Abstract
Purpose
To develop a robust T1ρ magnetic resonance imaging (MRI) sequence for assessment of myocardial disease in humans.
Materials and Methods
We developed a breath-held T1ρ mapping method using a single-shot, T1ρ-prepared balanced steady-state free-precession (bSSFP) sequence. The magnetization trajectory was simulated to identify sources of T1ρ error. To limit motion artifacts, an optical flow-based image registration method was used to align T1ρ images. The reproducibility and accuracy of these methods was assessed in phantoms and 10 healthy subjects. Results are shown in 1 patient with pre-ventricular contractions (PVCs), 1 patient with chronic myocardial infarction (MI) and 2 patients with hypertrophic cardiomyopathy (HCM).
Results
In phantoms, the mean bias was 1.0 ± 2.7 msec (100 msec phantom) and 0.9 ± 0.9 msec (60 msec phantom) at 60 bpm and 2.2 ± 3.2 msec (100 msec) and 1.4 ± 0.9 msec (60 msec) at 80 bpm. The coefficient of variation (COV) was 2.2 (100 msec) and 1.3 (60 msec) at 60 bpm and 2.6 (100 msec) and 1.4 (60 msec) at 80 bpm. Motion correction improved the alignment of T1ρ images in subjects, as determined by the increase in Dice Score Coefficient (DSC) from 0.76 to 0.88. T1ρ reproducibility was high (COV < 0.05, intra-class correlation coefficient (ICC) = 0.85–0.97). Mean myocardial T1ρ value in healthy subjects was 63.5 ± 4.6 msec. There was good correspondence between late-gadolinium enhanced (LGE) MRI and increased T1ρ relaxation times in patients.
Conclusion
Single-shot, motion corrected, spin echo, spin lock MRI permits 2D T1ρ mapping in a breath-hold with good accuracy and precision.
Introduction
There is increasing interest in quantitative, parametric mapping as a method for myocardial disease assessment in magnetic resonance imaging (MRI). One such method, called T1ρ (‘T1-rho’) mapping, quantitatively detects and spatially maps changes to water 1H T1ρ nuclear magnetic relaxation times and may provide useful information about myocardial structure. In vivo myocardial T1ρ mapping has detected relaxation time changes that correlate with histologically-confirmed scar in animal models of ischemic heart disease [1–3] and has been shown to detect disease in acute [4,5] and chronic ischemic patients [6].
Given the need for sensitive, non-invasive methods for myocardial tissue characterization, the development of robust T1ρ MRI pulse sequences is an important goal. Minimizing sources of relaxation time error, such as pulse sequence variability, sensitivity to myocardial and respiratory motion, and magnetic field heterogeneity, is essential to detect disease with high sensitivity. Unknown variability in pulse sequence parameters can contribute to significant error in T1ρ relaxation time mapping and overwhelm the intrinsic variation between normal and diseased myocardial tissue.
While T1ρ MRI pulse sequences have been previously reported for myocardial disease applications [5,7], cardiac T1ρ mapping in humans poses a few unique acquisition challenges that have not been addressed in detail. Although several methods have been established to reduce T1ρ sensitivity to field heterogeneity in heart muscle [8] and in other organs [9–11], cardiac and respiratory motion can reduce spatial resolution, T1ρ measurement accuracy, and introduce motion artifacts that impair quantification. Since cardiac T1ρ mapping methods acquire each T1ρ image in one (‘single-shot’) or more (‘multi-shot’) heartbeats, variation in myocardial or respiratory position alters voxel alignment and causes motion artifacts. While several methods have been tested for motion correction of cardiac parametric maps, such as T1 [12–14], T2 and arterial spin labeling (ASL) parametric maps [15,16], these methods have not been validated for T1ρ maps. Variations in myocardial and blood contrast, as well as signal intensity oscillations of off-resonance tissues may limit the success of conventional registration methods.
