs (Sexton and Cavalli 2015). Indeed, condensation and decondensation as well as “looping” out from chromosome territories have been correlated to the transcriptional activity of Hox loci during developmental transitions (Chambeyron and Bickmore 2004; Eskeland et al. 2010). Moreover, the positions of specific loci within the nuclear space have been recently demonstrated to be heritable over several cell divisions (Therizols et al. 2014).
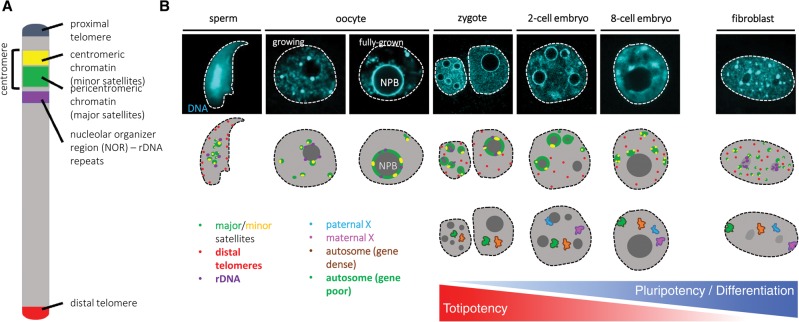
Figure 1.
Changes of chromatin organization during preimplantation development. (A) Schematic representation of the main building blocks and their organization on a mouse chromosome. rDNA repeats are present in only five of the 20 chromosomes in mice. (B) The differences in nuclear organization between mature gametes, preimplantation stages, and somatic cells are readily observable by a simple DNA (DAPI) staining. The top part of the scheme depicts the localization of the repetitive DNA elements studied by FISH: major satellites (green), minor satellites (yellow), rDNA (purple), and distal telomeres (red). In the mature sperm, rDNA and centromeric repeats cluster in the center, while distal telomeres are peripheral. During oocyte growth, centromeric repeats gradually assemble into a ring-like structure around the nucleolar precursor bodies (NPBs) and stay associated with them until the late two-cell stage, when they start reclustering into chromocenters, similar to somatic cells (fibroblast). rDNA is associated with the periphery of NPBs in oocytes and early embryos and gradually occupies nucleolar localization as somatic-like nucleoli form in place of NPBs. The localization of telomeres has not been determined in oocytes and, apart from the mature sperm, their localization has not been associated with nuclear compartments. The second scheme shows the localization of gene-rich (orange) and gene-poor (green) chromosomes as well as paternal (light blue) and maternal (pink) X chromosomes. Based on studies in bovine embryos, the gene density of a chromosome does not correlate with its radial position prior to embryonic genome activation (EGA, which occurs at the eight-cell stage in cattle and at the two-cell stage in mice). However, after EGA, gene-poor chromosomes occupy more peripheral localization compared with gene-dense ones. The imprinted inactive paternal X chromosome is preferably associated with the NPB in two- and four-cell stage mouse embryos but relocates from the NPB by the eight-cell stage. In the somatic cell depicted, due to random X inactivation, one of the X chromosomes forms the Barr body, preferentially located at the nuclear periphery.
Subnuclear compartments are structurally defined components constituted primarily of characteristic proteins. They constitute platforms for nuclear organization and, in some instances, attachment points for specific genomic loci. Indeed, the composition of the nuclear lamina, the position and architectural changes of the nucleolus, and the sequestration of genomic loci and regulatory proteins into promyelocytic leukemia (PML) bodies, nuclear speckles, histone locus bodies, transcription factories, Cajal bodies, and others have all been linked to gene regulation across a variety of cell types (Solovei et al. 2004; Peric-Hupkes et al. 2010; Sleeman and Trinkle-Mulcahy 2014).
Global changes in nuclear architecture are readily observable during tumorigenesis or differentiation or between different cell types. Rod cells from the retina of nocturnal mammals are a drastic example. They show global relocation of pericentromeric heterochromatin to the nuclear center during their post-mitotic terminal differentiation (Solovei et al. 2009). Studying this process in rod nuclei has yielded valuable information on the molecular mechanisms responsible for peripheral heterochromatin localization (Solovei et al. 2013).
The preimplantation mammalian embryo is another example. The nuclei of early embryos undergo an equally drastic yet very different global chromatin reorganization that occurs over just a couple of divisions after fertilization. These changes are visible when imaging the DNA directly and are dominated by the relocalization of AT-rich satellite sequences that are heavily labeled by, e.g., DAPI (Fig. 1B). The timing of this restructuring of the nuclear architecture coincides with the period of epigenetic reprogramming, the loss of totipotency, and the very first cell fate decisions. However, whether changes in nuclear organization are functionally required for these key biological processes is unknown. Notwithstanding, because similar general changes of chromatin organization are recapitulated in in vitro fertilized embryos and somatic cell nuclear transfer (SCNT)-derived embryos (Martin et al. 2006b; Popken et al. 2014), it is possible that this reorganization is a general feature of reprogramming to a totipotent state.
Here, we first review in general the protein composition of the main reference points of the nucleus and summarize how they are thought to serve as platforms for genome organization. We then collect the currently available literature on these subnuclear compartments and on how chromatin is organized in relation to mainly mouse preimplantation development
General overview of the main nuclear organizing platforms
Nuclear periphery: the nuclear lamina
The protein meshwork of intermediate-type filaments called the nuclear lamina provides the primary scaffold for the nuclear envelope. The main constituents of the nuclear lamina are A- and B-type lamins. Lamins B1 and B2 are transcribed from different genes, and at least one of them is present in every cell type (Dechat et al. 2010). The Lmna gene encodes two splice variants, lamins A and C, which differ in their C termini. Contrary to B-type lamins, lamin A has also been suggested to localize to the nuclear interior in some cell types analyzed, although the relevance of this finding is unclear (Legartova et al. 2014).
In higher eukaryotes, chromatin in the proximity of the nuclear lamina tends to be more heterochromatic and gene-poor and contains developmentally silenced and late-replicating domains (Guelen et al. 2008; Peric-Hupkes et al. 2010). This notion is supported by tethering experiments that show that recruitment of genetically modified loci using bacterial arrays to the nuclear periphery can result in their repression, albeit not in all cases analyzed (Finlan et al. 2008; Kumaran and Spector 2008; Reddy et al. 2008). Technically, the positional relationship between the genome and the nuclear lamina can be addressed using DNA adenine methyltransferase identification (DamID), in which the Dam enzyme of Escherichia coli is fused to components of the nuclear lamina and provokes 6A methylation in genomic regions that are in proximity. DamID allows the creation of maps of nuclear lamina contact probabilities across cell populations but also in single cells (Kind et al. 2015). Chromatin immunoprecipitation (ChIP) analysis of lamin B1 and lamin B1-DamID yield similar patterns of nuclear lamina associations (Handoko et al. 2011). Genome-wide mapping of nuclear lamina-proximal regions (referred to as lamina-associated domains [LADs]) in mouse embryonic stem cells (mESCs) and neural precursor cells revealed that the profile of LADs changes with cell differentiation (Peric-Hupkes et al. 2010). Interestingly however, the peripheral domain organization of chromatin does not seem to depend on lamins, at least in mESCs, indicating that other factors might be responsible for establishing a cell type-specific LAD composition in these cells (Amendola and van Steensel 2015). In some other more differentiated cell types, however, peripheral tethering of heterochromatin does depend on A-type lamins (Solovei et al. 2013).
