Abstract
Traditional cell-screening techniques such as FACS and MACS are better suited for large numbers of cells isolated from bulk tissue and cannot easily screen stem or progenitor cells from minute populations found in their physiological niches. Furthermore, these techniques rely upon irreversible antibody binding, potentially altering cell properties, including gene expression and regenerative capacity. To address these challenges, we have developed a novel, label-free stem-cell analysis and sorting platform capable of quantifying cell-surface marker expression of single functional organ stem cells directly isolated from their micro-anatomical niche. Using our unique platform, we have discovered a remarkable heterogeneity in both the regenerative capacity and expression of CXCR4, β1-integrin, Sca-1, M-cadherin, Syndecan-4, and Notch-1 in freshly isolated muscle stem (satellite) cells residing on different, single myofibers and have identified a small population of Sca-1+/Myf5+ myogenic satellite cells. Our results demonstrate the utility of our single-cell platform for uncovering and functionally characterizing stem-cell heterogeneity in the organ microniche.
Introduction
An understanding of both embryonic organogenesis and adult tissue regeneration relies on isolating and characterizing stem cells. In general, these cells are difficult to study because they constitute minute populations in organ niches and express multiple cell-surface markers, only some of which are known. Furthermore, their properties change quickly in vitro and possibly even during isolation procedures 1–3. The different combinations of markers used to identify muscle satellite cells [CD34 4,5, CXCR4 6, β1-integrin 6, Sca-1 5–6, M-cadherin 4, and Syndecan-4 7] illustrate the on-going debate as to which cell-surface markers do, indeed, identify particular rare stem cells. Fueling this debate are a number of important factors including: the known heterogeneity of muscle stem cells, variations introduced during sample isolation and processing, the difficulty in determining gene-expression accurately/quantitatively in the low starting stem-cell numbers within the micro-anatomical niche (i.e., a single myofiber), and the inability to functionally characterize single quiescent satellite cells. The skeletal muscle fiber (myofiber) has long been established as an undisputed microniche of satellite cells, first in the microscopy work pioneered by Mauro 8, then by the isolation and in vitro characterization of myofibers with their associated satellite cells pioneered by Schultz 9 and Bishoff 10, and finally through the serial transplantation of single myofibers with associated satellite cells by Collins 11. Comprehensively, this body of work determined that muscle stem cells reside beneath the basement membrane and on top of the sarcolemma of myofibers and that these cells are necessary and sufficient to account for the regenerative capacity of postnatal skeletal muscle 12–15.
Traditional quantitative methods of fluorescence-activated cell sorting (FACS) and magnetic-activated cell sorting (MACS) analysis are designed for large numbers of cells and cannot be easily applied to niche-specific characterization. In the case of muscle satellite cells, FACS and MACS cannot distinguish subsets of cells isolated from different myofibers or even separate hind-leg muscle groups. Microscopy, although capable of imaging stem cells in their niches, neither provides a straightforward means to quantify gene expression levels, nor allows further characterization of immunostained cells. Adding to the overall complexity is the fact that FACS, MACS, and fluorescence microscopy depend on irreversible antibody binding to stem-cell surface proteins that then become internalized, likely altering cell properties, including gene expression and regenerative capacity 16.
To address these challenges, we have developed a unique and label-free method for the objective, quantitative screening and characterization of single, functional organ stem cells. We demonstrate the power of our method by screening satellite cells freshly isolated from single fibers of non-injured extensor digitorum longus (EDL) muscle 17, 18 for heterogeneous cell-surface marker expression, and by subsequently sorting and characterizing these cells for their myogenic capacity. We show that our novel method combines the desired qualities of FACS, MACS, and microscopy, offering not only the high-resolution capability to quantitatively analyze rare stem cells residing in their undisputed organ micro-niche but also the ability to purify and subsequently characterize these cells without significantly altering their behavior through our screening and sorting processes.
Results
Description of the device, procedure, and control experiments
For our measurements, single satellite cells, freshly isolated from individual myofibers of uninjured EDL muscle (Figure 1A), are injected directly into a polydimethylsiloxane (PDMS) microfluidic channel (Figure 1B) that has been functionalized with a saturating concentration of either a specific or an isotype control antibody. A non-pulsatile pressure 19 is used to drive single satellite cells through filters, an inner reservoir, and finally through the functionalized microchannel for measurement. As individual satellite cells transit the microchannel, the flow of current is partially blocked, leading to a transient increase, or pulse, in the electrical resistance (Figure 1C) that is subsequently recorded and analyzed to characterize the cell 20–26. The pulse magnitude and width correspond to cell size and transit time, τ, across the microchannel, respectively. Because of specific interactions, cells transiting a microchannel functionalized with an antibody specific for an expressed surface marker have longer transit times than those transiting an isotype control microchannel. Figure 1D shows a typical current (I) vs. time measurement. Pulses 1 and 2 each correspond to a single cell transiting the microchannel. Pulse 3 corresponds to the rare occasion when two cells simultaneously transit the microchannel. As shown, the pulse has a characteristic profile that is easily recognized and discarded 19. Because we flow cells at a high rate through the channel (~106 s−1 shear rate), they only transiently interact with the functionalized antibody. As such, we have never observed cells adhering to our devices. Immediately after screening, each cell is collected and sorted in a separate well of 8-well slides using a pressurized flow of media in the outlet reservoir (Figure 1E). The single cells are then cultured for 14 days, fixed, and immunostained to determine their myogenic capacity, e.g. the ability to form colonies of Pax7+ and MyoD+ myogenic progenitors (SI Appendix).