The purpose of this work was to develop an approach for motion corrected T1ρ mapping of myocardial disease in humans, within a single breath-hold examination, and to assess pulse sequence parameters that influence the accuracy and reproducibility of the T1ρ map. Simulations of the magnetization trajectory, phantom experiments and in vivo experiments were performed under various conditions and the potential contributions to T1ρ map accuracy and reproducibility were quantified. The optimized sequence was evaluated in patients with PVCs, chronic MI or HCM pathology.
Materials and Methods
Pulse sequence design and Bloch simulations
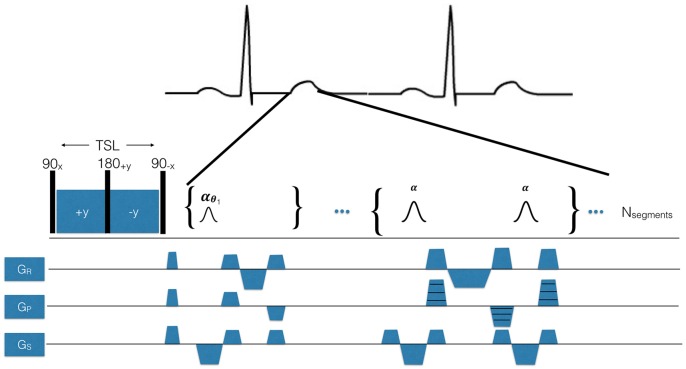
T1ρ MRI data was obtained using the pulse sequence shown in Fig 1. The main design criteria was to obtain a single slice T1ρ map in a single patient breath-hold with high accuracy and reproducibility. In general, the pulse sequence was composed of four periods: (1) T1ρ magnetization preparation, (2) bSSFP magnetization preparation (a flip angle ramp), (3) bSFFP spatial encoding, and (4) T1 recovery.
Fig 1. T1ρ pulse sequence designed for use in humans using breath-hold and electrocardiogram (ECG)-triggering.
The pulse sequence is synchronized to the R-wave using a MRI- compatible vector ECG. The spin lock pulse cluster shown here is a spin lock, spin echo (90x-SLy-180+y-SL-y-90-x), consisting of a pair of continuous RF spin locking pulses with opposite phase ±y to refocus magnetization in a heterogeneous B1 magnetic field and a refocusing pulse to refocus B0. TSL is total spin lock time. After the T1ρ pulse cluster, a crusher gradient is delivered to eliminate remaining transverse signal and then a second magnetization preparation period employing a bSSFP flip angle ramp to stabilize the transverse magnetization prior to spatial encoding. The readout sequence employed is a single-shot bSSFP.
During the first time period, the magnetization was T1ρ-prepared using a composite, radiofrequency (RF) pulse using a spin echo, spin lock (SL) (90x-SLy-180y-SL-y-90-x). The integration of a single refocusing pulse (spin-echo) with a pair of phase-reversed spin-locking pulses (spin echo, spin lock) has been previously shown to reduce some of the T1ρ artifacts associated with B0 and B1 magnetic field heterogeneity in the heart, as compared to rotary echo alone [9].
During the second time period, a second magnetization preparation employed a bSSFP flip angle ramp to stabilize the transverse magnetization prior to spatial encoding. The RF pulse flip angle ramp was composed of N = 10 pulses of increasing flip angle
where θi is the flip angle of the ith pulse (i ϵ 1..N) in the ramp and θ is the readout flip angle.