Association of specific chromatin domains with the nuclear periphery can occur by several lamin-associated transmembrane proteins. Among them, lamin-B receptor (LBR) can anchor genomic regions through the recognition of heterochromatic histone marks directly or through binding to heterochromatin protein 1 (HP1) (Ye and Worman 1996; Hirano et al. 2012). LBR deletion or overexpression leads to a global change in nuclear organization, mainly of peripheral heterochromatin (Solovei et al. 2013). Other integral proteins of the nuclear envelope that contain a LEM domain (such as Lap2, Emerin, and Man1) mainly interact with chromatin through indirect binding via the barrier to autointegration factor (BAF) (Wilson and Foisner 2010). Additionally, other components of the nuclear membrane integrate cytoplasmic signals to the nuclear interior. Proteins of the linker of nucleoskeleton and cytoskeleton (LINC) complex contain either SUN or KASH domains that enable them to span either the inner or outer nuclear membrane and thus can become mediators of mechanotransduction stimuli from the cytoskeleton (Osmanagic-Myers et al. 2015). Although little is known as to whether and how proteins of the LINC complex might determine genome organization, increasing evidence demonstrates that mechanical signals from the cytoplasm can regulate gene expression (Ghosh et al. 2012; Kumar et al. 2014). However, this has not yet been explored in the context of early development. Whether and how mechanotransduction can affect nuclear organization of development will be an important research avenue in the following years.
Proteins of the nuclear pore complex (NPC) exert their function on the genome at several scales (for a recent detailed review, see Ibarra and Hetzer 2015). Briefly, in addition to their function as NPC building blocks to create channels for nucleocytoplasmic transport, nucleoporins (Nups) are also regarded as key reference points for genome organization and as regulators of gene expression. In yeast, anchoring of actively transcribed genes to NPCs is well documented and widespread even though, in some cases, a repressive effect of the pores has also been observed. Nup-dependent chromatin anchoring also affects global nuclear organization in Drosophila oocytes (Breuer and Ohkura 2015). In mammals, Nups can bind chromatin when sequestered into the nuclear envelope, and soluble, nucleoplasmic Nups can regulate gene activity (Kalverda et al. 2010). For instance, in mESC cultures, Nup153 acts both in the nucleoplasm and when incorporated into the nuclear envelope to repress developmental genes associated with differentiation. Those Nup153 target genes that localize to the nuclear periphery shift toward the nuclear interior upon Nup153 depletion, accompanied by their transcriptional activation, indicating that Nups can serve as attachment points of genome organization (Jacinto et al. 2015).
Nuclear subcomparments: the nucleolus
The nucleolus forms around rDNA (clusters of the tandemly repeated ribosomal RNA genes). Somatic cell nucleoli show a typical tripartite morphology with defined functions. The transcription of rRNA by RNA polymerase I (Pol I) occurs at the central fibrillar regions and dense fibrillar components of the nucleolus, while the processing of rRNAs takes place in the distinct granular components. Genomic regions associated with nucleoli are referred to as nucleolus-associated domains (NADs). Active transcription within NADs arises from rRNA genes as well as genes transcribed by RNA Pol III. Interestingly, RNA Pol II-dependent genes within NADs tend to be silenced and labeled with repressive histone modifications (Nemeth et al. 2010; van Koningsbruggen et al. 2010). NADs and LADs are similar in size (0.1–10 Mb) and show a substantial overlap. The movement of genomic regions from the nuclear lamina to the nucleolus over cell division (Kind et al. 2013) suggests that the nucleolus might be a compartment that promotes silencing of Pol II-dependent transcription similarly to the nuclear lamina. Functional interactions between the nuclear lamina and the nucleoli are indeed documented, and lamin B1 physically interacts with one of the main components of the nucleolus, Nucleophosmin 1. Additionally, depletion of lamin B1 in HeLa cells causes rearrangement of nucleoli (Martin et al. 2009). Thus, both the nuclear lamina and the nucleolus have been correlated with gene silencing.
According to the definitions and items discussed above, we summarize the current knowledge of the changes in nuclear components and in the 3D organization of chromatin occurring during early mammalian development. While the nucleolus and, to some extent, the components of the nuclear lamina have been better studied, the literature regarding other nuclear compartments is scarce, and we therefore focus on the former.
Nuclear compartments of the preimplantation embryo
Nuclear envelope: making the first embryonic nuclei
Upon fertilization, the nuclear envelope of the sperm is rapidly degraded, and maternally derived nuclear membranes contribute to the formation of the nuclear envelope of both pronuclei (Usui et al. 1997). Specifically in zygotes and two-cell stage embryos, the inner membrane of the nuclear envelope forms “blebs” that seem to contain granular material and fold into the perinuclear space (Szollosi and Szollosi 1988). The investigators hypothesize that blebbing could constitute a form of material transport from the nucleus, as vesicles detaching toward the cytoplasm are seen in zygotes and two-cell embryos. Indeed, recent findings in Drosophila showing nuclear envelope-embedded nucleocytoplasmic export of signaling molecules have been documented (Speese et al. 2012). Importantly, blebbing is induced in somatic donor nuclei upon SCNT, suggesting that this process could be linked to reprogramming. Investigating the exact molecular composition of these blebs should provide answers to these questions.
From the two-cell stage onward, invaginations of the inner nuclear membrane toward the nucleoplasm become increasingly frequent (Szollosi and Szollosi 1988). Such invaginations have been observed in bovine preimplantation embryos as well, and the investigators of this study suggest that this folding might be related to the shrinking of nuclear size occurring over development (Popken et al. 2015). Because the degree of fluctuations in the folding and shape of the nuclear envelope and the turnover of lamin B1 are higher in ESCs compared with mouse embryonic fibroblasts (MEFs) (Bhattacharya et al. 2009), it is plausible that this higher plasticity of the nuclear periphery is related to a more undifferentiated state. However, in preimplantation stages, such measures have not been quantitatively compared. Current knowledge from the preimplantation embryo regarding the dynamics of the nucleus is restricted to the core mobility of histones. Histone mobility is higher in the totipotent two-cell stage embryo compared with the more pluripotent eight-cell stage nuclei (Boskovic et al. 2014), in line with a higher histone core mobility of ESCs compared with differentiated cells (Meshorer et al. 2006). Additionally, lamin B1 shows a higher turnover in ESCs than in MEFs as measured by fluorescence recovery after photobleaching (FRAP), suggesting a higher plasticity of the nuclear lamina in pluripotent cells as compared with somatic cells (Bhattacharya et al. 2009).
