Abstract
Context:
Polycystic ovary syndrome (PCOS) is a common complex genetic disease. It is characterized by hyperandrogenism, gonadotropin secretory changes, polycystic ovarian morphology, and insulin resistance. The etiology of PCOS remains unknown, but modern genetic approaches, such as genome-wide association studies (GWAS), Mendelian randomization, and next-generation sequencing, promise to identify the pathways that are primarily disrupted.
Evidence Acquisition:
The literature on PCOS, including the author's research, is discussed.
Evidence Synthesis:
Recent genetic analyses are reviewed.
Conclusions:
Considerable progress has been made mapping PCOS susceptibility genes. GWAS have implicated gonadotropin secretion and action as important primary defects in disease pathogenesis in European and Han Chinese PCOS cohorts, respectively. European women with the National Institutes of Health and Rotterdam phenotypes as well as those with self-reported PCOS have some gene regions in common, such as chromosome 11p14.1 region containing the FSH B polypeptide (FSHB) gene, suggesting shared genetic susceptibility. Several chromosomal signals are significant in both Han Chinese and European PCOS cohorts, suggesting that the susceptibility genes in these regions are evolutionarily conserved. In addition, GWAS have suggested that DENND1A, epidermal growth factor signaling, and DNA repair pathways play a role in PCOS pathogenesis. Only a small amount of the heritability of PCOS is accounted for by the common susceptibility variants mapped so far. Future studies should clarify the contribution of rare genetic variants and epigenetic factors to the PCOS phenotype. Furthermore, Mendelian randomization can be used to clarify causal relationships, and phenome-wide association studies can provide insight into health risks associated with PCOS susceptibility variants.
It is particularly timely to consider the disorder currently known as polycystic ovary syndrome (PCOS) because 2015 marked the 25th anniversary of the 1990 National Institute of Child Health and Human Development Conference on PCOS where the first diagnostic criteria for the syndrome were established (1). Moreover, the field has witnessed several important developments over the past few years. In December 2012, the expert panel of the National Institutes of Health (NIH) Office for Disease Prevention-sponsored Evidence-based Methodology Workshop on PCOS called for a new name for PCOS that better reflected its metabolic as well as reproductive features (2). Large genome-wide association studies (GWAS) in Han Chinese (3, 4) and in European ancestry (5, 6) PCOS cohorts have mapped PCOS susceptibility loci that promise to provide insight into the biological pathways that are primarily disrupted.
The core reproductive features of PCOS, disordered gonadotropin secretion (7) and increased androgen production (8), were described in the 1950s and 1960s (Figure 1). Insulin resistance was first noted to be associated with PCOS in the 1980s (9), and over the ensuing 30 years, the syndrome has become recognized as a major metabolic disorder (10). There is an extensive body of literature on the pathophysiology of PCOS that has been ably reviewed recently (10–12). However, the etiology or etiologies of PCOS remain unknown. There is experimental evidence that androgen administration (13), increasing GnRH or LH release (14, 15), or inducing insulin resistance (16, 17) can produce phenocopies of the syndrome (Figure 2). Thus, it is possible that primary derangements in androgen production, gonadotropin secretion, or insulin action could cause PCOS.
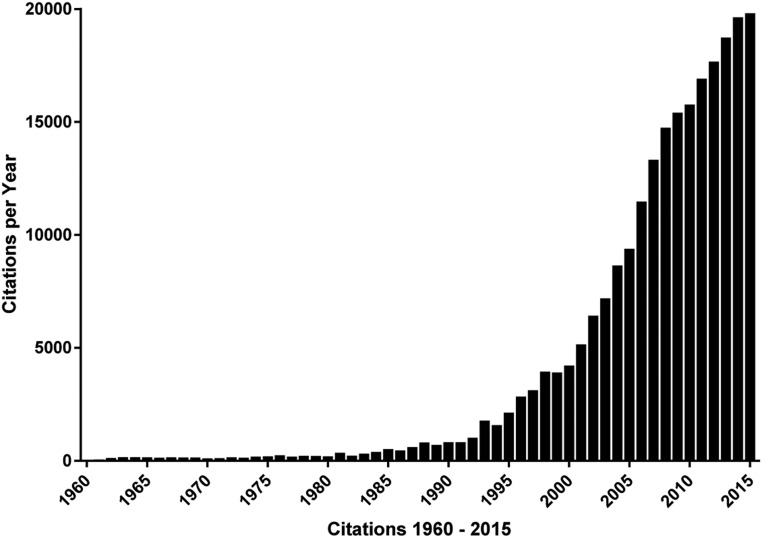
Figure 1.
Number of annual citations 1960–2015 in Web of Science to original research articles on Stein-Leventhal syndrome or PCOS (including name variants polycystic ovary/ovarian syndrome/disease or multicystic ovaries). There were very few citations before 1960, despite the fact that the disorder was first reported by Stein and Leventhal in 1935 (121). The number of citations was relatively stable at approximately 100 annually until 1980 when it began to in increase steadily.
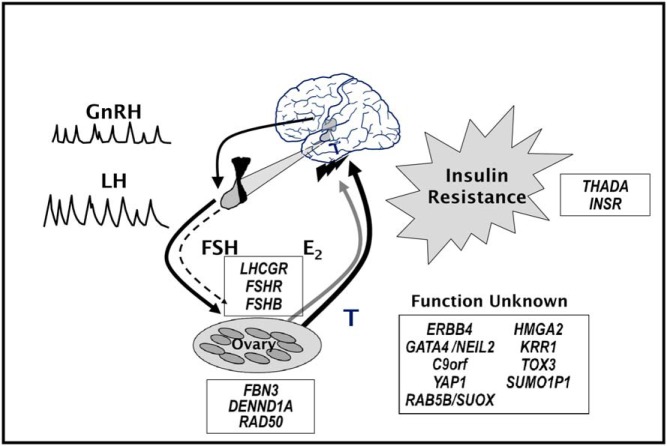
Figure 2.
