Thongthip et al. describe a FANCD2/FANCI-associated nuclease 1 (Fan1)-deficient mouse and show that FAN1 is required for cellular and organismal resistance to DNA interstrand cross-links. Karyomegaly becomes prominent in the kidneys and livers of Fan1-deficient mice with age, and mice develop liver dysfunction.
Keywords: DNA damage, interstrand cross-link repair, karyomegalic interstitial nephritis, Fanconi anemia, FAN1, SNM1A
Abstract
Deficiency of FANCD2/FANCI-associated nuclease 1 (FAN1) in humans leads to karyomegalic interstitial nephritis (KIN), a rare hereditary kidney disease characterized by chronic renal fibrosis, tubular degeneration, and characteristic polyploid nuclei in multiple tissues. The mechanism of how FAN1 protects cells is largely unknown but is thought to involve FAN1's function in DNA interstrand cross-link (ICL) repair. Here, we describe a Fan1-deficient mouse and show that FAN1 is required for cellular and organismal resistance to ICLs. We show that the ubiquitin-binding zinc finger (UBZ) domain of FAN1, which is needed for interaction with FANCD2, is not required for the initial rapid recruitment of FAN1 to ICLs or for its role in DNA ICL resistance. Epistasis analyses reveal that FAN1 has cross-link repair activities that are independent of the Fanconi anemia proteins and that this activity is redundant with the 5′–3′ exonuclease SNM1A. Karyomegaly becomes prominent in kidneys and livers of Fan1-deficient mice with age, and mice develop liver dysfunction. Treatment of Fan1-deficient mice with ICL-inducing agents results in pronounced thymic and bone marrow hypocellularity and the disappearance of c-kit+ cells. Our results provide insight into the mechanism of FAN1 in ICL repair and demonstrate that the Fan1 mouse model effectively recapitulates the pathological features of human FAN1 deficiency.
DNA interstrand cross-links (ICLs) impede the unwinding and separation of DNA by covalently linking its Watson and Crick strands. If unrepaired, ICLs block DNA replication and transcription. In humans, defects in the repair of ICLs are associated with the genetic disorders Fanconi anemia (FA) and karyomegalic interstitial nephritis (KIN) (for review, see Moldovan and D'Andrea 2009; Kottemann and Smogorzewska 2013). The clinical phenotypes of these two diseases are distinct. FA patients display congenital abnormalities, bone marrow failure, and predisposition to cancers (Fanconi 1967; Butturini et al. 1994; Alter 2003), whereas KIN patients develop early-onset end-stage kidney disease (Mihatsch et al. 1979; Spoendlin et al. 1995; Zhou et al. 2012) and may also exhibit elevated serum markers of hepatocyte damage, mild anemia, and recurrent upper respiratory infections (Bhandari et al. 2002; Palmer et al. 2007; Uz et al. 2011). Histologically, the kidneys, livers, and brains of KIN patients contain enlarged, polyploid (karyomegalic) nuclei (Spoendlin et al. 1995; Zhou et al. 2012), which have not been described in FA patients.
Mutations in at least 19 FANC genes have been identified in patients with FA, while FANCD2/FANCI-associated nuclease 1 (FAN1) is the only known gene that is found to be inactivated in KIN patients (Zhou et al. 2012; Rickman et al. 2015; Wang et al. 2015; Wang and Smogorzewska 2015). Proteins encoded by the FANC genes (FANCA to FANCT) are directly involved in the repair of the ICL lesions during DNA replication (for review, see Kottemann and Smogorzewska 2013; Walden and Deans 2014; Wang and Smogorzewska 2015). Eight of these proteins form the FA core complex, which is necessary for ubiquitination of the FANCD2–FANCI (ID2) complex. The ubiquitinated ID2 complex localizes to ICLs and is essential for downstream nucleolytic incisions and translesion synthesis repair events. FANCQ/XPF in a complex with FANCP/SLX4 is responsible for making the incisions in one of the parental strands to “unhook” the ICL. Unhooking allows for translesion polymerases to bypass the lesion on one strand of the replicated DNA, with the other strand being repaired using homology-directed repair (HDR) proteins, including FANCD1/BRCA2, FANCN/PALB2, and RAD51/FANCR.
FAN1 is associated with the FA pathway through its interaction with the ID2 complex. This interaction is mediated by the N-terminal ubiquitin-binding zinc finger (UBZ) domain of FAN1 and the monoubiquitinated lysine residue of FANCD2 in the ID2 complex (Kratz et al. 2010; Liu et al. 2010; MacKay et al. 2010; Smogorzewska et al. 2010). Given the direct association between FAN1 and the ID2 complex, it has been proposed that the intrinsic nuclease activities of FAN1 are required either downstream from ID2 monoubiquitination to contribute to ICL incision or later during HDR of DNA double-strand break intermediates after incision. However, the phenotypic differences between patients deficient in one of the FANC proteins and FAN1 prompted the question of whether the cross-link repair function of FAN1 is epistatic with the FA pathway (Zhou et al. 2012). The UBZ-mediated interaction with the ID2 complex was first shown to be necessary for the localization of FAN1 to ICL-induced DNA damage foci and psoralen plus UV laser-induced stripes, arguing that the ICL repair activity of FAN1 largely depends on the UBZ domain (Kratz et al. 2010; Liu et al. 2010; MacKay et al. 2010; Smogorzewska et al. 2010). The UBZ domain also appeared to be necessary for resistance to ICLs (Liu et al. 2010). However, this was recently challenged, as UBZ-deficient FAN1 could fully rescue the ICL repair defects of a KIN patient cell line completely lacking FAN1 protein (Zhou et al. 2012). This finding suggested a possibility that the FAN1 and ID2 complex interaction is not required for the ICL-processing activity of FAN1 and that the cross-link repair function of FAN1 can take place independently of the FA pathway. Evidence from studies in the chicken DT40 cell line also support this view, as FAN1−/−/FANCC−/− cells are more sensitive to cisplatin than wild-type, FAN1−/−, or FANCC−/− single-knockout cells (Yoshikiyo et al. 2010).
Regardless of the genetic interaction with the FA pathway, recent in vitro studies have highlighted a possible mode of FAN1 activity on the cross-linked DNA substrates (Wang et al. 2014; Zhao et al. 2014; Pizzolato et al. 2015). These studies showed that FAN1 nuclease activity is able to process the ICL-stalled replication fork-like structures by cleaving 5′ flaps and also by processing nicks made by other nucleases to digest 5′ to 3′ across the cross-link, de facto unhooking it. However, despite the convincing in vitro data, unhooking of a cisplatin ICL was unaffected in FAN1-immunodepleted Xenopus laevis extracts (Douwel et al. 2014). This DNA replication-dependent ICL repair system relies on the FA proteins, including the XPF/FANCQ-bound SLX4/FANCP, and the depletion of FAN1 or MUS81 alone did not have any effect on the repair of the ICL lesions (Raschle et al. 2008; Knipscheer et al. 2009; Douwel et al. 2014). This and the previous studies stress the role of the XPF–ERCC1–SLX4 complex as the essential nuclease for ICL unhooking (Bhagwat et al. 2009; Kim et al. 2013; Douwel et al. 2014; Hodskinson et al. 2014) but leave open the possibility that the other ICL repair nucleases, including SLX4-associated MUS81 and SLX1 as well as FAN1, may possess redundant ICL processing activities or act on structures other than the dual convergent fork during ICL repair (for review, see Zhang and Walter 2014).
The in vitro activity of FAN1 on substrates containing an ICL overlaps with the activity of SNM1A, one of the three human homologs of Pso2, a nuclease that functions in ICL repair in Saccharomyces cerevisiae (Henriques and Moustacchi 1980; Ruhland et al. 1981; Hejna et al. 2007; Wang et al. 2011). Mammalian SNM1A shares the most similarity with Pso2 and is the only homolog that can complement the ICL sensitivity of pso2Δ yeast (Hazrati et al. 2008). Nonetheless, SNM1A deficiency has been shown to confer very mild cross-link sensitivity in mouse and human cells (Dronkert et al. 2000; Ahkter et al. 2005; Wang et al. 2011). This suggests that SNM1A plays a minor role in mammalian ICL repair or that another ICL repair nuclease, such as FAN1, may compensate for the loss of SNM1A. Consistent with possible redundant roles of SNM1A and FAN1, the Schizosaccharomyces pombe Fan1 was recently shown to be nonepistatic with Pso2, as pso2Δfan1Δ displays greater cisplatin sensitivity than a single pso2Δ or fan1Δ mutant alone (Fontebasso et al. 2013).
Although the role of FAN1 in DNA ICL repair is now firmly established, how the loss of FAN1 function causes chronic kidney degeneration in humans remains unknown. It is also unclear whether the secondary phenotypes reported for KIN patients, including anemia, liver dysfunction, and upper respiratory infections, are a result of faulty DNA repair. In addition, recent reports implicate FAN1 as a colorectal cancer (CRC) and high-risk pancreatic cancer (PC) susceptibility gene (Segui et al. 2015; Smith et al. 2016).
To facilitate the study of FAN1 function, we created a mouse model of Fan1 deficiency and show that FAN1 is essential for ICL resistance at both the cellular and organismal levels. Cells lacking FAN1 are less sensitive to cross-link-inducing agents than cells lacking the FANC proteins, and FAN1 has functions in ICL repair outside of the FA pathway. In addition, we demonstrate that SNM1A partially compensates for lack of FAN1 activity. At the organismal level, FAN1 is required for the suppression of polyploidy and karyomegaly in the kidney and liver and to safeguard liver function with increasing age. FAN1 is also vital for the protection of the hematopoietic compartment when exogenous cross-linking agents are used.