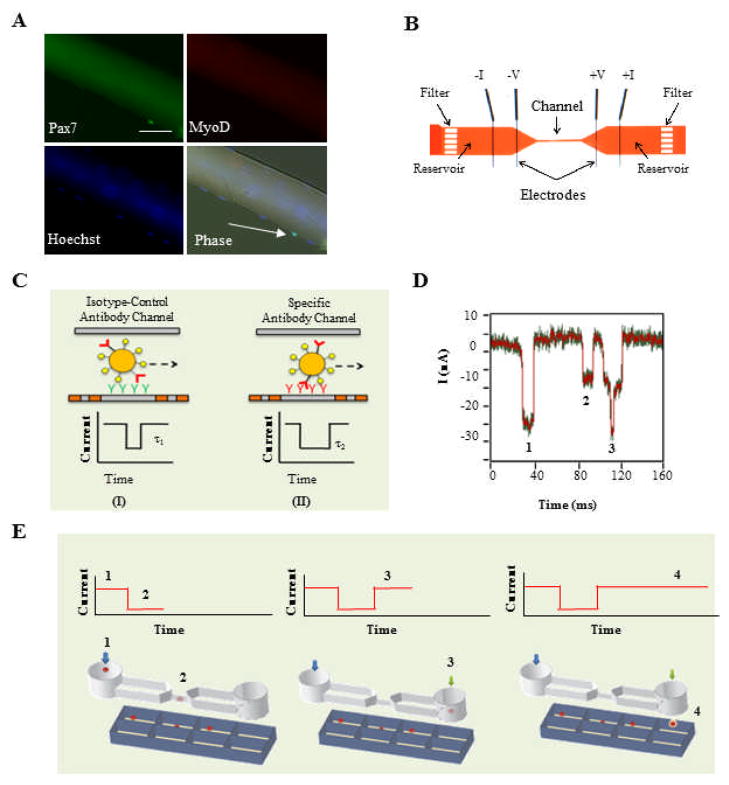
Figure 1. Detailed view and description of the stem-cell analysis platform.
(A) Freshly isolated single myofiber that has been immunostained. Satellite cells are sub-laminal and one expresses the conventional myogenic marker Pax7 (white arrow). No satellite cells express MyoD, indicating that they are not activated. Blue corresponds to Hoechst nuclear dye, and green to anti-Pax7 antibody. Scale bar corresponds to 100 μm. (B) Optical image of an actual device, consisting of two reservoirs connected by a single microchannel (800 μm x 25 μm x 25 μm, L x W x H), all embedded in a polydimethylsiloxane (PDMS) slab sealed to a glass substrate. (C) A four-terminal measurement is used to detect the current pulse generated when a cell transits the microchannel. Pulse magnitude and width correspond to cell size and transit time, τ, respectively. When the microchannel is functionalized with antibodies that have a high affinity to a particular cell-surface epitope, transient binding between the two leads to a longer transit time (II) than that due to non-specific interactions in an isotype control antibody microchannel (I). (D) A typical current (I) vs. time measurement as satellite cells flow through an anti-Sca-1 antibody microchannel. The green trace is the raw data, and the red trace is the analyzed data. Pulses 1 and 2 each correspond to a single cell passing through the microchannel. Pulse 3 corresponds to two cells passing through the pore simultaneously and is removed from any further analysis. (E) Schematic of cell sorting: a cell is initially injected into the microfluidic device (1), flows through the microchannel for measurement (2), and enters the exit chamber as a transverse flow of media is triggered (3) that enables collection in an individual well for culture (4).
To show that our method is capable of quantitatively determining both the expression and lack of expression of a particular surface antigen in a population, we screened primary-culture mouse myoblasts for Sca-1 and M-cadherin in prepared microchannels (SI Appendix). In agreement with previous reports that were based on FACS analysis, we found that 2.7% of the cultured myoblasts were Sca-1+ 27 and 93.0% were M-cadherin+ 23, 28, 29 (Figure S1). To demonstrate that our method could accurately analyze primary organ stem cells, we first used our published methods 30 to isolate a bulk population of satellite cells from uninjured muscle and then used both our functionalized microchannels and conventional FACS to screen this population for Sca-1 expression. As shown in Figure S2, the accuracy of our device and method are validated, as there is excellent correlation between the two analysis methods: 67.6% were determined to be Sca-1+ by our method and 66.1% by the gold standard, FACS. While both FACS and our device are capable of screening cells from bulk-tissue preparations, our method uniquely allows for the analysis of rare cells from a few single myofibers. Indeed, our next goal was to test this particular application of our novel single-cell analysis platform i.e. to assay single satellite cells associated with individual myofibers.
Niche-to-Niche Variation of Sca-1, CXCR4, β1-integrin, M-cadherin, Syndecan-4, and Notch-1 Expression
The presence, absence, and fiber-to-fiber heterogeneity of the surface markers Sca-1, CXCR4, β1-integrin, M-cadherin, Syndecan-4, and Notch-1 expressed by freshly isolated muscle satellite cells were examined. To ensure that our sample processing did not introduce artifacts, we hand-selected individual myofibers from bulk muscle for all our experiments. Furthermore, we sampled a subset of these single myofibers and immunostained them to ensure that all satellite cells were sub-laminal, expressed the conventional myogenic marker Pax7, and did not express MyoD or Ki67. In so doing, we confirmed that the satellite cells we studied were not activated 17, 18 (Figure 1A).
To determine which cells were Sca-1+, M-cadherin+, β1-integrin+, Syndecan-4+, CXCR4+, or Notch-1+, we statistically analyzed the transit-time data to assess whether a particular cell had an outlying slow transit time as compared to those cells passing through the isotype control microchannel (Figures 2A and S3). Because there is a direct correlation between the density of available epitopes and transit time (Figure S4), we can also determine expression levels of Sca-1, M-cadherin, β1-integrin, Syndecan-4, CXCR4, and Notch-1 in satellite cells (not just identify cells as positive or negative). Thus, cells whose transit times were significantly high (with raw p-values < 0.001 and Bonferroni corrected p-values < 0.05) were considered to have high levels of expression (Figures 2, 3A, and S3, red), while less stringent cutoffs on raw p-values of 0.01 and 0.05 were used to determine cells with medium (Figures 2A and S3, green) and low (Figures 2A and S3, blue) expression levels of these markers, respectively. In the case of Sca-1, M-cadherin, Syndecan-4, CXCR4, or Notch-1, the p-values were calculated directly from null distributions of transit time; the null distributions were estimated based on cells passing through the isotype control microchannel (SI Appendix). With the estimated null distributions, we further performed a False Discovery Rate (FDR) analysis to confirm our findings. The FDRs for cells with high levels of expression were all found to be < 0.01 (more than 80% of these cells have FDRs < 0.0001). The FDRs for cells with medium or low levels of expression were mostly found to be < 0.02 and 0.05, respectively. The FDR results provide a high confidence on the identified Sca-1+, M-cadherin+, Syndecan-4+, CXCR4+, or Notch-1+ cells. In the case of β1-integrin, the p-values were calculated from a Dixon’s Q Test, a robust statistical test that is used to identify values that appear diverging from a control sample and ideal for small sample sizes 31–33. Determination between the above two ways of p-value calculation was based on the number of cells measured in the control data.
