Abstract
Hepatitis C virus has been found to be associated with B-cell non-Hodgkin lymphomas, mostly marginal zone lymphomas and diffuse large B-cell lymphoma. Deregulation of signaling pathways involved in normal marginal zone development (NOTCH pathway, NF-κB, and BCR signaling) has been demonstrated in splenic marginal zone lymphoma. We studied mutations of NOTCH pathway signaling in 46 patients with hepatitis C virus-positive diffuse large B-cell lymphoma and in 64 patients with diffuse large B-cell lymphoma unrelated to HCV. NOTCH2 mutations were detected in 9 of 46 (20%) hepatitis C virus-positive patients, and NOTCH1 mutations in 2 of 46 (4%). By contrast, only one of 64 HCV-negative patients had a NOTCH1 or NOTCH2 mutation. The frequency of the NOTCH pathway lesions was significantly higher in hepatitis C virus-positive patients (P=0.002). The 5-year overall survival was 27% (95%CI: 5%–56%) for hepatitis C virus-positive diffuse large B-cell lymphoma patients carrying a NOTCH pathway mutation versus 62% (95%CI: 42%–77%) for those without these genetic lesions. By univariate analysis, age over 60 years, NOTCH2 mutation, and any mutation of the NOTCH pathway (NOTCH2, NOTCH1, SPEN) were associated with shorter overall survival. Mutation of the NOTCH pathway retained an independent significance (P=0.029). In conclusion, a subset of patients with hepatitis C virus-positive diffuse large B-cell lymphoma displays a molecular signature of splenic marginal zone and has a worse clinical outcome.
Introduction
Hepatitis C virus (HCV) HCV infection is a risk factor for the development of B-cell non-Hodgkin lymphomas (B-NHL), either indolent B-NHL, such as splenic marginal zone lymphoma (SMZL), or diffuse large B-cell lymphoma (DLBCL).1,2
Beside epidemiological evidence, the direct relationship between SMZL and HCV infection is also clinically supported by SMZL regression after antiviral treatments.3,4
Unbiased genomic studies have unraveled the typical coding genome of SMZL, which is characterized by mutations of genes involved in the physiological differentiation and homeostasis of marginal zone (MZ) B cells.5,6 Indeed, the commitment and retention of mature B cells to the MZ compartment requires NOTCH2 signaling activation. Consistently, the NOTCH pathway is affected in approximately 30%–40% of SMZL, including mutations of NOTCH2, the master regulator of MZ B-cell differentiation, in approximately 25% of cases. In addition to NOTCH2, other modulators or members of the NOTCH pathway are recurrently targeted by genetic lesions in SMZL, including NOTCH1, a paralog of NOTCH2, and SPEN, the main negative regulator of NOTCH signaling in MZ B cells. NOTCH2 mutations are also recurrent in clonal B-cell lymphocytosis with an MZ phenotype,7 a recently recognized entity8 that may represent a precursor of MZL.
HCV-positive DLBCL patients display specific presentation with respect to their HCV-negative counterparts, as documented by the preferential involvement of the spleen, and the occasional residual signs of low-grade lymphoma.9–11 These clinical and pathological features suggest that at least a fraction of HCV-positive DLBCL may represent the transformation of a pre-existent, though unrecognized MZL clone.
Though the genetics of DLBCL arising in HCV-negative patients has been extensively investigated, few data are currently available about the molecular mechanisms involved in the development of DLBCL in HCV subjects.
In this study we document that: i) NOTCH pathway mutations are a molecular clue associated to approximately 25% HCV-positive cases among DLBCL; and ii) among HCV-positive DLBCL, the molecular deregulation of NOTCH signaling associates with the co-existence of a low-grade component in the diagnostic biopsy and poor outcome. Overall, these data suggest that at least a fraction of HCV-positive DLBCL may represent the transformed phase of an MZL clone.
Methods
Patients
This study was approved by the institutional Ethics Committee (Comitato di Bioetica, Fondazione IRCCS Policlinico San Matteo, Pavia, Italy). The procedures followed were in accordance with the Declaration of Helsinki of 1975, as revised in 2000.
Fifty-six cases of newly diagnosed, previously untreated DLBCL arisen in HCV-positive subjects were retrieved from the files of the Departments of Hematology and Anatomic Pathology of the University of Pavia and the Amedeo Avogadro University of Eastern Piedmont, Italy. A total of 46 cases were included in the study based on the availability of adequate biological material, complete clinical data and final diagnosis after histological review. The cases arising in a setting of congenital or acquired (HIV-related or iatrogenic) immunodeficiency or with a history of previous low-grade NHL were excluded.
The 46 cases of DLBCL included in the study were differentiated into a discovery panel (n=19 cases) and an extension panel (n = 27 cases), based on the availability of frozen samples from pretreatment diagnostic biopsies. The full set of candidate genes was initially assessed in the discovery panel for which frozen material was available. The size of the discovery panel allowed a 90% statistical power to detect mutations represented in at least 10% of HCV-associated DLBCL. In order to refine the mutation frequency, genes that turned out to be mutated in the discovery panel were further investigated in the extension panel, for which only FFPE tissue samples were available.