The pathophysiology of PCOS and replicated genetic loci. PCOS is characterized by increased pulsatile GnRH release that selectively increases LH secretion while relatively suppressing FSH secretion. LH stimulates ovarian theca cell T production. T is not completely aromatized to estradiol (E2) because of insufficient FSH-stimulated granulosa cell aromatase activity. PCOS theca cells produce increased androgens basally as well as in response to LH. Circulating T produces masculinizing symptoms of PCOS. In addition, T feeds back on the hypothalamic-pituitary axis to decrease sensitivity to effects of E2 and progesterone (not noted in figure) to slow the GnRH pulse generator. These abnormalities are self-perpetuating, the so-called vicious cycle of PCOS. Insulin resistance is commonly associated with PCOS, and insulin amplifies the reproductive abnormalities. Seventeen genetic loci have been reproducibly mapped for PCOS, one by family-based association testing, fibrillin-3 (FBN3), and 16 by GWAS. LHCGR and FSHR modulate gonadotropin action, and FSHB modulates FSH secretion. FBN3 and DENND1A may regulate ovarian steroidogenesis. RAD50 may be involved in ovarian aging. THADA and INSR are T2D susceptibility genes. The function of the remaining genes in the pathogenesis of PCOS remains unknown. Used with permission of Andrea Dunaif.
Genetic Approaches to PCOS (Figure 2)
It has been proposed since the 1960s (18) that PCOS (then known as Stein-Leventhal syndrome) was heritable. In 1976, Kahn et al (19) described a new disorder in girls with virilization, acanthosis nigricans, polycystic ovaries, and extreme insulin resistance due to decreased insulin receptor number, which they designated the type A syndrome of insulin resistance. Subsequent studies (20) found that many affected girls had insulin receptor gene (INSR) mutations decreasing receptor number or function. The phenotypic similarities between type A syndrome and PCOS (21) suggested that PCOS might also be due to INSR mutations, although none were identified with the relatively primitive available analytic methods (22). Nevertheless, research into a possible genetic contribution to PCOS continued.
PCOS demonstrates non-Mendelian familial aggregation consistent with a complex genetic disease resulting from the interaction between susceptibility genes and environmental factors (23). Twin studies (24) have shown heritability of 79% consistent with a major influence of genetic factors in PCOS. Furthermore, investigating phenotypes in first-degree relatives has provided considerable insights into the features of the syndrome that are genetically determined. Hyperandrogenemia is a consistent reproductive phenotype (25–28), and insulin resistance (26, 28–32) is a consistent metabolic phenotype in male as well as female first-degree relatives of affected women. However, although these phenotypes are present before puberty, studies in the infants and children of affected women (33–36) have thus far failed to identify the initiating hormonal change.
As with other complex diseases (37), such as type 2 diabetes (T2D) and inflammatory bowel disease, genetic analyses hold considerable promise for elucidating the etiology of PCOS. More than 250 case-control studies of approximately 160 candidate genes implicated in androgen biosynthesis and action, gonadotropin secretion and action, folliculogenesis, insulin action and T2D, and energy homeostasis have been examined for association with PCOS (23, 38). These studies have a number of limitations. First, they are based on the assumption that the cases and controls are perfectly matched, except for the phenotype of interest. However, this assumption is rarely true due to population stratification because of the often subtle, racial and ethnic differences between cases and controls (39). Furthermore, failure to match for other phenotypes associated with PCOS, such as obesity, insulin resistance, or dysglycemia, could lead to the identification of susceptibility variants associated with these phenotypes rather than with PCOS (40, 41). Second, sample sizes in many studies have been small, resulting in a lack of adequate statistical power (23). This problem is further exacerbated by the frequent failure to adequately adjust for multiple testing, which increases the likelihood of false-positive results. Third, findings have rarely been replicated in additional case-control cohorts. Fourth, a limited number of single nucleotide polymorphisms (SNPs), usually selected based on their association with other phenotypes (42, 43) rather than SNPs spanning the entire gene, have been examined.
An alternative approach to mapping susceptibility genes for complex traits are family-based association tests (FBATs), such as the transmission disequilibrium test (44). These FBATs test for association in the presence of linkage (44). Because FBATs use within-family comparisons, they are not affected by population stratification (44). We found strong evidence by transmission disequilibrium test that an allele of a dinucleotide repeat D19S884 on chromosome 19p13.2 was linked and associated with the PCOS reproductive phenotype (45). These findings were replicated in an independent sample of PCOS families (46) and in a case-control study (47). Thus, this marker was the first replicated susceptibility locus for PCOS. D19S884 is a microsatellite marker that had been selected for mapping the INSR, but it mapped to intron 55 of the fibrillin-3 (FBN3) gene located approximately 1 Mb centromeric to the INSR on chromosome 19p13.2 (46, 48, 49). The allele was also associated with evidence for insulin resistance in women with PCOS and for pancreatic β-cell dysfunction in brothers, suggesting a sex difference in the associated metabolic phenotypes (49).
The 2005 publication of the human haplotype map (HapMap) (50) that cataloged common genetic variants enabled a revolution in case-control study design by permitting the mapping of the entire genome. These studies are known as “genome-wide association studies” (GWAS). Gene regions containing variations affecting a phenotype can be identified through the indirect relationship between the contributing variation and nearby variation that is in linkage disequilibrium with the contributing variation (51). The HapMap contains common haplotypes and SNPs that identify these haplotypes, so-called tag SNPs (50). GWAS map the genome by genotyping these tag SNPs to localize gene regions containing putative susceptibility genes for complex traits, such as obesity and T2D (52, 53). Importantly, GWAS permit an unbiased interrogation of the entire genome for novel disease susceptibility loci and are, unlike candidate gene approaches, hypothesis generating (39). Population stratification can be controlled for in GWAS by adjusting for axes of ancestry-specific variation (39, 54).
The first PCOS GWAS (3), which was published online in December 2010, was in Han Chinese PCOS cases and control women. The PCOS cases were diagnosed using the Rotterdam criteria. A second analysis published in 2012 (4) increased the original Han Chinese sample to a total of 8226 cases and 7578 controls. There was strong evidence for association with meta-analysis P values surpassing the proposed threshold for genome-wide significance of 5 × 10−8 (55, 56) between PCOS and 11 chromosomal loci (Table 1). Two loci, both on chromosome 2p16.3, contained the receptors for LH/human chorionic gonadotropin (LHCGR) and FSH (FSHR). Given the central role of LH in the pathogenesis of ovarian androgen excess in PCOS (10), LHCGR is a high-priority PCOS candidate gene (48, 57). The FSHR is another highly plausible candidate gene for PCOS, given its characteristic abnormalities of folliculogenesis (10, 58).
Table 1.