Results
FAN1 is required for resistance to DNA ICL-inducing agents in mouse embryonic fibroblasts (MEFs)
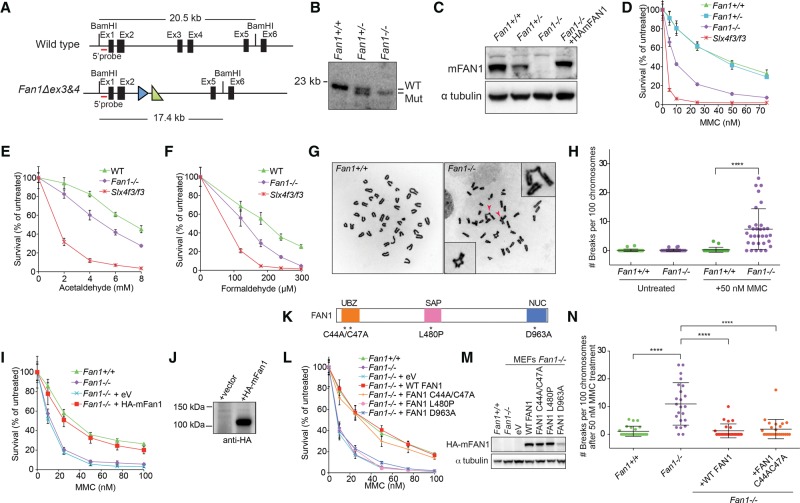
To investigate the cellular and organismal functions of FAN1, we generated Fan1-deficient mice. The Fan1 locus was targeted in embryonic stem cells using a conditional Fan1tm1a(KOMP)Wtsi (Fan1stop) construct (Knockout Mouse Project [KOMP]) (Supplemental Fig. S1A). The correctly targeted clone was used to create a mouse with a single Fan1stop allele (Supplemental Fig. S1B,C), which, through appropriate crosses (Supplemental Fig. S1A), gave rise to animals carrying the disrupted Fan1 allele Fan1Δex3&4, which, for ease of labeling, we refer to here as Fan1− (Fig. 1A; Supplemental Fig. S1D). Southern blotting and PCR genotyping verified the generation of Fan1−/− mice (Fig. 1B; Supplemental Fig. S1B–D). Removal of exons 3 and 4 from Fan1 resulted in low Fan1 transcript and no visible protein expression in MEFs obtained from homozygous Fan1−/− embryonic day 13.5 (E13.5) embryos (Fig. 1C; Supplemental Fig. S1E,F).
Figure 1.
FAN1 is necessary for resistance to DNA ICL damage in MEFs. (A) Schematic of the wild-type and Fan1Δex3&4 mutant alleles. The wild-type Fan1 gene contains a 20.5-kb BamHI restriction fragment that can be detected with a 5′ probe (red line). A correctly targeted Fan1 locus harbors a 3.1-kb deletion, including exons 3 and 4, resulting in a 17.4-kb fragment that hybridizes to the same probe. The FRT and LoxP sites are indicated as blue and green triangles, respectively. See Supplemental Figure. S1A for full Fan1 targeting strategy. (B) Southern blot analysis of BamHI-digested gDNA from Fan1+/+, Fan1+/−, and Fan1−/− MEFs. (WT) Wild type; (Mut) mutant. (C) Immunoblot of extracts from MEFs of the indicated genotypes using an antibody recognizing mouse FAN1. HAmFAN1 is N-terminally HA-tagged mouse FAN1 and is expressed in Fan1−/− MEFs. (D) Survival of MEFs after treatment with the indicated doses of mitomycin C (MMC). MEFs were treated in triplicate with increasing concentrations of MMC. After 6 d, cell numbers were determined and normalized to untreated control to calculate the percentage of survival. Error bars indicate SD. (E,F) Sensitivity to aldehydes of the indicated MEFs. MEFs were treated in triplicate with increasing concentrations of acetaldehyde (0–8 mM) (E) or formaldehyde (0–300 μM) (F) for 2 h before being washed three times with PBS and were cultured for an additional 6 d in non-drug-containing medium. After 6 d, cell numbers were determined and normalized to untreated control to calculate the percentage of survival. Error bars indicate SD. (G) Examples of metaphase spreads of Fan1+/+ (left) and Fan1−/− (right) after exposure to 50 nM MMC for 24 h. Arrowheads indicate radial chromosomes shown in the inset. (H) Quantification of chromosome breakage of Fan1+/+ and Fan1−/− MEFs shown in G. (****) P < 0.0001, calculated using a t-test. (I) Complementation of MMC sensitivity in Fan1−/− MEFs. Fan1−/− MEFs stably transduced with empty vector (eV) or vector expressing HA-tagged mouse FAN1 (HA-mFAN1) were exposed to different levels of MMC, as in D. Error bars indicate SD. (J) Immunoblot showing the expression of HA-mFAN1 used in I. (K) Schematic representation of the mouse FAN1 protein indicating the mutations in the key functional domains: C44A/C47A in the UBZ domain; p.L480P in the SAF-A/B, Acinus, and PIAS (SAP) domain; and p.D963A in (VRR-NUC) virus-type replication repair nuclease (NUC) domain. (L) MEFs stably transduced with empty vector or vector expressing Fan1 variants with mutations shown in K were MMC-treated as in D. Error bars indicate SD. (M) Immunoblot showing expression of mouse Fan1 mutant cDNAs in MEFs used in L. See also Supplemental Figure S1, I and J, for additional complementation assays. (N) Quantification of chromosome breakage analysis of the indicated MEFs treated with 50 nM MMC for 24 h.
To assess whether FAN1 deficiency recapitulates the cellular phenotypes seen in human cells devoid of FAN1, we studied MEFs treated with the ICL-inducing agent mitomycin C (MMC). Fan1−/− MEFs were hypersensitive to MMC when compared with Fan1+/+ or Fan1+/− MEFs (Fig. 1D). They were also more sensitive to treatment with acetaldehyde and formaldehyde, both of which have been implicated as endogenous sources of DNA ICLs (Fig. 1E,F). However, the level of sensitivity to all tested ICL-inducing agents was significantly lower than that of MEFs deficient in SLX4 (Slx4f3/f3), a scaffold for three nucleases, including XPF, which is the major effector of the FA pathway (Fig. 1D–F). Consistent with previous reports, Fan1-deficient MEFs did not show hypersensitivity to either camptothecin (CPT) or hydroxyurea (HU), suggesting that FAN1 is specifically required for the repair of DNA ICLs and not other S-phase-associated replication blocks (Supplemental Fig. S1G,H; Kratz et al. 2010; Liu et al. 2010; MacKay et al. 2010; Smogorzewska et al. 2010).
The inability to repair ICL damage in Fan1−/− MEFs was further illustrated by an increase in the level of chromosome breaks and radial chromosomes when cells were treated with MMC (Fig. 1G,H). Importantly, the cellular sensitivity and the increased breakage in Fan1−/− MEFs after MMC treatment could be fully complemented when cells were transduced with wild-type mouse Fan1 cDNA (Fig. 1C,I,J,L,N; Supplemental Fig. S1I,J). This shows that the cross-link repair defect in Fan1−/− MEFs is due to a lack of functional FAN1 protein.
The UBZ domain is not required for the cross-link repair function of FAN1 and is dispensable for the recruitment of FAN1 to sites of DNA damage
Our earlier work revealed that the UBZ domain was not essential for the cross-link resistance activity of human FAN1 (Zhou et al. 2012). In that study, the UBZ mutated Cys44AlaCys47Ala human FAN1 was shown to fully rescue the ICL repair defect in the KIN patient cells. To assess which domains of mouse FAN1 are important for ICL resistance, we expressed mouse FAN1 with mutated UBZ (p.Cys44Ala;Cys47Ala), SAP (p.Leu480Pro), or nuclease (p.Asp963Ala) domains in Fan1−/− MEFs and tested complementation of ICL sensitivity in a survival assay. Unlike expression of wild-type mouse Fan1 cDNA, which was able to fully complement the MMC sensitivity of Fan1-deficient MEFs, expression of cDNA carrying a mutation in the SAP or nuclease domain resulted in no rescue (Fig. 1K–M; Supplemental Fig. S1I,J). In agreement with our previous study, the Fan1 variant (p.Cys44Ala;Cys47Ala) behaved like the wild-type allele in this assay, indicating that FAN1-conferred ICL resistance is indeed independent of its FANCD2/FANCI interaction in mammalian cells (Fig. 1K–N). This result is perplexing, since the UBZ domain is critical for stable localization of FAN1 to the sites of DNA damage (Smogorzewska et al. 2010), and implies that a different domain of FAN1 might be important for localization of FAN1 to the ICLs.
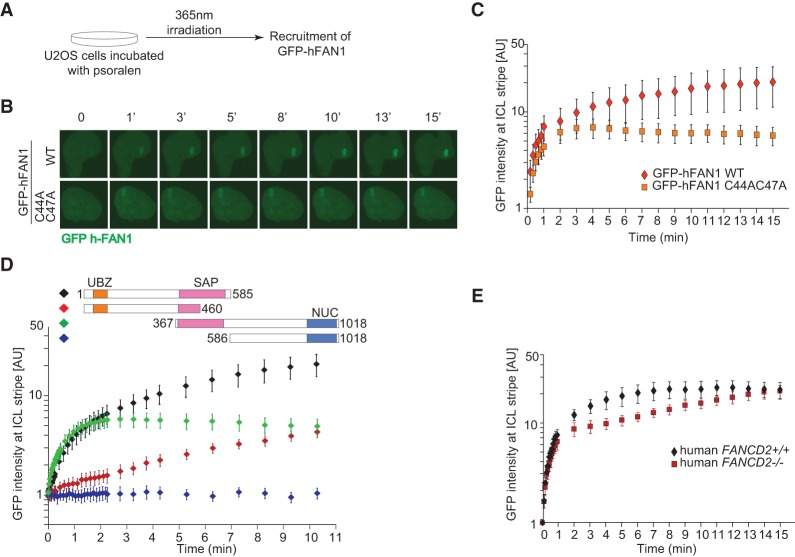
Recent crystallographic data revealed that the SAP domain interacts extensively with the DNA (Gwon et al. 2014; Wang et al. 2014; Zhang and Walter 2014), suggesting that direct DNA binding to the ICL might be responsible for the recruitment of FAN1 to sites of DNA damage. To investigate the contribution of the UBZ and the SAP domains to the localization of FAN1 at ICLs, we studied the recruitment of human GFP-tagged FAN1 (GFP-hFAN1) to psoralen-induced ICLs in U2OS cells (Fig. 2A; Yan et al. 2012). Accumulation of wild-type FAN1 was biphasic, with an initial rapid eightfold increase of the protein over the first 2 min followed by a slower but steady buildup over the next 13 min (Fig. 2B,C; Supplemental Fig. S2A,B). Cells expressing GFP tag alone or expressing GFP-hFAN1 but treated only with the UV laser without prior incubation in the presence of psoralen did not exhibit any recruitment to the laser stripe. The UBZ-deficient FAN1 displayed an intact initial rapid recruitment phase; however, its recruitment plateaued after the first 2 min (Fig. 2C). The SAP domain appeared to be necessary for the fast recruitment of FAN1 to ICLs, as the FAN1 variant with the UBZ domain but no SAP domain accumulated only very slowly at the sites of the cross-links (Fig. 2D). The C-terminal protein lacking both the UBZ and SAP domains did not accumulate at ICLs. Since the UBZ domain binds to FANCD2, we assessed recruitment of wild-type FAN1 in FANCD2-deficient cells. As predicted, the initial stage of recruitment was normal, but the second stage of recruitment was reduced (Fig. 2E). The effect was less pronounced than that seen with FAN1 lacking the UBZ domain, which could be due to residual levels of FANCD2 ubiquitination (Kalb et al. 2007) or another, unknown protein that is able to recruit FAN1 to sites of damage in a UBZ-dependent manner. These results lead us to conclude that FAN1 has two modes of recruitment to ICLs. One is very fast and depends on the SAP domain and presumably on the direct binding to the ICL. The other is a slower accumulation that depends on the UBZ domain and FANCD2.