All the diagnostic samples, were reviewed by 4 expert hematopathologists (MP, ML, MN, AR) according to the 2008 WHO Lymphoma Classification joining morphological, immunohistochemical and molecular information.12 Bone marrow biopsy was available in 36 patients.
For comparative purposes, 64 HCV-negative DLBCL were also included in the study. All of the 64 samples had been obtained at diagnosis from the involved site (lymph nodes or extranodal sites). HCV-positive and HCV-negative cohorts were matched for age.
Mutation analysis
The mutation hotspots of NOTCH1 (exon 34; RefSeq NM_017617.2), NOTCH2 (exon 34; RefSeq NM_024408.3), SPEN (exons 1–15; RefSeq NM_015001.2), MYD88 (exons 3, 5; RefSeq NM_001172567.1),5,13 BIRC3 (exons 6–9; RefSeq NM_001165.4),14,15 IKBKB (exon 7; RefSeq NM_001556.2),14 TNFAIP3 (exons 2–9; RefSeq NM_001270508.1), TRAF3 (exons 3–12, RefSeq NM_145725.2), CARD11 (exons 5–9; RefSeq NM_032415.5).16 CD79A (exons 4–5, RefSeq NM_001783.3) and CD79B (exons 5–6; RefSeq NM_001039933.1)17 genes were analyzed by PCR amplification and DNA direct sequencing of genomic DNA.
Statistical analysis
The non-parametric Wilcoxon rank-sum test was used to compare quantitative variables across groups of patients. Association between categorical variables was tested by Fisher exact test. Overall survival (OS) was measured using the Kaplan-Meier product limit method. Multivariate Cox regression model was also applied to evaluate the adjusted influence of mutations and clinical features significantly associated to OS in univariate analysis. P<0.05 was considered statistically significant. Statistical analyses were performed using Stata 12.1 (StataCorp. 2011. Stata Statistical Software: Release 12. College Station, TX, USA: StataCorp LP) software.
Results
Clinico-pathological features of HCV-positive DLBCL
Clinical and pathological features of 46 patients with HCV-positive DLBCL are summarized in Table 1. All patients, with the exception of 2, have been treated with CHOP or CHOP-like regimen (in 15 coupled with rituximab). All cases were HCV-RNA positive and HCV genotype was investigated in 11 patients: 5 had genotype 1b and 6 genotype 2a/2.
Table 1.
Clinical features at presentation of 46 cases of diffuse large B-cell lymphoma associated with hepatitis C virus infection
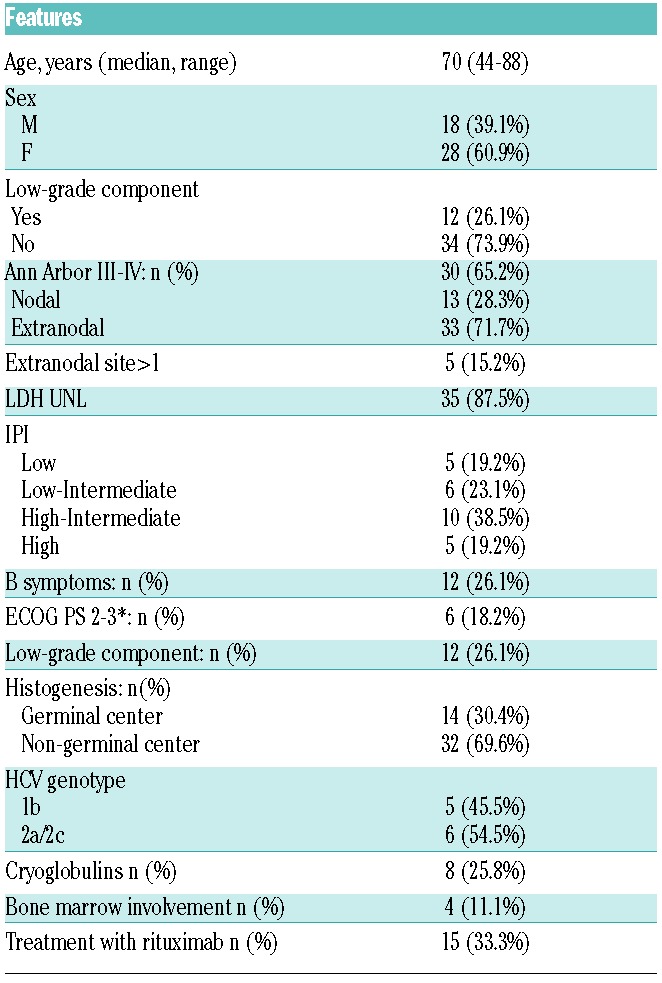
Extranodal disease was present in 72% (33 of 46) of patients and 17 patients had splenic involvement, including 32% (15 of 46) of patients with primary splenic disease. Cryoglobulins were detected in 26% (8 of 31) patients.