Sixteen PCOS GWAS Susceptibility Loci
| Chromosome | Han Chinese; Chen, 2011 (3), and Shi, 2012 (4) | European 1; Hayes, 2015 (5) | European 2; Day, 2015 (6) |
|---|---|---|---|
| 2p16.3 | LHCGR | ||
| 2p16.3 | FSHR | ||
| 2p21 | THADA | THADA | |
| 2q34 | ERBB4 | ||
| 5q13.1 | RAD50 | ||
| 8p32.1 | GATA4/NEIL2 | ||
| 9q22.32 | C9orf3 | C9orf3 | |
| 9q33.3 | DENND1A | ||
| 11p14.1 | FSHB | FSHB | |
| 11q22.1 | YAP1 | YAP1 | |
| 12q13.2 | RAB5B/SUOX | ||
| 12q14.3 | HMGA2 | ||
| 12q21.2 | KRR1 | ||
| 16q12.1 | TOX3 | ||
| 19q13.3 | INSR | ||
| 20q13.2 | SUMO1P1 |
A locus on chromosome 19q13.3 contained the INSR, a high-priority candidate gene for the PCOS metabolic phenotype. Another potential candidate for the PCOS metabolic phenotype was the strongest signal on chromosome 2p21, THADA, a gene originally identified in thyroid adenomas (3), but that was associated with T2D in a European GWAS (59). This association was not replicated in Chinese T2D, but the associated SNP is a rare variant in Asians (3). The region on chromosome 9q33.3 associated with PCOS was located within DENND1A, which encodes a domain differentially expressed in normal and neoplastic cells (DENN) that can bind to and negatively regulate endoplasmic reticulum aminopeptidase-1 (3). Recent studies have suggested that the DENND1A protein is involved in the PCOS theca cell phenotype (60). Several of the Han Chinese GWAS loci (DENND1A, THADA, YAP1, LHCGR, and FSHR) have been replicated in PCOS cohorts of European ancestry diagnosed by NIH (61–63) or Rotterdam criteria (64).
Another Asian ancestry GWAS in Korean women with PCOS diagnosed by Rotterdam criteria in 1249 PCOS cases and 1778 control women (65, 66) found no PCOS-associated signals reaching genome-wide significance levels of ≤ 5 × 10−8 (55, 56). The study was constrained not only by a relatively small sample size but also by the fact that 10.5% of the control women had hyperandrogenemia. A pathway analysis using these GWAS data from the discovery cohort of 1000 PCOS cases and 1000 controls (67) was limited by the lack of significant GWAS signals and the presence of hyperandrogenemia in the controls.
The first GWAS in European PCOS was published in August 2015 in 3000 PCOS cases diagnosed by NIH criteria and 5330 controls (5). Three loci reached genome-wide significance in the case-control analysis. Two loci were novel, chromosome 8p32.1 in the region of GATA4 and NEIL2 and chromosome 11p14.1 in the region of the FSH B polypeptide (FSHB) gene. One locus was found previously in Han Chinese PCOS, chromosome 9q22.32 in the region of c9orf3. Adjusting for body mass index (BMI) had little impact on the results, suggesting that the findings were independent of obesity. This study also included the first genome-wide analysis of quantitative traits; the same chromosome 11p14.1 SNP, rs11031006, in the region of FSHB was associated with LH levels at genome-wide significance.
The second European PCOS GWAS was published in September 2015 in a discovery cohort of 5184 self-reported cases of PCOS and 82 759 controls with replication in approximately 2000 cases diagnosed by NIH or Rotterdam criteria and approximately 100 000 controls (6). There were three novel loci reaching genome-wide significance, chromosome 2q34 in the region of ERBB4, chromosome 5q13.1 in the region of RAD50, and chromosome 12q21.2 in the region of KRR1. Importantly, the signal from the first European PCOS GWAS in the region of FSHB was replicated at genome-wide significance levels, as were the Han Chinese signals in the region of THADA and YAP1.
Several additional relevant genetic analyses were performed in this cohort (6). All GWAS SNPs in known biological pathways were examined, and enrichment was found in an ATP-binding cassette transporter pathway with PCOS-associated variants, including RAD50. Mendelian randomization uses genetic variants as proxies for putatively causal variables (68). For example, variants in the SHBG gene associated with circulating SHBG levels are also associated with T2D risk, suggesting that SHBG may play a causal role in T2D risk (69). In contrast, C-reactive protein gene variants associated with circulating levels are not associated with cardiovascular events, arguing against a causal role for C-reactive protein in cardiovascular disease (70). Mendelian randomization suggested causal associations with increasing BMI, more severe insulin resistance, lower SHBG, and later menopause in European PCOS (6). The PCOS susceptibility alleles in the second European PCOS GWAS were associated with higher anti-Müllerian hormone levels in girls, suggesting that these variants may be predictive of PCOS.
The European PCOS GWAS findings suggest that variation in FSHB plays an important role in the etiology of PCOS in this ancestry group. Furthermore, the FSHB association is likely mediated by LH because, in the quantitative trait analysis (5), adjusting for LH levels in the regression model between the FSHB region SNP rs11031006 and PCOS abolished the association. Taken together with the Chinese GWAS findings of associations with the genes encoding gonadotropin receptors (LHCGR and FSHR), these GWAS implicate genes regulating gonadotropin secretion (FSHB) and action (LHCGR and FSHR) in the etiology of PCOS. Thus, PCOS GWAS have provided important biological insights, much as the GWAS in T2D have implicated genes involved in insulin secretion (53) and those in obesity have implicated genes involved in the central nervous system regulation of food intake (52). Furthermore, finding the same signals in the region of THADA, YAP1, and c9orf3 in Chinese and European PCOS populations suggests that PCOS is an ancient trait that was present before humans migrated out of Africa (10, 71–73).
The potential role of genes at the other Han Chinese and European GWAS loci (Table 1) in the pathogenesis of PCOS remains unknown. It was suggested that the signal in the region of ERBB4 in the second European GWAS implicated epidermal growth factor receptors in the pathogenesis of PCOS because signals at two other genes in this family, ERBB3 and ERBB2, approached genome-wide significance (6). RAD50 is involved in DNA repair; other DNA repair genes have been associated with age at menopause (74). However, caution must be exercised in such speculations about putative disease mechanisms. Furthermore, even signals in the region of high-priority candidate genes may in fact reflect other genes or elements that regulate them. Indeed, because most GWAS loci have been intronic or intergenic, it has been exceptionally difficult to determine their biological relevance (75), although there has been some recent progress (76).