During the third time period, spatial encoding was performed using single-shot bSSFP and pulse sequence parameters were analyzed in simulations and in phantoms. These parameters included the flip angle (10, 30, 50, 70°), the number of Cartesian k-space lines acquired per shot (Nsegments), the number of heartbeats per image (Nshots = 1, 2, 4, 8), and the expected patient heart rate (HR = 60 beats-per-minute—bpm- or HR = 80 bpm). Imaging was performed every other heartbeat, so the time between T1ρ preparations was 2 sec (60 bpm) or 1.5 sec (80 bpm). Parameters held constant or not analyzed for their impact on relaxation times were the non-selective 90°/180° pulse (B1 = 1250 Hz, pulse duration = 200/400 us), TE = 1.11 msec, TR = 2.22 msec, k-space parallel imaging acceleration factor = 2, number of parallel imaging reference lines = 24, bandwidth = 898 Hz/pixel, spatial resolution = 1.6–2.2 mm2, rectangular field-of-view = 75%, matrix = 192x108, interpolated matrix = 192x144, frequency encode lines = 192, and slice thickness = 6 mm. The readout duration was TRO = NsegmentsTR. Parallel imaging reference lines were acquired in a separate heartbeat, without any T1ρ preparation. The total acquisition time was 17 heartbeats (17 sec at 60 bpm). The reference T1ρ sequence used the same parameters, but with θ = 90°, Nsegments = 1, Nshots = 54 and time between T1ρ preparations = 10 sec. While this scan was inappropriate for human use because of its total acquisition time, it served as a reference for relaxation times in phantoms. For all experiments, we used the maximum available spin lock amplitude (B1 = 500 Hz) within specific absorption rate (SAR) limits. Unless otherwise specified, all phantom and human scans used the same acquisition.
During the fourth period, a delay was introduced to permit recovery of longitudinal magnetization between consecutive shots. This recovery was primarily affected by heart rate (HR) and the duration of the desired breath-hold. In particular, elevated HR was expected to introduce undesired T1-weighting into the T1ρ-weighted images.
Simulations were performed to test the magnetization response to pulse sequence parameters. The time-dependent magnetization was solved using piecewise, time-independent matrix solution to the Bloch equations [17]
with the propagation matrix
where R1,2 were the longitudinal and transverse relaxation rates, Δω the resonance frequency shift, ω1 the RF pulse amplitude and θ the RF pulse phase. The augmented magnetization vector M = [Mx, My,Mz, 1]T. Simulations were performed in Matlab (The MathWorks, Natick, MA).
Magnetic Resonance Imaging
The accuracy and precision of different T1ρ MRI pulse sequences were determined in phantoms and human subjects. A phantom was prepared with multiple known nuclear relaxation properties, as determined by the reference T1ρ sequence, and differences between measured and true T1ρ relaxation times were quantified. The phantom contained 8 cylindrical samples prepared with solutions of H2O and MnCl2 in concentrations of 0.002, 0.004, 0.006, 0.008, 0.010, 0.012, 0.015, 0.017% solutions, providing a range of T1ρ relaxation times that would be expected in vivo. Mean bias was analyzed for two phantoms with relaxation times similar to normal myocardium (T1ρ ~ 60 msec, 0.012%) and scar tissue (T1ρ ~ 100 msec, 0.015%). Scan parameters were the same as in the previous section in a single slice intersecting all phantoms and with TSL = 50, 42, 34, 26, 18, 10, 2 msec.
T1ρ images were acquired in 10 normal subjects (7 men, 3 women, mean age = 29±7 years) without a previous history of cardiovascular events, 1 patient with chronic MI, 1 patient with PVCs, and 2 patients with HCM on a 1.5 T whole-body MRI system (Avanto Model, Siemens Healthcare, Erlangen, Germany) at the Hospital of the University of Pennsylvania. The University of Pennsylvania Institutional Review Board approved this study and all subjects gave written informed consent to participate. 5 of the subjects underwent 3 MRI scans to assess the reproducibility of T1ρ maps.
In humans, 2D multislice ECG-gated T1ρ-weighted images were acquired, during end-systole and end-expiration, to reduce the effects of cardiac and respiratory motion. The end-systolic cardiac phase was determined by first examining the cardiac motion on cine-SSFP short-axis images and then adjusting the beginning of spatial encoding with the period of maximum myocardial wall thickness. To further reduce respiratory motion artifacts, all T1ρ-weighted images were acquired within a single end-expiratory breath-hold using the same acquisition parameters as in the phantom experiments, and with flip angle = 70°, Nshots = 1, TSL = 50, 42, 34, 26, 18, 10, 2 msec, and number of slices = 7–10.