Histologically, most cases (37 of 46, 80%) had centroblastic cytology, with a minority of cases showing immunoblastic (1 of 46, 2%) or anaplastic (1 of 46, 2%) features. In 7 cases, the presence of some artifactual changes allowed a DLBCL diagnosis, but prevented any further subtyping. In 26% (12 of 46) of cases, a minor area of the diagnostic biopsy was composed of small to medium sized monocytoid B cells, suggesting a progression from an MZL that had not previously been recognized (Figure 1). According to the Hans algorithm, 30% (14 of 46) of cases had a germinal center (GC) phenotype while 70% (32 of 46) were of non-germinal center origin (non-GC). Median Ki67 expression was 70%. Bone marrow infiltration was documented in 11% (4 of 36) of cases, including one patient showing a small B-cell marrow infiltrate with features consistent with MZL. In 2 additional cases, a B-cell clone CD20+, CD5−, CD10− of small size was detected by flow cytometry in the bone marrow, despite the absence in these cases of lymphoma infiltration by morphology and the absence of a low-grade component in diagnostic biopsy. There was no difference in clinical and histological features among discovery and extension panels, except for more frequent detection of B symptoms in the discovery panel (Online Supplementary Table S1). Clinical features of a comparative cohort of HCV-negative DLBCL are reported in Online Supplementary Table S2.
Figure 1.
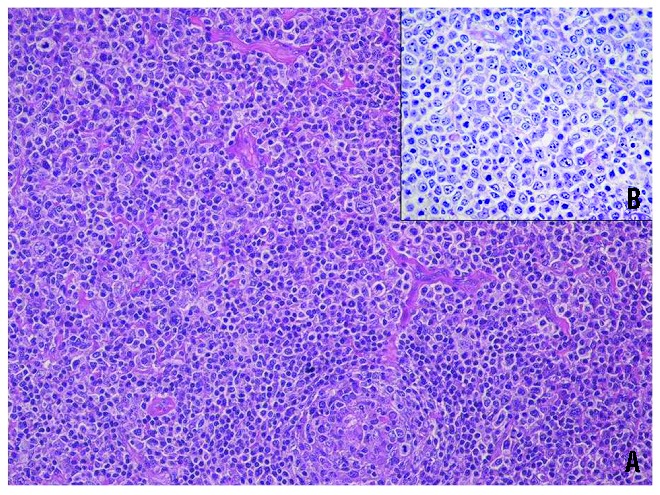
Histological picture of a case of diffuse large B-cell lymphoma exhibiting a perifollicular growth pattern (A) (HE 20X) and consisting of cells with abundant clear cytoplasm (B) (Giemsa 40x), resembling monocytoid B cells, that is reminiscent of a marginal zone origin.
The NOTCH pathway is recurrently mutated in HCV-positive DLBCL
Overall, the NOTCH pathway was recurrently mutated in 26% (12 of 46) HCV-positive DLBCL (Figure 2A). Among NOTCH pathway genes, NOTCH2 was mutated in 20% (9 of 46) of HCV-positive DLBCL, NOTCH1 in 4% (2 of 46), and SPEN in 2% (1 of 46); NOTCH1 and NOTCH2 mutations occur in a mutually exclusive manner (Figure 2A). In both NOTCH1 and NOTCH1 genes, mutations were represented by frame-shift indel or stop codons that truncated the PEST domain of the protein.
Figure 2.
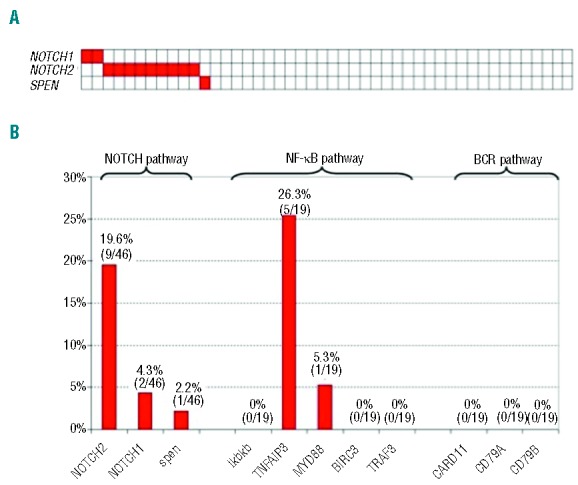
(A) Mutations of NOTCH genes in diffuse large B-cell lymphoma associated with hepatitis C virus infection (white: wild-type; red: mutations). (B) Percentages of diffuse large B-cell lymphoma cases harboring mutations in selected genes belonging to cellular pathways that are recurrently altered in splenic marginal zone lymphoma.
In our discovery panel of HCV-positive DLBCL, the prevalence and type of NF-κB pathway mutations was consistent with that of a cohort of unselected non-GC DLBCL,18 while BCR pathway genes (CARD11, CD79A and CD79B) were never affected by mutations in HCV-positive DLBCL (Figure 2B). A detailed description of mutations is reported in Online Supplementary Table S3.