It is of considerable interest with respect to functionality of PCOS GWAS susceptibility variants that DENND1A protein and mRNA levels are increased in PCOS theca cells (60). Furthermore, overexpression of a DENND1A mRNA splice variant in normal theca cells results in a PCOS phenotype (77). However, the DENND1A GWAS signal is intronic and does not affect DENND1A expression or splicing. No functional mutations were found in whole exome sequencing of DENND1A (60). Therefore, the mechanism by which genetic variation in the region of DENND1A contributes to PCOS remains unknown. Nevertheless, the discovery of a putative role for DENND1A in the etiology of PCOS is an excellent example of the novel biological insights that can be provided by GWAS (78).
Most recently, preliminary results of a meta-analysis from multiple GWAS in European ancestry PCOS (79), the PCOS Genetics Consortium, were presented at the 2015 meeting of the American Society of Human Genetics. There were three novel loci that reached genome-wide significance. The signals in the region of GATA4/NEIL2 and FSHB were replicated at genome-wide significance levels. There was adequate power to stratify the population by diagnostic criteria, and there was no significant difference in the signals in the NIH compared to the Rotterdam phenotypes or after adjustment for BMI. These findings, taken together with the consistency of the FSHB signal in cohorts diagnosed by NIH (5) or Rotterdam criteria (6) or by self-report (6), suggest shared genetic susceptibility to the various PCOS phenotypes. The findings challenge the alleged heterogeneity of PCOS. Indeed, the observation that there can be several phenotypes in affected sisters from the same family (25, 80) suggests that phenotypic heterogeneity could be accounted for by variable expression of shared genes.
The Future
Genetic architecture
We are using the European PCOS GWAS data to investigate whether the genetic architecture of PCOS metabolic phenotypes differs from that for these phenotypes in non-PCOS populations (40, 81). Neither of the major genes conferring increased risk for T2D, TCF7L2 (53), or obesity, FTO (52), has been significantly associated with PCOS in any of the GWAS studies performed to date (4–6), suggesting differences in genetic susceptibility for these metabolic traits in PCOS.
Phenome-wide association studies
We are also assessing whether any of our European PCOS-susceptibility SNPs are associated with other phenotypes, an analysis known as a phenome-wide association study, using electronic medical record codes linked to GWAS data (82). This analysis has replicated 66% of other GWAS associations tested in European ancestry individuals in the Electronic Medical Records and Genomics (eMERGE) network in which Feinberg School of Medicine is a member (83). It should allow us to interrogate whether other health risks, eg, cardiovascular disease and certain cancers, are associated with PCOS.
Rare genetic variants
The heritability of PCOS explained by the GWAS variants we have mapped is less than 5%, a substantial deficit from its estimated approximately 80% heritability (24). It is now clear that many of the replicated GWAS loci for complex traits confer very small increases in disease risk and that the loci discovered thus far, taken together, do not account for the observed heritability of T2D, obesity, and other complex traits (84–86). This so-called “missing” heritability (84) may reflect the fact that rare variants have a greater contribution to complex traits than previously anticipated (87, 88). GWAS are designed to detect common allelic variants with minor allele frequencies of 5% or greater (84, 87, 89). Common variants would be expected to have a modest effect on phenotype because they were not subjected to strong selective pressure (39). Variants with lower frequency and larger effect size that are not detected by GWAS may account for the deficit in heritability in complex traits (84). Recent estimates based on large-scale deep sequencing have predicted that each person carries rare single nucleotide variants that are predicted to affect protein function of approximately 300 genes per genome (90)! This large number of rare variants is postulated to be due to rapid human population expansion with a lack of selection pressure on such variants (91).
Support for this hypothesis comes from finding rare genetic variants with large effects on high-density lipoprotein (88), adiponectin (92), and triglyceride (93) levels in the general population. Perhaps the most compelling example of the clinical relevance of rare variants was the discovery in African Americans of inactivating mutations in the gene (PCSK9) encoding proprotein convertase subtilisin/kexin type 9 protein (PCSK9) that lower low-density lipoprotein (LDL) levels (94). PCSK9 plays a key role in LDL receptor degradation (95). Hepatic LDL receptor-mediated clearance modulates circulating LDL-cholesterol levels (95). Inactivating PSCK9 with monoclonal antibodies reduces LDL receptor degradation, resulting in decreased circulating cholesterol levels (95). This discovery has led to the development of a new class of cholesterol-lowering drugs, which have been approved recently by the U.S. Food and Drug Administration (96).
It is now becoming feasible to reliably identify rare allelic variants with the use of next-generation sequencing technologies (84, 87, 90). To detect less frequent or rare (but not private, ie, limited to one family) allelic variants, it is important to enrich the population for these variants (84, 87). This can be accomplished by investigating families with multiple affected individuals (97, 98), as was done to identify the rare variants in the adiponectin gene (92), and by investigating individuals with extreme phenotypes (99, 100).
We are currently applying whole genome sequencing in multiplex PCOS families coupled with robust bioinformatic analyses (101). Exonic low frequency/rare variants are anticipated to have much greater effects on phenotype (84). Indeed, the bioinformatic analyses of genome sequence data classify nonsynonymous protein-altering variants using algorithms that predict their likelihood to be deleterious (101–106). Therefore, traditional molecular biological approaches can be utilized to assess their functional significance (93, 107, 108).
We also plan to sequence the genome-wide significant PCOS GWAS loci to identify common as well as rare variants contributing to disease risk in these genomic regions. However, most GWAS loci are not exonic, making their functional impact more difficult to interrogate (109, 110). Genome-wide approaches to investigate these noncoding regions for regulatory elements have recently become available (110).
Epigenetics
It remains possible that some or all of the deficit in heritability in PCOS is accounted for by epigenetic factors. This hypothesis is particularly plausible because phenocopies of PCOS can be produced by androgen exposure at critical windows of development (10, 111). There have been a limited number of studies in PCOS suggesting that epigenetic changes are present (112–114). Furthermore, epigenetic mechanisms could contribute to the parent-of-origin effects on glucose homeostasis that we have found in PCOS (115). Epigenetic studies are challenging because of the difficulty of obtaining tissues and cells, eg, theca cells, relevant to the pathogenesis of PCOS. Nevertheless, such studies as well as those in animal models (111, 116, 117) will be critical for understanding the etiology of PCOS.