Late gadolinium enhanced (LGE) MRI was performed in patients using a 2D multislice, short-axis, phase-sensitive inversion recovery pulse sequence (TR = 850 msec, TE = 1.56 msec, acquisition matrix = 123x256, inversion time = 400–500 msec, flip angle = 20°, slice thickness = 8 mm). Approximately 15 minutes prior to LGE MRI, 0.15 mmol/kg gadolinium-based contrast agent (gadobenate dimeglumine, MultiHance, Bracco) was administered intravenously. In the patient with chronic scar, LGE and T1ρ MRI were performed 2 weeks apart in different imaging sessions.
Motion Correction
To mitigate cardiac motion, T1ρ images were acquired at the same end-systolic cardiac phase and during an end-expiratory breathhold. Even though this strategy reduced the influence of motion, residual myocardial motion was observed in images due to respiration and small variations in sinus rhythm. This issue was more problematic when the subjects were unable to hold their breath or were uncooperative.
To further reduce the effects of residual motion, we investigated image-based optical flow (OF) motion correction. The OF algorithm [18,19] was implemented in C++ [20] using a combination of the methods described in [21] and [22]. The motion correction scheme, T1ρ and R2 mapping were implemented into the online reconstruction software utilizing the Image Calculation Environment (ICE) framework of the clinical MRI System (Siemens Healthcare, Erlangen, Germany). Each T1ρ image series consisted of 8 images. Frame 4 (TSL = 34 msec) was chosen as the reference frame as it showed moderate T1ρ contrast and, in most cases, less motion compared to other frames. Two-frame image registration was performed between each of the 7 moving frames and the reference frame (frame 4).
To quantify the accuracy of motion correction, we measured the Dice similarity coefficient (DSC) between image pairs with large, moderate and no motion before and after motion correction. DSC was defined as:
where A and B are two segmented regions. A human rater was asked to classify the motion between each frame pair as large, moderate, or no motion and 133 image pairs (7 pairs per map, 19 total maps in 10 subjects) were classified. To measure DSC, the myocardium was manually segmented in each image frame. After motion correction, the segmented myocardium was propagated to the corrected images using OF-computed deformation fields. In the ideal case, the motion correction algorithm would perfectly align the segmentation of each image pair.
T1ρ Relaxation Mapping
We tested three models of T1ρ relaxation to assess their accuracy and precision (Table 1). The main differences between the models were linearity or nonlinearity and the number of free parameters. The 2-parameter linear model was solved using linear least-squares (‘\’ operator, MATLAB, version 2014a, Natick, MA). The 2- and 3-parameter nonlinear models were solved using constrained active-set minimization (MATLAB). The 3-parameter model included a steady-state magnetization parameter to account for the effects of the readout [23].
Table 1. Models of T1ρ relaxation.
| Known Parameters | xi ϵ ℝ2: ith image voxel location, where i = 1:Nvoxels |
| S0 ϵ ℂ: measured signal | |
| TSL: contrast evolution time (spin lock time) (msec) | |
| Unknown Parameters | η ϵ ℂ: steady state magnetization |
| S0 ϵ ℂ: initial signal | |
| T1ρ: relaxation time (msec) | |
| 3-parameter, nonlinear model (model I) | |
| 2-parameter, nonlinear model (model II) | |
| 2-parameter, linear model (model III) |
Statistics
T1ρ MRI accuracy was quantified as the percent error and precision was quantified as , where was the mean of N = 3 measurements and σ(T1ρ) the standard deviation. The intra-class correlation coefficient (ICC) was used to assess the T1ρ ROI measurement reproducibility between different scans (R software package). ICC values > 0.75 represent a good agreement [24].