NOTCH pathway mutations associate with co-existence of a low-grade component
Among pathological features, NOTCH pathway mutations were enriched in cases characterized by the co-existence in the diagnostic biopsy of a small to medium cell component along with the large cell component proper of DLBCL (NOTCH pathway mutations in 6 of 12, 50% of cases with low-grade component vs. 6 of 34, 18% in cases without low-grade component; P=0.052).
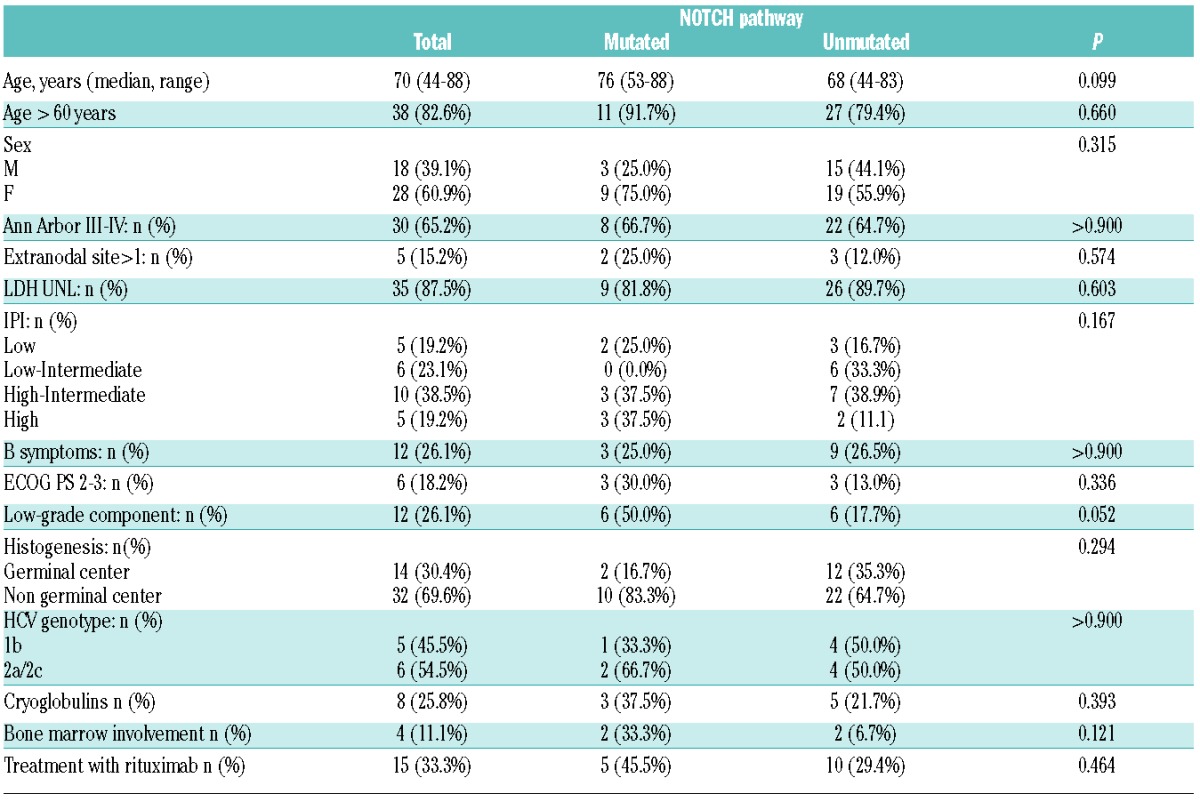
There was no difference in clinical features between NOTCH mutated and NOTCH wild-type patients (Table 2).
Table 2.
Clinical features of 46 cases of diffuse large B-cell lymphoma associated with hepatitis C virus infection according to NOTCH mutational status.
Among 9 patients with NOTCH2 mutation, 4 had extranodal disease (one spleen, one parotid gland, one meninges, salivary gland and liver, and one muscle).
The 2 cases harboring NOTCH1 mutation had splenic involvement and the trephine biopsy was negative. The single patient harboring a mutation of SPEN had involvement of Waldeyer’s ring and no involvement of bone marrow. One patient with a B-cell clone of MZL type in bone marrow carries mutations of NOTCH2 and TNFAIP3.
NOTCH pathway mutations are restricted to HCV-positive DLBCL
In order to define whether NOTCH pathway mutations are a genetic event specifically associated to DLBCL of the HCV infected patient, NOTCH2 and NOTCH1 mutations were investigated in a cohort of 64 HCV-negative DLBCL. NOTCH2 was rarely mutated in one of 64 (1.6%) of HCV-negative DLBCL (comparison with HCV-positive cases resulted statistically significant; P=0.002) while NOTCH1 was never affected (0 of 64) (comparison with HCV-positive cases; P=0.173) (Figure 3).