Summary
Modern genetic approaches have enabled substantial progress on elucidating the etiology of PCOS and should continue to do so. GWAS have implicated gonadotropin secretion and action as a key causal pathway. They have led to the discovery of an important role for DENNDA1 in the theca cell PCOS phenotype and suggested other pathways, such as epidermal growth factor signaling and DNA repair, of potential biological relevance. Importantly, GWAS suggest shared genetic susceptibility among the various PCOS phenotypes and across cohorts of different ancestry. These findings have major implications for diagnostic criteria, suggesting that self-report, NIH, or Rotterdam identify genetically similar phenotypes. Furthermore, the findings have evolutionary implications suggesting that the susceptibility loci shared between the Han Chinese and European PCOS are conserved because the ancestral Eurasian population migrated out of Africa approximately 100 000 years ago and these populations diverged > 40 000 years ago (71, 118).
Next-generation sequencing promises to identify rare genetic variants contributing to PCOS as well as to map the genetic variants contributing to the significant signals at the various PCOS GWAS loci. Other genetic analyses should also substantially advance the field. The PCOS literature is plagued by conclusions based on correlative findings. Mendelian randomization can be applied in many of these situations to determine whether such associations are causal, eg, increased inflammatory cytokines and PCOS. Phenome-wide association studies may enable identification of long-term health risks by assessing what diseases are associated with PCOS-susceptibility variants.
Finally, it may be possible to use PCOS-susceptibly variants to create disease-predictive genetic risk scores. This possibility is supported by the association of higher anti-Müllerian hormone levels with GWAS PCOS-susceptibility loci in a cohort of adolescent girls (6). The ability to predict PCOS would be extremely clinically relevant because there are already well-validated interventions, such as lifestyle modification and metformin, to prevent T2D in high-risk individuals (119). Furthermore, there are intriguing but limited data (120) to suggest that metformin begun in prepubertal girls at risk for PCOS, low birth weight girls with precocious pubarche, ameliorates the development of both metabolic and reproductive features of PCOS after menarche.
So much of the literature on PCOS contains variations on the statement that it is a complex and heterogeneous disorder that this description has become the “It was a dark and stormy night” of the field. It has always been arguable whether PCOS is any more complex or heterogeneous than other common disorders of unknown etiology, such as metabolic syndrome or T2D. Indeed, the core reproductive features of the PCOS are usually present: chronic anovulation, hyperandrogenism, and polycystic ovarian morphology. Genetic analyses should end the bickering over diagnostic criteria, and the emerging data suggest a remarkable degree of genetic homogeneity not only across diagnostic criteria but also across ethnicities. Clearly, there are many as yet to be discovered factors contributing to PCOS because the susceptibility loci identified so far contribute a relatively small amount of disease risk. Nevertheless, I believe we do the field a great disservice by emphasizing the so-called heterogeneity of PCOS rather than focusing on its unifying features because this creates the false impression that PCOS is somehow unknowable, which is a major barrier to broader engagement in the field. Current genetic analyses have markedly increased our knowledge of PCOS, and the future promises to make it much more “knowable.”
Acknowledgments
This research was supported by Grant P50 HD044405 (to A.D.) from the Eunice Kennedy Shriver National Institute of Child Health and Development.
Disclosure Summary: The author reports no potential conflicts of interest relevant to this article.
Footnotes
- BMI
- body mass index
- FBAT
- family-based association test
- GWAS
- genome-wide association studies
- LDL
- low-density lipoprotein
- PCOS
- polycystic ovary syndrome
- PCSK9
- proprotein convertase subtilisin/kexin type 9 protein
- SNP
- single nucleotide polymorphism
- T2D
- type 2 diabetes.
References
- 1. Dunaif A, Givens JR, Haseltine FP, Merriam GR. Polycystic Ovary Syndrome. Cambridge, MA: Blackwell Scientific Publications, Inc; 1992. [Google Scholar]
- 2. Dunaif A, Fauser BC. Renaming PCOS–a two-state solution. J Clin Endocrinol Metab. 2013;98:4325–4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen ZJ, Zhao H, He L, et al. Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Nat Genet. 2011;43:55–59. [DOI] [PubMed] [Google Scholar]
- 4. Shi Y, Zhao H, Shi Y, et al. Genome-wide association study identifies eight new risk loci for polycystic ovary syndrome. Nat Genet. 2012;44:1020–1025. [DOI] [PubMed] [Google Scholar]
- 5. Hayes MG, Urbanek M, Ehrmann DA, et al. Genome-wide association of polycystic ovary syndrome implicates alterations in gonadotropin secretion in European ancestry populations. Nat Commun. 2015;6:7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Day FR, Hinds DA, Tung JY, et al. Causal mechanisms and balancing selection inferred from genetic associations with polycystic ovary syndrome. Nat Commun. 2015;6:8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Keettel WC, Bradbury JT, Stoddard FJ. Observations on the polycystic ovary syndrome. Am J Obstet Gynecol. 1957;73:954–962; discussion, 962–955. [PubMed] [Google Scholar]
- 8. Lanthier A. Urinary 17-ketosteroids in the syndrome of polycystic ovaries and hyperthecosis. J Clin Endocrinol Metab. 1960;20:1587–1600. [DOI] [PubMed] [Google Scholar]
- 9. Burghen GA, Givens JR, Kitabchi AE. Correlation of hyperandrogenism with hyperinsulinism in polycystic ovarian disease. J Clin Endocrinol Metab. 1980;50:113–116. [DOI] [PubMed] [Google Scholar]
- 10. Diamanti-Kandarakis E, Dunaif A. Insulin resistance and the polycystic ovary syndrome revisited: an update on mechanisms and implications. Endocr Rev. 2012;33:981–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Legro RS, Arslanian SA, Ehrmann DA, et al. Diagnosis and treatment of polycystic ovary syndrome: an Endocrine Society Clinical Practice Guideline. J Clin Endocrinol Metab. 2013;98:4565–4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Conway G, Dewailly D, Diamanti-Kandarakis E, et al. The polycystic ovary syndrome: a position statement from the European Society of Endocrinology. Eur J Endocrinol. 2014;171:P1–P29. [DOI] [PubMed] [Google Scholar]