Lamins in the embryo
Zygotic deletion of lamins B1 and B2 simultaneously or lamin A alone results in lethality at birth or ∼2 wk after, respectively (Sullivan et al. 1999; Kim et al. 2011; Kim and Zheng 2013). However, lamin B1, B2, and A triple-knockout mESCs are capable of differentiation into all germ layers (Kim et al. 2013). Still, whether lamins are important for germline development or whether their maternal pools are necessary for progression through preimplantation stages has not been investigated. While mouse oocytes contain all of the canonical lamin isoforms (B1/B2, A, and C), mature spermatozoa are devoid of lamins (Schutz et al. 2005). In the oocyte, lamin A/C appear to be nonuniformly distributed along the nuclear envelope (Arnault et al. 2010). It was also observed that lamin A from oocytes shows decreased electrophoretic mobility compared with lamin A from fertilized embryos or any other mouse tissues, suggesting the existence of an oocyte-specific post-translational modification of lamin A (Houliston et al. 1988). Contrary to B-type lamins that are expressed at similar levels throughout preimplantation development, the level of lamin A/C decreases after fertilization, and several groups were unable to identify A-type lamins beyond the morula stage, the protein only becoming detectable again after embryonic day 9 (E9) (Schatten et al. 1985; Maul et al. 1987; Stewart and Burke 1987). Others have detected low levels of lamin A/C in all preimplantation stages, including blastocysts (Houliston et al. 1988; Eckersley-Maslin et al. 2013). These discrepancies may be due to antibody sensitivity. As translation of lamin A is seen in oocytes but only very weakly in eight-cell embryos (Houliston et al. 1988), it is most likely that the dilution of maternal lamin A over the first cleavages accounts for the very low A-type lamin immunoreactivity of the embryo.
Because the level of lamin A expression correlates with differentiation, with mESCs having significantly less A-type lamins when compared with differentiated cells (Constantinescu et al. 2006), it is plausible that the low A-type lamin level could be a prerequisite for the totipotent/pluripotent state. However, this does not seem to be so straightforward, as the overexpression of A-type lamins in ESCs does not affect pluripotency gene expression, differentiation potential, or growth (Peter and Nigg 1991; Melcer et al. 2012). Thus, whether lamin A/C levels play a role in cell plasticity and reprogramming in vivo still awaits functional testing. The early mouse embryo is clearly an ideal choice to directly address this. Some information on the potential role of lamin A during reprogramming stems from an SCNT study reporting that the reforming donor nuclei acquire strong perinuclear lamin A/C. This protein originates from newly transcribed RNA upon nuclear transfer, likely being the result of the somatic transcription program of the injected nucleus (Moreira et al. 2003). This raises the interesting possibility that high levels of A-type lamins in SCNT embryos might act as one of the barriers to efficient reprogramming. However, some interspecies differences may exist, as bovine and porcine SCNT embryos have A-type lamins present to an extent similar to and at the same stages of development as their IVF counterparts (Lee et al. 2007; Kelly et al. 2010).
It is plausible that early embryo-specific intermediate-type filaments might exist. Indeed, three proteins of the transcription-requiring complex (TRC) that are commonly used as markers for embryonic genome activation (EGA) were originally described as lamin-like proteins in the early 1990s based on their biochemical properties (Conover et al. 1991). However, whether these proteins are actual structural components of the nuclear lamina in the embryo and whether they contribute to organization or scaffolding of the nucleus have not been explored to date.
Phenotypes linked to nuclear envelope components and Nups
The maternal protein deposit of the oocyte remains present over several cell divisions after fertilization; thus, conventional zygotic gene knockout embryos derived from crosses between heterozygous parents are temporally not devoid of the targeted gene product. To address the function of a protein in early development, it is necessary to use maternal or maternal-zygotic deletion strategies; however, this is rarely pursued when knockout mice are generated. Zygotic LBR knockout embryos are born at sub-Mendelian ratios, with half of them likely dying before implantation, as no signs of a reabsorbed fetus are visible upon examination of heterozygous crosses (Shultz et al. 2003; Cohen et al. 2008). Moreover, two different LBR knockouts are either subfertile or sterile depending on the type of the mutation (Cohen et al. 2008), suggesting a role for LBR in gametogenesis and preimplantation development, but the exact cause of sterility and whether this is linked to defects in nuclear organization have not been explored. Zygotic deletion of Emerin does not affect development or fertility (Ozawa et al. 2006), and, although zygotic Man1-deficient embryos die at E10 due to defective TGFβ signaling (Cohen et al. 2007), conditional, tissue-specific mutants have not been created to address whether Man1 plays a role in germ cell development and/or after fertilization.
Recent 3D-SIM superresolution analyses have revealed that, during the first few cleavages of bovine embryos, NPCs are not detected in the nuclear envelope at regions where no chromatin accumulation is observed at the nuclear periphery, as assessed by DAPI (Popken et al. 2015). After EGA, the distribution of NPCs becomes uniform, suggesting that changes in the spatial composition of the nuclear envelope correlate with changes in de novo transcriptional activity. To address the functionality of NPC components in development, zygotic knockouts of several Nups have been generated in mice, which result in embryonic lethality with a variety of phenotypes. Nup214 and Rae1 mutants die before implantation, at the time when the maternally derived protein is completely exhausted (van Deursen et al. 1996; Babu et al. 2003). Deletion of other NPC subunits like Nup62/98/133/155 results in lethality at a later developmental stage, between E7 and E10 (Wu et al. 2001; Lupu et al. 2008; Zhang et al. 2008) Nup50 mutants die at late gestation (Smitherman et al. 2000), but the stage of embryonic lethality for Nup96 deletion was not determined (Faria et al. 2006).
Nucleolar precursor bodies (NPBs) are major organizers of embryonic nuclear architecture
Nucleoli in fully grown oocytes, zygotes, and two-cell stage embryos are compact, homogenous structures without distinctive fibrillar and granular components (Flechon and Kopecny 1998). Due to this unique nucleolar morphology, the nucleoli at these stages are commonly referred to as NPBs or nucleolar-like bodies (NLBs). Although little is known of their exact function, the number and distribution of NPBs in human pronuclei can serve as an indicator of embryonic developmental potential (Tesarik and Greco 1999; Gianaroli et al. 2003). The formation of NPBs from nucleoli occurs gradually during oocyte growth (Chouinard 1971) together with the changes in global chromatin reorganization of centromeric sequences (Fig. 1B). In fully grown oocytes, the NPB becomes transiently transcriptionally silent to become reactivated only after fertilization. Recently, rRNA transcription has been detected at the NPB as early as the zygote (Lin et al. 2014), contrary to previous data (Zatsepina et al. 2003), which suggested that rDNA transcription commences only at the late two-cell stage. The number of NPBs decreases during the cell cycle and over the progression from the zygote to the eight-cell stage, and the conventional tripartite morphology of somatic-type nucleoli gradually starts to appear forming around the NPB as a core (Flechon and Kopecny 1998; Aguirre-Lavin et al. 2012). Importantly, within 3 h after SCNT, NPBs morphologically identical to those in fertilized embryos form, and their number correlates with developmental potential, raising the possibility that NPB formation might be a critical component of reprogramming to the totipotent state (Martin et al. 2006b).
The role of NPBs has been investigated through their microsurgical removal (enucleolation) and reinjection at different developmental stages. During oocyte development, the “stage” of the nucleolus is crucial for meiotic resumption, as the transfer of the nucleolus from growing porcine oocytes into enucleolated fully grown oocytes compromises meiotic maturation. In addition, if the nucleolus of growing oocytes is removed, oocytes enter meiotic maturation more often, suggesting that the growing stage nucleolus hinders entry into meiosis I (Kyogoku et al. 2011). However, the NPB of the fully grown oocyte has no role in meiosis, as enucleolated fully grown oocytes show no defects in chromosome segregation or spindle assembly in the two meiotic divisions (Ogushi et al. 2008).