Figure 3.
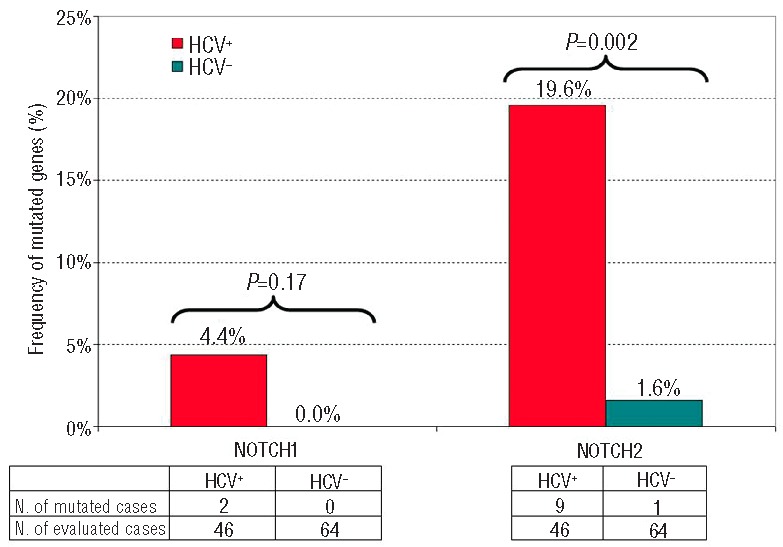
Comparison of NOTCH1 and NOTCH2 mutation diffuse large B-cell lymphoma with or without hepatitis C virus infection.
Impact of NOTCH pathway disruption on outcome of HCV-positive DLBCL
After a median follow up of 3.7 years for the entire series, the 5-year OS was 53% (95%CI: 37%–67%). In univariate analysis on HCV-positive DLBCL, the only factors negatively affecting OS are age, nodal disease with respect to extranodal disease and the mutation of NOTCH pathway (Table 3). More precisely, 5-year OS was 27% (95%CI: 5%–56%) for patients carrying NOTCH pathway mutation and 62% (95%CI: 42%–77%) for patients without these mutations (Figure 4).
Table 3.
Prognostic factors at univariate analyses in 46 patients with diffuse large B-cell lymphoma associated with hepatitis C virus infection.
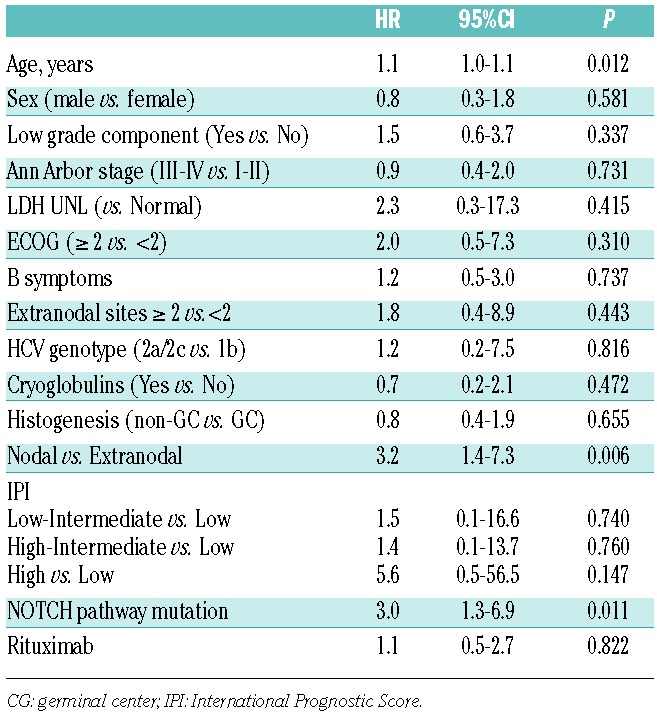
Figure 4.
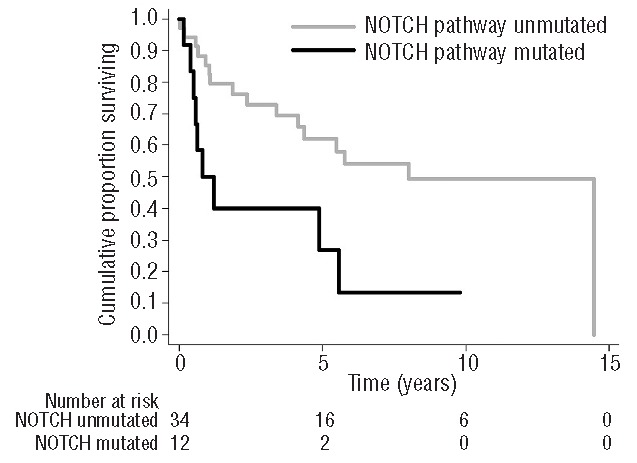
Overall survival of 46 patients with diffuse large B-cell lymphoma associated with hepatitis C virus infection according to mutational status of NOTCH pathway.