- 13. Dumesic DA, Goodarzi MO, Chazenbalk GD, Abbott DH. Intrauterine environment and polycystic ovary syndrome. Semin Reprod Med. 2014;32:159–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Spratt DI, Finkelstein JS, Butler JP, Badger TM, Crowley WF., Jr Effects of increasing the frequency of low doses of gonadotropin-releasing hormone (GnRH) on gonadotropin secretion in GnRH-deficient men. J Clin Endocrinol Metab. 1987;64:1179–1186. [DOI] [PubMed] [Google Scholar]
- 15. Risma KA, Clay CM, Nett TM, Wagner T, Yun J, Nilson JH. Targeted overexpression of luteinizing hormone in transgenic mice leads to infertility, polycystic ovaries, and ovarian tumors. Proc Natl Acad Sci USA. 1995;92:1322–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brüning JC, Gautam D, Burks DJ, et al. Role of brain insulin receptor in control of body weight and reproduction. Science. 2000;289:2122–2125. [DOI] [PubMed] [Google Scholar]
- 17. Wu S, Divall S, Nwaopara A, et al. Obesity-induced infertility and hyperandrogenism are corrected by deletion of the insulin receptor in the ovarian theca cell. Diabetes. 2014;63:1270–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cooper HE, Spellacy WN, Prem KA, Cohen WD. Hereditary factors in the Stein-Leventhal syndrome. Am J Obstet Gynecol. 1968;100:371–387. [DOI] [PubMed] [Google Scholar]
- 19. Kahn CR, Flier JS, Bar RS, et al. The syndromes of insulin resistance and acanthosis nigricans. Insulin-receptor disorders in man. N Engl J Med. 1976;294:739–745. [DOI] [PubMed] [Google Scholar]
- 20. Taylor SI, Cama A, Accili D, et al. Mutations in the insulin receptor gene. Endocr Rev. 1992;13:566–595. [DOI] [PubMed] [Google Scholar]
- 21. Dunaif A, Hoffman AR, Scully RE, et al. Clinical, biochemical, and ovarian morphologic features in women with acanthosis nigricans and masculinization. Obstet Gynecol. 1985;66:545–552. [PubMed] [Google Scholar]
- 22. Sorbara LR, Tang Z, Cama A, et al. Absence of insulin receptor gene mutations in three insulin-resistant women with the polycystic ovary syndrome. Metabolism. 1994;43:1568–1574. [DOI] [PubMed] [Google Scholar]
- 23. Kosova G, Urbanek M. Genetics of the polycystic ovary syndrome. Mol Cell Endocrinol. 2013;373:29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vink JM, Sadrzadeh S, Lambalk CB, Boomsma DI. Heritability of polycystic ovary syndrome in a Dutch twin-family study. J Clin Endocrinol Metab. 2006;91:2100–2104. [DOI] [PubMed] [Google Scholar]
- 25. Legro RS, Driscoll D, Strauss JF, 3rd, Fox J, Dunaif A. Evidence for a genetic basis for hyperandrogenemia in polycystic ovary syndrome. Proc Natl Acad Sci USA. 1998;95:14956–14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Legro RS, Kunselman AR, Demers L, Wang SC, Bentley-Lewis R, Dunaif A. Elevated dehydroepiandrosterone sulfate levels as the reproductive phenotype in the brothers of women with polycystic ovary syndrome. J Clin Endocrinol Metab. 2002;87:2134–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kashar-Miller M, Azziz R. Heritability and the risk of developing androgen excess. J Steroid Biochem Mol Biol. 1999;69:261–268. [DOI] [PubMed] [Google Scholar]
- 28. Yildiz BO, Yarali H, Oguz H, Bayraktar M. Glucose intolerance, insulin resistance, and hyperandrogenemia in first degree relatives of women with polycystic ovary syndrome. J Clin Endocrinol Metab. 2003;88:2031–2036. [DOI] [PubMed] [Google Scholar]
- 29. Sam S, Legro RS, Essah PA, Apridonidze T, Dunaif A. Evidence for metabolic and reproductive phenotypes in mothers of women with polycystic ovary syndrome. Proc Natl Acad Sci USA. 2006;103:7030–7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sam S, Sung YA, Legro RS, Dunaif A. Evidence for pancreatic β-cell dysfunction in brothers of women with polycystic ovary syndrome. Metabolism. 2008;57:84–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Coviello AD, Sam S, Legro RS, Dunaif A. High prevalence of metabolic syndrome in first-degree male relatives of women with polycystic ovary syndrome is related to high rates of obesity. J Clin Endocrinol Metab. 2009;94:4361–4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Baillargeon JP, Carpentier AC. Brothers of women with polycystic ovary syndrome are characterised by impaired glucose tolerance, reduced insulin sensitivity and related metabolic defects. Diabetologia. 2007;50:2424–2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sir-Petermann T, Angel B, Maliqueo M, Carvajal F, Santos JL, Pérez-Bravo F. Prevalence of type II diabetes mellitus and insulin resistance in parents of women with polycystic ovary syndrome. Diabetologia. 2002;45:959–964. [DOI] [PubMed] [Google Scholar]
- 34. Anderson H, Fogel N, Grebe SK, Singh RJ, Taylor RL, Dunaif A. Infants of women with polycystic ovary syndrome have lower cord blood androstenedione and estradiol levels. J Clin Endocrinol Metab. 2010;95:2180–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sir-Petermann T, Codner E, Pérez V, et al. Metabolic and reproductive features before and during puberty in daughters of women with polycystic ovary syndrome. J Clin Endocrinol Metab. 2009;94:1923–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Torchen LC, Fogel NR, Brickman WJ, Paparodis R, Dunaif A. Persistent apparent pancreatic β-cell defects in premenarchal PCOS relatives. J Clin Endocrinol Metab. 2014;99:3855–3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265:2037–2048. [DOI] [PubMed] [Google Scholar]
- 38. Welt CK, Duran JM. Genetics of polycystic ovary syndrome. Semin Reprod Med. 2014;32:177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. [DOI] [PubMed] [Google Scholar]
- 40. Thorleifsson G, Walters GB, Gudbjartsson DF, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. [DOI] [PubMed] [Google Scholar]
- 41. Zeggini E, Weedon MN, Lindgren CM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ewens KG, Jones MR, Ankener W, et al. Type 2 diabetes susceptibility single-nucleotide polymorphisms are not associated with polycystic ovary syndrome. Fertil Steril. 2011;95:2538–2541.e1–e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Barber TM, Bennett AJ, Groves CJ, et al. Association of variants in the fat mass and obesity associated (FTO) gene with polycystic ovary syndrome. Diabetologia. 2008;51:1153–1158. [DOI] [PubMed] [Google Scholar]
- 44. Spielman RS, McGinnis RE, Ewens WJ. The transmission/disequilibrium test detects cosegregation and linkage. Am J Hum Genet. 1994;54:559–560; author reply 560–563. [PMC free article] [PubMed] [Google Scholar]
