In this review, Huang and Bonni discuss the functions and mechanisms of the anaphase-promoting complex in neurogenesis; glial differentiation and migration; neuronal survival, metabolism, and morphogenesis; synapse formation and plasticity; and learning and memory.
Keywords: anaphase-promoting complex, E3 ubiquitin ligase, ubiquitin–proteasome system
Abstract
Control of protein abundance by the ubiquitin–proteasome system is essential for normal brain development and function. Just over a decade ago, the first post-mitotic function of the anaphase-promoting complex, a major cell cycle-regulated E3 ubiquitin ligase, was discovered in the control of axon growth and patterning in the mammalian brain. Since then, a large number of studies have identified additional novel roles for the anaphase-promoting complex in diverse aspects of neuronal connectivity and plasticity in the developing and mature nervous system. In this review, we discuss the functions and mechanisms of the anaphase-promoting complex in neurogenesis, glial differentiation and migration, neuronal survival and metabolism, neuronal morphogenesis, synapse formation and plasticity, and learning and memory. We also provide a perspective on future investigations of the anaphase-promoting complex in neurobiology.
Protein degradation regulates diverse biological processes in the developing and mature nervous system. Spatial and temporal control of protein turnover and abundance influences distinct stages of neural development and function from neurogenesis and neuronal morphogenesis to synapse formation and pruning to plasticity and learning (Hegde and Upadhya 2007; Yi and Ehlers 2007; Segref and Hoppe 2009; Yamada et al. 2013). Protein degradation by the proteasome requires ubiquitination, which is mediated by the sequential action of an E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzyme, and E3 ubiquitin ligase (Hershko and Ciechanover 1992; Scheffner et al. 1995; Ciechanover 2005; Hershko 2005; Weissman et al. 2011). Two E1, ∼40 E2, and >600 E3 enzymes are encoded in the human genome (Haas and Rose 1982; Pickart 2001; Ye and Rape 2009). E3 ubiquitin ligases are divided into two major classes, the HECT domain and RING domain families. RING domain E3 ubiquitin ligases, consisting of single subunit or multisubunit enzymes, represent the largest group and display wide-ranging functions within the cells. The large number of E3 ubiquitin ligases reflects their critical role in substrate specificity in the ubiquitination reaction (Deshaies and Joazeiro 2009; Rotin and Kumar 2009; Komander and Rape 2012).
The anaphase-promoting complex is a major E3 ubiquitin ligase and member of the RING domain family. First identified in genetic and biochemical studies of Cyclin B ubiquitination two decades ago (Irniger et al. 1995; King et al. 1995; Sudakin et al. 1995), the anaphase-promoting complex is a large complex of 15 subunits in vertebrates (Chang et al. 2014). Cdh1 and Cdc20 constitute two key regulatory subunits of this complex (Visintin et al. 1997). The association of Cdh1 or Cdc20 stimulates the catalytic activity of the anaphase-promoting complex and specifies substrate recognition (Harper et al. 2002; Peters 2006; Chang et al. 2015). Cdh1–anaphase-promoting complex (Cdh1–APC) and Cdc20–APC recognize substrates via a peptide motif, termed the destruction box (D box), within substrates (Glotzer et al. 1991; Zachariae and Nasmyth 1999; Burton and Solomon 2001). Cdh1–APC associates with substrates through additional recognition motifs, including the KEN and A boxes (Pfleger and Kirschner 2000; Littlepage and Ruderman 2002). The Cul1-related scaffold protein APC2 and the RING finger protein APC11 form the catalytic core of the anaphase-promoting complex (Harper et al. 2002; Peters 2006). Other subunits act as scaffold proteins, coordinating the juxtaposition of the catalytic and degron recognition modules of the complex (Passmore et al. 2005; Schreiber et al. 2011). Several scaffolding subunits, including APC3 (Cdc27), APC6 (Cdc16), APC8 (Cdc23), APC5, and APC7, contain tetratricopeptide repeat (TPR) motifs (Schreiber et al. 2011; Chang et al. 2015).
Although the anaphase-promoting complex in budding yeast collaborates with the E2s Ubc1 and Ubc4 that catalyze the assembly of K48-linked polyubiquitin chains of substrates (Rodrigo-Brenni and Morgan 2007), several ubiquitin chains of complex topology are generated through multiple lysine residues, including K11 and K63 in addition to K48 (Kirkpatrick et al. 2006). The human anaphase-promoting complex and its E2, UbcH10, preferentially assemble K11-linked branched ubiquitin chains on substrates to drive proteasomal degradation and mitotic exit (Jin et al. 2008). Branched ubiquitin chains that contain multiple blocks of K11-linked chains enhance substrate recognition by the proteasome (Meyer and Rape 2014). In a recent study using single-molecule kinetic analyses, Lu et al. (2015) characterized the role of ubiquitin configurations of substrates on proteasome binding and subsequent degradation. The total number of ubiquitin molecules rather than ubiquitin chain configuration on substrates of the anaphase-promoting complex determines the strength of binding of substrates with the proteasome (Lu et al. 2015). However, the configuration of ubiquitin chains on substrates determines degradation efficiency by affecting the translocation of substrates into the proteasome. A distributed array of short ubiquitin chains on substrates of the anaphase-promoting complex is a superior and optimal signal for degradation (Lu et al. 2015).
Structure of the anaphase-promoting complex
Elucidation of the structure of the anaphase-promoting complex is essential to understanding the function of this major E3 ubiquitin ligase. Crystallographic analyses of isolated subunits and small subcomplexes of the anaphase-promoting complex followed by cryo-electron microscopy (cryo-EM) studies have revealed the molecular architecture of this E3 ubiquitin ligase with its coactivators, subunit stoichiometry, and position of subunits as well as the coactivators Cdh1 and Cdc20 (Dube et al. 2005; Passmore et al. 2005; Ohi et al. 2007; Herzog et al. 2009). These studies have painted a picture of a triangular-shaped complex ∼1.7 MDa in mass and 19 × 17 × 15 nm in size (Passmore et al. 2005; Ohi et al. 2007).
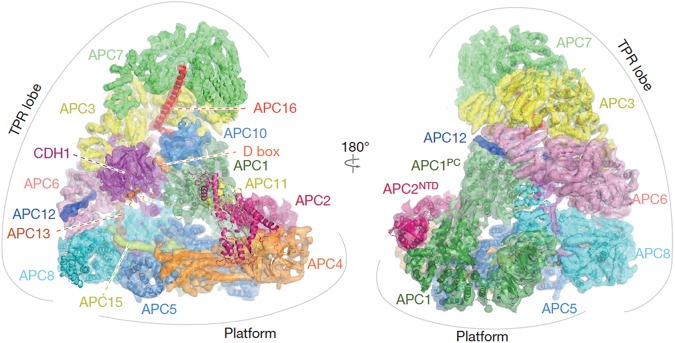
Recent cryo-EM studies have refined our understanding of the precise relationship of individual subunits within Cdh1–APC and how ubiquitination of substrates operates and inhibitors block ubiquitin ligase function (Frye et al. 2013; He et al. 2013; Chang et al. 2014, 2015; Yamaguchi et al. 2015). Reconstruction of Cdh1–APC by cryo-EM has largely confirmed the triangular shape of Cdh1–APC delineated by a lattice-like shell comprised of a catalytic “platform” and an “arc lamp” scaffolding structure (Fig. 1; Chang et al. 2014; Yamaguchi et al. 2015). Positioned at the periphery of the platform is the catalytic core, consisting of APC2 and APC11. The platform also includes APC1, APC4, APC5, and APC15. The arc lamp contains the subunits APC3, APC6, APC7, APC8, APC10, APC12, APC13, and APC16. The curved structure of the arc lamp, which serves as a scaffold for the recruitment of substrates, is established by layers of the TPR proteins APC8, APC6, APC3, and APC7 stacked on top of each other. Notably, APC10 contributes to the substrate D-box recognition module (da Fonseca et al. 2011; Chang et al. 2014). The homologous isoleucine–arginine “IR tail” sequences at the C terminus of APC10 and Cdh1 (or Cdc20) bind the APC3 homodimer. Thus, APC10 may act as a coreceptor for Cdh1–APC or Cdc20–APC substrates (Izawa and Pines 2011; Chang et al. 2014).
Figure 1.
EM reconstruction of the anaphase-promoting complex. The anaphase-promoting complex is a large complex of 15 subunits in vertebrates. It is triangular-shaped, ∼1.7 MDa in mass, and 19 × 17 × 15 nm in size. Cdh1 and Cdc20 constitute two key regulatory subunits of the anaphase-promoting complex. The Cul1-related scaffold protein APC2 and the RING finger protein APC11 form the catalytic core. The anaphase-promoting complex is delineated by a lattice-like shell comprised of a catalytic “platform” containing the subunits APC1, APC4, APC5, and APC15 and an “arc lamp” scaffolding structure of the TPR lobe containing the subunits APC3, APC6, APC7, APC8, APC10, APC12, APC13, and APC16. Together with Cdh1 or Cdc20, APC10 contributes to the substrate recognition module. This figure was adapted by permission from Macmillan Publishers Ltd.: Nature (Chang et al. 2014) © 2014.
The structural basis of Cdh1 stimulation of the catalytic activity of Cdh1–APC has also been revealed. The N-terminal domain of Cdh1 triggers displacement of the catalytic subunits APC11 and APC2 relative to the D-box recognition module of Cdh1 and APC10. This allosteric transition is accompanied by increased flexibility of the catalytic subunits and enhanced affinity for E2–ubiquitin, thus stimulating the catalytic activity of Cdh1–APC (Chang et al. 2014). The structural basis of inhibition of the anaphase-promoting complex by the protein interphase early mitotic inhibitor 1 (Emi1) has also been studied. Emi1 interacts with both the substrate recognition module of Cdh1 and the catalytic module of Cdh1–APC (Reimann et al. 2001; Frye et al. 2013; Chang et al. 2015), thereby inhibiting the substrate recruitment and catalytic activity of Cdh1–APC. Furthermore, the interaction of Emi1 with Cdh1–APC blocks the binding of the E2 UbcH10 with APC11 and thus inhibits ubiquitin chain elongation (Frye et al. 2013), suggesting that Emi1 directly inhibits the catalytic activity of the anaphase-promoting complex. Components of the mitotic checkpoint (MCC), including the proteins BubR1, Bub3, and Mad2, associate with the anaphase-promoting complex and thereby inhibit its activity (Hwang et al. 1998; Sudakin et al. 2001; Tang et al. 2001). Single-particle EM analyses have revealed that the MCC proteins interact with Cdc20–APC within a region that overlaps with the Cdc20-binding site (Herzog et al. 2009), thus inhibiting the binding of substrates of Cdc20–APC. The MCC proteins also directly contact the APC2–APC11 catalytic module (Herzog et al. 2009), suggesting that it may interfere with the catalytic core of Cdc20–APC.
In addition to endogenous Cdh1–APC and Cdc20–APC inhibitors, chemical inhibitors have been identified. The small molecule tosyl-L-arginine methyl ester (TAME) binds Cdh1–APC and Cdc20–APC and blocks ubiquitin ligase activation (Verma et al. 2004; Zeng et al. 2010; Zeng and King 2012). Because TAME structurally resembles the IR tail of Cdh1 and Cdc20, TAME binds the site of the IR tail-binding region within the anaphase-promoting complex, thereby interfering with the interaction with Cdc20 or Cdh1 (Zeng et al. 2010). Whereas TAME is a potent inhibitor of Cdh1–APC and Cdc20–APC in Xenopus extracts in vitro, a cell-permeable variant, proTAME, has been generated that is processed by intracellular esterases to yield the compound TAME (Zeng et al. 2010). Interestingly, proTAME may be activated selectively in a cell type-specific manner (Zeng et al. 2010). The small molecule apcin also inhibits the anaphase-promoting complex but shows more selective inhibition of Cdc20–APC (Sackton et al. 2014). Apcin occupies the D-box-binding pocket in the WD40 domain of Cdc20, thereby competitively blocking the binding of D-box-containing substrates with Cdc20 (Sackton et al. 2014). Although proteins that inhibit Cdh1–APC have been used in studies in the nervous system (Konishi et al. 2004; Huang et al. 2015), the potential utility of small molecule inhibitors of Cdh1–APC and Cdc20–APC in neurobiology has remained largely untapped.