In multivariate analysis, adjusting for clinical features significantly associated to OS in univariate analysis, the adverse prognostic impact of NOTCH pathway mutations was confirmed (HR=2.5; 95%CI: 1.0–5.8; P=0.041), while age lost its significance (HR=1.0; 95%CI: 0.9–1.1; P=0.101) and nodal disease became of border-line significance (HR=2.3; 95%CI: 1.0–5.5; P=0.050).
Discussion
The majority of clinical and biological investigations have focused on HCV-positive MZL,19,22 but less attention has been paid to HCV-positive DLBCL.23 In the available series of HCV-positive DLBCL, a predilection for extranodal presentation, including spleen tissue, has been observed.9–11,24
In the present study, we document that: i) NOTCH pathway mutations are a molecular clue associated to approximately 25% HCV-positive cases among DLBCL; and ii) among HCV-positive DLBCL, the molecular deregulation of NOTCH signaling associates with the co-existence of a low-grade component in the diagnostic biopsy and poorer outcome.
A key finding of this study was the identification of alterations on genes of the NOTCH pathway as highly recurrent in HCV-positive DLBCL. In particular, NOTCH2, known as a key regulator of MZ development, was the most frequently mutated gene (20%) This frequency is consistent with the prevalence of NOTCH2 mutations documented in SMZL5,6,18,25,26 and significantly higher than that observed in HCV-negative DLBCL (this study), and more in general, in unselected DLBCL cohorts27,28 (Online Supplementary Table S4). All NOTCH2 mutations observed in HCV-positive DLBCL cause disruption of the protein inhibitory PEST domain and are predicted to activate NOTCH signaling.27
In the general population, most cases of DLBCL arise de novo, but approximately 10% of cases are transformed from low-grade B-cell lymphomas, such as chronic lymphocytic leukemia29 and follicular lymphoma.30 Conversely, HCV-positive DLBCL seem to represent the transformation of a previous low-grade MZL in up to approximately 30% of cases.9 Consistent with this notion, our study documents that: i) 25% of HCV-positive DLBCL harbor a minor area of the diagnostic biopsy composed of small to medium sized monocytoid B cells consistent with an MZL; ii) SMZL-associated NOTCH pathway mutations are significantly enriched in HCV-positive DLBCL harboring both low- and high-grade components; and iii) HCV-positive DLBCL harboring NOTCH pathway mutations are characterized by worse survival in exploratory univariate and multivariate analyses, with an expected survival superimposable on that of transformed DLBCL from other indolent B-cell tumors.31,32
Overall, these data further support the notion that a subgroup of DLBCL arising in subjects carrying HCV infection represent the transformation of a previous, clinically unrecognized indolent MZL.
It is noteworthy that NOTCH signaling plays a role in marginal zone B-cell differentiation in the spleen,33 and that a lymphoid neoplasm such as splenic marginal zone lymphoma is frequently associated to cryoglobulinemia and HCV infection.34
From a clinical standpoint, the identification of NOTCH pathway mutations in HCV-positive DLBCL may have both prognostic and therapeutic implications. The identification of predictive prognostic markers in HCV-positive DLBCL is still a matter of debate. Indeed, at variance with unselected DLBCL, IPI and LDH are not predictive of survival in HCV-positive DLBCL, which is conceivable because, in these patients, LDH can be elevated not only due to lymphoma activity, but also because of the liver disease. Recently, the Fondazione Italiana Linfomi (FIL) proposed a new “HCV Prognostic Score” based on 3 risk factors (performance status, albumin level and HCV-RNA viral load), which is able to identify 3 risk categories with different survival, and may be a useful tool to predict the outcome of HCV-positive DLBCL.35 Considering that only a minority of our patients received rituximab, investigation of larger cohorts of rituximab-treated patients for NOTCH pathway mutations may be of help in integrating this molecular biomarker in a new comprehensive clinicobiological prognostic score useful for the stratification of HCV-positive DLBCL treated in the rituximab era.
From a therapeutical point of view, in DLBCL, we are facing a new era in which investigators are adding new drugs to standard RCHOP-21. Such new agents are designed to target the different specific oncogenic pathways disrupted in DLBCL.36,37 Regarding NOTCH pathway targeting, it has recently been reported that γ-secretase inhibitor PF-03084014 inhibits the constitutive NOTCH activation and induces selective apoptosis in CLL cells carrying NOTCH1 mutations.38
In conclusion, in this study, we document that NOTCH activation pathway mutations are a molecular clue associated to approximately 25% of HCV-positive cases among DLBCL and that among HCV-positive DLBCL, the molecular deregulation of NOTCH signaling associates with the co-existence of a low-grade component in the diagnostic biopsy and poor outcome. Overall, these data are consistent with the notion that at least a fraction of HCV-positive DLBCL represent the transformed phase of an MZL clone.