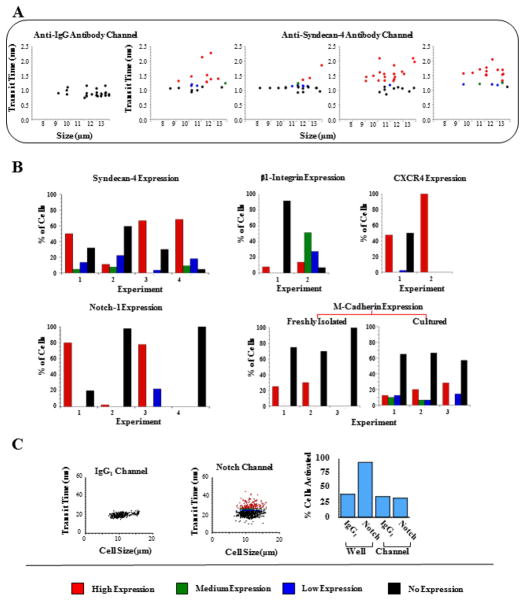
Figure 2. Screening analysis of satellite cells freshly derived from single muscle fibers.
(A) Syndecan-4 marker screening. Transit times of cells that passed through the isotype IgG1 control antibody microchannel were compared to those of 4 different experiments. Each experiment consisted of cells collected from 3 single muscle fibers and screened with a different anti-Syndecan-4 antibody microchannel. Based on an FDR analysis, cells were determined to have high (red), medium (green), low (blue), or no (black) Syndecan-4 expression. (B) Summary of the observed expression levels of Syndecan-4 (Experiments 1, 2, 3, and 4 with n=20, 26, 28, and 22 cells, respectively);β1-integrin (Experiments 1 and 2 with n=38 and 29 cells, respectively); CXCR4 (Experiments 1 and 2 with n=23 and 11 cells, respectively); Notch-1 (Experiments 1, 2, 3, and 4 with n=5, 43, 9, and 32 cells, respectively); and M-cadherin (Experiments 1, 2, and 3 with n=8, 10, and 3 cells, respectively). All markers screened show a wide range of heterogeneity from fiber to fiber. Heterogeneity and variance in expression levels of the studied markers increase upon muscle-stem-cell activation (M-cadherin freshly-isolated vs cultured, as shown. See also Figures 3A and S5). The bars correspond to the percentages calculated from the raw transit-time data in Figure S3. (C) Screening for Notch-1 protein expression on the surface of satellite cells. Transit time vs. cell size for satellite cells screened with a 2000 μm-long IgG1 or Notch-1 microchannel. τavg = 19.80 ± 2.36 ms for the IgG1 microchannel and τavg = 22.93 ± 4.83 ms for the Notch-1 microchannel Using an FDR calculation, with cutoffs of 0.1, 0.05, and 0.01, 21% of the 757 cells screened, had high levels of Notch1 expression (red), 8% had medium levels of expression (green), 9% had low levels (blue), respectively. The remaining cells (62%) did not express Notch1 (black). Percentage of cells showing high levels of nuclear Notch-1 after screening vs. control cultured conditions (See Figure S6). Cells were not activated when screened with our microchannels.
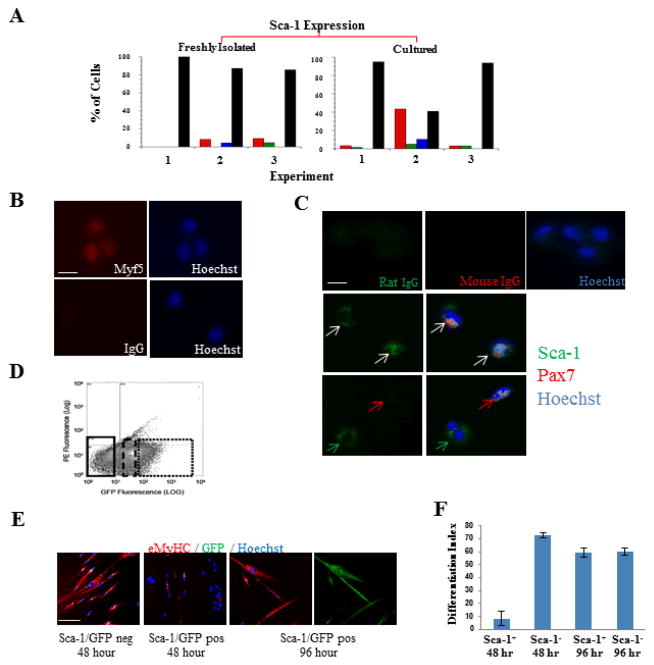
Figure 3.
A population of Sca-1+ cells is myogenic. (A) Summary of the observed expression levels of Sca-1 in freshly isolated (Experiments 1, 2, and 3 with n=5, 47, and 21 cells, respectively) and cultured satellite cells (Experiments 1, 2, and 3 with n = 60, 39, and 33 cells, respectively). As in Figure 2, red, green, blue, and black correspond to high, medium, low, and no expression, respectively. (B) Myofiber-associated cells that were Sca-1+ were analyzed after being captured by a microchannel functionalized with anti-Sca-1 antibody using a slow flow rate. The red and blue corresponds to Myf5 and Hoechst staining, respectively. Scale bar indicates 10 μm in all images. (C) Freshly isolated satellite cells were immunostained for Sca-1 and Pax7. The top row image shows antibody control, and the middle and bottom row shows Sca-1 staining. Green, red, and blue correspond to Sca-1, Pax7, and Hoechst staining, respectively. White, red, and green arrows indicate Sca-1+/Pax7+, Sca-1−/Pax7+, and Sca-1+/Pax7− cells, respectively. Scale bar indicates 10 μm in all images. (D) Myofiber-associated cells were derived from Sca-1-GFP Tg mice and live-sorted on GFP for three populations: low/no GFP (the lowest 21%, solid line region), an intermediate GFP (34%, dashed line region) and the highest GFP (17%, dotted line region). (E) The sorted cells were plated in DM for 48 hours or 96 hours, where myoblasts readily fuse into MyHC+ myotubes. Both Sca-1-GFP positive and Sca-1-GFP negative populations produced MyHC+ myotubes (red), demonstrating the myogenic nature of the Sca-1-expressing cells. Hoechst (blue) was used to label all nuclei. Scale bar indicates 100 μm. (F) Differentiation efficiency was quantified by percentage of nuclei in MyHC+ myotubes.