Results
Sources of measurement error
The magnetization was simulated to identify potential sources of T1ρ quantification error (Fig 2). Three types of error were identified:
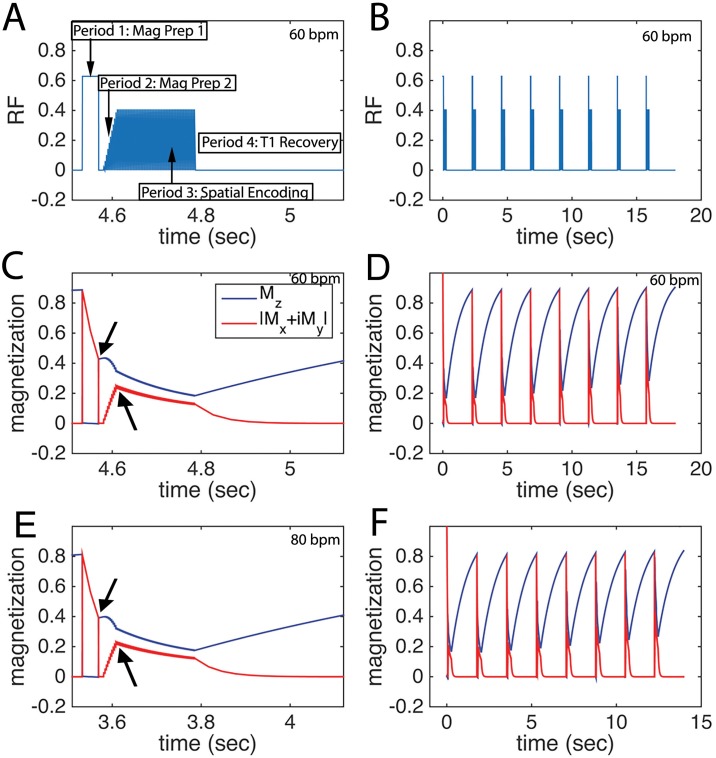
Fig 2. Pulse sequence and magnetization trajectory for single-shot cardiac T1ρ MRI in humans.
A, RF pulse diagram, showing magnetization preparation and spatial encoding periods with HR = 60 bpm. The spatial encoding period includes a flip angle ramp to stabilize the transverse magnetization, during which the magnetization is not spatially encoded. B, extended RF graph depicting 8 single-shot T1ρ-weighted images with varying spin lock duration to create a T1ρ map. C, longitudinal MZ and transverse |Mx + iMy| magnetization for the period in A. D, magnetization for 8 shots at 60 bpm. E, longitudinal MZ and transverse |Mx + iMy| magnetization at 80 bpm. and F, 80 bpm. The acquisition of parallel imaging reference data is not shown and occurs in a separate heartbeat. The arrows in C and D indicate that the transverse magnetization is different after the spin lock and after the ramp.
The RF pulse ramp (Fig 2A) disturbed the T1ρ magnetization prior to spatial encoding. The RF pulse ramp was designed to stabilize the magnetization prior to readout, so a balance between stabilization, achieved using a longer flip angle ramp, and perturbation of the T1ρ magnetization was desired.
While the ideal readout magnetization would be flat, the observed (red) magnetization trajectories (Fig 2C and 2E) showed a monotonic decay. The decay was expected to affect the voxel point spread function differently for centric and linear spatial encoding trajectories and introduce measurement bias. The behavior of the magnetization response was not significantly different at 60 bpm (Fig 2C) and 80 bpm (Fig 2E).
The inter-shot delay for both HR = 60 bpm (2 beats per shot, TR = 2 sec) and HR = 80 bpm (TR = 1.5 sec) was less than myocardial T1, so the magnetization did not fully recover. Fig 2B shows RF pulses for an 8 shot T1ρ map acquisition at 60 bpm and Fig 2D shows the magnetization trajectory. A comparison of Fig 2D to 2F showed minimal variation in the shot-to-shot magnetization, despite the increase in HR from 60 to 80 bpm.