When enucleolated oocytes are either fertilized, parthenogenetically activated, or used for SCNT experiments, the resulting embryos never develop to the blastocyst stage (Ogushi et al. 2008). Similarly, mice lacking the main protein component of the NPB (Npm2−/−) show no defects in meiosis, but the majority of zygotes arrest at the two-cell stage (Burns et al. 2003). A likely explanation for this developmental failure might lie in changes to global nuclear organization: Centromeres in zygotes derived from enucleolated oocytes become scattered around the nucleoplasm, contrary to their otherwise normal distribution in rings around the NPBs (Fig. 1B). This abnormal chromatin organization of zygotes derived from enucleolated oocytes can be restored and the developmental arrest can be rescued when the enucleolated oocytes are reinjected with NPBs from oocytes or two-cell embryos before fertilization (Ogushi et al. 2008; Kyogoku et al. 2012). Strikingly, the reinjection of NPBs after fertilization into early pronuclear stage zygotes does not restore development (Ogushi and Saitou 2010), indicating that the NPB might exert its main and perhaps only function in the very early zygote. This idea is further supported by experiments showing that the removal of NPBs from early zygotes does not abrogate development to term (Kyogoku et al. 2014). Although NPB transfer and enucleolation may result in the removal of a number of additional factors during the procedure, the fact that both zygotes derived from enucleolated oocytes and Npm2−/− embryos show a delay in the first mitosis (Burns et al. 2003; Ogushi and Saitou 2010) suggests that the surface of the NPB might provide a platform for the establishment of a centromeric chromatin state necessary for proper chromosome segregation. However, the role of NPBs seems to be critical only shortly after fertilization, most likely at the time when pronuclei are being formed.
PML and Cajal bodies in embryos
Information on other nuclear compartments is largely lacking with respect to preimplantation development. To our knowledge, the only two compartments investigated are the Cajal and PML bodies. The latter are defined primarily by the accumulation of the PML protein. Originally thought of as storage bodies, they have emerged as potential additional organizers of the nucleus required for heterochromatin integrity. Nevertheless, the number of studies documenting the presence of PML in mammalian preimplantation embryos is limited (Cho et al. 2011), and these studies have not addressed a potential function or structural role of these subnuclear bodies in early embryos. Cajal bodies are 0.5- to 1-µm-sized spherical structures present in the nucleus of most cell types. They are thought to play a role in the biogenesis of small nuclear ribonucleoproteins (snRNPs) that contribute to the pre-mRNA processing machinery (Cioce and Lamond 2005). However, the exact function of Cajal bodies still remains elusive, not only during preimplantation development. The main component of Cajal bodies, coilin, colocalizes with fibrillarin in numerous foci around the NPBs in zygotes and gradually accumulates into a single patch away from the NPB by the end of two-cell stage. It has been proposed that the Cajal body might serve as a site for preassembly of the rRNA transcription machinery, which becomes recruited to rDNA around the NPBs at the late two-cell stage (Zatsepina et al. 2003). However, in light of the recent identification of rRNA transcription at the zygote stage (Lin et al. 2014), these interpretations are questionable. Although mutant mice homozygous for coilin following zygotic or maternal-zygotic deletion of the p80 coilin protein are born at sub-Mendelian ratios and produce reduced litter sizes, this main component of the Cajal body appears to be dispensable for development (Walker et al. 2009).
Chromatin organization of the preimplantation embryo
Spectroscopic imaging studies have highlighted that nuclei of zygotes, and early two-cell stage embryos are devoid of highly compacted, dense chromatin, which is only poorly visible in proximity to the nuclear envelope or around nucleoli (Ahmed et al. 2010). These data suggest that the general idea of the nuclear periphery as a silencing compartment in somatic cells might not hold in these early developmental stages. In most cell types, gene-dense chromosomes localize internally compared with gene-poor ones (Mayer et al. 2005). However, one study applying 3D DNA-FISH to bovine embryos from the zygote to the blastocyst stage revealed that, in embryos prior to EGA, the gene density of a chromosome does not correlate with its radial distribution (Fig. 1B). However, this distribution is changed shortly after the zygotic genome initiates transcription, when gene-poor chromosomes start occupying more peripheral territories compared with those with more genes (Koehler et al. 2009).
Apart from these few studies on the spatial distribution of the autosomes, the relative position of the X chromosome across preimplantation stages has been studied in mice (Namekawa et al. 2010). During imprinted X inactivation, the paternal X chromosome (Xp), but not the maternal one (Xm), is closely associated with the NPBs in most two-cell and four-cell but not the eight-cell or later stage embryos, as demonstrated by combined chromosome painting and DNA-FISH for Xist (Fig. 1B). Thus, NPB association may underlie the epigenetic asymmetry of Xp and Xm at the onset of silencing. In addition, the repeats within the Xp translocate into the X territory during X inactivation.
Although the above-mentioned studies demonstrate some peculiarities of the global and, to some extent, chromosomal organization of embryos, until now, neither single locus nor genome-wide profiling of nuclear organization has been performed on preimplantation embryos. Thus, to date, the currently available information on specific genomic regions includes only repetitive sequences of centromeres, telomeres, and rDNA studied through 3D DNA-FISH.
In mice, one of the most visible changes in nuclear organization occurs at the transition from the two-cell to the four-cell stage, with the reorganization of the centromeres. The core of the centromere is organized around minor satellite repeats that are enriched in CENP-A (CenH3), a specific histone H3 variant crucial for kinetochore assembly. Minor satellites are flanked by pericentromeric heterochromatin that is built on major satellite repeats (Fig. 1A) and whose proper heterochromatin configuration is essential for chromosome segregation. In mouse interphase somatic nuclei, pericentromeric and centromeric sequences of several chromosomes cluster together, forming so-called chromocenters. Structurally, the pericentromeric sequences of different chromosomes are intermingled and surrounded by isolated pericentromeric DNA sequences of single chromosomes (Guenatri et al. 2004). In fully grown oocytes, drastic changes in nuclear organization occur, which are mainly observed by the relocation of pericentromeric repeats from chromocenters into ring-like structures around the NPBs (Zuccotti et al. 2005). This transition in nuclear architecture correlates with meiotic competence and preimplantation developmental potential (Zuccotti et al. 1998).
In the zygote, the formation of pericentromeric rings shows asymmetric parental dynamics. In the maternal pronucleus, the pericentromeres occupy the surface of the NPB already from a few hours after pronuclear formation. In contrast, the pericentromeric repeats in the male pronucleus first cluster in an unorganized mass at the center of the pronucleus and restructure around the surface of the NPBs later compared with the maternal pronucleus (Aguirre-Lavin et al. 2012). The chromatin structure of mature sperm provides a plausible explanation for this asymmetry, as pericentromeric domains are found in a single cluster in the center of the sperm nucleus (Haaf and Ward 1995). Of note, a minority of the pericentromeric signals does not localize to the NPB in either the male or female pronuclei but is positioned close to the nuclear periphery.