As shown in Figures 2B and 3A, we found significant heterogeneity between fibers in all studied cell-surface markers. In the summary of experiments we performed for each marker (Figure 2B and 3A), some fibers had almost no β1-integrin, M-cadherin, Notch-1, or Sca-1 expressing satellite cells. Other fibers had a significant number of satellite cells that express Syndecan-4, β1-integrin, CXCR4, or Notch-1 (>90% total expression). Significantly, almost no fibers had satellite cells with medium or low expression of CXCR4, Notch-1, or M-Cadherin. Notch-1 showed the greatest variability of expression between fibers, while Syndecan-4 showed the greatest variance in expression level of cells positive for this marker.
The high degree of variation of surface-marker expression levels between fibers that our method detected is surprising, and the first to be reported. To validate that this heterogeneity is an accurate reflection of the screened surface-markers, we compared freshly isolated and cultured satellite cells (SI Appendix) and found that, as expected, the heterogeneity and variance in expression levels of the studied markers increased upon in vitro culture (Figures 2B, 3A, and S5). Additionally, we confirmed that our device accurately reports virtually identical transit times in replicate runs of the same cell population (Figure S1C), hence providing another control for the uncovered phenomenon of fiber-dependent satellite-cell heterogeneity.
Following the discovery of microniche-specific heterogeneity of marker expression in satellite cells freshly isolated from single fibers, we investigated whether the high degree of cell-surface marker heterogeneity was due to the transient binding between the functionalized antibody and the cell-surface receptors that could activate receptor signaling and ultimately change cell properties 16. Since the anti-Notch-1 antibody (specific for the external part of Notch-1 receptor) employed in our microchannels has been shown to mimic the native ligand binding and activate Notch-1 robustly in satellite cells, resulting in high levels of the truncated intracellular portion of Notch that is localized to the cell nucleus 4, we analyzed whether Notch would become activated when detected in freshly harvested satellite cells (Figure 2C). As additional controls, unscreened satellite cells were plated on IgG1 and anti-Notch-1 antibody-coated culture wells overnight. To determine whether the transient interactions between the extracellular portion of the receptor and the functionalized antibodies activated the Notch pathway, we performed immunofluorescence on all cells using an antibody that specifically recognizes the truncated-activated form of Notch-1. In contrast to wells coated with anti-Notch-1 antibody, which induced robust nuclear-active Notch, cells from the IgG1 and anti-Notch-1 antibody microchannels and the IgG1-coated wells showed low levels of Notch activation (Figure 2C and Figure S6). Thus, the transient binding between functionalized antibody in our microchannel and specific receptors does not significantly contribute to Notch-pathway activation in satellite cells, thereby providing strong evidence that changes in signal transduction and cell behavior should not be expected when stem-cell populations are screened with our method.
Overall, our data suggest that individual myofibers from non-injured EDL muscle are heterogeneous with respect to the expression levels of Sca-1, M-cadherin, β1-integrin, Syndecan-4, CXCR4, and Notch-1 on their associated satellite cells. Our findings are highly consistent with previous reports, which have indicated that the muscle satellite-cell population is heterogeneous between different mice 34 and muscle groups 35. At the same time, our data are the first to reveal the variation of satellite cell subsets among single fibers in the same muscle and to establish quantitatively and directly this heterogeneity based on multiple surface markers.
A Significant Population of Sca-1+ Cells Are Myogenic
Of particular interest is the expression of Sca-1 in muscle satellite stem cells. While earlier studies reported that cells expressing Sca-1 protein and residing in skeletal muscle are indeed myogenic, i.e. they form myotubes in culture 36, later work seemed to exclude these cells as muscle precursors and even suggested that they give rise to adipose tissue 37–39. These reports recommended to eliminate Sca-1+ cells in bulk sorting of dissociated muscle tissue in order to obtain the myogenic precursor cells.
Since we found that some Sca-1+ cells reside in the muscle stem-cell micro-niche (Figure 3A), we first investigated the myogenicity of Sca-1+ cells through immunodetection. Freshly isolated satellite cells were either directly immunostained for Sca-1 and Pax7 (Figure 3C) or immunostained for the myogenic transcriptional factor, Myf5, after Sca-1+ cells were captured in a large microchannel (100 μm x 2000 μm x 40000 μm, H x W x L) functionalized with anti-Sca-1 antibody under a very slow flow rate (~103 s−1 shear rate) (Figure 3B, SI appendix). Through several methods of immunodetection, we identified a small, but significant population of Sca-1+/Myf5+ and Sca-1+/Pax7+ cells. This is in direct contrast to reports characterizing cells that express Sca-1 protein as non-myogenic 39, but in agreement with the previously reported expression of Sca-1 protein in single myofiber cultures 27.
To confirm the myogenicity of Sca-1-expressing cells, we isolated satellite cells from uninjured, intact myofibers of Sca-1-GFP transgenic mice, in which GFP expression signifies the presence and the levels of Sca-1 mRNA 40. The cells were cultured for 9 days, after which they were sorted on GFP mean fluorescence as high and low populations (Figure 3D), and plated in conventional differentiation medium, in which myoblasts fuse into eMyHC+ myotubes. As shown in Figures 3E and F, Sca-1-GFP+ cells differentiated into eMyHC+/GFP+ myotubes in culture, thus confirming their unambiguous myogenicity. Interestingly, Sca-1-GFP+ cells displayed a significant delay in differentiation: by 48 hours almost none of these cells fused into myotubes and very few expressed eMyHC, while myogenic differentiation was robust in Sca-1−-GFP− cells (Figures 3E and F). Nevertheless, by 96 hours, Sca-1-GFP+ satellite cells underwent terminal differentiation into multinucleated eMyHC+/GFP+ myotubes, demonstrating their capacity for myogenic lineage progression 41. These results suggest that Sca-1+ cells are myogenic and might have unique properties with respect to their terminal differentiation.