Accuracy and reproducibility in phantoms
Phantoms with different MR relaxation properties were scanned to assess accuracy and reproducibility of T1ρ relaxation times using a 3-parameter nonlinear model (Fig 3; 2-parameter model shown in S2 Dataset). The mean bias was 1.0 ± 2.7 msec (100 msec phantom) and 0.9 ± 0.9 msec (60 msec phantom) at 60 bpm and 2.2 ± 3.2 msec (100 msec) and 1.4 ± 0.9 msec (60 msec) at 80 bpm. COV = 2.2 (100 msec) and 1.3 (60 msec) at 60 bpm and 2.6 (100 msec) and 1.4 (60 msec) at 80 bpm for single shot-imaging at flip angle = 70°. Reproducibility in phantoms was very high (COV < 0.05 for all scans). Single-shot imaging with a readout flip angle = 70° achieved very low mean bias for all phantoms (Fig 3E, R2 = 0.99, p<0.01).
Fig 3. Mean bias of T1ρ MRI in phantoms of normal and ischemic myocardial tissue and dependence on number of shots and readout flip angle.
A, mean bias in normal myocardial tissue phantom (T1ρ = 60 msec) at 60 and B, 80 bpm. C, mean bias in scar tissue phantom (T1ρ = 100 msec) at 60 and D, 80 bpm. E, correlation of measured single-shot T1ρ to a standard T1ρ imaging sequence (readout flip angle = 70°).
Effects of motion on cardiac T1ρ maps
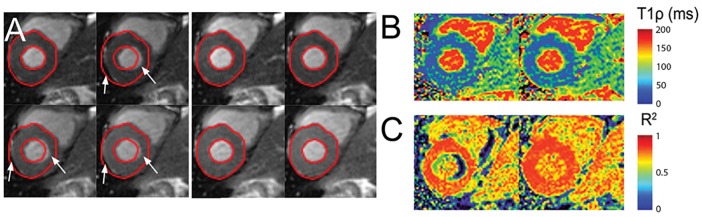
Large motion was observed in 25 image pairs (18.8%), moderate motion was present in 21 image pairs (15.8%), and no motion was found in 87 image pairs (65.4%). Fig 4 shows a T1ρ dataset containing several images exhibiting large motion. Myocardial regions with perceived large motion were labeled with white arrows in Fig 4A. There were overall improvements in the T1ρ map fit (Fig 4B) and in the R2 map at the myocardial free wall and subendocardium (Fig 4C).
Fig 4. Motion correction of a T1ρ dataset in the large motion category.
A, Top: Original images showing large motion. Bottom: Results of motion correction by applying optical flow registration. B, T1ρ maps before (left) and after (right) motion correction. C, R2 maps before (left) and after (right) motion correction.
Fig 5A shows the results of motion correction for a dataset containing T1ρ images with both large motion and moderate motion. OF successfully corrected most of the image frames. However, large motion of the basal myocardium below the aortic valve was difficult to correct, since the myocardial wall partially moved out of the image frame during respiration. Nevertheless, a visual comparison of T1ρ and R2 maps after registration showed an improvement in these areas (Fig 5B and 5C) and reduced motion artifacts.
Fig 5. Motion correction of a T1ρ dataset in the moderate motion category.
A, Top: Original images showing large motion. Bottom: Results of motion correction by applying optical flow registration. B, T1ρ maps before (left) and after (right) motion correction. C, R2 maps before (left) and after (right) motion correction.
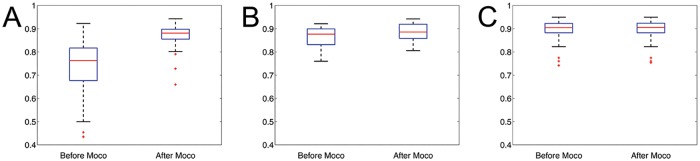
DSC values for different motion categories are shown (Fig 6). For large motion datasets, there was a significant increase in DSC (p << 0.01). For moderate and no motion datasets, the improvement in DSC was not significant (p > 0.05).