The functions of the anaphase-promoting complex were first characterized in proliferating cells. It promotes the transition of cells from metaphase to anaphase during cell division (Irniger et al. 1995; King et al. 1995), which is mediated by the ordered degradation of cyclins, securin, mitotic kinases, and microtubule motors and assembly factors (Harper et al. 2002; Peters 2006; Sivakumar and Gorbsky 2015). Cdc20–APC and Cdh1–APC control distinct temporal phases of the cell cycle. While Cdc20–APC drives anaphase in early mitosis, Cdh1–APC plays key roles in mitotic exit and G1 phase of the cell cycle (Peters 2006; Sivakumar and Gorbsky 2015). The activity of Cdh1–APC and Cdc20–APC is tightly controlled by post-translational modifications, including phosphorylation and ubiquitination as well as interactions with inhibitors in cycling cells (Reimann et al. 2001; Kraft et al. 2003; Peters 2006; Pesin and Orr-Weaver 2008). Cyclin-dependent kinases (CDKs) phosphorylate the anaphase-promoting complex, which promotes its association with Cdc20 (Kotani et al. 1999; Kramer et al. 2000), whereas phosphorylation of Cdh1 triggers its dissociation from the anaphase-promoting complex (Zachariae et al. 1998; Kramer et al. 2000). Characterization of the functions and regulation of Cdh1–APC and Cdc20–APC in dividing cells has provided invaluable clues for our understanding of the novel functions and mechanisms of the anaphase-promoting complex in the nervous system.
Identification of functions of the anaphase-promoting complex in the nervous system
While studies of Cdc20–APC and Cdh1–APC have occupied the leading edge of cell cycle research since its identification in the mid 1990s, early clues suggested that the anaphase-promoting complex might also have functions in post-mitotic tissues beyond the control of the cell cycle. Components of Cdh1–APC were found ubiquitously expressed in post-mitotic neurons, including in adult mouse and human brains (Gieffers et al. 1999). However, it was not until 2004 that the first neuronal function of the anaphase-promoting complex was identified in the mammalian brain (Konishi et al. 2004). Konishi et al. (2004) discovered that Cdh1–APC plays a critical role in the control of axon growth and patterning in the rodent cerebellar cortex. Other studies identified functions for Cdh1–APC in synapse development and function in flies and nematodes (Juo and Kaplan 2004; Van Roessel et al. 2004). Since these early reports over a decade ago, a large number of studies have uncovered additional functions and mechanisms of the anaphase-promoting complex in the nervous system (Fig. 2). The picture emerging from these investigations is that Cdh1–APC and Cdc20–APC play pleiotropic roles in distinct stages of nervous system development and function.
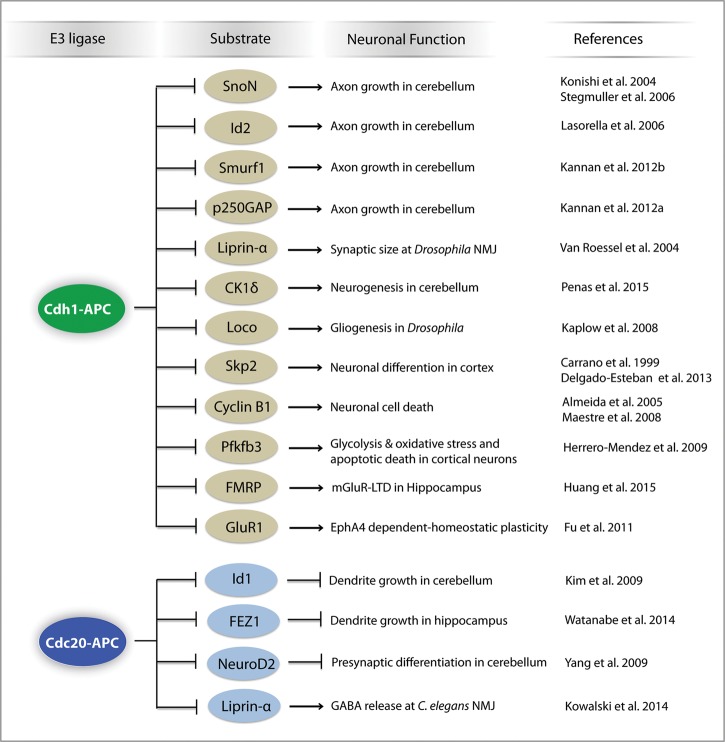
Figure 2.
Substrates of the anaphase-promoting complex in the nervous system. Substrates of Cdh1–APC and Cdc20–APC and their roles in the nervous system are shown. SnoN, Id2, Smurf1, and p250GAP are substrates of Cdh1–APC in the regulation of axon growth in the rodent cerebellum. Liprin-α is a substrate of Cdh1–APC in the regulation of synaptic size and transmission at the Drosophila neuromuscular junction (NMJ). Casein kinase 1δ (CK1δ) is a substrate of Cdh1–APC in the regulation of neurogenesis in the rodent cerebellum. Loco is a substrate of Cdh1–APC in the regulation of gliogenesis in Drosophila. Skp2 is a substrate of Cdh1–APC in the regulation of cortical neuronal differentiation. Cyclin B1 and 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-3 (Pfkfb3) are substrates of Cdh1–APC in the regulation of neuronal cell death. FMRP is a substrate of Cdh1–APC in the regulation of hippocampal mGluR-dependent long-term depression (mGluR-LTD). GluR1 is a substrate of Cdh1–APC in the regulation of EphA4-dependent homeostatic plasticity in cortical neurons. Id1 and FEZ1 are substrates of Cdc20–APC in the regulation of dendrite growth in the cerebellum and hippocampus, respectfully. NeuroD2 is a substrate of Cdc20–APC in the regulation of presynaptic differentiation in the cerebellum. Liprin-α is a substrate of Cdc20–APC in the regulation of GABA release at the Caenorhabditis elegans NMJ.
Control of neurogenesis and gliogenesis by Cdh1–APC
The function of Cdh1–APC in neural precursors was first characterized in Cdh1 heterozygous mutant mice. Although Cdh1+/– mice have little or no defects in the structure of the nervous system, a specific set of neural progenitor cells in the subventricular zone show increased proliferation in the mutant mice (Garcia-Higuera et al. 2008). The function of Cdh1–APC in neurogenesis in the cerebral cortex was further characterized in conditional Cdh1 knockout mice using the Nestin-Cre driver as well as in embryo-restricted Cdh1 knockout mice using the Sox2-Cre driver (Delgado-Esteban et al. 2013; Eguren et al. 2013). Conditional knockout of Cdh1 in neural progenitor cells in both of these models impairs cortical neurogenesis due to delay of mitotic exit, accelerated entry into the S phase, replicative stress, and p53-induced apoptotic cell death. Thus, conditional knockout of Cdh1 reduces the number of cortical neurons and hence cortical size and thickness. Conditional knockout of Cdh1 induced by Nestin-Cre in neural progenitors leads to increased abundance of previously identified Cdh1–APC substrates, including Aurora A, Aurora B, Tpx2, and Cyclin B1 (Fig. 3A; Eguren et al. 2013), which consequently results in shorter G1 and premature entry into the S phase of the progenitors. Cdh1 depletion-induced replicative stress and proliferative defects can be rescued by moderate inhibition of Cdks in Cdh1 knockout neural progenitors, suggesting that inappropriate activation of Cdks may account for the Cdh1 depletion-induced phenotypes in cortical progenitor cells (Eguren et al. 2013).
Figure 3.
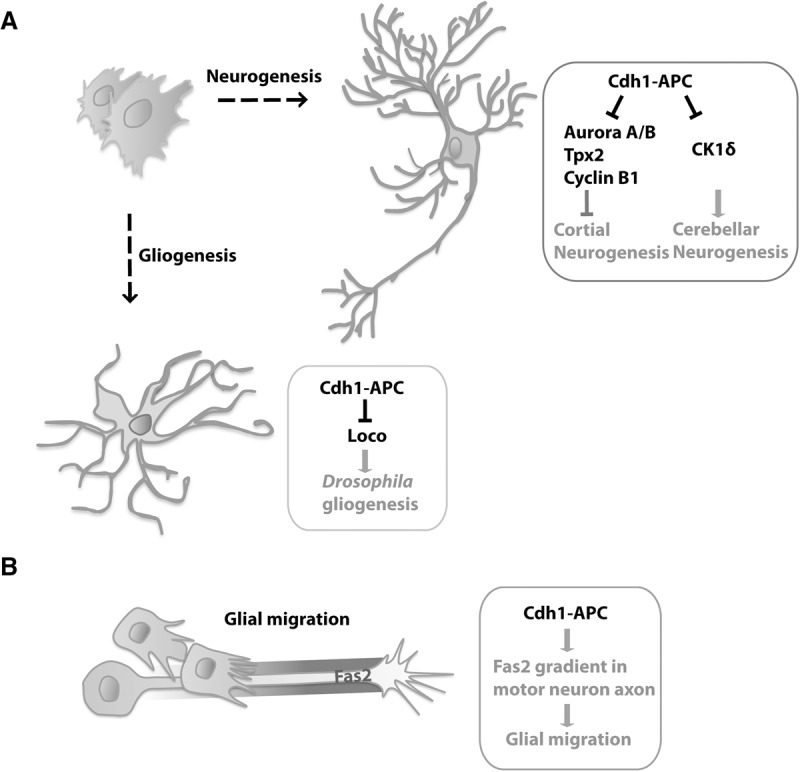
The anaphase-promoting complex regulates neurogenesis, gliogenesis, and glial migration. (A) Cdh1–APC controls neurogenesis in the cerebral cortex and cerebellum and gliogenesis in Drosophila. Cdh1–APC promotes cortical neurogenesis via degradation of the cell cycle proteins Aurora A/B, Tpx2, and Cyclin B1 in neural precursor cells. In the cerebellum, Cdh1–APC may control neurogenesis via degradation of the protein kinase CK1δ. In Drosophila, Cdh1–APC limits gliogenesis via degradation of Loco, a regulator of G-protein signaling protein. (B) In Drosophila, Cdh1–APC controls peripheral glial migration along motor neuron axons. Cdh1–APC acts non-cell-autonomously in the establishment of a gradient of the immunoglobulin superfamily cell adhesion molecule Fas2 in motor neuron axons, leading to low levels of Fas2 proximally and high levels distally, facilitating glial migration along motor neuron axons.
Cdh1–APC has also been reported to control neurogenesis in the cerebellum via degradation of the protein kinase casein kinase 1δ (CK1δ) (Fig. 3A; Penas et al. 2015). Knockdown of CK1δ by siRNA or pharmacologically by small molecule inhibitors leads to cell cycle arrest in granule neuron progenitors, suggesting that CK1δ promotes the proliferation of granule neuron progenitors. Cdh1–APC triggers the ubiquitination and consequent degradation of CK1δ in granule neuron progenitors (Penas et al. 2015), suggesting that Cdh1–APC might promote cell cycle exit and consequent granule neuron differentiation. Surprisingly, however, conditional knockout of Cdh1 in granule neuron progenitors fails to alter the development of the mouse cerebellum even though this leads to increased levels of CK1δ in these cells (Penas et al. 2015).
Cdh1 is up-regulated in primary neural stem cells upon neuronal differentiation in response to retinoic acid (Yao et al. 2010), suggesting a potential role for Cdh1–APC in neuronal differentiation. Consistent with these results, ablation of Cdh1 in neural progenitors in conditional Cdh1 knockout mice using the Sox2-Cre driver impairs neuronal differentiation (Delgado-Esteban et al. 2013). Primary cortical neuron cultures prepared from Sox2-Cre-induced conditional Cdh1 knockout mice contain a higher proportion of precursor cells and a lower proportion of post-mitotic neurons compared with control cultures. Correspondingly, the ventricular and subventricular zones in conditional Cdh1 knockout mice contain a higher proportion of precursor cells and fewer post-mitotic neurons (Delgado-Esteban et al. 2013), suggesting that Cdh1 promotes neuronal differentiation in the cerebral cortex. Cdh1–APC regulates neuronal differentiation through rapid degradation of the protein Skp2, leading to the stabilization of the Cdk inhibitor p27 (Carrano et al. 1999; Cuende et al. 2008), which in turn is thought to promote neuronal differentiation of cortical progenitor cells (Cuende et al. 2008; Harmey et al. 2009; Delgado-Esteban et al. 2013). The phosphorylation status of Cdh1 is critical in regulating Cdh1–APC function in neuronal differentiation. Cdh1–APC activity increases in primary cortical neurons during the first 2 d in culture, which is associated with dephosphorylation of Cdh1 by the protein phosphatase 2A (PP2A) (Delgado-Esteban et al. 2013). Incubation of cortical neurons with the PP2A inhibitor okadaic acid or transfecting neurons with a phosphomimetic form of Cdh1 inhibits neurite extension (Delgado-Esteban et al. 2013), suggesting that dephosphorylation of Cdh1 by PP2A promotes Cdh1–APC function in neuronal differentiation.