Figure 2.
Two modes of FAN1 recruitment to sites of DNA–psoralen cross-links. (A) U2OS cells stably transduced with a vector expressing wild-type or a UBZ mutant (C44A/C47A) GFP-hFAN1 were subjected to psoralen/UV laser-induced ICLs in a localized region of the nucleus and subsequently imaged to monitor the recruitment of the GFP-tagged protein to the cross-link stripe. (B) Illustrative images showing an accumulation of wild-type or a UBZ mutant (C44A/C47A) GFP-hFAN1 at the targeted site as a function of time. (WT) Wild type. (C) A recruitment dynamics for wild-type or a UBZ mutant (C44A/C47A) GFP-hFAN1. The graph depicts GFP intensity at the targeted site as a function of time in arbitrary units (AU). Ten cells were analyzed and averaged for each cell line. n = 10. Error bars indicate SD. (D) Schematic of GFP-FAN1 deletion mutants used to define the domains involved in FAN1 recruitment to ICL sites. U2OS cells stably transduced with the GFP-tagged FAN1 deletion mutants depicted were treated as shown in A. The graph depicts GFP intensity at the targeted site as a function of time in arbitrary units (AU). Five to 10 cells were analyzed and averaged for each cell line. n = 5–10. Error bars indicate SD. (E) PD20 cells complemented with human FANCD2 (human FANCD2+/+) or the empty vector (human FANCD2−/−) and stably transduced with a vector expressing wild-type GFP-hFAN1 were treated as in A. The graph depicts GFP intensity at the targeted site as a function of time in arbitrary units (AU). Ten cells were analyzed and averaged for each cell line. n = 10. Error bars indicate SD.
FAN1 can function independently of the FA pathway
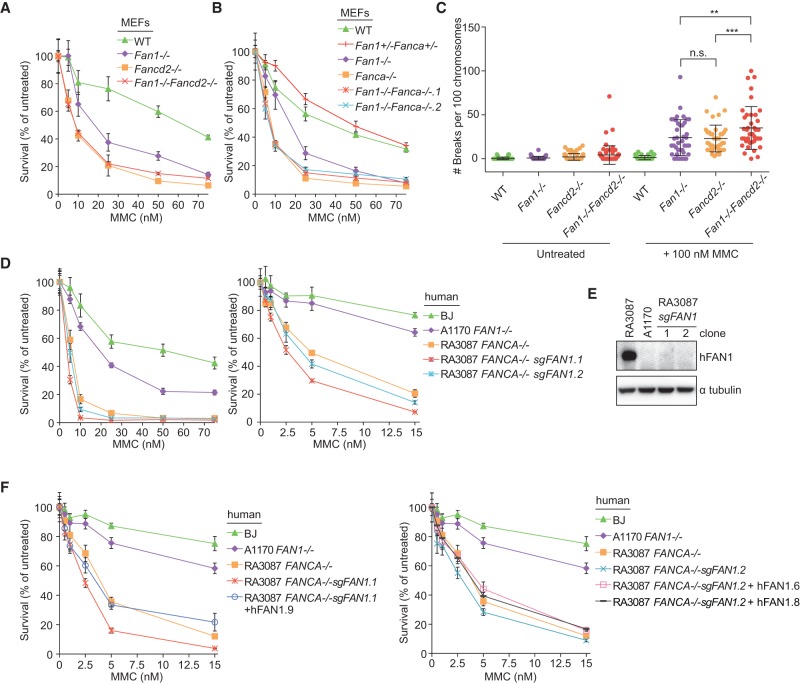
Having established that FAN1 can repair ICL damage independently of its binding to FANCI/D2, we sought to determine whether FAN1 is epistatic with the FA pathway. To elucidate this genetic interaction, we generated Fan1−/− Fancd2−/− and Fan1−/−Fanca−/− double-knockout MEFs by crossing Fan1+/− mice with Fancd2+/− (Houghtaling et al. 2003) or Fanca−/− (Noll et al. 2002) mice. Fan1−/− Fancd2−/− animals generated from the intercrosses between Fan1−/−Fancd2+/− mice were born at an observed ratio of 0.21, which is not significantly different from the expected ratio of 0.25. Fancd2−/− and Fanca−/− MEFs were more sensitive to MMC than the Fan1−/− cells (Fig. 3A,B). No further MMC-induced proliferation defect was apparent in the double-mutant MEF cell lines compared with cells lacking Fancd2 or Fanca alone (Fig. 3A,B). However, a deficiency in Fan1 enhanced the degree of MMC-induced chromosomal abnormalities in Fancd2−/− MEFs (Fig. 3C). To assess the epistasis in human cells, we knocked out FAN1 in a patient cell line with biallelic deletion of FANCA (Kim et al. 2013) using CRISPR/Cas9. In this setting, FAN1 deficiency further sensitized cells to MMC, which was complemented back to levels of MMC resistance seen in FANCA−/− cells by exogenously expressing wild-type FAN1 (Fig. 3D–F; Supplemental Fig. S2D). Collectively, the analysis of the UBZ domain mutant and the epistasis analysis support that FAN1 has a cross-link resistance function that is independent of the FA pathway. This FAN1-specific repair function is more subtle compared with the activity of the FA-dependent pathway in mammalian cells.
Figure 3.
Fan1 and FA genes are nonepistatic. (A) Survival of single Fan1−/− and Fancd2−/− and double Fan1−/−Fancd2−/− MEFs following single treatment with MMC. After 6 d, cell numbers were determined and normalized to the untreated control to calculate the percentage of survival. (WT) Wild type. Error bars indicate SD. (B) Survival of single Fan1−/−-deficient, Fanca−/−-deficient, and double Fan1−/−Fanca−/−- deficient MEFs following single treatment with MMC as described in A. Error bars indicate SD. (C) Quantification of chromosome breakage analysis of the indicated MEFs collected 24 h after 100 nM MMC treatment or left untreated. (**) P < 0.01; (***) P < 0.001, calculated by one-way ANOVA. (D) Survival of wild-type fibroblasts (BJ) and A1170-22 FAN1−/−, RA3087 FANCA−/−, and two clones (1 and 2) of RA3087 FANCA−/− FAN1−/− cells obtained by CRISPR/Cas9 gene editing. Cells were treated with MMC as described in A. Error bars indicate SD. (E) Immunoblot assessing expression of human FAN1 in human fibroblasts used in D. (F) Survival of RA3087 FANCA−/− FAN1−/− clone 1 and clone 2 complemented with HA-hFAN1. Cells were treated with MMC as described in A.
Genetic interactions of Fan1 with Mus81
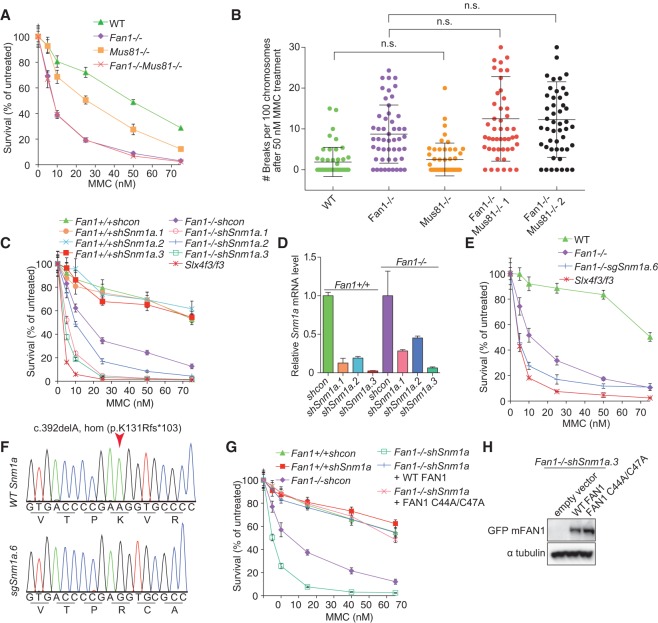
To determine the genetic interaction between Fan1 and Mus81, which contributes to ICL resistance, we generated Fan1−/−Mus81−/− MEFs. Mus81−/− cells were significantly more sensitive to MMC than wild-type cells but less sensitive compared with Fan1−/− cells (Fig. 4A). Notably, lack of Mus81 did not induce a remarkable increase in MMC-induced chromosomal breakages, unlike that seen in Fan1 deficiency (Fig. 4B). The double-knockout Fan1−/−Mus81−/− MEFs generated through mouse intercrosses were as sensitive to MMC as Fan1−/− cells, suggesting that Fan1 and Mus81 are epistatic. Furthermore, Mus81 knockout also did not significantly change the extent of chromosomal aberrations in MEFs lacking Fan1 (Fig. 4B), although there was a trend of increased abnormalities in the double-deficient cells.
Figure 4.
FAN1 and SNM1a are redundant for sensitivity to ICL-inducing agents. (A) Survival of Fan1−/−-deficient, Mus81−/−-deficient, and double-deficient MEFs treated with the indicated levels of MMC. After 6 d, cell numbers were determined and normalized to the untreated control to calculate the percentage of survival. (WT) Wild type. Error bars indicate SD. (B) Quantification of chromosome breakage analysis of the indicated MEFs treated with 50 nM MMC for 24 h. (n.s.) Not significant, as determined by one-way ANOVA. (C) Survival of Fan1+/+ or Fan1−/− stably transduced with control shRNA or shRNA targeting the Snm1a gene after treatment with MMC treatment as described in A. Slx4f3/f3 was used as a control. Error bars indicate SD. (D) Quantitative RT–PCR to assess the level of Snm1a transcript depletion in cell lines used in D. (E) Survival of wild-type, Fan1−/−, Fan1−/−sgSnm1a.6, and Slx4f3/f3 MEFs after MMC treatment as described in A. Error bars indicate SD. Fan1−/−sgSnm1a.6 is a clonal cell line with a biallelic frameshift mutation in Snm1a generated by CRISPR/Cas9 genome editing. (F) Sequencing of the Fan1−/−sgSnm1a.6 MEF clone used in E. (G) Survival of the indicated MEFs after MMC treatment. Fan1−/− MEFs depleted of Snm1a were stably transduced with wild-type GFP-FAN1 or GFP-Fan1 C44A/C47A. Error bars indicate SD. (H) Immunoblot showing the expression of wild-type GFP-FAN1 or GFP-FAN1 C44A/C47A in Fan1−/−shSnm1a.3 MEFs used in G.