As the NORs of most Mus musculus subspecies are located on six autosomes and as are all centromerically located (Britton-Davidian et al. 2012; Gibbons et al. 2015), it was initially suggested that rDNA might be a player for the peripheral NPB localization of pericentromeres. However, 3D mapping of rDNA by DNA-FISH in the zygote revealed that chromosomes without NORs also associate with NPBs, and not all rDNA-bearing chromosomes have their centromeres on the NPB surface, but some sit close to the nuclear periphery (Aguirre-Lavin et al. 2012). Thus, rDNA alone does not seem to cause the peculiar organization of pericentromeres around the NPB. Recent experimental evidence indicates that the 3D location of centromeres around the NPB is a prerequisite for embryonic development. When major satellite sequences were artificially tethered away from the NPB, toward the nuclear envelope, in two-cell stage embryos, development to blastocyst was compromised, possibly due to insufficient centromeric heterochromatin organization and defective silencing (Jachowicz et al. 2013). Importantly, these observations indicate that embryonic nuclei are functionally regionalized from the earliest stages of development. In addition, these experiments suggest that the nuclear envelope is not de facto a silent compartment. The molecular mechanism of how the NPB serves as a platform for genome organization in the embryo remains to be determined.
Coinciding with the time of EGA, the most apparent chromatin reorganization starts at the late two-cell stage, when most pericentromeric sequences, which still localize in a “shell” around NPBs similarly to zygotes, start to recluster into patches yet still associate with the NPBs (Martin et al. 2006a; Probst et al. 2007; Aguirre-Lavin et al. 2012). In the four-cell stage, pericentromeric sequences become dissociated from the NPBs and, from the eight-cell stage onward, appear as conventional chromocenters distributed over the nucleoplasm. The timing of this large-scale genome reorganization is similar in fertilized, parthenogenetically activated, and SCNT embryos, suggesting that the formation of chromocenters is linked to loss of totipotency (Martin et al. 2006b; Merico et al. 2007). Indeed, the interference with major satellite transcripts results in incomplete chromocenter formation and a developmental arrest at the two-cell stage (Probst et al. 2010; Casanova et al. 2013). However, the molecular mechanism behind and the exact role of this large-scale chromatin reorganization remain to be deciphered.
Analysis of the localization of telomeres, also heterochromatic in nature, has also been performed in both the early embryo and the germline. The positioning of telomeres during germ cell development shows a clear patterning, as the recruitment of telomeres to the nuclear periphery is necessary during meiotic recombination in both sexes (Ding et al. 2007). Although this configuration seems to be kept even in the mature sperm head (Haaf and Ward 1995), germinal vesicle (GV) oocytes have not been systematically analyzed for the position of telomeres. From the zygote stage onward, the distal telomeres are found either close to the nuclear periphery or free in the nucleoplasm without any obvious patterning (Aguirre-Lavin et al. 2012). Telomeres of zygotes and two-cell stage embryos undergo alternative lengthening of telomeres (ALT) to reset the shortened gametic telomeres to somatic length (Liu et al. 2007). However, whether the process of ALT in the early embryo is associated with specific subnuclear compartments (such as PML bodies) has not been investigated.
From all of the above together, it is evident that the embryo at the zygote and two-cell stages displays major differences in global nuclear organization as compared with embryos at later stages. This is mostly defined by the localization of pericentromeres but also by the changes in the relative position of chromosome territories of gene-rich and gene-poor regions. Therefore, we can distinguish two “global” patterns of nuclear organization in preimplantation embryos: one atypical pattern with central NPBs and few electron-dense regions in proximity to the nuclear membrane and the second one, which is more similar to that of mESCs and differentiated cells, characterized by the presence of chromocentres and a clear accumulation of heterochromatin in proximity to the nuclear lamina. Biologically, we postulate that these two states might delineate totipotent versus pluripotent nuclei.
Conclusions and perspectives
In conclusion, the preimplantation embryo offers a valuable system to study how changes in genome organization might play a role in cell differentiation and cellular state. While many studies pertaining to nuclear organization have been performed using ESCs, nuclear organization of ESCs must not be extrapolated to nuclear organization, at least in its global topological and functional features, in preimplantation embryos. The advent of single-cell techniques to study nuclear organization will begin to shed light on a more refined analysis of embryonic nuclear organization, in particular to define potential changes of specific genomic regions across development. The adaptation of approaches such as formaldehyde-assisted isolation of regulatory elements (FAIRE) sequencing (FAIRE-seq), DamID, or DNase sequencing (DNase-seq) to single cells of the early embryo will be a first step toward this goal and a necessary requirement to address a potential function of nuclear organization in regulating cellular plasticity.
Acknowledgments
We thank A. Burton for critical reading of the manuscript. M.-E.T.-P. acknowledges funding from EpiGeneSys NoE, ERC-Stg “NuclearPotency,” EMBO Young Investigator Programme, and the Schlumberger Foundation for Research and Education. M.B. is a recipient of a fellowship from Boehringer Ingelheim Fonds.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.273805.115.
References
- Aguirre-Lavin T, Adenot P, Bonnet-Garnier A, Lehmann G, Fleurot R, Boulesteix C, Debey P, Beaujean N. 2012. 3D-FISH analysis of embryonic nuclei in mouse highlights several abrupt changes of nuclear organization during preimplantation development. BMC Dev Biol 12: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed K, Dehghani H, Rugg-Gunn P, Fussner E, Rossant J, Bazett-Jones DP. 2010. Global chromatin architecture reflects pluripotency and lineage commitment in the early mouse embryo. PLoS One 5: e10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiken CE, Swoboda PP, Skepper JN, Johnson MH. 2004. The direct measurement of embryogenic volume and nucleo-cytoplasmic ratio during mouse pre-implantation development. Reproduction 128: 527–535. [DOI] [PubMed] [Google Scholar]
- Amendola M, van Steensel B. 2015. Nuclear lamins are not required for lamina-associated domain organization in mouse embryonic stem cells. EMBO Rep 16: 610–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrey G, Montavon T, Mascrez B, Gonzalez F, Noordermeer D, Leleu M, Trono D, Spitz F, Duboule D. 2013. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science 340: 1234167. [DOI] [PubMed] [Google Scholar]
- Arnault E, Doussau M, Pesty A, Lefevre B, Courtot AM. 2010. Review: lamin A/C, caspase-6, and chromatin configuration during meiosis resumption in the mouse oocyte. Reprod Sci 17: 102–115. [DOI] [PubMed] [Google Scholar]