Whereas Cdh1–APC promotes neurogenesis in the rodent cortex (Delgado-Esteban et al. 2013; Eguren et al. 2013), it limits gliogenesis in flies (Fig. 3A; Kaplow et al. 2008). Drosophila Cdh1 mutants have a significantly higher number of glial cells in the brain. Conversely, Cdh1 overexpression in flies markedly reduces the number of glial cells. The protein Loco, a regulator of G-protein signaling, has been identified as a substrate of Cdh1–APC in the regulation of gliogenesis (Kaplow et al. 2008). Cdh1 interacts with Loco in the Drosophila larva brain, leading to its ubiquitination and consequent degradation. Loss-of-function Loco mutant flies harbor a reduced number of glial cells (Kaplow et al. 2008). Whether Cdh1–APC suppresses gliogenesis in the mammalian nervous system remains to be determined. Together, studies of Cdh1–APC in precursor cell differentiation suggest that Cdh1–APC promotes neurogenesis and limits gliogenesis. Whether Cdh1–APC plays an instructive cell-intrinsic role in cell fate specification in the same population of neural precursors is unknown.
The function of Cdh1–APC in glia extends beyond the control of their differentiation to influencing their migration in Drosophila (Fig. 3B). In genetic screens using ethyl methanesulfonate (EMS) mutagenesis, Cdh1 mutants show markedly reduced peripheral glial migration along motor neuron axons, suggesting that Cdh1–APC is required for glial migration (Silies and Klambt 2010). Remarkably, Cdh1 acts non-cell autonomously in regulating glial cell migration. Cdh1 is required for the establishment of a gradient of the immunoglobulin superfamily cell adhesion molecule Fasciclin 2 (Fas2) in motor neuron axons, leading to low levels of Fas2 proximally and high levels distally (Silies and Klambt 2010). Loss of Fas2 graded distribution in motor neuron axons of Cdh1 mutants causes impaired migration of peripheral glia cells (Silies and Klambt 2010). However, whether Cdh1–APC plays a direct role in ubiquitinating Fas2 remains to be determined, and a role for Cdh1–APC in the regulation of glial migration in vertebrates still remains to be explored.
Although Cdh1–APC functions have been characterized in neurogenesis and gliogenesis, the potential roles of Cdc20–APC in the biology of neural precursors in the developing nervous system have remained largely unexplored. Loss-of-function Cdc20 mutant mice have been generated from a gene trap embryonic stem cell clone containing a splicing acceptor and β-geo sequence inserted into intron 9 of the Cdc20 gene (Li et al. 2007). The predicted Cdc20 gene trap allele (Cdc20gt) encodes a fusion of 440 amino acids of the N-terminal portion of Cdc20 with β-geo. The loss-of-function Cdc20 mutant in Cdc20gt/gt mice triggers metaphase arrest at the two-cell embryonic stage (Li et al. 2007). It is presumed that the essential role of Cdc20–APC in cell proliferation extends to neural precursor cells, although demonstration of this function awaits generation of conditional Cdc20 knockout mice using precursor cell-specific drivers. In recent studies, a key function for Cdc20–APC has been uncovered in the biology of human glioblastoma brain tumor stem cells (BTSCs) (Mao et al. 2015). Using both knockdown and gain-of-function approaches, Mao et al. (2015) found that Cdc20–APC promotes the self-renewal and invasiveness of patient-derived glioblastoma BTSCs. In a manner distinct from its characteristic control of the cell cycle, Cdc20 interacts with the transcription factor Sox2 in the nucleus through the WD40 domain of Cdc20. Surprisingly, Cdc20–APC stabilizes Sox2 and hence enhances Sox2-dependent transcription in human BTSCs, leading to stem cell renewal and invasiveness (Mao et al. 2015). The mechanism by which Cdc20–APC promotes the accumulation of Sox2 in glioblastoma BTSCs and whether regulation of Sox2 extends to other stem cells during normal development remain important questions for future studies.
Role of the anaphase-promoting complex in neuronal survival and metabolism
Studies of Cdh1–APC in rat cortical neurons have pointed to a role for this major E3 ubiquitin ligase in the control of cell survival and metabolism. Knockdown of Cdh1 in rat cortical neurons by siRNAs leads to the accumulation of Cyclin B1 and aberrant entry into S phase and apoptotic cell death (Almeida et al. 2005). In subsequent studies, Cdk5 phosphorylation of Cdh1 has been shown to inhibit Cdh1–APC function in NMDA- or glutamate-induced neural excitotoxicity in cortical neurons, leading to Cyclin B1 accumulation, p27 depletion, aberrant cell cycle entry, and apoptotic neuronal cell death (Maestre et al. 2008; Veas-Perez de Tudela et al. 2015a,b). These studies suggest that Cdh1–APC may promote the survival of cortical neurons.
Interestingly, Cdh1–APC may control neuronal survival via a second substrate that plays a key role in cell metabolism (Herrero-Mendez et al. 2009; Rodriguez-Rodriguez et al. 2012). The glycolytic enzyme 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-3 (Pfkfb3) has been identified as a substrate of Cdh1–APC in rat cortical neurons. Pfkfb3 generates fructose-2,6-bisphosphate, which activates 6-phophofruto-1-kinase (Pfk1), the rate-limiting enzyme in glycolysis (Rapoport et al. 1976; Hue and Rider 1987). Under baseline conditions in primary cortical neurons, the levels of Pfkfb3 are low. However, upon Cdh1 knockdown, Pfkfb3 protein accumulates in neurons (Herrero-Mendez et al. 2009). Conversely, overexpression of Cdh1 in astrocytes, which have high levels of Pfkfb3, leads to down-regulation of Pfkfb3. Cdh1 interacts with Pfkfb3 via a KEN box, and a Pfkfb3 mutant protein in which the KEN box is mutated accumulates in neurons (Herrero-Mendez et al. 2009). Notably, knockdown of Cdh1 or up-regulation of Pfkfb3 activates glycolysis in neurons. The up-regulation of Pfkfb3 also reduces the consumption of glucose via the pentose phosphate pathway, leading to the accumulation of reactive oxygen species and apoptotic cell death. Thus, Cdh1–APC-mediated degradation of Pfkfb3 in neurons may inhibit glycolysis and maintain the antioxidant status of neurons (Herrero-Mendez et al. 2009). In subsequent studies, stimulation of the glutamate receptor NMDAR has been reported to stabilize Pfkfb3 secondary to Cdk5-mediated inhibition of Cdh1–APC activity (Rodriguez-Rodriguez et al. 2012). It will be important in future studies to determine the circumstances in which Cdh1–APC regulates neuronal survival via the accumulation of Cyclin B or Pfkfb3 in the mammalian brain in vivo.
The connection between neuronal survival and the anaphase-promoting complex has also been demonstrated in Drosophila larva brain neural stem cells in which Cdc20–APC promotes proliferation and concomitantly suppresses necrosis (Kuang et al. 2014). In a genetic screen, loss-of-function mutations of fzy (Cdc20) trigger premature brain neuroblast loss through necrosis, which occurs independently of Cdc20's function in cell proliferation. Cdc20–APC suppresses necrosis in larva brain neural stem cells by antagonizing the oxidoreductase Aif and the stress-activated protein kinase JNK signaling. In addition, p53 appears to promote cell death in these cells via inhibition of Cdc20–APC-dependent survival (Kuang et al. 2014). The Cdc20–APC substrate that regulates necrosis remains to be identified, and whether Cdc20–APC regulates necrosis in neural stem cells in the mammalian brain is unknown.
Regulation of neuronal morphogenesis by Cdh1–APC and Cdc20–APC
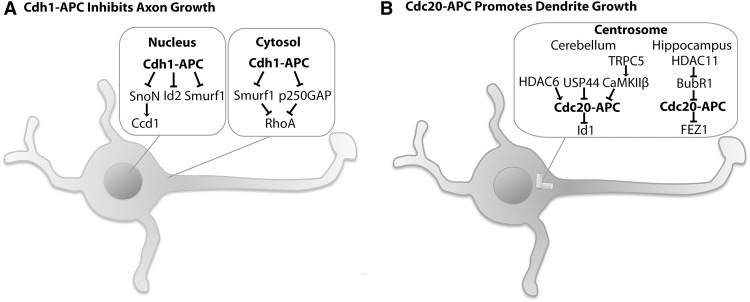
Although the biological roles of the anaphase-promoting complex in neural precursor cells might reflect functions and mechanisms of this complex in proliferating cells, the greatest surprises have come from studies of this complex in post-mitotic neurons (Juo and Kaplan 2004; Konishi et al. 2004; Van Roessel et al. 2004). The first insights into the role of the anaphase-promoting complex in the nervous system came from studies of Cdh1–APC in neuronal morphogenesis (Fig. 4A; Konishi et al. 2004). Using a knockdown approach in granule neurons of the rodent cerebellar cortex, Konishi et al. (2004) discovered that Cdh1–APC selectively limits the growth of axons but not dendrites. Expression of the Cdh1–APC inhibitor Emi1 or expression of a dominant interfering form of the catalytic subunit APC11 in granule neurons phenocopied the Cdh1 knockdown-induced increase in axon length in granule neurons (Konishi et al. 2004). To characterize the role of Cdh1–APC in the control of granule neuron axon growth in the brain, Konishi et al. (2004) developed and used an in vivo RNAi approach in postnatal rat pups. These studies revealed that Cdh1–APC controls the growth and patterning of granule neuron parallel fiber axons in the rodent cerebellum in vivo (Konishi et al. 2004).
Figure 4.
The anaphase-promoting complex regulates neuronal morphogenesis. (A) Cdh1–APC regulates axon growth in the cerebellum granule neurons. Cdh1–APC operates in the nucleus to limit axon growth by targeting the transcriptional regulators SnoN and Id2 and the E3 ubiquitin ligase Smurf1 for degradation. Cdh1–APC also operates in the cytosol to limit axon growth by targeting Smurf1 and the RhoGAP p250GAP for degradation and disinhibiting the negative effect of the small GTPase RhoA on axon growth. (B) In granule neurons of the cerebellum, Cdc20–APC operates at the centrosome to drive elaboration of dendrite arbors by targeting the helix–loop–helix (HLH) protein Id1 for degradation. At the centrosome, histone deacetylase 6 (HDAC6) promotes the polyubiquitination of Cdc20, whereas the deubiquitinase USP44 deubiquitinates Cdc20. Polyubiquitination of Cdc20 enhances Cdc20–APC activity, whereas the protein kinase CaMKIIβ inhibits centrosomal Cdc20–APC signaling, and the activity of CaMKIIβ is stimulated in turn by the calcium channel TRPC5. In hippocampal dentate gyrus granule neurons, Cdc20–APC also operates at the centrosome to drive elaboration of dendrite arbors by targeting the schizophrenia-linked protein FEZ1 for degradation. At the centrosome in the hippocampal neurons, HDAC11 deacetylates and inhibits BubR1, and BubR1 suppresses Cdc20–APC activity for dendrite growth.
To identify the mechanisms by which Cdh1–APC regulates axon growth in post-mitotic neurons, Stegmuller et al. (2006) characterized the subcellular locale of Cdh1 function in granule neurons. In structure–function analyses of an RNAi-resistant form of Cdh1 in the background of Cdh1 RNAi, Cdh1–APC operates in the nucleus rather than the cytoplasm to control axon growth. The transcriptional regulator SnoN has been identified as a substrate of neuronal Cdh1–APC in granule neurons (Fig. 4A). Cdh1–APC interacts with SnoN and stimulates the ubiquitination and consequent degradation of SnoN in granule neurons. Consistent with a critical role for SnoN in axon growth, knockdown of SnoN reduces axon length in granule neurons and triggers substantial loss of parallel fiber axons in the rodent cerebellum in vivo (Stegmuller et al. 2006). SnoN knockdown suppresses the Cdh1 knockdown-induced axon phenotype in granule neurons, consistent with the conclusion that Cdh1–APC operates upstream of SnoN in the regulation of axon growth in the mammalian brain (Stegmuller et al. 2006).
How the Cdh1–APC substrate SnoN promotes axon growth has also been characterized. Surprisingly, whereas SnoN typically operates as a transcriptional corepressor (Luo 2004; Pot and Bonni 2008), SnoN acts as a transcriptional activator in promoting axon growth (Ikeuchi et al. 2009). SnoN forms a complex with the transcriptional coactivator p300 and stimulates expression of the signaling scaffold protein Ccd1 (Fig. 4A), which is enriched at the axon terminal. Ccd1 activates the axon growth-promoting kinase JNK (Ikeuchi et al. 2009). Ccd1 knockdown mimics the effects of SnoN knockdown, including reducing the formation of parallel fiber formation in vivo, and suppresses SnoN-mediated axon growth (Ikeuchi et al. 2009). Beyond promoting the growth of axons in the developing cerebellum (Stegmuller et al. 2006), SnoN may also promote the regeneration of axons following injury (Do et al. 2013). Consistent with this result, inhibition of Cdh1–APC overrides the axon inhibitory effect of myelin (Konishi et al. 2004; Lasorella et al. 2006).