- 45. Urbanek M, Woodroffe A, Ewens KG, et al. Candidate gene region for polycystic ovary syndrome on chromosome 19p13.2. J Clin Endocrinol Metab. 2005;90:6623–6629. [DOI] [PubMed] [Google Scholar]
- 46. Stewart DR, Dombroski BA, Urbanek M, et al. Fine mapping of genetic susceptibility to polycystic ovary syndrome on chromosome 19p13.2 and tests for regulatory activity. J Clin Endocrinol Metab. 2006;91:4112–4117. [DOI] [PubMed] [Google Scholar]
- 47. Tucci S, Futterweit W, Concepcion ES, et al. Evidence for association of polycystic ovary syndrome in Caucasian women with a marker at the insulin receptor gene locus. J Clin Endocrinol Metab. 2001;86:446–449. [DOI] [PubMed] [Google Scholar]
- 48. Urbanek M, Legro RS, Driscoll DA, et al. Thirty-seven candidate genes for polycystic ovary syndrome: strongest evidence for linkage is with follistatin. Proc Natl Acad Sci USA. 1999;96:8573–8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Urbanek M, Sam S, Legro RS, Dunaif A. Identification of a polycystic ovary syndrome susceptibility variant in fibrillin-3 and association with a metabolic phenotype. J Clin Endocrinol Metab. 2007;92:4191–4198. [DOI] [PubMed] [Google Scholar]
- 50. International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang WY, Barratt BJ, Clayton DG, Todd JA. Genome-wide association studies: theoretical and practical concerns. Nat Rev Genet. 2005;6:109–118. [DOI] [PubMed] [Google Scholar]
- 52. Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Saxena R, Elbers CC, Guo Y, et al. Large-scale gene-centric meta-analysis across 39 studies identifies type 2 diabetes loci. Am J Hum Genet. 2012;90:410–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Price AL, Butler J, Patterson N, et al. Discerning the ancestry of European Americans in genetic association studies. PLoS Genet. 2008;4:e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hoggart CJ, Clark TG, De Iorio M, Whittaker JC, Balding DJ. Genome-wide significance for dense SNP and resequencing data. Genet Epidemiol. 2008;32:179–185. [DOI] [PubMed] [Google Scholar]
- 57. Urbanek M. The genetics of the polycystic ovary syndrome. Nat Clin Pract Endocrinol Metab. 2007;3:103–111. [DOI] [PubMed] [Google Scholar]
- 58. Franks S, Stark J, Hardy K. Follicle dynamics and anovulation in polycystic ovary syndrome. Hum Reprod Update. 2008;14:367–378. [DOI] [PubMed] [Google Scholar]
- 59. Zeggini E, Scott LJ, Saxena R, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. McAllister JM, Modi B, Miller BA, et al. Overexpression of a DENND1A isoform produces a polycystic ovary syndrome theca phenotype. Proc Natl Acad Sci USA. 2014;111:E1519–E1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Goodarzi MO, Jones MR, Li X, et al. Replication of association of DENND1A and THADA variants with polycystic ovary syndrome in European cohorts. J Med Genet. 2012;49:90–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Welt CK, Styrkarsdottir U, Ehrmann DA, et al. Variants in DENND1A are associated with polycystic ovary syndrome in women of European ancestry. J Clin Endocrinol Metab. 2012;97:E1342–E1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mutharasan P, Galdones E, Peñalver Bernabé B, et al. Evidence for chromosome 2p16.3 polycystic ovary syndrome susceptibility locus in affected women of European ancestry. J Clin Endocrinol Metab. 2013;98:E185–E190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Louwers YV, Stolk L, Uitterlinden AG, Laven JS. Cross-ethnic meta-analysis of genetic variants for polycystic ovary syndrome. J Clin Endocrinol Metab. 2013;98:E2006–E2012. [DOI] [PubMed] [Google Scholar]
- 65. Hwang JY, Lee EJ, Jin Go M, et al. Genome-wide association study identifies GYS2 as a novel genetic factor for polycystic ovary syndrome through obesity-related condition. J Hum Genet. 2012;57:660–664. [DOI] [PubMed] [Google Scholar]
- 66. Lee H, Oh JY, Sung YA, et al. Genome-wide association study identified new susceptibility loci for polycystic ovary syndrome. Hum Reprod. 2015;30:723–731. [DOI] [PubMed] [Google Scholar]
- 67. Shim U, Kim HN, Lee H, Oh JY, Sung YA, Kim HL. Pathway analysis based on a genome-wide association study of polycystic ovary syndrome. PLoS One. 2015;10:e0136609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ebrahim S, Davey Smith G. Mendelian randomization: can genetic epidemiology help redress the failures of observational epidemiology? Hum Genet. 2008;123:15–33. [DOI] [PubMed] [Google Scholar]
- 69. Ding EL, Song Y, Manson JE, et al. Sex hormone-binding globulin and risk of type 2 diabetes in women and men. N Engl J Med. 2009;361:1152–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Casas JP, Shah T, Cooper J, et al. Insight into the nature of the CRP-coronary event association using Mendelian randomization. Int J Epidemiol. 2006;35:922–931. [DOI] [PubMed] [Google Scholar]
- 71. Li H, Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475:493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gronau I, Hubisz MJ, Gulko B, Danko CG, Siepel A. Bayesian inference of ancient human demography from individual genome sequences. Nat Genet. 2011;43:1031–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Azziz R, Dumesic DA, Goodarzi MO. Polycystic ovary syndrome: an ancient disorder? Fertil Steril. 2011;95:1544–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Stolk L, Perry JR, Chasman DI, et al. Meta-analyses identify 13 loci associated with age at menopause and highlight DNA repair and immune pathways. Nat Genet. 2012;44:260–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Freedman ML, Monteiro AN, Gayther SA, et al. Principles for the post-GWAS functional characterization of cancer risk loci. Nat Genet. 2011;43:513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Claussnitzer M, Dankel SN, Kim KH, et al. FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med. 2015;373:895–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. McAllister JM, Legro RS, Modi BP, Strauss JF., 3rd Functional genomics of PCOS: from GWAS to molecular mechanisms. Trends Endocrinol Metab. 2015;26:118–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hirschhorn JN. Genomewide association studies–illuminating biologic pathways. N Engl J Med. 2009;360:1699–1701. [DOI] [PubMed] [Google Scholar]
- 79. Meun C, Karaderi T, Magi R, et al. Genome-wide meta-analysis of polycystic ovary syndrome in women of European ancestry identifies new loci in hormone pathways. In: Proceedings from the American Society of Human Genetics; October 6–10, 2015; Baltimore, MD Poster 933T. [Google Scholar]