- Babu JR, Jeganathan KB, Baker DJ, Wu X, Kang-Decker N, van Deursen JM. 2003. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J Cell Biol 160: 341–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya D, Talwar S, Mazumder A, Shivashankar GV. 2009. Spatio–temporal plasticity in chromatin organization in mouse cell differentiation and during Drosophila embryogenesis. Biophys J 96: 3832–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boskovic A, Eid A, Pontabry J, Ishiuchi T, Spiegelhalter C, Raghu Ram EV, Meshorer E, Torres-Padilla ME. 2014. Higher chromatin mobility supports totipotency and precedes pluripotency in vivo. Genes Dev 28: 1042–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle S, Rodesch MJ, Halvensleben HA, Jeddeloh JA, Bickmore WA. 2011. Fluorescence in situ hybridization with high-complexity repeat-free oligonucleotide probes generated by massively parallel synthesis. Chromosome Res 19: 901–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer M, Ohkura H. 2015. A negative loop within the nuclear pore complex controls global chromatin organization. Genes Dev 29: 1789–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton-Davidian J, Cazaux B, Catalan J. 2012. Chromosomal dynamics of nucleolar organizer regions (NORs) in the house mouse: micro-evolutionary insights. Heredity 108: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KH, Viveiros MM, Ren Y, Wang P, DeMayo FJ, Frail DE, Eppig JJ, Matzuk MM. 2003. Roles of NPM2 in chromatin and nucleolar organization in oocytes and embryos. Science 300: 633–636. [DOI] [PubMed] [Google Scholar]
- Casanova M, Pasternak M, El Marjou F, Le Baccon P, Probst AV, Almouzni G. 2013. Heterochromatin reorganization during early mouse development requires a single-stranded noncoding transcript. Cell Rep 4: 1156–1167. [DOI] [PubMed] [Google Scholar]
- Chambeyron S, Bickmore WA. 2004. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev 18: 1119–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Park JS, Kang YK. 2011. Dual functions of histone-lysine N-methyltransferase Setdb1 protein at promyelocytic leukemia-nuclear body (PML-NB): maintaining PML-NB structure and regulating the expression of its associated genes. J Biol Chem 286: 41115–41124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouinard LA. 1971. A light- and electron-microscope study of the nucleolus during growth of the oocyte in the prepubertal mouse. J Cell Sci 9: 637–663. [DOI] [PubMed] [Google Scholar]
- Cioce M, Lamond AI. 2005. Cajal bodies: a long history of discovery. Annu Rev Cell Dev Biol 21: 105–131. [DOI] [PubMed] [Google Scholar]
- Cohen TV, Kosti O, Stewart CL. 2007. The nuclear envelope protein MAN1 regulates TGFβ signaling and vasculogenesis in the embryonic yolk sac. Development 134: 1385–1395. [DOI] [PubMed] [Google Scholar]
- Cohen TV, Klarmann KD, Sakchaisri K, Cooper JP, Kuhns D, Anver M, Johnson PF, Williams SC, Keller JR, Stewart CL. 2008. The lamin B receptor under transcriptional control of C/EBPε is required for morphological but not functional maturation of neutrophils. Hum Mol Genet 17: 2921–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conover JC, Temeles GL, Zimmermann JW, Burke B, Schultz RM. 1991. Stage-specific expression of a family of proteins that are major products of zygotic gene activation in the mouse embryo. Dev Biol 144: 392–404. [DOI] [PubMed] [Google Scholar]
- Constantinescu D, Gray HL, Sammak PJ, Schatten GP, Csoka AB. 2006. Lamin A/C expression is a marker of mouse and human embryonic stem cell differentiation. Stem Cells 24: 177–185. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer M, Dietzel S, Muller S, Solovei I, Fakan S. 2006. Chromosome territories—a functional nuclear landscape. Curr Opin Cell Biol 18: 307–316. [DOI] [PubMed] [Google Scholar]
- Dechat T, Adam SA, Taimen P, Shimi T, Goldman RD. 2010. Nuclear lamins. Cold Spring Harb Perspect Biol 2: a000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Xu R, Yu J, Xu T, Zhuang Y, Han M. 2007. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev Cell 12: 863–872. [DOI] [PubMed] [Google Scholar]
- Eckersley-Maslin MA, Bergmann JH, Lazar Z, Spector DL. 2013. Lamin A/C is expressed in pluripotent mouse embryonic stem cells. Nucleus 4: 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan Y, Skoultchi AI, Wutz A, et al. 2010. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell 38: 452–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria AM, Levay A, Wang Y, Kamphorst AO, Rosa ML, Nussenzveig DR, Balkan W, Chook YM, Levy DE, Fontoura BM. 2006. The nucleoporin Nup96 is required for proper expression of interferon-regulated proteins and functions. Immunity 24: 295–304. [DOI] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA. 2008. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet 4: e1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flechon JE, Kopecny V. 1998. The nature of the ‘nucleolus precursor body’ in early preimplantation embryos: a review of fine-structure cytochemical, immunocytochemical and autoradiographic data related to nucleolar function. Zygote 6: 183–191. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Gardner JM, Smoyer CJ, Friederichs JM, Unruh JR, Slaughter BD, Alexander R, Chisholm RD, Lee KK, Workman JL, et al. 2012. Acetylation of the SUN protein Mps3 by Eco1 regulates its function in nuclear organization. Mol Biol Cell 23: 2546–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianaroli L, Magli MC, Ferraretti AP, Fortini D, Grieco N. 2003. Pronuclear morphology and chromosomal abnormalities as scoring criteria for embryo selection. Fertil Steril 80: 341–349. [DOI] [PubMed] [Google Scholar]
- Gibbons JG, Branco AT, Godinho SA, Yu S, Lemos B. 2015. Concerted copy number variation balances ribosomal DNA dosage in human and mouse genomes. Proc Natl Acad Sci 112: 2485–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453: 948–951. [DOI] [PubMed] [Google Scholar]
- Guenatri M, Bailly D, Maison C, Almouzni G. 2004. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J Cell Biol 166: 493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaf T, Ward DC. 1995. Higher order nuclear structure in mammalian sperm revealed by in situ hybridization and extended chromatin fibers. Exp Cell Res 219: 604–611. [DOI] [PubMed] [Google Scholar]
- Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CW, Ye C, Ping JL, Mulawadi F, et al. 2011. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet 43: 630–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson AS, Warburton D, Atwood KC. 1972. Location of ribosomal DNA in the human chromosome complement. Proc Natl Acad Sci 69: 3394–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano Y, Hizume K, Kimura H, Takeyasu K, Haraguchi T, Hiraoka Y. 2012. Lamin B receptor recognizes specific modifications of histone H4 in heterochromatin formation. J Biol Chem 287: 42654–42663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houliston E, Guilly MN, Courvalin JC, Maro B. 1988. Expression of nuclear lamins during mouse preimplantation development. Development 102: 271–278. [DOI] [PubMed] [Google Scholar]
- Ibarra A, Hetzer MW. 2015. Nuclear pore proteins and the control of genome functions. Genes Dev 29: 337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jachowicz JW, Santenard A, Bender A, Muller J, Torres-Padilla ME. 2013. Heterochromatin establishment at pericentromeres depends on nuclear position. Genes Dev 27: 2427–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto FV, Benner C, Hetzer MW. 2015. The nucleoporin Nup153 regulates embryonic stem cell pluripotency through gene silencing. Genes Dev 29: 1224–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. 2012. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotechnol 30: 90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalverda B, Pickersgill H, Shloma VV, Fornerod M. 2010. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell 140: 360–371. [DOI] [PubMed] [Google Scholar]