Cdh1–APC regulates axon growth via additional substrates besides SnoN (Lasorella et al. 2006). Immunoprecipitation followed by mass spectrometry analyses led to the identification of the helix–loop–helix (HLH) protein Id2 as an interactor of Cdh1–APC (Fig. 4A; Lasorella et al. 2006). Like SnoN, Id2 contains the Cdh1 recognition motif, the D box, and undergoes APC-induced ubiquitination and consequent proteasome-dependent degradation. Expression of Id2 with a D-box mutation in granule neurons promotes the growth of axons (Lasorella et al. 2006). As a negative regulator of basic HLH (bHLH)-dependent transcription (Sun et al. 1991), Id2 inhibits the expression of a number of genes encoding proteins with roles in axon growth inhibition (Lasorella et al. 2006). These results suggest that Cdh1–APC-induced degradation of Id2 induces the expression of a program of axon growth inhibitory signals. Like SnoN, Id2 may also promote the regeneration of axons following injury (Yu et al. 2011). Expression of the D-box mutant Id2 using adenovirus-based gene delivery in DRG neurons stimulates axon growth in the lesioned dorsal ascending axons following dorsal spinal cord hemisection (Yu et al. 2011), suggesting that inhibiting Cdh1–APC-mediated degradation of Id2 facilitates axon regeneration after spinal cord injury. By inducing the degradation of SnoN and Id2, Cdh1–APC therefore acts as a pivotal cell-intrinsic regulator of axon growth by controlling the responsiveness of axons to extrinsic cues and intracellular signaling proteins.
Cdh1–APC limits axon growth via the ubiquitination and consequent degradation of a third substrate, the E3 ubiquitin ligase Smurf1 (Fig. 4A; Kannan et al. 2012b). Cdh1 interacts with Smurf1 and targets it for ubiquitination and degradation in a D-box-dependent manner. Smurf1 stimulates axon growth in granule neurons by promoting the turnover of the small GTPase RhoA (Wang et al. 2003; Kannan et al. 2012b), which characteristically inhibits axon growth (Fournier et al. 2003; Govek et al. 2005). These findings suggest that the Cdh1–APC/Smurf1 pathway limits axon growth via activation of RhoA signaling. The RhoGAP p250GAP represents another target of Cdh1–APC that operates upstream of RhoA in the control of axon growth (Fig. 4A; Kannan et al. 2012a). Accordingly, the two targets of Cdh1–APC, Smurf1 and p250GAP, synergistically promote axon growth by disinhibiting the effect of RhoA on axon growth (Kannan et al. 2012a). Whereas Cdh1–APC targets Smurf1 for degradation, Cdh1–APC induction of p250GAP ubiquitination does not lead to its degradation (Kannan et al. 2012a).
How Cdh1–APC function is regulated in the control of axon growth has also been the subject of scrutiny. Thus far, these investigations have benefited from studies of Cdh1–APC regulation in proliferating cells. The TGFβ–Smad signaling pathway promotes the ability of Cdh1–APC to promote the degradation of SnoN in the G1 phase of the cell cycle in dividing cells (Bonni et al. 2001; Stroschein et al. 2001; Wan et al. 2001). Analogously, TGFβ–Smad2 signaling operates upstream of the Cdh1–APC/SnoN pathway in the control of axon growth in granule neurons, where knockdown of Smad2 stimulates axon growth (Stegmuller et al. 2008). In epistasis analyses, Smad2 acts upstream of SnoN and Cdh1–APC in the regulation of axon growth. Phosphorylation of Cdh1 figures prominently in the regulation of Cdh1–APC function of the cell cycle, whereby phosphorylation of Cdh1 on Cdk sites disrupts its interaction with the anaphase-promoting complex and inhibits Cdh1–APC function during S phase and early mitosis (Zachariae et al. 1998; Kramer et al. 2000; Peters 2006). In post-mitotic granule neurons of the rodent cerebellum, phosphorylation of Cdh1 at Cdk sites inhibits the ability of Cdh1–APC to limit axon growth (Huynh et al. 2009). Interestingly, a dephosphorylated mimic of Cdh1 in which nine sites of Cdk-induced phosphorylation are replaced with alanine (9A) is found in the nucleus, whereas the phosphorylated mimic in which the nine sites are replaced with aspartate (9D) accumulates in the cytoplasm. Moreover, the 9A mutant is found in low amounts, whereas the 9D mutant is stabilized and found in higher amounts. In structure–function analyses, the dephosphorylated Cdh1 mimic (9A), but not the phosphorylated Cdh1 mimic (9D), limits the growth of axons in granule neurons (Huynh et al. 2009). Collectively, these studies suggest that Cdh1–APC is subject to control by both extrinsic cues and intracellular signaling events in the control of axon growth.
While studies of Cdh1–APC in the nervous system began over a decade ago, studies of Cdc20–APC in the nervous system lagged behind, perhaps owing to the observation that, although Cdh1 is highly expressed in post-mitotic neurons in the adult brain, little or no Cdc20 appears to be expressed in adult brain neurons (Gieffers et al. 1999). However, the situation changed when Kim et al. (2009) discovered that Cdc20 is expressed in post-mitotic neurons during development. In contrast to Cdh1–APC regulation of axon growth (Konishi et al. 2004), Cdc20–APC drives the elaboration of dendrite arbors in the rodent cerebellum in vivo (Fig. 4B; Kim et al. 2009). Remarkably, Cdc20 is enriched at the centrosome in granule neurons, and centrosomal localization is critical for the ability of Cdc20–APC to promote dendrite arbor elaboration (Kim et al. 2009).
The HLH protein Id1 has been identified as a substrate of Cdc20–APC in the control of dendrite growth in granule neurons (Fig. 4B; Kim et al. 2009). A proportion of Id1 is localized at the centrosome in neurons. Cdc20 interacts with Id1 via a D box within Id1. Knockdown of Id1 in granule neurons promotes the growth of dendrites and suppresses the Cdc20 knockdown-induced dendrite phenotype, suggesting that Cdc20–APC operates upstream of Id1 in neurons. Notably, although Id1 is an inhibitor of bHLH-dependent transcription, Id1 operates at the centrosome to control dendrite growth in granule neurons (Kim et al. 2009). Remarkably, Id1 regulates dendrite growth by interacting with the proteasome lid protein S5a and thereby inhibiting proteasome function at the centrosome in granule neuron (Puram et al. 2013). Together, these studies suggest an intriguing interplay between the function of the E3 ubiquitin ligase Cdc20–APC and neuronal proteasomes at the centrosome in the nervous system.
Cdc20–APC function in dendrite elaboration is regulated by activating and inhibitory signaling mechanisms (Fig. 4B). Using a rational approach, Kim et al. (2009) found that the centrosomally localized protein histone deacetylase 6 (HDAC6) interacts with and stimulates the activity of Cdc20–APC in neurons. Surprisingly, rather than regulating Cdc20–APC through deacetylation of Cdc20, HDAC6 promotes via its C-terminal ZnF-UBP domain the polyubiquitinated state of Cdc20 and hence activates Cdc20–APC function in dendrite arbor morphogenesis in granule neurons. Consistent with the conclusion that the polyubiquitination of Cdc20 per se stimulates the activity of Cdc20–APC in dendrite growth, knockdown of the Cdc20-specific deubiquitinase USP44 (Stegmeier et al. 2007) increases the polyubiquitinated state of Cdc20 in granule neurons and stimulates dendrite growth in a Cdc20-dependent manner. These results suggest that the polyubiquitination of Cdc20 stimulates centrosomal Cdc20–APC signaling and consequent dendrite elaboration.
In contrast to HDAC6 activation of centrosomal Cdc20–APC signaling, the protein kinase CaMKIIβ inhibits centrosomal Cdc20–APC signaling in granule neurons (Fig. 4B; Puram et al. 2011a). Remarkably, CaMKIIβ localizes to the centrosome via a centrosomal targeting sequence (CTS) and operates independently of CaMKIIα, the other major brain CaMKII isoform (Hudmon and Schulman 2002), to promote granule neuron dendrite retraction and pruning (Puram et al. 2011a). CaMKIIβ catalyzes the phosphorylation of Cdc20 at a distinct site, Ser51, and thereby promotes the dispersion of Cdc20 away from the centrosome, leading to the inhibition of centrosomal Cdc20–APC activity in granule neurons. Consequently, CaMKIIβ induces the transition from granule neuron dendrite elaboration to dendrite pruning in the rodent brain (Puram et al. 2011a). The activity of CaMKIIβ is stimulated in turn in granule neurons of the cerebellum by the calcium channel TRPC5 (Puram et al. 2011b). Collectively, these studies suggest that a TRPC5/CaMKIIβ pathway triggers the phosphorylation of Cdc20 and consequently suppresses centrosomal Cdc20–APC signaling in neurons.
Cdc20–APC has also been implicated in promoting dendrite growth in the postnatal mouse hippocampus (Fig. 4B; Watanabe et al. 2014). Expression of Cdc20 stimulates, whereas knockdown of Cdc20 impairs, dendrite growth in dentate gyrus granule neurons. In these studies, the schizophrenia-linked protein FEZ1 has been identified as a substrate of Cdc20–APC. Upstream, the protein kinase BubR1 inhibits the function of Cdc20–APC in dendrite growth. Further upstream, the centrosomally localized HDAC11 triggers the deacetylation of BubR1 and thereby stimulates the activity of Cdc20–APC in dendrite growth. Interestingly, HDAC11 forms a complex with HDAC6, and these centrosomally localized proteins synergistically promote dendrite growth (Watanabe et al. 2014). These results suggest that centrosomal Cdc20–APC signaling is a key pathway in the regulation of dendrite morphogenesis in the mammalian brain.
The anaphase-promoting complex regulates synapse formation and function
Beyond the regulation of axon and dendrite morphogenesis, the anaphase-promoting complex has been implicated directly in neuronal connectivity by controlling synapse formation (Fig. 5). Early clues to the role of the anaphase-promoting complex in synapse development came from investigations in flies and nematodes (Juo and Kaplan 2004; Van Roessel et al. 2004). Flies with loss-of-function mutations of APC2 in which the entire cullin domain of APC2 is removed display expanded neuromuscular junction (NMJ) with an increase in the number of presynaptic boutons (Van Roessel et al. 2004). Expression of APC2 in motor neurons but not muscle reverses the NMJ overgrowth phenotype. The protein Liprin-α represents a downstream target of Cdh1–APC in the regulation of synaptic size in the presynaptic site (Fig. 5). Independently of its effects on the presynaptic side, Cdh1–APC also controls synaptic function on the postsynaptic side of the NMJ in Drosophila (Van Roessel et al. 2004). The APC2 mutation-induced postsynaptic phenotype is rescued by expression of APC2 in muscle cells but not motor neurons. Associated with this phenotype, APC2 mutant flies have higher levels of the glutamate receptor GluRIIa at the NMJ (Fig. 5; Van Roessel et al. 2004). In immunofluorescence analyses, APC2, Cdc27, and Cdh1, but not Cdc20, immunoreactivity is present at the NMJ synapse (Van Roessel et al. 2004), suggesting that Cdh1–APC regulates the size of Drosophila NMJ synapses. Collectively, these results suggest that Cdh1–APC regulates presynaptic size and postsynaptic function at the NMJ in Drosophila.
Figure 5.
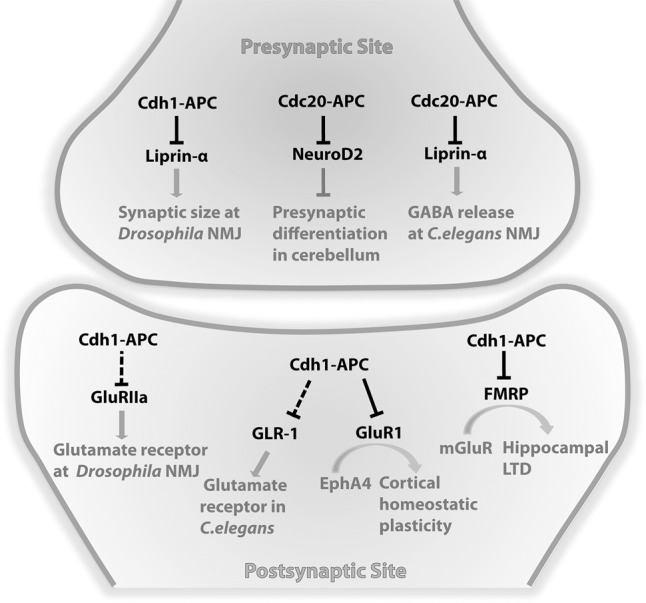
The anaphase-promoting complex regulates synapse development and function. At presynaptic sites, Cdh1–APC regulates synapse size at Drosophila NMJ via degradation of the protein Liprin-α. Cdc20–APC regulates GABA release and inhibitory synaptic transmission at C. elegans NMJ also by targeting Liprin-α for degradation. Cdc20–APC promotes presynaptic differentiation in granule neurons of the cerebellum via degradation of the transcription factor NeuroD2. At postsynaptic sites, Cdh1–APC limits expression of glutamate receptors GluRIIa at Drosophila NMJ and controls the endocytosis or recycling of GLR-1 in C. elegans ventral cord interneurons, thus regulating excitatory synaptic strength in flies and nematodes. In the regulation of synaptic plasticity, Cdh1–APC facilitates EphA4-dependent homeostatic plasticity by targeting the glutamate receptor GluR1 for degradation in cortical neurons. Cdh1–APC drives mGluR-LTD by targeting the fragile X mental retardation protein FMRP for degradation in the hippocampus.