Correlating Myogenicity with the Heterogeneity of Cell-Surface Markers
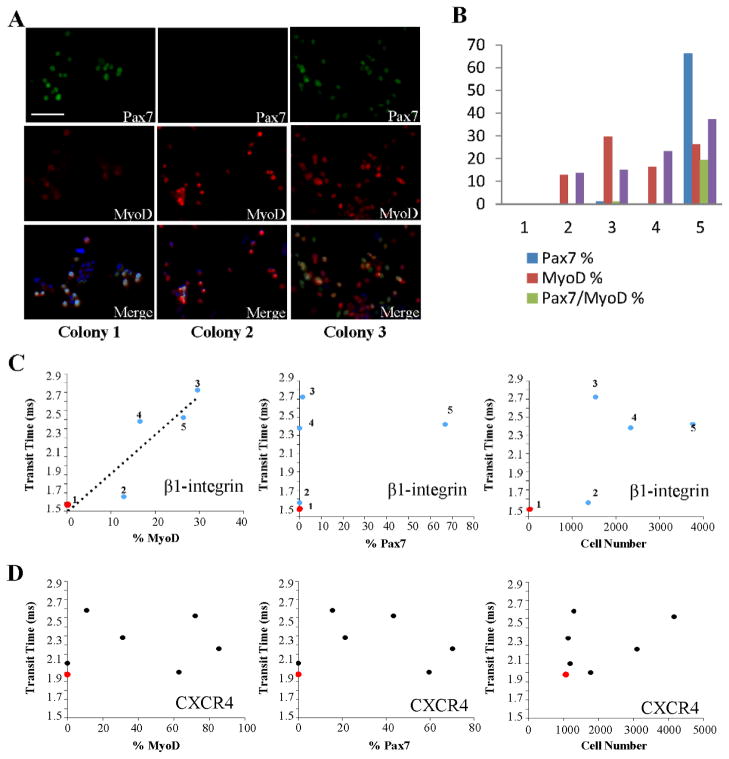
Satellite cells were isolated as above from single myofibers of non-injured EDL muscle and screened and sorted by our microfluidic channels functionalized with anti-β1-integrin and anti-CXCR4 antibodies. Single cells with known levels of β1-integrin and CXCR4 were then plated and cultured for 14 days (the time-frame during which quiescent satellite cells are typically activated in culture and form myogenic colonies 42). The capacity to form colonies of myogenic progenitor cells (Pax7+ and/or MyoD+) was assessed for each satellite cell that was isolated from each myofiber and the myogenicity was correlated with the levels of cell-surface markers. As shown in Figures 4A and B, satellite cells analyzed and sorted by our device formed myogenic colonies and very interestingly, the size of the colonies, as well as the percent of Pax7+/MyoD−, Pax7+/MyoD+, and Pax7−/MyoD+ cells, varied significantly, revealing the functional heterogeneity of muscle stem cells that associate with myofibers of the same EDL muscle. We correlated such functional heterogeneity with the expression of β1-integrin and CXCR4 in these sorted single cells and found a positive correlation between the levels of β1-integrin and the percent of differentiated MyoD+ cells in the colonies 43 that were formed by single satellite cells (Figure 4C). Interestingly, we found no correlation between the levels of CXCR4 and either Pax7+, MyoD+, or total cell numbers in the colonies; moreover, one CXCR4+ satellite cell (out of 16 clonally plated cells) formed a non-myogenic colony that had neither Pax7+ nor MyoD+ myogenic cells (Figure 4D, red dot). One cell screened with the β1-integrin channel also formed a non-myogenic colony (Figure 4C, red dot), but in contrast to the CXCR4 case, this cell did not have detectable levels of β1-integrin. Incidents of satellite-cell lineage conversion to non-myogenic lineage have been well documented 44, and we have confirmed this phenomenon at a single-cell level. Overall, these data indicate that: (1) the levels of β1-integrin can predict the differentiation capacity of satellite cells; and (2) some CXCR4+ cells might not be myogenic. Furthermore, with respect to the colony size and expression of the self-renewal marker, Pax7, these data show that a combination of multiple markers (rather than a single marker) would be a better predictor of such functional properties of satellite cells. More importantly, these results demonstrate that our device is capable of sorting single satellite cells and analyzing the correlation between their cell surface-marker expression and their myogenicity.
Figure 4.
Functional heterogeneity of single satellite cells. (A) Single cells with different β1-integrin expression levels were cultured at clonal density in growth medium for 2 weeks before being immunostained for Pax7 and MyoD. Three representative colonies are presented. Scale bar indicates 100 μm. (B) Cell number, Pax7+, MyoD+, and Pax7+/MyoD+ cell percentages were quantified in 5 different colonies grown from single satellite cells that were analyzed for β1-integrin expression. (C) Transit time of individual cells screened with an anti-β1-integrin antibody microchannel vs. MyoD+ cell percentage, Pax7+ cell percentage, and total cell number of colonies derived from the individual cells analyzed. Each dot and number label (1–5) corresponds to a particular cell that gave rise to one of the five colonies analyzed in Figure 4B. Cells 1 and 2 were β1-integrin− and cells 3–5 were β1-integrin+. (D) Transit time of individual cells screened with an anti-CXCR4 antibody channel vs. MyoD+ cell percentage, Pax7+ cell percentage, and total cell number of colonies derived from the individual cells analyzed. All cells were CXCR4+. Blue and black dots indicate cells that gave rise to myogenic colonies, and red dots indicate cells that gave rise to non-myogenic colonies.
Discussion
This work describes a novel, label-free microfluidic approach for accurately measuring the cell-surface marker expression in rare subsets of stem cells that have been freshly isolated from their organ micro-niche and for sorting these cells for subsequent analysis. Using this method, we have discovered a significant heterogeneity in all studied cell-surface markers (Sca-1, M-cadherin, β1-integrin, Syndecan-4, CXCR4, and Notch-1) in satellite cells freshly isolated from different single myofibers of EDL. Furthermore, our data suggest that this heterogeneity is functionally important and that there is a correlation with the myogenic capacity of muscle stem cells. These results provide the first direct demonstration of a microniche-specific variation in gene expression in stem cells of the same lineage and suggest that different individual myofibers in the same muscle might not be equal with respect to the efficiency of their repair by resident satellite cells and to their developmental and/or postnatal myogenesis 45. Additionally, a variation among the different fiber types (e.g. fast versus slow) 46,47 can potentially contribute to the heterogeneity of associated satellite cells. The presence, absence, and niche-to-niche variation of muscle stem-cell markers was defined quantitatively through a statistical comparison of time-of-flight data for single, live cells that had been derived from their undisputed organ niche and that had transited across a surface functionalized with a saturating concentration of antibodies.