FAN1 deficiency can be partially compensated for by SNM1A
Recent biochemical analyses revealed that in vitro FAN1 can incise close to a cross-link embedded in a variety of DNA substrates. In some cases, the processing of cross-link substrates by FAN1 occurred at both the 5′ and 3′ sides of the cross-link, leading to complete unhooking of the DNA cross-link (Wang et al. 2014; Pizzolato et al. 2015). To date, SNM1A, a homolog of yeast Pso2, is the only other known ICL-processing nuclease that is capable of digesting past the ICL, leading to the insight that FAN1 may be functionally redundant with SNM1A in DNA ICL repair. To test this hypothesis, we used three different shRNAs to deplete Snm1a in both wild-type and Fan1−/− MEFs. As previously reported, Snm1a depletion alone did not cause significant sensitivity to MMC in mouse cells (Fig. 4C,D). However, depletion of Snm1a further sensitized Fan1-deficient cells to MMC, and the level of MMC sensitization correlated with the level of Snm1a mRNA knockdown (Fig. 4C,D; Supplemental Fig. S2E). To confirm these results in a cell line that completely lacks both FAN1 and SNM1A, we knocked out Snm1a using CRISPR/Cas9. The double-deficient cells were more sensitive than Fan1−/− cells and almost as sensitive as the SLX4-deficient cells (Fig. 4E,F). FAN1 with an inactivated UBZ domain was able to complement an ICL repair defect in Fan1−/−shSnm1a MEFs to the level of wild-type cells, indicating that the UBZ domain is also dispensable for ICL resistance in this setting. (Fig. 4G,H).
Lack of Fan1 results in progressive kidney and liver karyomegaly and liver dysfunction
Fan1−/− mice were born at the expected Mendelian ratio with no overt developmental abnormalities (Supplemental Fig. S3A). The growth of Fan1−/− mice from birth until 12 wk of age was comparable with wild-type and heterozygous littermates (Supplemental Fig. S3B). Furthermore, Fan1−/− mice were fertile, producing on average eight pups per cross, a number comparable with those produced in crosses of heterozygous mice (Supplemental Fig. S3C).
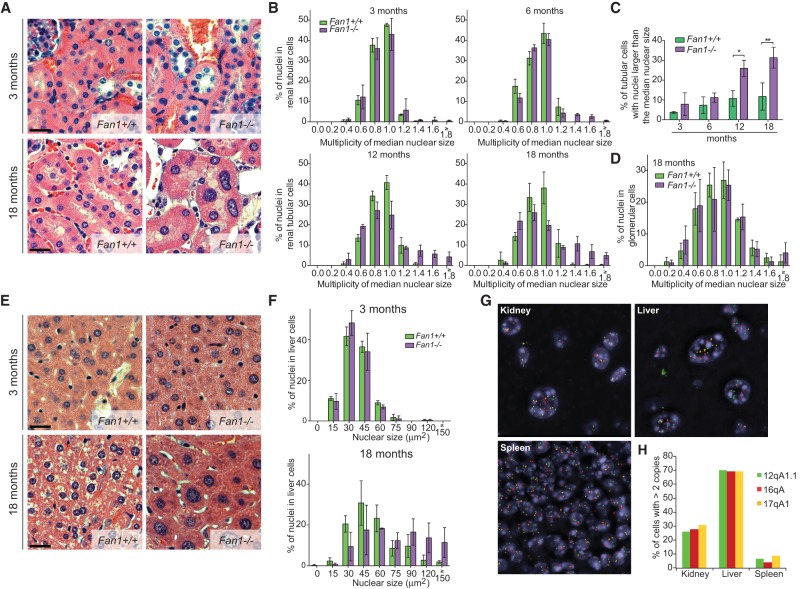
Since one of the most pronounced characteristics of human KIN is the presence of karyomegalic nuclei in cells of multiple tissues, we monitored the histology of kidney and liver tissues in an aging cohort of mice. At 3 mo, we did not observe any remarkable differences between the distribution of nuclear sizes in renal tubular cells of Fan1−/− and wild-type animals (Fig. 5A,B). Increased nuclear size became detectable in Fan1 mice at 6 mo of age (Fig. 5B). The level of renal karyomegaly further intensified with age, as the proportion of cells with larger than median nuclear size rose more than twofold to ∼29% in 12-mo-old Fan1 mice (Fig. 5B,C). Notably, the number of tubular cells with nuclei larger than the median size also went up to ∼10% in 12-mo-old wild-type animals. However, the nuclear size in this cohort did not exceed 1.2 times the median, unlike Fan1−/− animals at an equivalent age, which displayed a significant fraction (17%) that were >1.2 of the median. Significant karyomegaly was absent from glomeruli of Fan1−/− mice (Fig. 5D)
Figure 5.
Fan1 mice develop karyomegaly. (A) Hematoxylin and eosin (H&E) staining of kidneys from Fan1+/+ and Fan1−/− mice at 3 and 18 mo. (B) Quantification of the nuclear size of tubular epithelial cells in the cortex of kidneys from Fan1+/+ and Fan1−/− mice at 3, 6, 12, and 18 mo. The size of each nucleus was normalized to the median nuclear size present in the section to control for fixation and embedding conditions. Data were then plotted and grouped according to size. n = 3. Error bars indicate SD. (C) Percentage of karyomegalic nuclei larger than the median nuclear size in Fan1+/+ and Fan1−/− mice at 3, 6, 12, and 18 mo. (*) P < 0.05; (**) P < 0.01, calculated by one-way ANOVA. n = 3. Error bars indicate SD. (D) Quantification of the nuclear size of glomerular cells in Fan1+/+ and Fan1−/− mice at 18 mo. The size of each nucleus was normalized to the median nuclear size of each animal. Data were then plotted and grouped according to size. n = 3. Error bars indicate SD. (E) H&E staining of liver sections from Fan1+/+ and Fan1−/− animals at 3 and 18 mo. (F) Quantification of the size of nuclei in the liver isolated from Fan1+/+ and Fan1−/− mice at 3 and 18 mo. n = 3. Error bars indicate SD. (G) Representative images of fluorescent in situ hybridization (FISH) performed with probes against the indicated loci in the kidneys, livers, and spleens of 18-mo-old Fan1−/− animals. (H) Quantification of the cells with more than two signals per nucleus in 18-mo-old Fan1−/− animals. The analysis was restricted to tubular epithelial cells in the kidney and liver hepatocytes.
Despite the presence of karyomegaly, we detected no significant elevation in the level of blood urea nitrogen, creatinine, phosphorus, and magnesium in Fan1−/− mice throughout the experiment (Supplemental Fig. S4A–D). These results indicated that, even with apparent karyomegaly, the kidneys of the 18-mo-old Fan1−/− animals still retain sufficient function to support homeostasis. Additional experimental manipulations inducing kidney injury may be necessary to elicit overt kidney failure.
Similar to the kidney, there was no discernable difference between the hepatocyte nuclear size distribution of 3-mo-old wild-type and Fan1−/− mice. However, in 18-mo-old mice without functional FAN1, a large fraction (>50%) of nuclei became significantly enlarged (Fig. 5E,F). Consistent with the polyploidization of the cells in KIN patients, the enlarged cells in the kidney and liver tissue had increased numbers of hybridization signals of probes recognizing three different chromosomes. In contrast, the spleen, which did not manifest karyomegaly, had very few cells with more than two signals (Fig. 5G,H; Supplemental Fig. S4E).
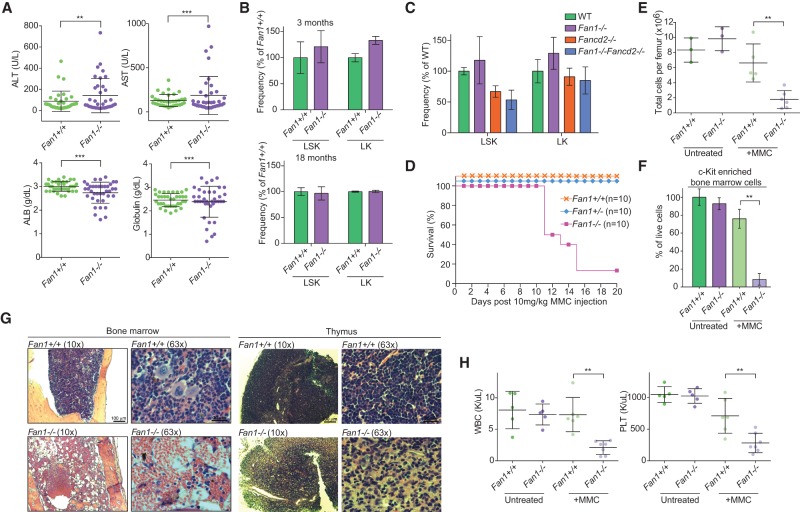
In order to determine whether the liver morphology had an impact on liver function, we analyzed liver enzymes and serum proteins in 3- to 18-mo-old animals. Fan1−/− animals displayed elevated alanine transaminase and aspartate transaminase while exhibiting lowered albumin and globulin levels (Fig. 6A; Supplemental Fig. S4F–I). The rise in serum liver enzyme and concurrent reduction in serum proteins indicate that the increase in karyomegalic hepatocytes coincides with the deterioration of liver function. Taken together, these results strongly supported that FAN1 is required to suppress age-dependent development of karyomegaly in the kidney and liver. The Fan1−/− mouse model thus accurately phenocopies the KIN conditions observed in humans and will be instrumental in understanding human disease and potentially the aging-associated changes in these tissues.
Figure 6.