- Kelly RD, Alberio R, Campbell KH. 2010. A-type lamin dynamics in bovine somatic cell nuclear transfer embryos. Reprod Fertil Dev 22: 956–965. [DOI] [PubMed] [Google Scholar]
- Kim Y, Zheng Y. 2013. Generation and characterization of a conditional deletion allele for Lmna in mice. Biochem Biophys Res Commun 440: 8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Sharov AA, McDole K, Cheng M, Hao H, Fan CM, Gaiano N, Ko MS, Zheng Y. 2011. Mouse B-type lamins are required for proper organogenesis but not by embryonic stem cells. Science 334: 1706–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Zheng X, Zheng Y. 2013. Proliferation and differentiation of mouse embryonic stem cells lacking all lamins. Cell Res 23: 1420–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, van Steensel B. 2013. Single-cell dynamics of genome-nuclear lamina interactions. Cell 153: 178–192. [DOI] [PubMed] [Google Scholar]
- Kind J, Pagie L, de Vries SS, Nahidiazar L, Dey SS, Bienko M, Zhan Y, Lajoie B, de Graaf CA, Amendola M, et al. 2015. Genome-wide maps of nuclear lamina interactions in single human cells. Cell 163: 134–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler D, Zakhartchenko V, Froenicke L, Stone G, Stanyon R, Wolf E, Cremer T, Brero A. 2009. Changes of higher order chromatin arrangements during major genome activation in bovine preimplantation embryos. Exp Cell Res 315: 2053–2063. [DOI] [PubMed] [Google Scholar]
- Kumar A, Mazzanti M, Mistrik M, Kosar M, Beznoussenko GV, Mironov AA, Garre M, Parazzoli D, Shivashankar GV, Scita G, et al. 2014. ATR mediates a checkpoint at the nuclear envelope in response to mechanical stress. Cell 158: 633–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran RI, Spector DL. 2008. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol 180: 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyogoku H, Ogushi S, Miyano T, Fulka J Jr. 2011. Nucleoli from growing oocytes inhibit the maturation of enucleolated, full-grown oocytes in the pig. Mol Reprod Dev 78: 426–435. [DOI] [PubMed] [Google Scholar]
- Kyogoku H, Ogushi S, Miyano T. 2012. Nucleoli from two-cell embryos support the development of enucleolated germinal vesicle oocytes in the pig. Biol Reprod 87: 113. [DOI] [PubMed] [Google Scholar]
- Kyogoku H, Fulka J Jr, Wakayama T, Miyano T. 2014. De novo formation of nucleoli in developing mouse embryos originating from enucleolated zygotes. Development 141: 2255–2259. [DOI] [PubMed] [Google Scholar]
- Lee K, Fodor WL, Machaty Z. 2007. Dynamics of lamin A/C in porcine embryos produced by nuclear transfer. Mol Reprod Dev 74: 1221–1227. [DOI] [PubMed] [Google Scholar]
- Legartova S, Stixova L, Laur O, Kozubek S, Sehnalova P, Bartova E. 2014. Nuclear structures surrounding internal lamin invaginations. J Cell Biochem 115: 476–487. [DOI] [PubMed] [Google Scholar]
- Lin CJ, Koh FM, Wong P, Conti M, Ramalho-Santos M. 2014. Hira-mediated H3.3 incorporation is required for DNA replication and ribosomal RNA transcription in the mouse zygote. Dev Cell 30: 268–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Bailey SM, Okuka M, Munoz P, Li C, Zhou L, Wu C, Czerwiec E, Sandler L, Seyfang A, et al. 2007. Telomere lengthening early in development. Nat Cell Biol 9: 1436–1441. [DOI] [PubMed] [Google Scholar]
- Lupianez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al. 2015. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161: 1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupu F, Alves A, Anderson K, Doye V, Lacy E. 2008. Nuclear pore composition regulates neural stem/progenitor cell differentiation in the mouse embryo. Dev Cell 14: 831–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Beaujean N, Brochard V, Audouard C, Zink D, Debey P. 2006a. Genome restructuring in mouse embryos during reprogramming and early development. Dev Biol 292: 317–332. [DOI] [PubMed] [Google Scholar]
- Martin C, Brochard V, Migne C, Zink D, Debey P, Beaujean N. 2006b. Architectural reorganization of the nuclei upon transfer into oocytes accompanies genome reprogramming. Mol Reprod Dev 73: 1102–1111. [DOI] [PubMed] [Google Scholar]
- Martin C, Chen S, Maya-Mendoza A, Lovric J, Sims PF, Jackson DA. 2009. Lamin B1 maintains the functional plasticity of nucleoli. J Cell Sci 122: 1551–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul GG, Schatten G, Jimenez SA, Carrera AE. 1987. Detection of nuclear lamin B epitopes in oocyte nuclei from mice, sea urchins, and clams using a human autoimmune serum. Dev Biol 121: 368–375. [DOI] [PubMed] [Google Scholar]
- Mayer R, Brero A, von Hase J, Schroeder T, Cremer T, Dietzel S. 2005. Common themes and cell type specific variations of higher order chromatin arrangements in the mouse. BMC Cell Biol 6: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister P, Mango SE, Gasser SM. 2011. Locking the genome: nuclear organization and cell fate. Curr Opin Genet Dev 21: 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcer S, Hezroni H, Rand E, Nissim-Rafinia M, Skoultchi A, Stewart CL, Bustin M, Meshorer E. 2012. Histone modifications and lamin A regulate chromatin protein dynamics in early embryonic stem cell differentiation. Nat Commun 3: 910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merico V, Barbieri J, Zuccotti M, Joffe B, Cremer T, Redi CA, Solovei I, Garagna S. 2007. Epigenomic differentiation in mouse preimplantation nuclei of biparental, parthenote and cloned embryos. Chromosome Res 15: 341–360. [DOI] [PubMed] [Google Scholar]
- Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. 2006. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell 10: 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira PN, Robl JM, Collas P. 2003. Architectural defects in pronuclei of mouse nuclear transplant embryos. J Cell Sci 116: 3713–3720. [DOI] [PubMed] [Google Scholar]
- Namekawa SH, Payer B, Huynh KD, Jaenisch R, Lee JT. 2010. Two-step imprinted X inactivation: repeat versus genic silencing in the mouse. Mol Cell Biol 30: 3187–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth A, Conesa A, Santoyo-Lopez J, Medina I, Montaner D, Peterfia B, Solovei I, Cremer T, Dopazo J, Langst G. 2010. Initial genomics of the human nucleolus. PLoS Genet 6: e1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordermeer D, Leleu M, Splinter E, Rougemont J, De Laat W, Duboule D. 2011. The dynamic architecture of Hox gene clusters. Science 334: 222–225. [DOI] [PubMed] [Google Scholar]
- Ogushi S, Saitou M. 2010. The nucleolus in the mouse oocyte is required for the early step of both female and male pronucleus organization. J Reprod Dev 56: 495–501. [DOI] [PubMed] [Google Scholar]
- Ogushi S, Palmieri C, Fulka H, Saitou M, Miyano T, Fulka J Jr. 2008. The maternal nucleolus is essential for early embryonic development in mammals. Science 319: 613–616. [DOI] [PubMed] [Google Scholar]
- Osmanagic-Myers S, Dechat T, Foisner R. 2015. Lamins at the crossroads of mechanosignaling. Genes Dev 29: 225–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa R, Hayashi YK, Ogawa M, Kurokawa R, Matsumoto H, Noguchi S, Nonaka I, Nishino I. 2006. Emerin-lacking mice show minimal motor and cardiac dysfunctions with nuclear-associated vacuoles. Am J Pathol 168: 907–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. 2010. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell 38: 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M, Nigg EA. 1991. Ectopic expression of an A-type lamin does not interfere with differentiation of lamin A-negative embryonal carcinoma cells. J Cell Sci 100: 589–598. [DOI] [PubMed] [Google Scholar]