- 80. Franks S, Webber LJ, Goh M, et al. Ovarian morphology is a marker of heritable biochemical traits in sisters with polycystic ovaries. J Clin Endocrinol Metab. 2008;93:3396–3402. [DOI] [PubMed] [Google Scholar]
- 81. Shungin D, Winkler TW, Croteau-Chonka DC, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Denny JC, Bastarache L, Ritchie MD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31:1102–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. McCarty CA, Chisholm RL, Chute CG, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360:1696–1698. [DOI] [PubMed] [Google Scholar]
- 86. Wheeler E, Barroso I. Genome-wide association studies and type 2 diabetes. Brief Funct Genomics. 2011;10:52–60. [DOI] [PubMed] [Google Scholar]
- 87. Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–425. [DOI] [PubMed] [Google Scholar]
- 88. Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. [DOI] [PubMed] [Google Scholar]
- 89. McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. [DOI] [PubMed] [Google Scholar]
- 90. Tennessen JA, Bigham AW, O'Connor TD, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Coventry A, Bull-Otterson LM, Liu X, et al. Deep resequencing reveals excess rare recent variants consistent with explosive population growth. Nat Commun. 2010;1:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Bowden DW, An SS, Palmer ND, et al. Molecular basis of a linkage peak: exome sequencing and family-based analysis identify a rare genetic variant in the ADIPOQ gene in the IRAS Family Study. Hum Mol Genet. 2010;19:4112–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Rees MG, Ng D, Ruppert S, et al. Correlation of rare coding variants in the gene encoding human glucokinase regulatory protein with phenotypic, cellular, and kinetic outcomes. J Clin Invest. 2012;122:205–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Cohen J, Pertsemlidis A, Kotowski IK, Graham R, Garcia CK, Hobbs HH. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet. 2005;37:161–165. [DOI] [PubMed] [Google Scholar]
- 95. Lambert G, Sjouke B, Choque B, Kastelein JJ, Hovingh GK. The PCSK9 decade. J Lipid Res. 2012;53:2515–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Everett BM, Smith RJ, Hiatt WR. Reducing LDL with PCSK9 inhibitors–the clinical benefit of lipid drugs. N Engl J Med. 2015;373:1588–1591. [DOI] [PubMed] [Google Scholar]
- 97. Curtis D. Assessing the contribution family data can make to case-control studies of rare variants. Ann Hum Genet. 2011;75:630–638. [DOI] [PubMed] [Google Scholar]
- 98. Zhu X, Feng T, Li Y, Lu Q, Elston RC. Detecting rare variants for complex traits using family and unrelated data. Genet Epidemiol. 2010;34:171–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Guey LT, Kravic J, Melander O, et al. Power in the phenotypic extremes: a simulation study of power in discovery and replication of rare variants. Genet Epidemiol. 2011;35:236–246. [DOI] [PubMed] [Google Scholar]
- 100. Li D, Lewinger JP, Gauderman WJ, Murcray CE, Conti D. Using extreme phenotype sampling to identify the rare causal variants of quantitative traits in association studies. Genet Epidemiol. 2011;35:790–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. [DOI] [PubMed] [Google Scholar]
- 104. Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. [DOI] [PubMed] [Google Scholar]
- 106. Shihab HA, Gough J, Cooper DN, et al. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat. 2013;34:57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Johansen CT, Wang J, Lanktree MB, et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat Genet. 2010;42:684–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Majithia AR, Flannick J, Shahinian P, et al. Rare variants in PPARG with decreased activity in adipocyte differentiation are associated with increased risk of type 2 diabetes. Proc Natl Acad Sci USA. 2014;111:13127–13132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Maurano MT, Humbert R, Rynes E, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Vockley CM, Guo C, Majoros WH, et al. Massively parallel quantification of the regulatory effects of noncoding genetic variation in a human cohort. Genome Res. 2015;25:1206–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Abbott DH, Nicol LE, Levine JE, Xu N, Goodarzi MO, Dumesic DA. Nonhuman primate models of polycystic ovary syndrome. Mol Cell Endocrinol. 2013;373:21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Hickey TE, Legro RS, Norman RJ. Epigenetic modification of the X chromosome influences susceptibility to polycystic ovary syndrome. J Clin Endocrinol Metab. 2006;91:2789–2791. [DOI] [PubMed] [Google Scholar]
- 113. Xu N, Azziz R, Goodarzi MO. Epigenetics in polycystic ovary syndrome: a pilot study of global DNA methylation. Fertil Steril. 2010;94:781–783.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Wang P, Zhao H, Li T, et al. Hypomethylation of the LH/choriogonadotropin receptor promoter region is a potential mechanism underlying susceptibility to polycystic ovary syndrome. Endocrinology. 2014;155:1445–1452. [DOI] [PubMed] [Google Scholar]
- 115. Kobaly K, Vellanki P, Sisk RK, et al. Parent-of-origin effects on glucose homeostasis in polycystic ovary syndrome. J Clin Endocrinol Metab. 2014;99:2961–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Xu N, Chua AK, Jiang H, Liu NA, Goodarzi MO. Early embryonic androgen exposure induces transgenerational epigenetic and metabolic changes. Mol Endocrinol. 2014;28:1329–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Xu N, Kwon S, Abbott DH, et al. Epigenetic mechanism underlying the development of polycystic ovary syndrome (PCOS)-like phenotypes in prenatally androgenized rhesus monkeys. PLoS One. 2011;6:e27286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Fu Q, Meyer M, Gao X, et al. DNA analysis of an early modern human from Tianyuan Cave, China. Proc Natl Acad Sci USA. 2013;110:2223–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Knowler WC, Barrett-Connor E, Fowler SE, et al. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med. 2002;346:393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Ibáñez L, López-Bermejo A, Díaz M, Marcos MV, de Zegher F. Metformin treatment for four years to reduce total and visceral fat in low birth weight girls with precocious pubarche. J Clin Endocrinol Metab. 2008;93:1841–1845. [DOI] [PubMed] [Google Scholar]
- 121. Stein IF, Leventhal ML. Amenorrhea associated with bilateral polycystic ovaries. Am J Obstet Gynecol. 1935;29:181–191. [Google Scholar]