Acknowledgments
MA is recipient of a fellowship from Anatomic Pathology Section, IRCCS Fondazione Policlinico “San Matteo,” Pavia, Italy; RR is a recipient of a grant (project #580) from the Italian Ministry of Health (5 per Mille) of Fondazione IRCCS Policlinico San Matteo, Pavia, Italy.
We thank dr. Laura Vanelli for providing flow cytometry data.
Footnotes
The online version of this article has a Supplementary Appendix.
Funding
This work was supported by grants from AIRC, Milan, Italy (My First AIRC Grant 2011 #11415 to LA; AIRC Special Program Molecular Clinical Oncology, 5 × 1000, No. 10007 to GG); Fondazione IRCCS Policlinico San Matteo, Pavia, Italy to LA); Ricerca Finalizzata, Ministero della Salute, Rome, Italy (to GG).
Authorship and Disclosures
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.de Sanjose S, Benavente Y, Vajdic CM, et al. Hepatitis C and non-Hodgkin lymphoma among 4784 cases and 6269 controls from the International Lymphoma Epidemiology Consortium. Clin Gastroenterol Hepatol. 2008;6(4):451–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mele A, Pulsoni A, Bianco E, et al. Hepatitis C virus and B-cell non-Hodgkin lymphomas: an Italian multicenter case-control study. Blood. 2003;102(3):996–999. [DOI] [PubMed] [Google Scholar]
- 3.Saadoun D, Suarez F, Lefrere F, et al. Splenic lymphoma with villous lymphocytes, associated with type II cryoglobulinemia and HCV infection: a new entity? Blood. 2005; 105(1):74–76. [DOI] [PubMed] [Google Scholar]
- 4.Hermine O, Lefrere F, Bronowicki JP, et al. Regression of splenic lymphoma with villous lymphocytes after treatment of hepatitis C virus infection. N Engl J Med. 2002;347(2):89–94. [DOI] [PubMed] [Google Scholar]
- 5.Rossi D, Trifonov V, Fangazio M, et al. The coding genome of splenic marginal zone lymphoma: activation of NOTCH2 and other pathways regulating marginal zone development. J Exp Med. 2012; 209(9):1537–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiel MJ, Velusamy T, Betz BL, et al. Whole-genome sequencing identifies recurrent somatic NOTCH2 mutations in splenic marginal zone lymphoma. J Exp Med. 2012;209(9):1553–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruscaggin A, Monti S, Arcaini L, et al. Molecular lesions of signalling pathway genes in clonal B-cell lymphocytosis with marginal zone features. Br J Haematol. 2014;167(5):718–720. [DOI] [PubMed] [Google Scholar]
- 8.Xochelli A, Kalpadakis C, Gardiner A, et al. Clonal B-cell lymphocytosis exhibiting immunophenotypic features consistent with a marginal-zone origin: is this a distinct entity? Blood. 2014;123(8):1199–1206. [DOI] [PubMed] [Google Scholar]
- 9.Besson C, Canioni D, Lepage E, et al. Characteristics and outcome of diffuse large B-cell lymphoma in hepatitis C virus-positive patients in LNH 93 and LNH 98 Groupe d’Etude des Lymphomes de l’Adulte programs. J Clin Oncol. 2006;24(6):953–960. [DOI] [PubMed] [Google Scholar]
- 10.Visco C, Arcaini L, Brusamolino E, et al. Distinctive natural history in hepatitis C virus positive diffuse large B-cell lymphoma: analysis of 156 patients from northern Italy. Ann Oncol. 2006; 17(9):1434–1440. [DOI] [PubMed] [Google Scholar]
- 11.Ennishi D, Maeda Y, Niitsu N, et al. Hepatic toxicity and prognosis in hepatitis C virus-infected patients with diffuse large B-cell lymphoma treated with rituximab-containing chemotherapy regimens: a Japanese multicenter analysis. Blood. 2010;116(24):5119–5125. [DOI] [PubMed] [Google Scholar]
- 12.Swerdlow SHCE, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon: IARC Press, 2008. [Google Scholar]
- 13.Ngo VN, Young RM, Schmitz R, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470(7332):115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rossi D, Deaglio S, Dominguez-Sola D, et al. Alteration of BIRC3 and multiple other NF-kappaB pathway genes in splenic marginal zone lymphoma. Blood. 2011; 118(18):4930–4934. [DOI] [PubMed] [Google Scholar]
- 15.Rossi D, Rasi S, Spina V, et al. Integrated mutational and cytogenetic analysis identifies new prognostic subgroups in chronic lymphocytic leukemia. Blood. 2013;121(8): 1403–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lenz G, Davis RE, Ngo VN, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319(5870): 1676–1679. [DOI] [PubMed] [Google Scholar]
- 17.Davis RE, Ngo VN, Lenz G, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463(7277):88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Compagno M, Lim WK, Grunn A, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459(7247):717–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peveling-Oberhag J, Crisman G, Schmidt A, et al. Dysregulation of global microRNA expression in splenic marginal zone lymphoma and influence of chronic hepatitis C virus infection. Leukemia. 2012;26(7):1654–1662. [DOI] [PubMed] [Google Scholar]