- Popken J, Brero A, Koehler D, Schmid VJ, Strauss A, Wuensch A, Guengoer T, Graf A, Krebs S, Blum H, et al. 2014. Reprogramming of fibroblast nuclei in cloned bovine embryos involves major structural remodeling with both striking similarities and differences to nuclear phenotypes of in vitro fertilized embryos. Nucleus 5: 555–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popken J, Graf A, Krebs S, Blum H, Schmid VJ, Strauss A, Guengoer T, Zakhartchenko V, Wolf E, Cremer T. 2015. Remodeling of the nuclear envelope and lamina during bovine preimplantation development and its functional implications. PLoS One 10: e0124619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst AV, Santos F, Reik W, Almouzni G, Dean W. 2007. Structural differences in centromeric heterochromatin are spatially reconciled on fertilisation in the mouse zygote. Chromosoma 116: 403–415. [DOI] [PubMed] [Google Scholar]
- Probst AV, Okamoto I, Casanova M, El Marjou F, Le Baccon P, Almouzni G. 2010. A strand-specific burst in transcription of pericentric satellites is required for chromocenter formation and early mouse development. Dev Cell 19: 625–638. [DOI] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H. 2008. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 452: 243–247. [DOI] [PubMed] [Google Scholar]
- Roix JJ, McQueen PG, Munson PJ, Parada LA, Misteli T. 2003. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat Genet 34: 287–291. [DOI] [PubMed] [Google Scholar]
- Schatten G, Maul GG, Schatten H, Chaly N, Simerly C, Balczon R, Brown DL. 1985. Nuclear lamins and peripheral nuclear antigens during fertilization and embryogenesis in mice and sea urchins. Proc Natl Acad Sci 82: 4727–4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutz W, Alsheimer M, Ollinger R, Benavente R. 2005. Nuclear envelope remodeling during mouse spermiogenesis: postmeiotic expression and redistribution of germline lamin B3. Exp Cell Res 307: 285–291. [DOI] [PubMed] [Google Scholar]
- Sexton T, Cavalli G. 2015. The role of chromosome domains in shaping the functional genome. Cell 160: 1049–1059. [DOI] [PubMed] [Google Scholar]
- Shultz LD, Lyons BL, Burzenski LM, Gott B, Samuels R, Schweitzer PA, Dreger C, Herrmann H, Kalscheuer V, Olins AL, et al. 2003. Mutations at the mouse ichthyosis locus are within the lamin B receptor gene: a single gene model for human Pelger-Huet anomaly. Hum Mol Genet 12: 61–69. [DOI] [PubMed] [Google Scholar]
- Sleeman JE, Trinkle-Mulcahy L. 2014. Nuclear bodies: new insights into assembly/dynamics and disease relevance. Curr Opin Cell Biol 28: 76–83. [DOI] [PubMed] [Google Scholar]
- Smitherman M, Lee K, Swanger J, Kapur R, Clurman BE. 2000. Characterization and targeted disruption of murine Nup50, a p27(Kip1)-interacting component of the nuclear pore complex. Mol Cell Biol 20: 5631–5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovei I, Grandi N, Knoth R, Volk B, Cremer T. 2004. Positional changes of pericentromeric heterochromatin and nucleoli in postmitotic Purkinje cells during murine cerebellum development. Cytogenet Genome Res 105: 302–310. [Google Scholar]
- Solovei I, Kreysing M, Lanctot C, Kosem S, Peichl L, Cremer T, Guck J, Joffe B. 2009. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell 137: 356–368. [DOI] [PubMed] [Google Scholar]
- Solovei I, Wang AS, Thanisch K, Schmidt CS, Krebs S, Zwerger M, Cohen TV, Devys D, Foisner R, Peichl L, et al. 2013. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 152: 584–598. [DOI] [PubMed] [Google Scholar]
- Speese SD, Ashley J, Jokhi V, Nunnari J, Barria R, Li Y, Ataman B, Koon A, Chang YT, Li Q, et al. 2012. Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell 149: 832–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart C, Burke B. 1987. Teratocarcinoma stem cells and early mouse embryos contain only a single major lamin polypeptide closely resembling lamin B. Cell 51: 383–392. [DOI] [PubMed] [Google Scholar]
- Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B. 1999. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol 147: 913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szollosi MS, Szollosi D. 1988. ‘Blebbing’ of the nuclear envelope of mouse zygotes, early embryos and hybrid cells. J Cell Sci 91(Pt 2): 257–267. [DOI] [PubMed] [Google Scholar]
- Talamas JA, Capelson M. 2015. Nuclear envelope and genome interactions in cell fate. Front Genet 6: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesarik J, Greco E. 1999. The probability of abnormal preimplantation development can be predicted by a single static observation on pronuclear stage morphology. Hum Reprod 14: 1318–1323. [DOI] [PubMed] [Google Scholar]
- Therizols P, Illingworth RS, Courilleau C, Boyle S, Wood AJ, Bickmore WA. 2014. Chromatin decondensation is sufficient to alter nuclear organization in embryonic stem cells. Science 346: 1238–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui N, Ogura A, Kimura Y, Yanagimachi R. 1997. Sperm nuclear envelope: breakdown of intrinsic envelope and de novo formation in hamster oocytes or eggs. Zygote 5: 35–46. [DOI] [PubMed] [Google Scholar]
- van Deursen J, Boer J, Kasper L, Grosveld G. 1996. G2 arrest and impaired nucleocytoplasmic transport in mouse embryos lacking the proto-oncogene CAN/Nup214. EMBO J 15: 5574–5583. [PMC free article] [PubMed] [Google Scholar]
- van Koningsbruggen S, Gierlinski M, Schofield P, Martin D, Barton GJ, Ariyurek Y, den Dunnen JT, Lamond AI. 2010. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol Biol Cell 21: 3735–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker MP, Tian L, Matera AG. 2009. Reduced viability, fertility and fecundity in mice lacking the cajal body marker protein, coilin. PLoS One 4: e6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KL, Foisner R. 2010. Lamin-binding Proteins. Cold Spring Harb Perspect Biol 2: a000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Kasper LH, Mantcheva RT, Mantchev GT, Springett MJ, van Deursen JM. 2001. Disruption of the FG nucleoporin NUP98 causes selective changes in nuclear pore complex stoichiometry and function. Proc Natl Acad Sci 98: 3191–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Q, Worman HJ. 1996. Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. J Biol Chem 271: 14653–14656. [DOI] [PubMed] [Google Scholar]
- Zatsepina O, Baly C, Chebrout M, Debey P. 2003. The step-wise assembly of a functional nucleolus in preimplantation mouse embryos involves the cajal (coiled) body. Dev Biol 253: 66–83. [DOI] [PubMed] [Google Scholar]
- Zhang X, Chen S, Yoo S, Chakrabarti S, Zhang T, Ke T, Oberti C, Yong SL, Fang F, Li L, et al. 2008. Mutation in nuclear pore component NUP155 leads to atrial fibrillation and early sudden cardiac death. Cell 135: 1017–1027. [DOI] [PubMed] [Google Scholar]
- Zhang Y, McCord RP, Ho YJ, Lajoie BR, Hildebrand DG, Simon AC, Becker MS, Alt FW, Dekker J. 2012. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell 148: 908–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccotti M, Giorgi Rossi P, Martinez A, Garagna S, Forabosco A, Redi CA. 1998. Meiotic and developmental competence of mouse antral oocytes. Biol Reprod 58: 700–704. [DOI] [PubMed] [Google Scholar]
- Zuccotti M, Garagna S, Merico V, Monti M, Alberto Redi C. 2005. Chromatin organisation and nuclear architecture in growing mouse oocytes. Mol Cell Endocrinol 234: 11–17. [DOI] [PubMed] [Google Scholar]