In nematodes, synapses in ventral cord interneurons have been analyzed using a fusion of the glutamate receptor GLR-1 and GFP (GLR-1::GFP) (Rongo et al. 1998). Temperature-sensitive mutations of several core components of the anaphase-promoting complex lead to increased width, but not density or amplitude, of GLR-1::GFP puncta in these interneurons (Juo and Kaplan 2004). This phenotype is mimicked in transgenic nematodes expressing dominant-negative Cdh1 but not dominant-negative Cdc20 (Juo and Kaplan 2004). These results suggest that Cdh1–APC regulates the abundance of GLR-1 receptors in ventral cord interneurons in nematodes (Fig. 5). GLR-1 is unlikely to be the direct target of Cdh1–APC; rather, Cdh1–APC appears to regulate endocytosis or recycling of GLR-1, although the direct substrate in this process remains to be identified. Notably, in primary cortical neurons in vertebrates, Cdh1–APC has been suggested to directly induce the ubiquitination and consequent degradation of the glutamate receptor GluR1 during EphA4-dependent homeostatic plasticity (Fu et al. 2011).
Although most studies of the anaphase-promoting complex in synapse biology have focused on the role of Cdh1–APC, Yang et al. (2009) uncovered a critical role for Cdc20–APC in presynaptic differentiation in the mammalian brain. Knockdown of Cdc20 dramatically reduces the number of granule neuron parallel fiber active zone protein-containing boutons that are contiguous with PSD95-positive postsynaptic densities in the rodent cerebellum. This effect is not mimicked by knockdown of Cdh1, suggesting a selective role for Cdc20–APC in promoting the number of presynaptic sites in the cerebellar cortex. In primary granule neurons, Cdc20–APC promotes the formation of functional presynaptic sites. Cdc20–APC represents the first identified E3 ubiquitin ligase that drives presynaptic development in the mammalian brain.
How does Cdc20–APC regulate presynaptic differentiation? The transcription factor NeuroD2 has been identified as a substrate of neuronal Cdc20–APC (Fig. 5; Yang et al. 2009). Cdc20 interacts with NeuroD2 via a conserved D box. NeuroD2 is ubiquitinated in granule neurons, and knockdown of Cdc20 increases the levels of endogenous NeuroD2 in granule neurons. Importantly, knockdown of NeuroD2 leads to an increase in the number of presynaptic boutons in granule neurons and suppresses the Cdc20 knockdown-induced impairment of presynaptic differentiation. NeuroD2 suppresses the formation of presynaptic boutons in granule neurons via the synaptic regulator Complexin 2 (Yang et al. 2009). Collectively, these results suggest that Cdc20–APC and NeuroD2 form components of a ubiquitin signaling pathway that regulates presynaptic development. Several questions remain to be addressed in studies of Cdc20–APC regulation of presynaptic differentiation. Among these questions, one of the most interesting is the subcellular locale of Cdc20–APC action in presynaptic differentiation. In the regulation of neuronal morphogenesis, Cdh1–APC acts in the nucleus to control axon growth, whereas Cdc20–APC operates at the centrosome to drive dendrite arbor elaboration. Whether Cdc20–APC acts in the nucleus, centrosome, or elsewhere within the cell to promote the ubiquitination of the transcription factor NeuroD2 remains an open question.
Whereas much of the focus on synaptic roles of the anaphase-promoting complex has been on excitatory synapses, intriguing findings by Kowalski et al. (2014) suggest that Cdc20–APC may also play a crucial role in the regulation of inhibitory synapses by GABAergic motor neurons in C. elegans (Fig. 5). In nematodes, body wall muscles receive excitatory synaptic inputs by cholinergic motor neurons as well as inhibitory synaptic inputs by GABAergic motor neurons (Richmond and Jorgensen 1999; Gao and Zhen 2011). Temperature-sensitive mutations of APC4, Cdc16 (APC6), APC1, and Cdc23 (APC8) as well as knockdown of Cdc20, but not a Cdh1 mutant, impair GABA release at GABAergic motor neurons in C. elegans, as measured by inhibitory postsynaptic currents (IPSCs) at the NMJ and behavioral sensitivity to a cholinesterase inhibitor (Kowalski et al. 2014). Cdc20–APC mutants lead to increased levels of the protein Liprin-α presynaptically in GABAergic neurons (Kowalski et al. 2014), but how Cdc20–APC regulates presynaptic release of GABA remains to be fully elucidated.
Cdh1–APC promotes synaptic plasticity and learning and memory
In view of functions of the anaphase-promoting complex in synapse formation, the high level of expression of Cdh1–APC in the brain into adulthood suggests that this major E3 ubiquitin ligase may have roles in adaptive responses of the nervous system to experience. Consistent with this prediction, Cdh1–APC has been implicated in several forms of synaptic plasticity, including long-term potentiation (LTP), mGluR-dependent long-term depression (mGluR-LTD), and homeostatic synaptic plasticity (Fig. 5; Li et al. 2008; Fu et al. 2011; Pick et al. 2013a,b; Huang et al. 2015). Cdh1–APC regulates synaptic plasticity in several areas of the brain, including the hippocampus, amygdala, and cortex (Li et al. 2008; Fu et al. 2011; Pick et al. 2013a; Huang et al. 2015). Consistent with these results, conditional knockout of Cdh1 or APC2 leads to cognitive impairments in spatial learning and fear memory (Kuczera et al. 2011; Pick et al. 2013a,b).
The first description of Cdh1–APC's function in synaptic plasticity came from studies of mice generated from a gene trap embryonic stem cell clone containing a splicing acceptor and β-geo sequence inserted into intron 5 of the Cdh1 gene (Li et al. 2008). The predicted Cdh1 gene trap allele (Cdh1gt) encodes a fusion of 125 amino acids of the N-terminal portion of Cdh1 with β-geo. Although constitutive Cdh1gt/gt mice die in early development, mice heterozygous for the Cdh1gt allele have little or no gross brain morphological abnormalities despite reduction of Cdh1 protein levels by ∼50% in the brain. Synapses in the heterozygous mice show no abnormalities in basal transmission or presynaptic short-term plasticity. However, late-phase LTP (L-LTP) evoked by multiple trains of high-frequency stimulation (HFS) at 100 Hz, but not early-phase LTP (E-LTP) evoked by a single train of HFS, is impaired in acute hippocampal slices from Cdh1 heterozygous mice. L-LTP in slices from Cdh1 heterozygous mice decays to baseline levels with more rapid kinetics when compared with slices from control littermates (Li et al. 2008). To date, substrates of Cdh1–APC that mediate regulation of L-LTP remain unknown.
In contrast to heterozygous Cdh1+/gt mice (Li et al. 2008), conditional Cdh1 knockout mice generated using a CaMKII-Cre driver have no abnormalities of LTP in the hippocampus (Pick et al. 2013a), suggesting that impairment of L-LTP in the Cdh1 heterozygous mice may reflect a requirement for Cdh1 in the development of the hippocampus. On the other hand, the CaMKII-Cre-induced conditional Cdh1 knockout mice display modest impairment of L-LTP in the amygdala (Pick et al. 2013a). This phenotype is associated with increased abundance of the postsynaptic density protein Shank1 and glutamate receptor NR2A in the amygdala of conditional Cdh1 knockout mice. However, whether ubiquitination and degradation of these proteins mediate the effect of Cdh1–APC on L-LTP in the amygdala remains unknown. Notably, Shank1 contains a D box, and its levels are also increased in APC2-deficient primary hippocampal neurons (Hung et al. 2010), suggesting that Shank1 may represent a Cdh1–APC substrate.
Recent studies have uncovered a novel role for Cdh1–APC in driving mGluR-LTD (Fig. 5; Huang et al. 2015). Huang et al. (2015) found that conditional knockout of Cdh1 in forebrain-specific knockout mice using the Emx-Cre driver specifically impairs mGluR-LTD induced in the hippocampus by the group 1 mGluR agonist DHPG or a paired-pulse low-frequency stimulation paradigm. Expression of the Cdh1–APC inhibitor Emi1 in mice by in utero electroporation phenocopies the conditional Cdh1 knockout-induced impairment in mGluR-LTD in the hippocampus, suggesting that Cdh1 acts in concert with the anaphase-promoting complex to promote mGluR-LTD. Structure–function analyses of Cdh1 expressed in the hippocampus in vivo by in utero electroporation in the background of conditional Cdh1 knockout mice reveal that Cdh1 operates in the cytoplasm rather than the nucleus to drive mGluR-LTD (Huang et al. 2015).
The fragile X syndrome protein FMRP has been identified as a novel substrate of Cdh1–APC in the control of mGluR-LTD (Huang et al. 2015). The mGluR activator DHPG stimulates the ubiquitination and consequent degradation of FMRP in the hippocampus (Hou et al. 2006; Nalavadi et al. 2012). FMRP contains a conserved D box, and Cdh1 interacts with FMRP via the D box. DHPG-induced ubiquitination and degradation of FMRP is blocked in the hippocampus of conditional Cdh1 knockout mice (Huang et al. 2015). Accordingly, the levels of FMRP increase in the hippocampus in conditional Cdh1 knockout mice. Importantly, FMRP knockout, which enhances mGluR-LTD (Huber et al. 2002), suppresses the conditional Cdh1 knockout-induced mGluR-LTD phenotype (Huang et al. 2015). Collectively, these findings define a Cdh1–APC/FMRP ubiquitin signaling pathway that drives mGluR-dependent synaptic plasticity in the mammalian brain.
Cdh1–APC has also been implicated in homeostatic synaptic plasticity (Fig. 5; Fu et al. 2011), which refers to the capacity of neurons to regulate their own excitability relative to network activity (Turrigiano 2008; Pozo and Goda 2010). In Ephrin-A1/EphA4-mediated homeostatic plasticity at excitatory synapses, activation of the receptor EphA4 in primary cortical neurons in response to the ligand Ephrin-A1 induces chronic elevation of neural activity and results in down-regulation of excitatory synaptic transmission with decreased synaptic and surface GluR1 expression and reduced mEPSC frequency (Fu et al. 2007, 2011). Beginning with an interaction of EphA4 with a truncated fragment of APC2 in a yeast-two hybrid assay, Fu et al. (2007, 2011) found that Cdh1–APC mediates EphA4-dependent down-regulation of AMPA receptors in cortical neurons in response to chronic elevation of synaptic activity. EphA4 interacts with APC2 in 293T cells and cortical neurons, an interaction that is enhanced upon exposure to Ephrin-A1. The glutamate receptor GluR1 appears to operate as a substrate of Cdh1–APC in the regulation of homeostatic plasticity. The ubiquitination and degradation of GluR1 is dependent on Cdh1–APC in 293T cells. Knockdown of Cdh1 suppresses Ephrin-A1-induced down-regulation of GluR1 in cortical neurons. These findings suggest a role for Cdh1–APC in the turnover of postsynaptic AMPAR during EphA4-dependent homeostatic synaptic plasticity in cortical neurons.
Because memory traces are thought to be encoded by alterations of synaptic connectivity among neurons (Martin et al. 2000; Neves et al. 2008), the role of the anaphase-promoting complex in the regulation of neuronal connectivity and synaptic plasticity would be expected to impact the cognitive behavior of animals and, in particular, learning and memory. Several studies have probed the function of Cdh1–APC in spatial learning and fear memory (Li et al. 2008; Kuczera et al. 2011; Pick et al. 2013a,b). In APC2 conditional knockout mice generated using the CaMKIIα-Cre driver in which APC2 is depleted in adult forebrain excitatory neurons, Kuczera et al. (2011) characterized the role of the anaphase-promoting complex in learning and memory. Conditional APC2 knockout mice show normal brain morphology, home cage behavior, motor coordination, basal anxiety, and depressive-like behavior, making them suitable for studies of the anaphase-promoting complex in learning and memory. In a Pavlovian fear conditioning paradigm of associative fear memory in rodents (Anagnostaras et al. 2000), animals are placed in a fear conditioning apparatus, and an aversive stimulus (a foot shock) is paired with an auditory conditioned stimulus within the novel environment. After training, freezing behavior of the animals is measured in response to presentation of either the context (contextual fear conditioning) or auditory stimulus in a different context (cued fear conditioning). Female mice with conditional knockout of APC2 display increased freezing behavior during contextual memory assays performed 1 d after the electric shock. Interestingly, when animals are subjected to a fear extinction paradigm in which animals are subjected to repetitive presentation of the conditioned stimulus in the absence of the unconditioned stimulus after they have already been conditioned to associate the two stimuli, both male and female conditional APC2 knockout mice show higher freezing on all extinction trials (Kuczera et al. 2011). In addition to impaired fear extinction, conditional knockout of APC2 also display impaired hippocampal-dependent spatial memory using the Morris water maze (MWM) test of spatial learning and memory behavior in rodents, in which animals are placed in a large circular pool and trained to find an invisible platform that allows them to escape the water by using various spatial cues (Kuczera et al. 2011). Because APC2 is depleted in mature excitatory neurons rather than during development in these mice, the function of the anaphase-promoting complex in fear extinction and spatial memory may reflect its role in the regulation of synaptic strength in the hippocampus or amygdala in the adult brain.