With respect to the myogenic capacity, we have shown that a number of Sca-1+ cells express Myf5, in contrast to recent reports that suggest Sca-1+ cells are not myogenic. Our work prompts further studies on the properties of Sca-1+/Myf5+ cells, which support prior research demonstrating that single Sca-1+/Pax7+ cells can give rise to myogenic progenitors through asymmetric division 7. Furthermore, we demonstrated that we could screen for the Notch-1 marker in muscle satellite cells without triggering the cleavage of Notch-1, thereby suggesting that our label-free method minimizes the activation of the molecules which are being analyzed, a concern for conventional techniques that rely on irreversible antibody binding. Based on this result for Notch-1, our microchannels have the potential to be used for the analysis of cells without altering cell properties to the extent that permanent antibody binding does. However, further testing on a per-cell type, per-marker basis would be necessary to confirm this characteristic of our platform in other applications.
To demonstrate the strong potential of our method for analyzing stem cells from their microniche, we chose to screen muscle (satellite) stem cells from single muscle fibers. While we manually isolated satellite cells from their respective fibers and then injected them into our device for screening, the modular nature of our microfluidics platform allows for the incorporation of a series of gated microfluidic valves, reservoirs to introduce enzymes for cell disassociation from tissue, and mixers that ultimately would enable isolation and subsequent screening in situ. Such an in situ method and device could be generally applied to other stem-cell systems in which both sample and cells are rare.
Currently, our label-free method screens for a single surface antigen; however, multiple surface-marker screening could be accomplished by expanding our device into multiple microchannels, each functionalized with a different antibody corresponding to a different complementary antigen. Our current studies have already determined that the levels of β1-integrin expression in satellite cells positively correlate with the percent of newly formed MyoD+ progenitor cells. The development of our next-generation devices will enable an analysis of the correlation between multiple cell-surface markers and the capacity of satellite cells to self-renew, proliferate, and differentiate, thereby allowing us to predict more accurately and comprehensively how the heterogeneous profile of cell-surface markers translates into the heterogeneous regenerative capacity of single, skeletal-muscle stem cells.
The focus of this work was the quantitative analysis of satellite cells from their microanatomical niche and thus throughput was not an issue. For applications requiring higher throughput, e.g. deriving stem cells from a larger pool of cells, we have successfully used this platform to screen cells as fast as ~1,000 cells per minute with neither loss in pulse-width resolution nor ability to detect specific ligand-receptor interactions with a specific affinity. For significantly larger sample sizes, multiple screening devices could be easily integrated on a single chip for parallel processing and further increased throughput. Beyond muscle stem cells, our method could be broadly applied to the quantitative analysis of single stem cells in other adult and developing organs, such as hair follicles or the sub-granular zone of the hippocampus, potentially leading to profound new discoveries on stem-cell properties and regenerative potential.
Experimental Procedures
Harvesting of satellite cells from single muscle fibers
EDL muscle was dissected from the hind leg of a 3-month old C57/black-6 mouse and incubated at 37°C in digestion medium for 1 hour with gentle agitation. Digested muscle was gently triturated and single fibers were handpicked under a microscope. Satellite cells were liberated from three single fibers by digestion with 1 U/mL Dispase and 40 U/mL Collagenase type II in medium for one hour before analysis. See SI Materials/Methods. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The procedures and animal protocols used were approved by the Office of Laboratory Animal Care, UC Berkeley.
Preparation of microchannel devices
Negative-relief masters of the final microfluidic channel devices were fabricated on polished silicon wafers using standard soft lithography 48. The electrodes were lithographically patterned onto RCA-cleaned glass substrates using Microposit S1813 resist. A 75/250 Å Ti/Pt thin film was deposited using an electron-gun evaporator. Before sealing to the PDMS slab, the glass substrates were functionalized. See SI Materials/Methods.
Data acquisition and analysis
Cells were injected into the microchannel device, and a non-pulsatile pressure of 17.4 kPa was used to drive the cells through the device without permanent retention in the channel. As cells passed through the microchannel, a four-point measurement of the current was performed using a constant applied AC voltage (typically 0.2–0.4 V), as previously published 23, 24, 26, 49, 50. The magnitude and width of each pulse were analyzed to determine cell size and transit time, respectively. See SI Materials/Methods.
Statistical analysis of transit-time data
Two different methods, Dixon’s Q Test 32, 33, 51 and FDR, were employed to compare the transit times of cells passing through isotype control channels to those functionalized with an antibody specific for expressed surface markers (e.g. Sca-1, CXCR4, M-cadherin, β1-integrin, Notch-1, and Syndecan-4). For larger quasi-bulk samples (i.e. screening Notch-1 expression in satellite cells), an FDR was used to control for false positives 52. We analyzed only those data that corresponded to cells of size 8–13 μm, the size range of satellite cells 30. See SI Materials/Methods.
Analysis of Sca-1 expression through immunofluorescence staining and microfluidic channel capture
Freshly isolated satellite cells were captured in a large microfluidic channel (100 μm x 2000 μm x 40000 μm, H x W x L) functionalized with anti-Sca-1 antibody using an extremely slow fluid flow rate (~103 s−1 shear rate) or attached to Matrigel plates before being co-stained for Sca-1 with the myogenic marker Pax7 or Myf5. See SI Materials/Methods.
Clonal analysis of single satellite cells
Single myofibers were isolated and analyzed with the devices. Immediately after screening for a pulse corresponding to a single satellite cell, the cell was collected in an individual well in an 8-well slide. Isolated cells were cultured in myoblast growth medium for 14 days before being immunostained for Pax7 and MyoD. Cell number, Pax7+, MyoD+, and Pax7+/MyoD+ cell percentages were quantified for each cell colony.
Supplementary Material
Acknowledgments
The authors wish to thank Vicki Plaks and Zena Werb for Sca-1-GFP mice, and Evan Lyall and Anand Kesavaraju for assistance with image analysis and cell-size determination, respectively. This work was supported in part by the W. M. Keck Foundation (L. L. S. and I. M. C.), the California Institute of Regenerative Medicine through a postdoctoral fellowship (S. K. M.) and a young faculty award RN1-00532 (I. M. C.), NIH EY019094 (H. H.), and NIH/NIA AG 027252 (I. M. C.).