FAN1-deficient mice develop liver dysfunction, are sensitive to MMC, and develop hypocellular bone marrow. (A) Analysis of liver function from 12- to 18-mo-old Fan1+/+ and Fan1−/− mice. Alanine transaminase (ALT), aspartate transaminase (AST), albumin (ALB), and globulin (GLOB) were tested in the serum. Bars represent mean ± SD. (***) P < 0.001; (**) P < 0.01, calculated using F-test. (B) Quantification of hematopoietic stem cells (HSCs) isolated from Fan1+/+ and Fan1−/− mice assessed by FACS. n = 3 per genotype. Error bars indicate SD. (C) Quantification of HSCs isolated from mice of the indicated genotype, assessed by FACS. n = 3 per genotype. (WT) Wild type. Error bars indicate SD. (D) Survival of Fan1+/+, Fan1+/−, and Fan1−/− mice (n = 10 per genotype) after a single treatment with 10 mg/kg MMC. Mice from each genotype were monitored up to 20 d following MMC intraperitoneal injection. (E) Total number of cells per femur of Fan1+/+ and Fan1−/− mice untreated or treated 1 wk prior with 10 mg/kg MMC. Bars represent mean ± SD. (**) P < 0.01, calculated using unpaired t-test. (F) Percentage of live bone marrow cells that were enriched for c-Kit+ population. The bone marrow cells were isolated from Fan1+/+ and Fan1−/− mice untreated or treated 1 wk prior with 10 mg/kg MMC . Bars represent mean ± SD. (**) P < 0.01, calculated using unpaired t-test. (G) Bone marrow and thymus histology from Fan1+/+ and Fan1−/− mice treated 1 wk prior with 10 mg/kg MMC. Representative images at 10× and 63× are shown. (H) White blood cell (WBC) and platelet (PLT) counts of Fan1+/+ and Fan1−/− mice untreated or treated 1 wk prior with 10 mg/kg MMC. Bars represent mean ± SD. (**)P < 0.01, calculated using unpaired t-test.
Fan1−/− mice are susceptible to cross-linking agent-induced bone marrow failure
Anemia has been reported in a number of human KIN cases, prompting us to investigate the possible hematological dysfunction in Fan1-deficient mice. At the peripheral blood level, Fan1−/− mice did not show obvious signs of cytopenia at a young age (Supplemental Fig. S5). To explore the possibility of bone marrow abnormality in Fan1 animals, we analyzed the hematopoietic stem cell (HSC) population in the bone marrow of young (3 mo) and old (18 mo) mice with or without Fan1 deficiency. We could not detect any remarkable changes in the level of the Lineage (Lin)− Sca-1+ c-Kit+ (LSK) or Lin− Sca-1− c-Kit+ (LK) populations between Fan1+/+ and Fan1−/− mice (Fig. 6B; Supplemental Fig. S6A). The numbers of long-term, short-term, and multipotent HSCs as well as the more differentiated progenitor cells—megakaryocyte/erythroid progenitors, granulocyte/monocyte progenitors, and common myeloid progenitors—in Fan1−/− animals were also similar to that of the wild-type control in the same age groups (Supplemental Fig. S6B,C). Also, inactivation of Fan1 in mice with Fancd2 deficiency did not significantly reduce the level of hematopoietic progenitor cells compared with Fancd2 single-mutant mice (Fig. 6C; Supplemental Fig. S6D,E). Collectively, these results indicated that the activity of FAN1 is not required for the protection of early HSCs under unstressed conditions.
Although the bone marrow was not functionally compromised in Fan1−/− mice, we asked whether FAN1 was required upon exogenous treatment with ICL-inducing agents. Fan1−/− and control mice were treated with a single intraperitoneal injection of 10 mg/kg MMC and were followed over the next 3 wk. Some wild-type and heterozygous Fan1+/− mice showed a minor drop in body weight after receiving the MMC; nevertheless, most eventually regained the initial weight and tolerated the dose of MMC given until the end of the experiment (20 d) (Supplemental Fig. S7A). In contrast, all Fan1−/− mice reacted acutely to MMC, displaying a significant drop in body weight (10%–30%) following the treatment (Supplemental Fig. S7A). Ninety percent of Fan1−/− mice tested died (Fig. 6D). On day 7 after treatment with MMC, a parallel cohort of Fan1−/− mice displayed depletion of the hematopoietic progenitor cells, as shown by a decrease in total cell number in the femurs and a complete disappearance of c-Kit+ bone marrow cells following MMC treatment (Fig. 6E,F; Supplemental Fig. S7B,C). In agreement, the histological analysis revealed pronounced hypocellularity in the bone marrow and the thymus of Fan1−/− mice (Fig. 6G). The effect of ICL repair deficiency on the hematopoietic system of Fan1−/− animals was further supported by significant neutropenia and thrombocytopenia. Although the red blood cell (RBC) count was largely unaltered due to the long half-life of RBCs, the reticulocyte counts of Fan1−/− mice was significantly lower than Fan1+/+ mice at day 7 after MMC treatment (Fig. 6H; Supplemental Fig. S7D). These results illustrate the necessity of FAN1 for the maintenance of hematopoiesis and survival of animals exposed to high levels of exogenous cross-link damage.
Discussion
We created a Fan1-deficient mouse model and demonstrated the significance of FAN1 in the protection of cells and organs from DNA ICL damage. On the cellular level, FAN1 is necessary for resistance to DNA ICL agents, with FAN1-deficient cells showing decreased survival after various ICL-inducing agents—MMC, acetaldehyde, and formaldehyde—when compared with FAN1-proficient cells. The absence of FAN1 in mice reveals karyomegaly in the liver associated with liver dysfunction as well as karyomegaly in the kidney. In addition, the mice are sensitive to MMC and develop bone marrow dysfunction when challenged. Overall, our mouse model recapitulates the pathology of human KIN in which FAN1 mutations have been identified (Zhou et al. 2012). Although FAN1 mutations in the general population are rare, patients with such mutations are predicted to have adverse reaction if they were to be treated with ICL-inducing chemotherapeutics.
FAN1 and organ dysfunction
Fan1-deficient mice display karyomegaly in epithelial cells lining the renal tubules and in the liver as early as at 6 mo of age, and the number of enlarged nuclei as well as the average size of the nuclei increase with time, suggesting a continuous and chronic process. The fluorescent in situ hybridization (FISH) analysis of 18-mo-old mice revealed increased ploidy at three different chromosomes, consistent with reduplication of the genome in the karyomegalic nuclei. This implies that these cells induce replication cycles, which are not followed by mitoses. It is unknown at this time whether the whole genome is reduplicated in the karyomegalic cells or whether there remain portions of the genome that are underreplicated in comparison with the bulk of the genome. It will be of interest to examine whether underreplicated regions exist and, if so, where they occur. The extensive karyomegaly seen in the liver in this mouse model might also be useful for understanding of the mechanism of polyploidization in normal hepatic tissues during aging.
Similar to human KIN patients, Fan1-deficient mice exhibit liver function abnormality, as determined by an increase in aspartate transaminase and alanine transaminase and a reduction in the level of liver-produced albumin and globulin at 18 mo of age. Hepatocytes are especially prone to increasing their ploidy during development and in response to insults, including DNA damage (for review, see Gentric and Desdouets 2014). The progressive nature of polyploidization and the correlation of ploidy increase and functional decline suggest that polyploidization is a pathologic and not adaptive event in the FAN1-deficient animal.
Although both the livers and kidneys of the Fan1-deficient mice display karyomegaly at 18 mo, we observed only liver, but no kidney, dysfunction, as assessed by blood serum levels of urea nitrogen, creatinine, and electrolytes. However, the blood level of these metabolites are more specifically measures of glomerular function and are not sensitive markers of the function of proximal tubules, the primary subtype of kidney cells affected in the Fan1-deficient mice. Furthermore, the kidney is a very resilient organ, and usually only loss of the majority of kidney tissue results in a visible functional decline. The C57BL/6J background may also be protective for the kidney in our model. Functional decline of the kidney may become detectable with age or might have to be elicited by nephrectomy or an increase of endogenous DNA damage load (see below).
The major unanswered question in the understanding of the pathophysiology of KIN is the nature of the lesion that FAN1 repairs. The progressive nature of the disease and our knowledge that FAN1 participates in ICL repair suggest that an endogenous ICL-inducing agent may be a culprit in this disease. However, it is possible that different DNA lesions may play a role in KIN pathogenesis. Generally, cases of KIN described in the literature exclude exposure to environmental genotoxins such as ochratoxin A, heavy metals, or herbal medicines (Mihatsch et al. 1979; Bhandari et al. 2002; Uz et al. 2011; Radha et al. 2014), leaving endogenously derived toxins as more likely candidates. Given the recent data implicating acetaldehyde and formaldehyde in the pathogenesis of FA (Langevin et al. 2011; Garaycoechea et al. 2012; Hira et al. 2013; Pontel et al. 2015), the contribution of aldehydes to the pathogenesis of KIN needs to be examined. Recent work demonstrates that an inactivation of alcohol dehydrogenase 5 (ADH5/GSNOR), which is responsible for formaldehyde detoxification, results in karyomegaly, kidney failure, and abnormal liver function when combined with Fancd2 deficiency (Pontel et al. 2015). Although, the glomerular damage seen in the Fancd2−/−Adh5−/− mice is not observed in Fan1−/− animals, implying differences in the pathology of the underlying disease, the increased levels of endogenous formaldehyde present in the Adh5−/− background will provide a good model to test whether formaldehyde-inflicted damage necessitates FAN1 activity in the kidney or liver. We showed that FAN1 is essential for cellular resistance to acetaldehyde and formaldehyde; thus, it is possible that the double-mutant mice with deficiency in Fan1 and formaldehyde catabolism will expose the kidney phenotype at a faster rate. Endogenous aldehydes could also be linked to chronic kidney disease in humans.
In contrast to patients with FA, KIN patients do not develop pancytopenia at a young age (Zhou et al. 2012). However, when diagnosed with kidney failure, some KIN patients exhibit mild anemia of a presently unknown cause. The analyses of the bone marrow histology and HSCs in Fan1−/− mice do not reveal a discernable change from the wild-type level in either young or old animals. Furthermore, Fan1 deficiency does not significantly enhance the HSC defect observed in Fancd2−/− animals. These data suggest that the activity of FAN1 is dispensable for the protection of blood stem cells from DNA damage under unperturbed conditions. Only one Fan1−/− mouse in our study exhibited signs of blood cytopenia during the first 18 mo of life. Similar to the kidney function, it remains possible that the expression of a mild anemia phenotype observed in humans requires a latency period >18 mo in mice. If that proves to be true, the question of whether the origin of such a phenotype is due to an intrinsic bone marrow stem cell insufficiency or is the consequence of a kidney defect needs to be addressed.
Despite not being required under unstressed conditions, FAN1 is essential for the maintenance of hematopoiesis when animals are challenged with doses of cross-linking agents that can be tolerated by the wild-type animals. The bone marrow and thymus were particularly prone to ICL-induced toxicity in Fan1-deficient animals, and the treatment with MMC resulted in 90% mortality. Defects in the kidney and liver were not detected under the acute MMC treatment used in this study. More chronic, low-dose treatments might be necessary to induce kidney and liver dysfunction in young Fan1−/− animals.