In view of the high expression of Cdh1 in the adult brain, the role of the anaphase-promoting complex in learning and memory has been investigated on the basis of Cdh1–APC activity. Constitutive heterozygous Cdh1 knockout mice (Cdh1+/gt) generated by Li et al. (2008) have been subjected to learning and memory tests in contextual and cued fear conditioning. Whereas no difference in cued fear conditioning have been observed, heterozygous Cdh1 knockout mice display impaired contextual fear conditioning. These results suggest that Cdh1–APC specifically regulates hippocampus-dependent memory.
To better elucidate the function of Cdh1–APC in learning and memory, Pick et al. (2013a) generated conditional Cdh1 knockout mice using the CaMKIIα-Cre driver in which Cdh1 is depleted in adult forebrain excitatory neurons. Interestingly, phenotypes in conditional Cdh1 knockout and conditional APC2 knockout mice differ even though similar CaMKIIα-Cre drivers have been used in both studies (Kuczera et al. 2011; Pick et al. 2013a). Whereas conditional APC2 knockout mice do not show abnormalities in explorative and locomotor behavior in an open field paradigm (Kuczera et al. 2011), conditional Cdh1 knockout mice spend less time in the center and more time in the periphery of the open field arena (Pick et al. 2013a), suggesting that knockout of Cdh1, but not APC2, impairs exploration in a novel context. Likewise, whereas conditional APC2 knockout mice display impaired spatial learning in the MWM paradigm (Kuczera et al. 2011), conditional knockout of Cdh1 does not show such impairment (Pick et al. 2013a). However, in a reversal learning paradigm of the MWM, in which the hidden platform is moved to a different quadrant, conditional Cdh1 knockout mice show enhanced ability to reverse the previously consolidated spatial memory and establish new memory faster than control animals (Pick et al. 2013a). These conditional Cdh1 knockout mice also show enhanced reversal learning in a water-based Y maze (Pick et al. 2013a), another spatial learning paradigm. Consistent with the constitutive heterozygous Cdh1 knockout mice but inconsistent with the conditional APC2 knockout mice (Li et al. 2008; Kuczera et al. 2011), conditional Cdh1 knockout mice show reduction in their freezing behavior in a contextual fear conditioning paradigm 24 h after training (Pick et al. 2013a), suggesting that knockout of Cdh1 impairs contextual fear memory.
Another line of conditional Cdh1 knockout mice has been generated using a neuron-specific enolase (Nse)-Cre driver to delete Cdh1 in all neurons from the onset of differentiation (Pick et al. 2013b). These conditional Cdh1 knockout mice display normal spatial memory in both the MWM and water-based Y maze as well as normal contextual fear memory. However, these mice show deficits in learning flexibility. Although conditional Cdh1 knockout mice under CaMKIIα-Cre in another study show enhanced reversal learning in a water-based Y maze (Pick et al. 2013a), conditional Cdh1 knockout mice under the Nse-Cre driver display reduced reversal learning in the same learning paradigm (Pick et al. 2013b). These mice also have a diminished ability to extinguish previously consolidated fear memory in the associative auditory fear memory paradigm. Thus, the picture that emerges is that Cdh1–APC may have a specific role in memory flexibility.
Perspectives
More than a decade after the first description of a function for the anaphase-promoting complex in the nervous system (Konishi et al. 2004), Cdh1–APC and Cdc20–APC have emerged as major E3 ubiquitin ligases with pleiotropic roles in neurobiology. Several key themes and principles have emerged from studies of Cdh1–APC and Cdc20–APC in the nervous system, which should guide future investigations in this field.
The anaphase-promoting complex contributes to the regulation of diverse aspects of neuronal development and plasticity, from the control of neurogenesis and gliogenesis to neuronal survival and metabolism to axon and dendrite morphogenesis to synaptic connectivity and plasticity to the control of learning and memory. How does the specificity of the anaphase-promoting complex function arise? At least part of the answer may lie in the principle that spatial localization of Cdh1–APC and Cdc20–APC dictates its function (Yamada et al. 2013). For example, nuclear localization of Cdh1–APC in granule neurons of the developing cerebellum plays a critical role in the ability of Cdh1–APC to regulate axon growth owing to targeting transcriptional regulators in the nucleus for ubiquitination and consequent degradation (Konishi et al. 2004; Lasorella et al. 2006; Stegmuller et al. 2006). On the other hand, Cdc20–APC operates at the centrosome to drive granule neuron dendrite arbor elaboration by triggering the ubiquitination and degradation of a centrosomally localized substrate (Kim et al. 2009; Puram et al. 2011a,b; Watanabe et al. 2014). Cdc20–APC function in dendrite elaboration is also regulated by centrosomally localized signals in granule neurons. Thus, the centrosome provides a hub for Cdc20–APC ubiquitin signaling and hence specificity for its function in the control of dendrite morphogenesis. In hippocampal neurons, Cdh1–APC acts outside the nucleus, in the cytoplasm, to drive mGluR-LTD (Huang et al. 2015). Whether Cdh1–APC and Cdc20–APC operate at additional subcellular locales to specify other functions in the nervous system remains to be determined.
Whereas both Cdh1–APC and Cdc20–APC have been demonstrated to control neuronal morphogenesis and synaptic connectivity during development, only Cdh1–APC has been implicated in the control of synaptic plasticity and learning and memory in the adult brain (Almeida et al. 2005; Li et al. 2008; Fu et al. 2011; Kuczera et al. 2011; Pick et al. 2013a,b; Huang et al. 2015). This is perhaps not surprising in view of the high level of expression of Cdh1 in the adult brain (Gieffers et al. 1999). However, it will be interesting to determine whether Cdc20–APC is expressed or activated in specific cell types or under specific circumstances in the adult brain. In addition to identifying novel roles for Cdc20–APC in the adult brain, it will be also interesting to probe the functions of the anaphase-promoting complex regulated by Cdh1 or Cdc20 in other cognitive functions and behaviors beyond the control of learning and memory in the hippocampus and amygdala.
The identification of Cdh1–APC and Cdc20–APC functions in the nervous system has expanded the list of substrates that are ubiquitinated by the anaphase-promoting complex in cells. This trend is likely to continue in the years ahead as additional roles for Cdh1–APC and Cdc20–APC are uncovered in neurobiology. The use of conditional knockout strategies for Cdh1 and Cdc20 will be especially useful in this regard, as these regulators are knocked out in distinct populations of neurons, and glial cells in the nervous system should provide novel insights into the functions and mechanisms of Cdh1–APC and Cdc20–APC in the brain. For instance, while much of the work on the anaphase-promoting complex in neurons has focused on excitatory neurons and the control of excitatory synapses (Konishi et al. 2004; Kim et al. 2009; Yang et al. 2009; da Fonseca et al. 2011; Fu et al. 2011; Huang et al. 2015), studies in C. elegans suggest that Cdc20–APC may also play important roles in GABAergic motor neurons and the regulation of inhibitory synapses at the NMJ (Kowalski et al. 2014). Thus, it will be interesting to probe the role of Cdc20–APC in GABAergic interneurons and in the control of inhibitory synapse formation and function in the mammalian brain. Additional substrates of Cdh1–APC and Cdc20–APC will be identified, starting with new biological roles for the anaphase-promoting complex. Alternatively, advanced proteomics approaches also may be used to identify new substrates of Cdh1–APC and Cdc20–APC as a starting point in these studies.
Characterization of novel functions of the anaphase-promoting complex in neuronal connectivity and plasticity has raised prospects that deregulation of Cdh1–APC and Cdc20–APC ubiquitin signaling may also contribute to neurological diseases. The recent identification of the Cdh1–APC/FMRP ubiquitin signaling pathway in the control of mGluR-LTD provides an example of how a novel link between Cdh1–APC and a protein whose mutations cause fragile X syndrome might lead to new insights into intellectual disability and autism (Huang et al. 2015). Fragile X syndrome patients harbor a large expansion (>200) of the CGG trinucleotide repeat within the 5′ untranslated region of the FMR1 gene, which leads to the hypermethylation and consequent silencing of the FMR1 gene (Bhakar et al. 2012; Santoro et al. 2012). Importantly, in a substantial proportion of fragile X syndrome patients as well as carriers of the FMR1 gene in whom the CGG repeats expand to an intermediate level (55–200), the FMR1 gene is not silenced but is reduced in expression. Such fragile X syndrome patients and a substantial proportion of the fragile X syndrome carriers with the premutation have neuropsychological symptoms, including manifestation of autism spectrum disorder (Tassone et al. 2000; Farzin et al. 2006). If levels of FMRP can be increased in these patients upon inhibition of Cdh1–APC, such measures might be anticipated to reduce cognitive impairments in these patients. It will be important to test this hypothesis by inhibiting Cdh1–APC in mouse models that express premutation of the FMR1 gene (Entezam et al. 2007; Qin et al. 2011). Beyond the implications of Cdh1–APC/FMRP ubiquitin signaling in neurological disease, characterization of functions of the anaphase-promoting complex may provide clues for additional roles in disease. For instance, the ability of Cdh1–APC to inhibit axon growth may be harnessed to generate means of promoting axon growth upon inhibition of Cdh1–APC. In addition, the role of Cdh1–APC in synaptic plasticity and learning and memory raises the possibility that deregulation of Cdh1–APC signaling may contribute to disorders of memory. In future studies, it will be exciting to define the extent of anaphase-promoting complex ubiquitin signaling in brain disorders and explore the potential for small molecule inhibitors of the anaphase-promoting complex in the treatment of these disorders.
Acknowledgments
We thank the members of the Bonni laboratory for helpful advice and critical reading of the manuscript. The authors are supported by National Institutes of Health grant NS051255 (to A.B.), the Mathers Foundation (to A.B.), and a post-doctoral fellowship award from McDonnell Center for Cellular and Molecular Neurobiology (to J.H.).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.274324.115.