Footnotes
Author Contributions: M. R. C. designed experiments, performed experiments, analyzed data, and was involved in writing the paper. K. B. performed experiments, analyzed data, and was involved in writing the manuscript. J. L. helped design experiments, performed experiments and was involved in writing the paper. M. J. C. helped design experiments, provided cells for experiments, performed and analyzed the FACS and immunofluorescence experiments, and was involved in writing the paper. H. H. analyzed data and was involved in writing the paper. S. K. M. performed the pilot experiments. E. J. performed experiments and was involved in writing the manuscript. J. H. performed experiments and analyzed data. I. M. C. and L. L. S conceived the idea, directed the study, and wrote the manuscript.
References
- 1.Slack JM. Origin of stem cells in organogenesis. Science. 2008;322:1498–501. doi: 10.1126/science.1162782. [DOI] [PubMed] [Google Scholar]
- 2.Shefer G, Yablonka-Reuveni Z. Isolation and culture of skeletal muscle myofibers as a means to analyze satellite cells. Methods Mol Biol. 2005;290:281–304. doi: 10.1385/1-59259-838-2:281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuci S, Kuci Z, Latifi-Pupovci H, Niethammer D, Handgretinger R, Schumm M, Bruchelt G, Bader P, Klingebiel T. Adult stem cells as an alternative source of multipotential (pluripotential) cells in regenerative medicine. Curr Stem Cell Res Ther. 2009;4:107–17. doi: 10.2174/157488809788167427. [DOI] [PubMed] [Google Scholar]
- 4.Conboy IM, Conboy MJ, Smythe GM, Rando TA. Notch-mediated restoration of regenerative potential to aged muscle. Science. 2003;302:1575–7. doi: 10.1126/science.1087573. [DOI] [PubMed] [Google Scholar]
- 5.Zammit PS, Partridge TA, Yablonka-Reuveni Z. The skeletal muscle satellite cell: the stem cell that came in from the cold. J Histochem Cytochem. 2006;54:1177–91. doi: 10.1369/jhc.6R6995.2006. [DOI] [PubMed] [Google Scholar]
- 6.Sherwood RI, Christensen JL, Conboy IM, Conboy MJ, Rando TA, Weissman IL, Wagers AJ. Isolation of adult mouse myogenic progenitors: functional heterogeneity of cells within and engrafting skeletal muscle. Cell. 2004;119:543–54. doi: 10.1016/j.cell.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka KK, Hall JK, Troy AA, Cornelison DD, Majka SM, Olwin BB. Syndecan-4-expressing muscle progenitor cells in the SP engraft as satellite cells during muscle regeneration. Cell stem cell. 2009;4:217–25. doi: 10.1016/j.stem.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mauro A. Satellite cell of skeletal muscle fibers. J Biophys Biochem Cytol. 1961;9:493–5. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schultz E. Changes in the satellite cells of growing muscle following denervation. The Anatomical record. 1978;190:299–311. doi: 10.1002/ar.1091900212. [DOI] [PubMed] [Google Scholar]
- 10.Bischoff R. Proliferation of muscle satellite cells on intact myofibers in culture. Dev Biol. 1986;115:129–39. doi: 10.1016/0012-1606(86)90234-4. [DOI] [PubMed] [Google Scholar]
- 11.Collins CA, Olsen I, Zammit PS, Heslop L, Petrie A, Partridge TA, Morgan JE. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell. 2005;122:289–301. doi: 10.1016/j.cell.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 12.Morgan JE, Partridge TA. Muscle satellite cells. Int J Biochem Cell Biol. 2003;35:1151–6. doi: 10.1016/s1357-2725(03)00042-6. [DOI] [PubMed] [Google Scholar]
- 13.Buckingham M. Myogenic progenitor cells and skeletal myogenesis in vertebrates. Curr Opin Genet Dev. 2006;16:525–32. doi: 10.1016/j.gde.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 14.Collins CA. Satellite cell self-renewal. Curr Opin Pharmacol. 2006;6:301–6. doi: 10.1016/j.coph.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 15.Zammit PS. All muscle satellite cells are equal, but are some more equal than others? J Cell Sci. 2008;121:2975–82. doi: 10.1242/jcs.019661. [DOI] [PubMed] [Google Scholar]
- 16.Tarnok A, Ulrich H, Bocsi J. Phenotypes of stem cells from diverse origin. Cytometry A. 2010;77:6–10. doi: 10.1002/cyto.a.20844. [DOI] [PubMed] [Google Scholar]
- 17.Chakkalakal JV, Jones KM, Basson MA, Brack AS. The aged niche disrupts muscle stem cell quiescence. Nature. 2012;490:355–60. doi: 10.1038/nature11438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheung TH, Quach NL, Charville GW, Liu L, Park L, Edalati A, Yoo B, Hoang P, Rando TA. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature. 2012;482:524–8. doi: 10.1038/nature10834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carbonaro A, Mohanty SK, Huang H, Godley LA, Sohn LL. Cell characterization using a protein-functionalized pore. Lab Chip. 2008;8:1478–85. doi: 10.1039/b801929k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Counter WH. U.S.P. Office; 2,656,508 Means for counting particles suspeded in a fluid. 1953
- 21.Kubitschek HE. Electronic counting and sizing of bacteria. Nature. 1958;182:234–5. doi: 10.1038/182234a0. [DOI] [PubMed] [Google Scholar]
- 22.Deblois RW, Bean CP. Counting and Sizing of Submicron Particles by Resistive Pulse Technique. Review of Scientific Instruments. 1970;41:909. [Google Scholar]
- 23.Moore R, Walsh FS. The cell adhesion molecule M-cadherin is specifically expressed in developing and regenerating, but not denervated skeletal muscle. Development. 1993;117:1409–20. doi: 10.1242/dev.117.4.1409. [DOI] [PubMed] [Google Scholar]
- 24.Saleh OA, Sohn LL. Quantitative sensing of nanoscale colloids using a microchip Coulter counter. Review of Scientific Instruments. 2001;72:4449–4451. [Google Scholar]
- 25.Saleh OA, Sohn LL. Direct detection of antibody-antigen binding using an on-chip artificial pore. P Natl Acad Sci USA. 2003;100:820–824. doi: 10.1073/pnas.0337563100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saleh OA, Sohn LL. An artificial nanopore for molecular sensing. Nano Letters. 2003;3:37–38. [Google Scholar]