FAN1 and ICL repair
Our studies using MEFs recapitulate the behavior of human cells deficient for FAN1, with the nuclease activity of FAN1 being necessary for protection against ICLs. Resistance in MEFs heterozygous for the Fan1-deficient allele suggests that FAN1 is not haploinsufficient for ICL repair, consistent with KIN being a recessive disease in humans. Our data clearly indicate that FAN1 acts during ICL repair independently of its UBZ domain. The interaction between FAN1 and the ID2 complex has generally been thought to be crucial for the recruitment of FAN1 to the sites of cross-linked DNA and its involvement in ICL repair (Kratz et al. 2010; Liu et al. 2010; MacKay et al. 2010; Smogorzewska et al. 2010). However, the current study provides strong evidence to support our previous observation that the UBZ domain is not essential for ICL resistance (Zhou et al. 2012). FAN1 can travel to the ICLs independently of the UBZ domain and in cells lacking FANCD2. The SAP domain is critical for the rapid recruitment of FAN1 to the ICLs, and the UBZ-dependent phase of ICL recruitment coincides with the localization of FANCD2 at the cross-link that slowly peaks following an earlier enrichment of the FA core complex (Yan et al. 2012). These experiments indicate that the direct DNA–FAN1 interaction that brings the nuclease to the ICL is the important step during ICL repair. Its function could be used in the context where the FA proteins are unable to act efficiently; for example, in the G0 or G1 stage of the cell cycle or at a subset of lesions that have structures other than the convergent replication fork.
This still leaves the ICL repair functions, if any, of the UBZ domain and the interaction between FAN1 and the ID2 complex unexplained. It is possible that the UBZ domain is required for the functions of FAN1 and ubiquitinated FANCD2/FANCI that are unrelated to the repair of the ICLs. The UBZ domain is a more recent evolutionary extension of FAN1 protein architecture (MacKay et al. 2010; Smogorzewska et al. 2010). In yeast, Fan1 is capable of conferring ICL repair activity in the absence of the UBZ domain (Fontebasso et al. 2013). One potential role of the interaction between FAN1 and FANCD2/FANCI outside of ICL repair includes the processing of replication forks under normal or replicative stress conditions (Schlacher et al. 2012; Chaudhury et al. 2014; Chen et al. 2015). Most recently, it has been shown that the UBZ domain of FAN1 is essential for the processing of replication forks and protection against chromosome instability after treatment with HU and MMC (Lachaud et al. 2016). Although our experimental conditions did not reveal the necessity of the functional UBZ domain for the maintenance of chromosomal stability after MMC treatment, it remains to be determined whether the fork-protective function of FAN1 is also responsible for organ-protective functions.
Consistent with the UBZ not being necessary for ICL resistance, we showed that FAN1 has functions outside of the FA repair pathway. Our data here are consistent with studies in DT40 chicken cells, which report that FAN1 is not epistatic with FANCC and FANCJ in ICL resistance (Yoshikiyo et al. 2010). Based on the MMC-induced proliferation defect, the FA protein network represents the predominant ICL repair pathway in mammalian cells (Zhou et al. 2012). However, FAN1 inactivation can additively enhance the ICL-induced proliferation defect in FANCA-deficient cells and chromosomal aberration in cells lacking FANCD2 at high loads of cross-links. These results firmly show the presence of an FA-independent function of FAN1 that can contribute to the protection of cells from accruing extensive genomic alterations inflicted by ICL agents.
FAN1 vs. other nucleases
Processing of ICL lesions is complex. Recently, in vitro biochemical analyses of SLX4/FANCP- and XPF/FANCQ-dependent processing of the cross-links and the ICL unhooking studies from Xenopus egg extracts strongly support the major involvement of the 3′ flap endonuclease activity of XPF/FANCQ in initiating and completing ICL incisions (Douwel et al. 2014; Hodskinson et al. 2014). However, it is evident from the published biochemical work and our unpublished data that FAN1, through its endonuclease and exonuclease activities, is able to incise and completely unhook different types of cross-link substrates in vitro (Zhao et al. 2014; Pizzolato et al. 2015; A Wang, L Timashev, and A Smogorzewska, unpubl.). The discrepancy may come from the fact that FAN1 may be involved at stalled forks that do not look like the divergent replication forks formed on plasmids in the Xenopus egg extracts or that FAN1 predominantly works in G1 and not in S phase. The other possibility as to why FAN1 depletion in the egg extracts does not lead to a discernable ICL unhooking defect is the presence of redundant nucleases in the extracts. In the present study, we identified SNM1A as being redundant with FAN1 during the ICL damage response. Unlike FAN1, SNM1A deficiency by itself exhibits only a very mild cross-link sensitivity in mammalian cells (Dronkert et al. 2000; Ahkter et al. 2005). In fact, in our hands, depletion of SNM1A in fibroblasts elicited no significant sensitivity to ICLs. On the other hand, cells lacking both FAN1 and SNM1A were almost as sensitive to cross-linking agents as cells lacking the FA proteins such as SLX4 or FANCD2, suggesting that the ICL incision could be severely compromised when both SNM1A and FAN1 are absent. The analysis of the ICL incision and repair after a double depletion of both FAN1 and SNM1A in the Xenopus egg extracts will be helpful to address whether these nucleases have redundant functions in this setting. The long-term effects of SNM1A deficiency have not been studied in mice, and it will be of interest to determine whether the Snm1a−/− animals develop KIN. If Fan1−/−Snm1a−/− mice show exacerbation of the KIN phenotype, that would confirm the redundant function of the two nucleases in vivo.
Materials and methods
Generation of Fan1-deficient mouse strains
All of the animals were handled according to the Rockefeller University Institutional Animal Care and Use Committee protocols. The Fan1 gene targeting construct [Fan1tm1a(KOMP)Wtsi; Mouse Genome Informatics (MGI) code 4940765] was generated by the KOMP. The construct was electroporated into albino B6 mouse-derived embryonic stem cells, and the targeted embryonic stem cells were injected into C57BL/6J blastocyst to generate chimeric animals. Germline transmission of the targeted allele Fan1(stop) was verified by the 5′ and 3′ probes on Southern blots (Supplemental Fig. S1A–C) and used to generate Fan1lox from the cross with the FLPe deleter mouse stain (Jackson Laboratories). The Fan1Δex3&4 allele was generated from the cross between Fan1+/lox and E2a-Cre mice (Jackson Laboratories). Fanca- and Fancd2-deficient mice of the 129SV background were obtained from Marcus Grompe (Noll et al. 2002; Houghtaling et al. 2003). Slx4- and Mus81-deficient mice of the C57/Bl6 background were obtained from Paula Cohen (Dendouga et al. 2005; Holloway et al. 2011). Genotyping was carried out by PCR on DNA isolated from mouse tail biopsy samples digested overnight in 0.4 mg/mL Proteinase K supplemented with Direct PCR lysis reagent (yolk sac; Viagenbiotech). Genotyping primers are shown in Supplemental Table S1.
Cell culture
Primary MEFs were obtained from E13.5 embryos using standard techniques and cultured in DMEM containing 15% heat-inactivated FBS, 100 U/mL penicillin, 0.1 μg/mL streptomycin, 0.1 mM nonessential amino acids, and glutamax (Life Technologies). MEFs were immortalized at passage 2 using pMSCVNeo HPV16E6E7.
DNA damage sensitivity assays
Cells were plated in a six-well cell culture plate in triplicate at a density of 3.5 × 104 to 4 × 104 cells per well. Twenty-four hours later, drugs (MMC, HU, and CPT) were added. After 6–8 d of culture, cell numbers were determined using the Z2 Coulter Counter (Beckman Coulter). Cell numbers after treatment were normalized to cell numbers in the untreated sample to give the percentage of survival.
Chromosome breakage analysis
Cells were exposed to 50 or 100 nM MMC for 24 h before 2 h of treatment with 0.167 μg of colcemid per milliliter of medium. Cells were harvested, incubated for 15 min at 37°C in 0.075 M KCl, and fixed in freshly prepared methanol:glacial acidic acid (3:1). Metaphase spreads were prepared by dropping cells onto wet microscope slides. Slides were air-dried for 12 h at 40°C before staining with 6% Karyomax Giemsa (Life Technologies) in Gurr buffer (Life Technologies) for 3 min. After rinsing with fresh Gurr buffer and distilled water, slides were fully dried at room temperature and scanned using the Metasystems Metafer. Chromosome breakage is reported as breaks per 100 chromosomes to remove any biases that come from polyploidization of immortalized MEFs. One-way ANOVA or t-test was used to determine the statistical significance. The quantification was performed in a blinded manner.
Mutagenesis
Mouse Fan1 cDNA was cloned from wild-type MEFs into pENTR/D-TOPO (Life Technologies). Mutagenesis was performed using the QuikChange multisite-directed mutagenesis kit (Agilent) per the manufacturer's instructions using the oligonucleotides listed in Supplemental Table S1.
RNAi and CRISPR/Cas9-driven genome editing
Snm1a mRNA level was stably knocked down in MEFs using pZIP (mCMV) lentiviruses (Transomic). MEFs were transduced with shRNA-expressing lentiviruses and selected for 1 wk with 3 μg/mL puromycin. RA3087 FANCA−/− human fibroblasts and Fan1−/− MEFs were transduced with lentivirus expressing pCW-Cas9 vector encoding a doxycycline-inducible Cas9 expression cassette (plasmid 50661; Addgene; originally from the Lander and Sabatini laboratories, Massachusetts Institute of Technology). After selection with 50 μg/mL hygromycin (for MEFs) or 3 μg/mL puromycin (for human fibroblasts), a single clone with an undetected basal level of pCW-Cas9 but showing increased expression following doxycycline induction was selected before being transduced with lentivirus expressing single guide RNA (sgRNA) against target sequence. Following selection in 3 μg/mL puromycin (for MEFs) or 5 μg/mL blasticidin (for human fibroblasts), cells were treated with 100 ng/mL doxycycline for 48 h before single cells were plated in 96-well plates. Isolated clones were screened for CRISPR-mediated genome editing at the target region by sequencing of genomic DNA. The reduction of the corresponding mRNA transcripts or protein level was used to validate knockout. Sequences of shRNAs and sgRNAs are listed in Supplemental Table S1.