References
- Almeida A, Bolanos JP, Moreno S. 2005. Cdh1/Hct1–APC is essential for the survival of postmitotic neurons. J Neurosci 25: 8115–8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anagnostaras SG, Josselyn SA, Frankland PW, Silva AJ. 2000. Computer-assisted behavioral assessment of Pavlovian fear conditioning in mice. Learn Mem 7: 58–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhakar AL, Dolen G, Bear MF. 2012. The pathophysiology of fragile X (and what it teaches us about synapses). Annu Rev Neurosci 35: 417–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonni S, Wang HR, Causing CG, Kavsak P, Stroschein SL, Luo K, Wrana JL. 2001. TGF-β induces assembly of a Smad2–Smurf2 ubiquitin ligase complex that targets SnoN for degradation. Nat Cell Biol 3: 587–595. [DOI] [PubMed] [Google Scholar]
- Burton JL, Solomon MJ. 2001. D box and KEN box motifs in budding yeast Hsl1p are required for APC-mediated degradation and direct binding to Cdc20p and Cdh1p. Genes Dev 15: 2381–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrano AC, Eytan E, Hershko A, Pagano M. 1999. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol 1: 193–199. [DOI] [PubMed] [Google Scholar]
- Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. 2014. Molecular architecture and mechanism of the anaphase-promoting complex. Nature 513: 388–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. 2015. Atomic structure of the APC/C and its mechanism of protein ubiquitination. Nature 522: 450–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A. 2005. Intracellular protein degradation: from a vague idea, through the lysosome and the ubiquitin–proteasome system, and onto human diseases and drug targeting (Nobel lecture). Angew Chem Int Ed Engl 44: 5944–5967. [DOI] [PubMed] [Google Scholar]
- Cuende J, Moreno S, Bolanos JP, Almeida A. 2008. Retinoic acid downregulates Rae1 leading to APC(Cdh1) activation and neuroblastoma SH-SY5Y differentiation. Oncogene 27: 3339–3344. [DOI] [PubMed] [Google Scholar]
- da Fonseca PC, Kong EH, Zhang Z, Schreiber A, Williams MA, Morris EP, Barford D. 2011. Structures of APC/C(Cdh1) with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature 470: 274–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Esteban M, Garcia-Higuera I, Maestre C, Moreno S, Almeida A. 2013. APC/C–Cdh1 coordinates neurogenesis and cortical size during development. Nat Commun 4: 2879. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. 2009. RING domain E3 ubiquitin ligases. Annu Rev Biochem 78: 399–434. [DOI] [PubMed] [Google Scholar]
- Do JL, Bonni A, Tuszynski MH. 2013. SnoN facilitates axonal regeneration after spinal cord injury. PLoS One 8: e71906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube P, Herzog F, Gieffers C, Sander B, Riedel D, Muller SA, Engel A, Peters JM, Stark H. 2005. Localization of the coactivator Cdh1 and the cullin subunit Apc2 in a cryo-electron microscopy model of vertebrate APC/C. Mol Cell 20: 867–879. [DOI] [PubMed] [Google Scholar]
- Eguren M, Porlan E, Manchado E, Garcia-Higuera I, Canamero M, Farinas I, Malumbres M. 2013. The APC/C cofactor Cdh1 prevents replicative stress and p53-dependent cell death in neural progenitors. Nat Commun 4: 2880. [DOI] [PubMed] [Google Scholar]
- Entezam A, Biacsi R, Orrison B, Saha T, Hoffman GE, Grabczyk E, Nussbaum RL, Usdin K. 2007. Regional FMRP deficits and large repeat expansions into the full mutation range in a new Fragile X premutation mouse model. Gene 395: 125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzin F, Perry H, Hessl D, Loesch D, Cohen J, Bacalman S, Gane L, Tassone F, Hagerman P, Hagerman R. 2006. Autism spectrum disorders and attention-deficit/hyperactivity disorder in boys with the fragile X premutation. J Dev Behav Pediatr 27: S137–S144. [DOI] [PubMed] [Google Scholar]
- Fournier AE, Takizawa BT, Strittmatter SM. 2003. Rho kinase inhibition enhances axonal regeneration in the injured CNS. J Neurosci 23: 1416–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye JJ, Brown NG, Petzold G, Watson ER, Grace CR, Nourse A, Jarvis MA, Kriwacki RW, Peters JM, Stark H, et al. 2013. Electron microscopy structure of human APC/C(CDH1)–EMI1 reveals multimodal mechanism of E3 ligase shutdown. Nat Struct Mol Biol 20: 827–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu WY, Chen Y, Sahin M, Zhao XS, Shi L, Bikoff JB, Lai KO, Yung WH, Fu AK, Greenberg ME, et al. 2007. Cdk5 regulates EphA4-mediated dendritic spine retraction through an ephexin1-dependent mechanism. Nat Neurosci 10: 67–76. [DOI] [PubMed] [Google Scholar]
- Fu AK, Hung KW, Fu WY, Shen C, Chen Y, Xia J, Lai KO, Ip NY. 2011. APC(Cdh1) mediates EphA4-dependent downregulation of AMPA receptors in homeostatic plasticity. Nat Neurosci 14: 181–189. [DOI] [PubMed] [Google Scholar]
- Gao S, Zhen M. 2011. Action potentials drive body wall muscle contractions in Caenorhabditis elegans. Proc Natl Acad Sci 108: 2557–2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera I, Manchado E, Dubus P, Canamero M, Mendez J, Moreno S, Malumbres M. 2008. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat Cell Biol 10: 802–811. [DOI] [PubMed] [Google Scholar]
- Gieffers C, Peters BH, Kramer ER, Dotti CG, Peters JM. 1999. Expression of the CDH1-associated form of the anaphase-promoting complex in postmitotic neurons. Proc Natl Acad Sci 96: 11317–11322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW. 1991. Cyclin is degraded by the ubiquitin pathway. Nature 349: 132–138. [DOI] [PubMed] [Google Scholar]
- Govek EE, Newey SE, Van Aelst L. 2005. The role of the Rho GTPases in neuronal development. Genes Dev 19: 1–49. [DOI] [PubMed] [Google Scholar]
- Haas AL, Rose IA. 1982. The mechanism of ubiquitin activating enzyme. A kinetic and equilibrium analysis. J Biol Chem 257: 10329–10337. [PubMed] [Google Scholar]
- Harmey D, Smith A, Simanski S, Moussa CZ, Ayad NG. 2009. The anaphase promoting complex induces substrate degradation during neuronal differentiation. J Biol Chem 284: 4317–4323. [DOI] [PubMed] [Google Scholar]
- Harper JW, Burton JL, Solomon MJ. 2002. The anaphase-promoting complex: it's not just for mitosis any more. Genes Dev 16: 2179–2206. [DOI] [PubMed] [Google Scholar]
- He J, Chao WC, Zhang Z, Yang J, Cronin N, Barford D. 2013. Insights into degron recognition by APC/C coactivators from the structure of an Acm1-Cdh1 complex. Mol Cell 50: 649–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde AN, Upadhya SC. 2007. The ubiquitin–proteasome pathway in health and disease of the nervous system. Trends Neurosci 30: 587–595. [DOI] [PubMed] [Google Scholar]
- Herrero-Mendez A, Almeida A, Fernandez E, Maestre C, Moncada S, Bolanos JP. 2009. The bioenergetic and antioxidant status of neurons is controlled by continuous degradation of a key glycolytic enzyme by APC/C–Cdh1. Nat Cell Biol 11: 747–752. [DOI] [PubMed] [Google Scholar]
- Hershko A. 2005. The ubiquitin system for protein degradation and some of its roles in the control of the cell-division cycle (Nobel lecture). Angew Chem Int Ed Engl 44: 5932–5943. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. 1992. The ubiquitin system for protein degradation. Annu Rev Biochem 61: 761–807. [DOI] [PubMed] [Google Scholar]
- Herzog F, Primorac I, Dube P, Lenart P, Sander B, Mechtler K, Stark H, Peters JM. 2009. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science 323: 1477–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Antion MD, Hu D, Spencer CM, Paylor R, Klann E. 2006. Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression. Neuron 51: 441–454. [DOI] [PubMed] [Google Scholar]
- Huang J, Ikeuchi Y, Malumbres M, Bonni A. 2015. A Cdh1–APC/FMRP ubiquitin signaling link drives mGluR-dependent synaptic plasticity in the mammalian brain. Neuron 86: 726–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber KM, Gallagher SM, Warren ST, Bear MF. 2002. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc Natl Acad Sci 99: 7746–7750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudmon A, Schulman H. 2002. Neuronal CA2+/calmodulin-dependent protein kinase II: the role of structure and autoregulation in cellular function. Annu Rev Biochem 71: 473–510. [DOI] [PubMed] [Google Scholar]
- Hue L, Rider MH. 1987. Role of fructose 2,6-bisphosphate in the control of glycolysis in mammalian tissues. Biochem J 245: 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung AY, Sung CC, Brito IL, Sheng M. 2010. Degradation of postsynaptic scaffold GKAP and regulation of dendritic spine morphology by the TRIM3 ubiquitin ligase in rat hippocampal neurons. PLoS One 5: e9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh MA, Stegmuller J, Litterman N, Bonni A. 2009. Regulation of Cdh1–APC function in axon growth by Cdh1 phosphorylation. J Neurosci 29: 4322–4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang LH, Lau LF, Smith DL, Mistrot CA, Hardwick KG, Hwang ES, Amon A, Murray AW. 1998. Budding yeast Cdc20: a target of the spindle checkpoint. Science 279: 1041–1044. [DOI] [PubMed] [Google Scholar]
- Ikeuchi Y, Stegmuller J, Netherton S, Huynh MA, Masu M, Frank D, Bonni S, Bonni A. 2009. A SnoN–Ccd1 pathway promotes axonal morphogenesis in the mammalian brain. J Neurosci 29: 4312–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irniger S, Piatti S, Michaelis C, Nasmyth K. 1995. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell 81: 269–278. [DOI] [PubMed] [Google Scholar]
- Izawa D, Pines J. 2011. How APC/C–Cdc20 changes its substrate specificity in mitosis. Nat Cell Biol 13: 223–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Williamson A, Banerjee S, Philipp I, Rape M. 2008. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell 133: 653–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juo P, Kaplan JM. 2004. The anaphase-promoting complex regulates the abundance of GLR-1 glutamate receptors in the ventral nerve cord of C. elegans. Curr Biol 14: 2057–2062. [DOI] [PubMed] [Google Scholar]
- Kannan M, Lee SJ, Schwedhelm-Domeyer N, Nakazawa T, Stegmuller J. 2012a. p250GAP is a novel player in the Cdh1–APC/Smurf1 pathway of axon growth regulation. PLoS One 7: e50735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan M, Lee SJ, Schwedhelm-Domeyer N, Stegmuller J. 2012b. The E3 ligase Cdh1-anaphase promoting complex operates upstream of the E3 ligase Smurf1 in the control of axon growth. Development 139: 3600–3612. [DOI] [PubMed] [Google Scholar]
- Kaplow ME, Korayem AH, Venkatesh TR. 2008. Regulation of glia number in Drosophila by Rap/Fzr, an activator of the anaphase-promoting complex, and Loco, an RGS protein. Genetics 178: 2003–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim AH, Puram SV, Bilimoria PM, Ikeuchi Y, Keough S, Wong M, Rowitch D, Bonni A. 2009. A centrosomal Cdc20–APC pathway controls dendrite morphogenesis in postmitotic neurons. Cell 136: 322–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Peters JM, Tugendreich S, Rolfe M, Hieter P, Kirschner MW. 1995. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell 81: 279–288. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi SP. 2006. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol 8: 700–710. [DOI] [PubMed] [Google Scholar]
- Komander D, Rape M. 2012. The ubiquitin code. Annu Rev Biochem 81: 203–229. [DOI] [PubMed] [Google Scholar]
- Konishi Y, Stegmuller J, Matsuda T, Bonni S, Bonni A. 2004. Cdh1–APC controls axonal growth and patterning in the mammalian brain. Science 303: 1026–1030. [DOI] [PubMed] [Google Scholar]
- Kotani S, Tanaka H, Yasuda H, Todokoro K. 1999. Regulation of APC activity by phosphorylation and regulatory factors. J Cell Biol 146: 791–800. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kowalski JR, Dube H, Touroutine D, Rush KM, Goodwin PR, Carozza M, Didier Z, Francis MM, Juo P. 2014. The anaphase-promoting complex (APC) ubiquitin ligase regulates GABA transmission at the C. elegans neuromuscular junction. Mol Cell Neurosci 58: 62–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C, Herzog F, Gieffers C, Mechtler K, Hagting A, Pines J, Peters JM. 2003. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J 22: 6598–6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer ER, Scheuringer N, Podtelejnikov AV, Mann M, Peters JM. 2000. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol Biol Cell 11: 1555–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang C, Golden KL, Simon CR, Damrath J, Buttitta L, Gamble CE, Lee CY. 2014. A novel fizzy/Cdc20-dependent mechanism suppresses necrosis in neural stem cells. Development 141: 1453–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczera T, Stilling RM, Hsia HE, Bahari-Javan S, Irniger S, Nasmyth K, Sananbenesi F, Fischer A. 2011. The anaphase promoting complex is required for memory function in mice. Learn Mem 18: 49–57. [DOI] [PubMed] [Google Scholar]
- Lasorella A, Stegmuller J, Guardavaccaro D, Liu G, Carro MS, Rothschild G, de la Torre-Ubieta L, Pagano M, Bonni A, Iavarone A. 2006. Degradation of Id2 by the anaphase-promoting complex couples cell cycle exit and axonal growth. Nature 442: 471–474. [DOI] [PubMed] [Google Scholar]
- Li M, York JP, Zhang P. 2007. Loss of Cdc20 causes a securin-dependent metaphase arrest in two-cell mouse embryos. Mol Cell Biol 27: 3481–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Shin YH, Hou L, Huang X, Wei Z, Klann E, Zhang P. 2008. The adaptor protein of the anaphase promoting complex Cdh1 is essential in maintaining replicative lifespan and in learning and memory. Nat Cell Biol 10: 1083–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlepage LE, Ruderman JV. 2002. Identification of a new APC/C recognition domain, the A box, which is required for the Cdh1-dependent destruction of the kinase Aurora-A during mitotic exit. Genes Dev 16: 2274–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Lee BH, King RW, Finley D, Kirschner MW. 2015. Substrate degradation by the proteasome: a single-molecule kinetic analysis. Science 348: 1250834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K. 2004. Ski and SnoN: negative regulators of TGF-β signaling. Curr Opin Genet Dev 14: 65–70. [DOI] [PubMed] [Google Scholar]