- 27.Mitchell PO, Mills T, O’Connor RS, Graubert T, Dzierzak E, Pavlath GK. Sca-1 negatively regulates proliferation and differentiation of muscle cells. Developmental Biology. 2005;283:240–252. doi: 10.1016/j.ydbio.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 28.Irintchev A, Zeschnigk M, Starzinski-Powitz A, Wernig A. Expression pattern of M-cadherin in normal, denervated, and regenerating mouse muscles. Dev Dyn. 1994;199:326–37. doi: 10.1002/aja.1001990407. [DOI] [PubMed] [Google Scholar]
- 29.Kaufmann U, Martin B, Link D, Witt K, Zeitler R, Reinhard S, Starzinski-Powitz A. M-cadherin and its sisters in development of striated muscle. Cell Tissue Res. 1999;296:191–8. doi: 10.1007/s004410051280. [DOI] [PubMed] [Google Scholar]
- 30.Conboy MJ, Conboy IM. Preparation of adult muscle fiber-associated stem/precursor cells. Methods Mol Biol. 2010;621:149–63. doi: 10.1007/978-1-60761-063-2_10. [DOI] [PubMed] [Google Scholar]
- 31.Dean RB, Dixon WJ. Simplified Statistics for Small Numbers of Observations. Analytical Chemistry. 1951;23:636–638. [Google Scholar]
- 32.Rorabacher DB. Statistical Treatment for Rejection of Deviant Values - Critical-Values of Dixon Q Parameter and Related Subrange Ratios at the 95-Percent Confidence Level. Analytical Chemistry. 1991;63:139–146. [Google Scholar]
- 33.Dixon WJ. Analysis of Extreme Values. Ann Math Stat. 1950;21:488–506. [Google Scholar]
- 34.Zammit PS, Golding JP, Nagata Y, Hudon V, Partridge TA, Beauchamp JR. Muscle satellite cells adopt divergent fates: a mechanism for self-renewal? J Cell Biol. 2004;166:347–357. doi: 10.1083/jcb.200312007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ono Y, Boldrin L, Knopp P, Morgan JE, Zammit PS. Muscle satellite cells are a functionally heterogeneous population in both somite-derived and branchiomeric muscles. Dev Biol. 2010;337:29–41. doi: 10.1016/j.ydbio.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Royer CL, Howell JC, Morrison PR, Srour EF, Yoder MC. Muscle-derived CD45-SCA-1+c-kit- progenitor cells give rise to skeletal muscle myotubes in vitro. In Vitro Cell Dev Biol Anim. 2002;38:512–7. doi: 10.1290/1071-2690(2002)038<0512:MCPCGR>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 37.Schulz TJ, Huang TL, Tran TT, Zhang HB, Townsend KL, Shadrach JL, Cerletti M, McDougall LE, Giorgadze N, Tchkonia T, Schrier D, Falb D, Kirkland JL, Wagers AJ, Tseng YH. Identification of inducible brown adipocyte progenitors residing in skeletal muscle and white fat. P Natl Acad Sci USA. 2011;108:143–148. doi: 10.1073/pnas.1010929108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherwood RI, Christensen JL, Conboy IM, Conboy MJ, Rando TA, Weissman IL, Wagers AJ. Isolation of adult mouse myogenic progenitors: Functional heterogeneity of cells within and engrafting skeletal muscle. Cell. 2004;119:543–554. doi: 10.1016/j.cell.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 39.Cerletti M, Jurga S, Witczak CA, Hirshman MF, Shadrach JL, Goodyear LJ, Wagers AJ. Highly efficient, functional engraftment of skeletal muscle stem cells in dystrophic muscles. Cell. 2008;134:37–47. doi: 10.1016/j.cell.2008.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanson P, Mathews V, Marrus SH, Graubert TA. Enhanced green fluorescent protein targeted to the Sca-1 (Ly-6A) locus in transgenic mice results in efficient marking of hematopoietic stem cells in vivo. Exp Hematol. 2003;31:159–167. doi: 10.1016/s0301-472x(02)01021-4. [DOI] [PubMed] [Google Scholar]
- 41.Kirillova I, Gussoni E, Goldhamer DJ, Yablonka-Reuveni Z. Myogenic reprogramming of retina-derived cells following their spontaneous fusion with myotubes. Dev Biol. 2007;311:449–63. doi: 10.1016/j.ydbio.2007.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conboy IM, Rando TA. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Developmental cell. 2002;3:397–409. doi: 10.1016/s1534-5807(02)00254-x. [DOI] [PubMed] [Google Scholar]
- 43.Kuang S, Kuroda K, Le Grand F, Rudnicki MA. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brack AS, Conboy MJ, Roy S, Lee M, Kuo CJ, Keller C, Rando TA. Increased Wnt signaling during aging alters muscle stem cell fate and increases fibrosis. Science. 2007;317:807–10. doi: 10.1126/science.1144090. [DOI] [PubMed] [Google Scholar]
- 45.Biressi S, Rando TA. Heterogeneity in the muscle satellite cell population. Semin Cell Dev Biol. 2010;21:845–54. doi: 10.1016/j.semcdb.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dimario JX, Fernyak SE, Stockdale FE. Myoblasts Transferred to the Limbs of Embryos Are Committed to Specific Fiber Fates. Nature. 1993;362:165–167. doi: 10.1038/362165a0. [DOI] [PubMed] [Google Scholar]
- 47.Kalhovde JM, Jerkovic R, Sefland I, Cordonnier C, Calabria E, Schiaffino S, Lomo T. ‘Fast’ and ‘slow’ muscle fibres in hindlimb muscles of adult rats regenerate from intrinsically different satellite cells. J Physiol-London. 2005;562:847–857. doi: 10.1113/jphysiol.2004.073684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xia YN, Whitesides GM. Soft lithography. Annu Rev Mater Sci. 1998;28:153–184. [Google Scholar]
- 49.Saleh OA, Sohn LL. Direct detection of antibody-antigen binding using an on-chip artificial pore. Proc Natl Acad Sci U S A. 2003;100:820–4. doi: 10.1073/pnas.0337563100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carbonaro A, Mohanty SK, Huang HY, Godley LA, Sohn LL. Cell characterization using a protein-functionalized pore. Lab on a Chip. 2008;8:1478–1485. doi: 10.1039/b801929k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dixon WJ. Ratios involving extreme values. Ann Math Stat. 1951;21:488–506. [Google Scholar]
- 52.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B (Methodological) 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.