Localization of GFP-FAN1 to psoralen ICLs
We followed a published protocol to detect proteins recruited at laser-induced localized ICLs (Thazhathveetil et al. 2007). Cells seeded in a 35-mm glass-bottomed culture dish (MatTek) were incubated with 5 μM psoralen for 20 min at 37°C prior to laser photoactivation. Localized irradiation was performed using the Nikon Eclipse TE2000 confocal microscope equipped with an SRS NL100 nitrogen laser-pumped dye laser (Photonics Instruments) that fires 5-nsec pulses with a repetition rate of 10 Hz at 365 nm and a power of 0.7 nW, measured at the back aperture of the 60× objective. The diffraction limited spot size was ∼300 nm. The laser, controlled by Volocity 5 software (Improvision, Perkin Elmer), was directed to deliver pulses to a specified rectangular region of interest (ROI) within the nucleus of a cell (4 × 20 pixels, 0.16 µm per pixel) visualized with a Plan Fluor 60×/N.A. 1.25 oil objective. The laser beam fired randomly throughout the ROI until the entire region was exposed, after which the photoactivation of the ROI was repeated. Throughout an experiment, cells were maintained at 37°C, 5% CO2, and 80% humidity using a Live Cell environmental chamber. At different time intervals, cells from different areas of the dish were treated with the laser to generate a time course on a single plate. After the final time point, cells were fixed immediately in freshly prepared 4% formaldehyde in PBS for 10 min at room temperature. The GFP signal was imaged using a Hamamatsu EM-CCD digital camera attached to the Nikon Eclipse TE2000 confocal microscope.
Antibodies
Immunoblotting and immunofluorescence were performed using the following antibodies: mouse FAN1 (S101D; a gift from John Rouse), mouse FANCD2 (Abcam, EPR2302), HA (Covance, MMS-101R), human FAN1 (antibody raised in-house, RC394), GFP (Roche), and γH2AX (Upstate Biotechnology).
RT–PCR
RNA was isolated using the RNeasy Plus extraction kit (Qiagen). cDNA was synthesized using the SuperScript III first strand synthesis system (Life Technologies), and RT–PCR was performed using the Platinum SYBR Green qPCR SuperMix-UDG (Life Technologies). All steps were carried out per the manufacturers’ instructions. Mouse actin was used as an internal control. Primers used for RT–PCR are listed in Supplemental Table S1.
Histology
Tissues were harvested and fixed in neutral buffered formalin for 20 h at 4°C. They were then embedded in paraffin, sectioned, and hematoxylin and eosin (H&E)-stained. Quantification of nuclear size in tissues was performed blinded to the genotype of the sample. Images were taken on a Zeiss Axiovert 40 microscope and analyzed in ImageJ. At least 150 cells from each tissue were included in the analysis.
FISH
FISH analysis was performed on formalin-fixed paraffin sections using an in-house three-color probe for chromosomal regions 12qA1.1, 16qA, and 17qA1. The probe mix consisted of the following BAC clones: 12qA1.1 (clones RP23-168O9, RP23-54G4, and RP23-41E22, labeled with green dUTP), 16qA (clones RP23-290E4, RP23-356A24, and RP23-258J4, labeled with red dUTP), and 17qA1 (clones RP23-73N16, RP23-354J18, and RP23-202G20, labeled with orange dUTP). Probe labeling, hybridization, post-hybridization washing, and fluorescence detection were performed according to standard procedures. Slides were scanned using a Zeiss Axioplan 2i epifluorescence microscope equipped with a megapixel CCD camera (CV-M4+CL, JAI) controlled by Isis 5.5.9 imaging software (MetaSystems Group, Inc.).
Whole-animal MMC sensitivity studies
Twelve-week-old mice were injected intraperitoneally with a single dose of 10 mg of MMC per kilogram of body weight. Survival and body weight measurements were recorded daily. The percentage of survival was calculated according to the Kaplan and Meier method (Kaplan and Meier 1958). Differences in survival between the different genotypes were tested for significance by the log-rank test. The hematopoietic analyses were done 7 d following MMC treatment in an independent cohort of mice.
LSK analysis
Bone marrow cells were isolated from the femurs and tibiae of mice by crushing the bone in FACs buffer (2% FBS in 1× PBS) before filtering through a 40-mm cell strainer. The early progenitor cells (c-Kit+) were enriched by a 30-min incubation with CD117 (c-Kit) microbeads (Miltenyi Biotec) before being stained for 1 h at 4°C with the following antibodies: PerCP-cyanine5.5-conjugated lineage cocktail with antibodies anti-CD4 (clone RM4-5; eBioscience), CD3e (clone 145-2C11; eBioscience), Ly-6G/Gr-1 (RB6-8C5; eBioscience), CD11b/Mac-1 (clone M1/70; eBioscience), CD45R/B220 (RA3-6B2; eBioscience), CD8a (clone 53-6.7; eBioscience), TER-119 (eBioscience), anti-c-Kit (APC, clone 2B8; eBioscience), anti-Sca-1 (BV421, clone D7; Biolegend), anti-CD150 (SLAM; PE, clone TC15-12F12.2 biolegend), anti-CD34 (FITC, clone RAM34; eBioscience), and anti- CD16/32 (AF700, clone 93; eBioscience) (Frascoli et al. 2012). Samples were run on a LSRII flow cytometer (BD Pharmingen), and the data were analyzed with FlowJo software.
Supplementary Material
Acknowledgments
We thank the members of the Smogorzewska laboratory for comments on the manuscript, and Johannes Walter for comments on the manuscript and sharing unpublished results. We thank Manish Ponda for helpful discussion, Markus Grompe for sharing Fancd2- and Fanca-deficient mice, and Paula Cohen for sharing Slx4- and Mus81-deficient mice. We thank Charlotte Cockram for the cloning of Snm1a CRISPR guide RNA plasmid. We are grateful to Chingwen Yang and members of the Gene Targeting Center, the Transgenic Services Center, the Comparative Bioscience Center, the Genomics Center, and the Flow Cytometry Resource Center at The Rockefeller University, and Michelle Lepherd and members of the Center for Comparative Medicine and Pathology at Memorial Sloan Kettering Cancer Center (MSKCC). FISH analysis was performed at the MSKCC Molecular Cytogenetics Core supported by National Institutes of Health Cancer Center support grant P30 CA008748. We thank John Rouse for antibodies against mouse FAN1. We thank John Rouse and Friedhelm Hildebrandt for sharing unpublished data. The mouse Fan1 targeting vector was generated by the trans-National Institutes of Health KOMP and obtained from the KOMP Repository (http://www.komp.org). This research was supported in part by the Intramural Research Program of the National Institutes of Health, the National Institute on Aging (M.S.), and grants from the Rita Allen Foundation Scholars Program (A.S.) and the National Institutes of Health (RO1 HL120922 to A.S.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.276261.115.
References
- Ahkter S, Richie CT, Zhang N, Behringer RR, Zhu C, Legerski RJ. 2005. Snm1-deficient mice exhibit accelerated tumorigenesis and susceptibility to infection. Mol Cell Biol 25: 10071–10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter BP. 2003. Cancer in Fanconi anemia, 1927–2001. Cancer 97: 425–440. [DOI] [PubMed] [Google Scholar]
- Bhagwat N, Olsen AL, Wang AT, Hanada K, Stuckert P, Kanaar R, D'Andrea A, Niedernhofer LJ, McHugh PJ. 2009. XPF–ERCC1 participates in the Fanconi anemia pathway of cross-link repair. Mol Cell Biol 29: 6427–6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari S, Kalowski S, Collett P, Cooke BE, Kerr P, Newland R, Dowling J, Horvath J. 2002. Karyomegalic nephropathy: an uncommon cause of progressive renal failure. Nephrol Dial Transplant 17: 1914–1920. [DOI] [PubMed] [Google Scholar]
- Butturini A, Gale RP, Verlander PC, Adler-Brecher B, Gillio AP, Auerbach AD. 1994. Hematologic abnormalities in Fanconi anemia: an International Fanconi Anemia Registry study. Blood 84: 1650–1655. [PubMed] [Google Scholar]
- Chaudhury I, Stroik DR, Sobeck A. 2014. FANCD2-controlled chromatin access of the Fanconi-associated nuclease FAN1 is crucial for the recovery of stalled replication forks. Mol Cell Biol 34: 3939–3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-H, Jones MJK, Yin Y, Crist SB, Colnaghi L, Sims I, Robert J, Rothenberg E, Jallepalli PV, Huang TT. 2015. ATR-mediated phosphorylation of FANCI regulates dormant origin firing in response to replication stress. Mol Cell 58: 323–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dendouga N, Gao H, Moechars D, Janicot M, Vialard J, McGowan CH. 2005. Disruption of murine Mus81 increases genomic instability and DNA damage sensitivity but does not promote tumorigenesis. Mol Cell Biol 25: 7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douwel DK, Boonen RACM, Long DT, Szypowska AA, Räschle M, Walter JC, Knipscheer P. 2014. XPF–ERCC1 acts in unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Mol Cell 54: 460–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dronkert ML, de Wit J, Boeve M, Vasconcelos ML, van Steeg H, Tan TL, Hoeijmakers JH, Kanaar R. 2000. Disruption of mouse SNM1 causes increased sensitivity to the DNA interstrand cross-linking agent mitomycin C. Mol Cell Biol 20: 4553–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanconi G. 1967. Familial constitutional panmyelocytopathy, Fanconi's anemia (F.A.). I. Clinical aspects. Semin Hematol 4: 233–240. [PubMed] [Google Scholar]
- Fontebasso Y, Etheridge TJ, Oliver AW, Murray JM, Carr AM. 2013. The conserved Fanconi anemia nuclease Fan1 and the SUMO E3 ligase Pli1 act in two novel Pso2-independent pathways of DNA interstrand crosslink repair in yeast. DNA Repair 12: 1011–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frascoli M, Proietti M, Grassi F. 2012. Phenotypic analysis and isolation of murine hematopoietic stem cells and lineage-committed progenitors. J Vis Exp 10.3791/3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaycoechea JI, Crossan GP, Langevin F, Daly M, Arends MJ, Patel KJ. 2012. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature 489: 571–575. [DOI] [PubMed] [Google Scholar]
- Gentric G, Desdouets C. 2014. Polyploidization in liver tissue. Am J Pathol 184: 322–331. [DOI] [PubMed] [Google Scholar]