- Maestre C, Delgado-Esteban M, Gomez-Sanchez JC, Bolanos JP, Almeida A. 2008. Cdk5 phosphorylates Cdh1 and modulates cyclin B1 stability in excitotoxicity. EMBO J 27: 2736–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao DD, Gujar AD, Mahlokozera T, Chen I, Pan Y, Luo J, Brost T, Thompson EA, Turski A, Leuthardt EC, et al. 2015. A CDC20–APC/SOX2 signaling axis regulates human glioblastoma stem-like cells. Cell Rep 11: 1809–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SJ, Grimwood PD, Morris RG. 2000. Synaptic plasticity and memory: an evaluation of the hypothesis. Annu Rev Neurosci 23: 649–711. [DOI] [PubMed] [Google Scholar]
- Meyer HJ, Rape M. 2014. Enhanced protein degradation by branched ubiquitin chains. Cell 157: 910–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalavadi VC, Muddashetty RS, Gross C, Bassell GJ. 2012. Dephosphorylation-induced ubiquitination and degradation of FMRP in dendrites: a role in immediate early mGluR-stimulated translation. J Neurosci 32: 2582–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves G, Cooke SF, Bliss TV. 2008. Synaptic plasticity, memory and the hippocampus: a neural network approach to causality. Nat Rev Neurosci 9: 65–75. [DOI] [PubMed] [Google Scholar]
- Ohi MD, Feoktistova A, Ren L, Yip C, Cheng Y, Chen JS, Yoon HJ, Wall JS, Huang Z, Penczek PA, et al. 2007. Structural organization of the anaphase-promoting complex bound to the mitotic activator Slp1. Mol Cell 28: 871–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore LA, Booth CR, Venien-Bryan C, Ludtke SJ, Fioretto C, Johnson LN, Chiu W, Barford D. 2005. Structural analysis of the anaphase-promoting complex reveals multiple active sites and insights into polyubiquitylation. Mol Cell 20: 855–866. [DOI] [PubMed] [Google Scholar]
- Penas C, Govek EE, Fang Y, Ramachandran V, Daniel M, Wang W, Maloof ME, Rahaim RJ, Bibian M, Kawauchi D, et al. 2015. Casein kinase 1δ is an APC/C(Cdh1) substrate that regulates cerebellar granule cell neurogenesis. Cell Rep 11: 249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesin JA, Orr-Weaver TL. 2008. Regulation of APC/C activators in mitosis and meiosis. Annu Rev Cell Dev Biol 24: 475–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM. 2006. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol 7: 644–656. [DOI] [PubMed] [Google Scholar]
- Pfleger CM, Kirschner MW. 2000. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev 14: 655–665. [PMC free article] [PubMed] [Google Scholar]
- Pick JE, Malumbres M, Klann E. 2013a. The E3 ligase APC/C–Cdh1 is required for associative fear memory and long-term potentiation in the amygdala of adult mice. Learn Mem 20: 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick JE, Wang L, Mayfield JE, Klann E. 2013b. Neuronal expression of the ubiquitin E3 ligase APC/C–Cdh1 during development is required for long-term potentiation, behavioral flexibility, and extinction. Neurobiol Learn Mem 100: 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM. 2001. Mechanisms underlying ubiquitination. Annu Rev Biochem 70: 503–533. [DOI] [PubMed] [Google Scholar]
- Pot I, Bonni S. 2008. SnoN in TGF-β signaling and cancer biology. Curr Mol Med 8: 319–328. [DOI] [PubMed] [Google Scholar]
- Pozo K, Goda Y. 2010. Unraveling mechanisms of homeostatic synaptic plasticity. Neuron 66: 337–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puram SV, Kim AH, Ikeuchi Y, Wilson-Grady JT, Merdes A, Gygi SP, Bonni A. 2011a. A CaMKIIβ signaling pathway at the centrosome regulates dendrite patterning in the brain. Nat Neurosci 14: 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puram SV, Riccio A, Koirala S, Ikeuchi Y, Kim AH, Corfas G, Bonni A. 2011b. A TRPC5-regulated calcium signaling pathway controls dendrite patterning in the mammalian brain. Genes Dev 25: 2659–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puram SV, Kim AH, Park HY, Anckar J, Bonni A. 2013. The ubiquitin receptor S5a/Rpn10 links centrosomal proteasomes with dendrite development in the mammalian brain. Cell Rep 4: 19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin M, Entezam A, Usdin K, Huang T, Liu ZH, Hoffman GE, Smith CB. 2011. A mouse model of the fragile X premutation: effects on behavior, dendrite morphology, and regional rates of cerebral protein synthesis. Neurobiol Dis 42: 85–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport TA, Heinrich R, Rapoport SM. 1976. The regulatory principles of glycolysis in erythrocytes in vivo and in vitro. A minimal comprehensive model describing steady states, quasi-steady states and time-dependent processes. Biochem J 154: 449–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimann JD, Freed E, Hsu JY, Kramer ER, Peters JM, Jackson PK. 2001. Emi1 is a mitotic regulator that interacts with Cdc20 and inhibits the anaphase promoting complex. Cell 105: 645–655. [DOI] [PubMed] [Google Scholar]
- Richmond JE, Jorgensen EM. 1999. One GABA and two acetylcholine receptors function at the C. elegans neuromuscular junction. Nat Neurosci 2: 791–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo-Brenni MC, Morgan DO. 2007. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell 130: 127–139. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Rodriguez P, Fernandez E, Almeida A, Bolanos JP. 2012. Excitotoxic stimulus stabilizes PFKFB3 causing pentose-phosphate pathway to glycolysis switch and neurodegeneration. Cell Death Differ 19: 1582–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongo C, Whitfield CW, Rodal A, Kim SK, Kaplan JM. 1998. LIN-10 is a shared component of the polarized protein localization pathways in neurons and epithelia. Cell 94: 751–759. [DOI] [PubMed] [Google Scholar]
- Rotin D, Kumar S. 2009. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol 10: 398–409. [DOI] [PubMed] [Google Scholar]
- Sackton KL, Dimova N, Zeng X, Tian W, Zhang M, Sackton TB, Meaders J, Pfaff KL, Sigoillot F, Yu H, et al. 2014. Synergistic blockade of mitotic exit by two chemical inhibitors of the APC/C. Nature 514: 646–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro MR, Bray SM, Warren ST. 2012. Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Annu Rev Pathol 7: 219–245. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Nuber U, Huibregtse JM. 1995. Protein ubiquitination involving an E1–E2–E3 enzyme ubiquitin thioester cascade. Nature 373: 81–83. [DOI] [PubMed] [Google Scholar]
- Schreiber A, Stengel F, Zhang Z, Enchev RI, Kong EH, Morris EP, Robinson CV, da Fonseca PC, Barford D. 2011. Structural basis for the subunit assembly of the anaphase-promoting complex. Nature 470: 227–232. [DOI] [PubMed] [Google Scholar]
- Segref A, Hoppe T. 2009. Think locally: control of ubiquitin-dependent protein degradation in neurons. EMBO Rep 10: 44–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silies M, Klambt C. 2010. APC/C(Fzr/Cdh1)-dependent regulation of cell adhesion controls glial migration in the Drosophila PNS. Nat Neurosci 13: 1357–1364. [DOI] [PubMed] [Google Scholar]
- Sivakumar S, Gorbsky GJ. 2015. Spatiotemporal regulation of the anaphase-promoting complex in mitosis. Nat Rev Mol Cell Biol 16: 82–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F, Rape M, Draviam VM, Nalepa G, Sowa ME, Ang XL, McDonald ER III, Li MZ, Hannon GJ, Sorger PK, et al. 2007. Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature 446: 876–881. [DOI] [PubMed] [Google Scholar]
- Stegmuller J, Konishi Y, Huynh MA, Yuan Z, Dibacco S, Bonni A. 2006. Cell-intrinsic regulation of axonal morphogenesis by the Cdh1–APC target SnoN. Neuron 50: 389–400. [DOI] [PubMed] [Google Scholar]
- Stegmuller J, Huynh MA, Yuan Z, Konishi Y, Bonni A. 2008. TGFβ–Smad2 signaling regulates the Cdh1–APC/SnoN pathway of axonal morphogenesis. J Neurosci 28: 1961–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroschein SL, Bonni S, Wrana JL, Luo K. 2001. Smad3 recruits the anaphase-promoting complex for ubiquitination and degradation of SnoN. Genes Dev 15: 2822–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, Luca FC, Ruderman JV, Hershko A. 1995. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell 6: 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V, Chan GK, Yen TJ. 2001. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol 154: 925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XH, Copeland NG, Jenkins NA, Baltimore D. 1991. Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix–loop–helix proteins. Mol Cell Biol 11: 5603–5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Bharadwaj R, Li B, Yu H. 2001. Mad2-Independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev Cell 1: 227–237. [DOI] [PubMed] [Google Scholar]
- Tassone F, Hagerman RJ, Taylor AK, Mills JB, Harris SW, Gane LW, Hagerman PJ. 2000. Clinical involvement and protein expression in individuals with the FMR1 premutation. Am J Med Genet 91: 144–152. [DOI] [PubMed] [Google Scholar]
- Turrigiano GG. 2008. The self-tuning neuron: synaptic scaling of excitatory synapses. Cell 135: 422–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Roessel P, Elliott DA, Robinson IM, Prokop A, Brand AH. 2004. Independent regulation of synaptic size and activity by the anaphase-promoting complex. Cell 119: 707–718. [DOI] [PubMed] [Google Scholar]
- Veas-Perez de Tudela M, Delgado-Esteban M, Maestre C, Bobo-Jimenez V, Jimenez-Blasco D, Vecino R, Bolanos JP, Almeida A. 2015a. Regulation of Bcl-xL–ATP synthase interaction by mitochondrial cyclin B1–cyclin-dependent kinase-1 determines neuronal survival. J Neurosci 35: 9287–9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veas-Perez de Tudela M, Maestre C, Delgado-Esteban M, Bolanos JP, Almeida A. 2015b. Cdk5-mediated inhibition of APC/C–Cdh1 switches on the cyclin D1–Cdk4–pRb pathway causing aberrant S-phase entry of postmitotic neurons. Sci Rep 5: 18180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Peters NR, D'Onofrio M, Tochtrop GP, Sakamoto KM, Varadan R, Zhang M, Coffino P, Fushman D, Deshaies RJ, et al. 2004. Ubistatins inhibit proteasome-dependent degradation by binding the ubiquitin chain. Science 306: 117–120. [DOI] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A. 1997. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science 278: 460–463. [DOI] [PubMed] [Google Scholar]
- Wan Y, Liu X, Kirschner MW. 2001. The anaphase-promoting complex mediates TGF-β signaling by targeting SnoN for destruction. Mol Cell 8: 1027–1039. [DOI] [PubMed] [Google Scholar]
- Wang HR, Zhang Y, Ozdamar B, Ogunjimi AA, Alexandrova E, Thomsen GH, Wrana JL. 2003. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science 302: 1775–1779. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Khodosevich K, Monyer H. 2014. Dendrite development regulated by the schizophrenia-associated gene FEZ1 involves the ubiquitin proteasome system. Cell Rep 7: 552–564. [DOI] [PubMed] [Google Scholar]
- Weissman AM, Shabek N, Ciechanover A. 2011. The predator becomes the prey: regulating the ubiquitin system by ubiquitylation and degradation. Nat Rev Mol Cell Biol 12: 605–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Yang Y, Bonni A. 2013. Spatial organization of ubiquitin ligase pathways orchestrates neuronal connectivity. Trends Neurosci 36: 218–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Yu S, Qiao R, Weissmann F, Miller DJ, VanderLinden R, Brown NG, Frye JJ, Peters JM, Schulman BA. 2015. Structure of an APC3–APC16 complex: insights into assembly of the anaphase-promoting complex/cyclosome. J Mol Biol 427: 1748–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Kim AH, Yamada T, Wu B, Bilimoria PM, Ikeuchi Y, de la Iglesia N, Shen J, Bonni A. 2009. A Cdc20–APC ubiquitin signaling pathway regulates presynaptic differentiation. Science 326: 575–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao W, Qian W, Zhu C, Gui L, Qiu J, Zhang C. 2010. Cdh1–APC is involved in the differentiation of neural stem cells into neurons. Neuroreport 21: 39–44. [DOI] [PubMed] [Google Scholar]
- Ye Y, Rape M. 2009. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol 10: 755–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi JJ, Ehlers MD. 2007. Emerging roles for ubiquitin and protein degradation in neuronal function. Pharmacol Rev 59: 14–39. [DOI] [PubMed] [Google Scholar]
- Yu P, Zhang YP, Shields LB, Zheng Y, Hu X, Hill R, Howard R, Gu Z, Burke DA, Whittemore SR, et al. 2011. Inhibitor of DNA binding 2 promotes sensory axonal growth after SCI. Exp Neurol 231: 38–44. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Nasmyth K. 1999. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev 13: 2039–2058. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Schwab M, Nasmyth K, Seufert W. 1998. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science 282: 1721–1724. [DOI] [PubMed] [Google Scholar]
- Zeng X, King RW. 2012. An APC/C inhibitor stabilizes cyclin B1 by prematurely terminating ubiquitination. Nat Chem Biol 8: 383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Sigoillot F, Gaur S, Choi S, Pfaff KL, Oh DC, Hathaway N, Dimova N, Cuny GD, King RW. 2010. Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer Cell 18: 382–395. [DOI] [PMC free article] [PubMed] [Google Scholar]