- 20.Zibellini S, Capello D, Forconi F, et al. Stereotyped patterns of B-cell receptor in splenic marginal zone lymphoma. Haematologica. 2010; 95(10):1792–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paulli M, Arcaini L, Lucioni M, et al. Subcutaneous ‘lipoma-like’ B-cell lymphoma associated with HCV infection: a new presentation of primary extranodal marginal zone B-cell lymphoma of MALT. Ann Oncol. 2010;21(6):1189–1195. [DOI] [PubMed] [Google Scholar]
- 22.Mollejo M, Menarguez J, Guisado-Vasco P, et al. Hepatitis C virus-related lymphoproliferative disorders encompass a broader clinical and morphological spectrum than previously recognized: a clinicopathological study. Mod Pathol. 2014;27(2):281–293. [DOI] [PubMed] [Google Scholar]
- 23.Peveling-Oberhag J, Arcaini L, Hansmann ML, Zeuzem S. Hepatitis C-associated B-cell non-Hodgkin lymphomas. Epidemiology, molecular signature and clinical management. J Hepatol. 2013; 59(1):169–177. [DOI] [PubMed] [Google Scholar]
- 24.Ennishi D, Maeda Y, Niitsu N, et al. Hepatic toxicity and prognosis in hepatitis C virus-infected patients with diffuse large B-cell lymphoma treated with rituximab-containing chemotherapy regimens: a Japanese multicenter analysis. Blood. 2010; 116(24):5119–5125. [DOI] [PubMed] [Google Scholar]
- 25.Martinez N, Almaraz C, Vaque JP, et al. Whole-exome sequencing in splenic marginal zone lymphoma reveals mutations in genes involved in marginal zone differentiation. Leukemia. 2014;28(6):1334–1340. [DOI] [PubMed] [Google Scholar]
- 26.Parry M, Rose-Zerilli MJ, Gibson J, et al. Whole exome sequencing identifies novel recurrently mutated genes in patients with splenic marginal zone lymphoma. PloS one. 2013. 8(12):e83244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee SY, Kumano K, Nakazaki K, et al. Gain-of-function mutations and copy number increases of Notch2 in diffuse large B-cell lymphoma. Cancer science. 2009; 100(5):920–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lohr JG, Stojanov P, Lawrence MS, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci USA. 2012; 109(10):3879–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rossi D, Gaidano G. Richter syndrome. Adv Exp Med Biol. 2013;792:173–191. [DOI] [PubMed] [Google Scholar]
- 30.Lossos IS, Gascoyne RD. Transformation of follicular lymphoma. Best Pract Res Clin Haematol. 2011;24(2):147–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer AH, Stroux A, Lerch K, et al. Transformation and additional malignancies are leading risk factors for an adverse course of disease in marginal zone lymphoma. Ann Oncol. 2014;25(1):210–215. [DOI] [PubMed] [Google Scholar]
- 32.Ghesquieres H, Berger F, Felman P, et al. Clinicopathologic characteristics and outcome of diffuse large B-cell lymphomas presenting with an associated low-grade component at diagnosis. J Clin Oncol. 2006;24(33):5234–5241. [DOI] [PubMed] [Google Scholar]
- 33.Lobry C, Oh P, Mansour MR, Look AT, Aifantis I. Notch signaling: switching an oncogene to a tumor suppressor. Blood. 2014;123(16):2451–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suarez F, Lortholary O, Hermine O, Lecuit M. Infection-associated lymphomas derived from marginal zone B cells: a model of antigen-driven lymphoproliferation. Blood. 2006;107(8):3034–3044. [DOI] [PubMed] [Google Scholar]
- 35.Merli M, Visco C, Spina M, et al. Outcome prediction of diffuse large B-cell lymphomas associated with hepatitis C virus infection: a study on behalf of the Fondazione Italiana Linfomi. Haematologica. 2014;99(3):489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruan J, Martin P, Furman RR, et al. Bortezomib plus CHOP-rituximab for previously untreated diffuse large B-cell lymphoma and mantle cell lymphoma. J Clin Oncol. 2011;29(6):690–697. [DOI] [PubMed] [Google Scholar]
- 37.Chiappella A, Tucci A, Castellino A, et al. Lenalidomide plus cyclophosphamide, doxorubicin, vincristine, prednisone and rituximab is safe and effective in untreated, elderly patients with diffuse large B-cell lymphoma: a phase I study by the Fondazione Italiana Linfomi. Haematologica. 2013;98(11):1732–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopez-Guerra M, Xargay-Torrent S, Rosich L, et al. The gamma-secretase inhibitor PF-03084014 combined with fludarabine antagonizes migration, invasion and angiogenesis in NOTCH1-mutated CLL cells. Leukemia. 2015;29(1):96–106. [DOI] [PubMed] [Google Scholar]