- Gwon GH, Kim Y, Liu Y, Watson AT, Jo A, Etheridge TJ, Yuan F, Zhang Y, Kim Y, Carr AM, et al. 2014. Crystal structure of a Fanconi anemia-associated nuclease homolog bound to 5′ flap DNA: basis of interstrand cross-link repair by FAN1. Genes Dev 28: 2276–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazrati A, Ramis-Castelltort M, Sarkar S, Barber LJ, Schofield CJ, Hartley JA, McHugh PJ. 2008. Human SNM1A suppresses the DNA repair defects of yeast pso2 mutants. DNA Repair 7: 230–238. [DOI] [PubMed] [Google Scholar]
- Hejna J, Philip S, Ott J, Faulkner C, Moses R. 2007. The hSNM1 protein is a DNA 5′-exonuclease. Nucleic Acids Res 35: 6115–6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques JA, Moustacchi E. 1980. Isolation and characterization of pso mutants sensitive to photo-addition of psoralen derivatives in Saccharomyces cerevisiae. Genetics 95: 273–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hira A, Yabe H, Yoshida K, Okuno Y, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Nakamura J, Kojima S, et al. 2013. Variant ALDH2 is associated with accelerated progression of bone marrow failure in Japanese Fanconi anemia patients. Blood 122: 3206–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodskinson MRG, Silhan J, Crossan GP, Garaycoechea JI, Mukherjee S, Johnson CM, Schärer OD, Patel KJ. 2014. Mouse SLX4 is a tumor suppressor that stimulates the activity of the nuclease XPF–ERCC1 in DNA crosslink repair. Mol Cell 54: 472–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway JK, Mohan S, Balmus G, Sun X, Modzelewski A, Borst PL, Freire R, Weiss RS, Cohen PE. 2011. Mammalian BTBD12 (SLX4) protects against genomic instability during mammalian spermatogenesis. PLoS Genet 7: e1002094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghtaling S, Timmers C, Noll M, Finegold MJ, Jones SN, Meyn MS, Grompe M. 2003. Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev 17: 2021–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalb R, Neveling K, Hoehn H, Schneider H, Linka Y, Batish SD, Hunt C, Berwick M, Callen E, Surralles J, et al. 2007. Hypomorphic mutations in the gene encoding a key Fanconi anemia protein, FANCD2, sustain a significant group of FA-D2 patients with severe phenotype. Am J Hum Genet 80: 895–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan EL, Meier P. 1958. Nonparametric-estimation from incomplete observations. J Am Stat Assoc 53: 457–481. [Google Scholar]
- Kim Y, Spitz GS, Veturi U, Lach FP, Auerbach AD, Smogorzewska A. 2013. Regulation of multiple DNA repair pathways by the Fanconi anemia protein SLX4. Blood 121: 54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC. 2009. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science 326: 1698–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottemann MC, Smogorzewska A. 2013. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature 493: 356–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratz K, Schöpf B, Kaden S, Sendoel A, Eberhard R, Lademann C, Cannavó E, Sartori AA, Hengartner MO, Jiricny J. 2010. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell 142: 77–88. [DOI] [PubMed] [Google Scholar]
- Lachaud C, Moreno A, Marchesi F, Toth R, Blow JJ, Rouse J. 2016. Ubiquitinated Fancd2 recruits Fan1 to stalled replication forks to prevent genome instability. Science 10.1126/science.aad5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ. 2011. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature 475: 53–58. [DOI] [PubMed] [Google Scholar]
- Liu T, Ghosal G, Yuan J, Chen J, Huang J. 2010. FAN1 acts with FANCI–FANCD2 to promote DNA interstrand cross-link repair. Science 329: 693–696. [DOI] [PubMed] [Google Scholar]
- MacKay C, Déclais A-C, Lundin C, Agostinho A, Deans AJ, MacArtney TJ, Hofmann K, Gartner A, West SC, Helleday T, et al. 2010. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell 142: 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihatsch MJ, Gudat F, Zollinger HU, Heierli C, Tholen H, Reutter FW. 1979. Systemic karyomegaly associated with chronic interstitial nephritis. A new disease entity? Clin Nephrol 12: 54–62. [PubMed] [Google Scholar]
- Moldovan G-L, D'Andrea AD. 2009. How the Fanconi anemia pathway guards the genome. Annu Rev Genet 43: 223–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M, Battaile KP, Bateman R, Lax TP, Rathbun K, Reifsteck C, Bagby G, Finegold M, Olson S, Grompe M. 2002. Fanconi anemia group A and C double-mutant mice: functional evidence for a multi-protein Fanconi anemia complex. Exp Hematol 30: 679–688. [DOI] [PubMed] [Google Scholar]
- Palmer D, Lallu S, Matheson P, Bethwaite P, Tompson K. 2007. Karyomegalic interstitial nephritis: a pitfall in urine cytology. Diagn Cytopathol 35: 179–182. [DOI] [PubMed] [Google Scholar]
- Pizzolato J, Mukherjee S, Schärer OD, Jiricny J. 2015. FANCD2-associated nuclease 1, but not exonuclease 1 or flap endonuclease 1, is able to unhook DNA interstrand cross-links in vitro. J Biol Chem 290: 22602–22611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontel LB, Rosado IV, Burgos-Barragan G, Garaycoechea JI, Yu R, Arends MJ, Chandrasekaran G, Broecker V, Wei W, Liu L, et al. 2015. Endogenous formaldehyde is a hematopoietic stem cell genotoxin and metabolic carcinogen. Mol Cell 60: 177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radha S, Tameem A, Rao BS. 2014. Karyomegalic interstitial nephritis with focal segmental glomerulosclerosis: a rare association. Indian J Nephrol 24: 117–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschle M, Knipscheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Scharer OD, Walter JC. 2008. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell 134: 969–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickman KA, Lach FP, Abhyankar A, Donovan FX, Sanborn EM, Kennedy JA, Sougnez C, Gabriel SB, Elemento O, Chandrasekharappa SC, et al. 2015. Deficiency of UBE2T, the E2 ubiquitin ligase necessary for FANCD2 and FANCI ubiquitination, causes FA-T subtype of Fanconi anemia. Cell Rep 12: 35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhland A, Haase E, Siede W, Brendel M. 1981. Isolation of yeast mutants sensitive to the bifunctional alkylating agent nitrogen mustard. Mol Gen Genet 181: 346–351. [DOI] [PubMed] [Google Scholar]
- Schlacher K, Wu H, Jasin M. 2012. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51–BRCA1/2. Cancer Cell 22: 106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segui N, Mina LB, Lazaro C, Sanz-Pamplona R, Pons T, Navarro M, Bellido F, Lopez-Doriga A, Valdes-Mas R, Pineda M, et al. 2015. Germline mutations in FAN1 cause hereditary colorectal cancer by impairing DNA repair. Gastroenterology 149: 563–566. [DOI] [PubMed] [Google Scholar]
- Smith AL, Alirezaie N, Connor A, Chan-Seng-Yue M, Grant R, Selander I, Bascunana C, Borgida A, Hall A, Whelan T, et al. 2016. Candidate DNA repair susceptibility genes identified by exome sequencing in high-risk pancreatic cancer. Cancer Lett 370: 302–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Desetty R, Saito TT, Schlabach M, Lach FP, Sowa ME, Clark AB, Kunkel TA, Harper JW, Colaiácovo MP, et al. 2010. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell 39: 36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoendlin M, Moch H, Brunner F, Brunner W, Burger HR, Kiss D, Wegmann W, Dalquen P, Oberholzer M, Thiel G, et al. 1995. Karyomegalic interstitial nephritis: further support for a distinct entity and evidence for a genetic defect. Am J Kidney Dis 25: 242–252. [DOI] [PubMed] [Google Scholar]
- Thazhathveetil AK, Liu ST, Indig FE, Seidman MM. 2007. Psoralen conjugates for visualization of genomic interstrand cross-links localized by laser photoactivation. Bioconjug Chem 18: 431–437. [DOI] [PubMed] [Google Scholar]
- Uz E, Bayram Y, Haltas H, Bavbek N, Kanbay M, Guz G, Akçay A. 2011. Karyomegalic tubulointerstitial nephritis: a rare cause of chronic kidney disease. Nephro Urol Mon 3: 201–203. [Google Scholar]
- Walden H, Deans AJ. 2014. The Fanconi anemia DNA repair pathway: structural and functional insights into a complex disorder. Annu Rev Biophys 43: 257–278. [DOI] [PubMed] [Google Scholar]
- Wang AT, Smogorzewska A. 2015. SnapShot: Fanconi anemia and associated proteins. Cell 160: 354–354.e1. [DOI] [PubMed] [Google Scholar]
- Wang AT, Sengerova B, Cattell E, Inagawa T, Hartley JM, Kiakos K, Burgess-Brown NA, Swift LP, Enzlin JH, Schofield CJ, et al. 2011. Human SNM1A and XPF–ERCC1 collaborate to initiate DNA interstrand cross-link repair. Genes Dev 25: 1859–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Persky NS, Yoo B, Ouerfelli O, Smogorzewska A, Elledge SJ, Pavletich NP. 2014. DNA repair. Mechanism of DNA interstrand cross-link processing by repair nuclease FAN1. Science 346: 1127–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AT, Kim T, Wagner JE, Conti BA, Lach FP, Huang AL, Molina H, Sanborn EM, Zierhut H, Cornes BK, et al. 2015. A dominant mutation in human RAD51 reveals its function in DNA interstrand crosslink repair independent of homologous recombination. Mol Cell 59: 478–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Guo R, Paramasivam M, Shen W, Ling C, Fox D III, Wang Y, Oostra AB, Kuehl J, Lee DY, et al. 2012. A ubiquitin-binding protein, FAAP20, links RNF8-mediated ubiquitination to the Fanconi anemia DNA repair network. Mol Cell 47: 61–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikiyo K, Kratz K, Hirota K, Nishihara K, Takata M, Kurumizaka H, Horimoto S, Takeda S, Jiricny J. 2010. KIAA1018/FAN1 nuclease protects cells against genomic instability induced by interstrand cross-linking agents. Proc Natl Acad Sci 107: 21553–21557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Walter JC. 2014. Mechanism and regulation of incisions during DNA interstrand cross-link repair. DNA Repair 19: 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Xue X, Longerich S, Sung P, Xiong Y. 2014. Structural insights into 5′ flap DNA unwinding and incision by the human FAN1 dimer. Nat Commun 5: 5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Otto EA, Cluckey A, Airik R, Hurd TW, Chaki M, Diaz K, Lach FP, Bennett GR, Gee HY, et al. 2012. FAN1 mutations cause karyomegalic interstitial nephritis, linking chronic kidney failure to defective DNA damage repair. Nat Genet 44: 910–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.