Fig 6. DSC quantification of motion correction for large, moderate and no motion T1ρ datasets.
A, Overall DSC for large motion datasets. B, Overall DSC for moderate motion datasets. C, Overall DSC for moderate motion datasets. For the large motion category, the DSC values are significantly improved after correction. In the case of moderate motion datasets, DSC values improved slightly after the registration. In the datasets without heart motion, the DSC values were similar before and after registration.
Effects of heart rate, linear and centric encoding, and model fitting
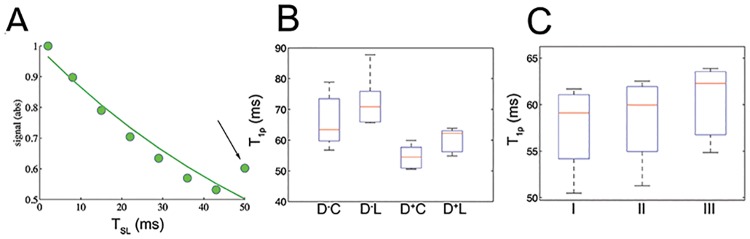
The magnetization for the first shot was significantly different from subsequent shots and biased the model estimate (Fig 7A, black arrow). This was consistent with simulations in which the initial shot-to-shot magnetization varied for the first shot in a heart rate-dependent fashion. The effect was eliminated with an initial ‘dummy’ shot with TSL = 50 msec. The dummy pulse (Fig 7B, D+) significantly reduced variability and bias.
Fig 7. T1ρ relaxation times in normal myocardial tissue and dependence on pulse sequence parameters.
A, heart rate variability causes the first shot (black arrow) to be inconsistent with our signal intensity measurements and overestimation of T1ρ relaxation times. B, Reproducibility and bias with and without an initial ‘dummy’ shot (D+ or D-) and for centric and linear encoding (C,L). C, Reproducibility and bias of 3 models for T1ρ relaxation.
Linear spatial encoding introduced a small bias compared to centric encoding, but had reduced artifacts arising from initial perturbations of the T1ρ magnetization by the readout pulses.
A small bias was observed between model types in normal myocardium, but these differences were not significant (Fig 7C).
Reproducibility in humans
The T1ρ values from region of interests (ROIs) in the septum for each of the 3 models are shown in Table 2. The COVs for each subject were highly reproducible (COV < 0.05). The ICC values for T1ρ measurements between 3 different scans in subjects 1 to 5 were reproducible (ICC = 0.85–0.97).
Table 2. T1ρ (msec) of healthy volunteers in 3 different trials.
| Subject 1 | Subject 2 | Subject 3 | Subject 4 | Subject 5 | Mean | |
|---|---|---|---|---|---|---|
| Trial 1 | 62.3±6.9 | 66.1±3.0 | 68.3±8.1 | 60.3±4.2 | 68.3±4.5 | |
| Trial 2 | 58.4±6.2 | 64.7±4.5 | 69.3±5.9 | 55.2±6.8 | 69.3±5.2 | |
| Trial 3 | 59.1±8.0 | 64.9±4.9 | 64.0±7.2 | 57.5±5.1 | 64.9±5.8 | |
| Mean | 59.9 | 65.3 | 67.2 | 57.7 | 67.5 | 64.5 |
| SD | 2.1 | 0.8 | 2.8 | 2.6 | 2.3 | 2.1 |
| COV | 0.03 | 0.01 | 0.04 | 0.04 | 0.03 | 0.03 |
Patient imaging
Fig 8 shows T1ρ maps and LGE images from patients with PVCs (Fig 8A), chronic MI (8B), and in two patients with HCM (Fig 8C and 8D). In all patients, there was moderate correspondence of increased T1ρ relaxation times with injury, as confirmed by signal enhancement on LGE MRI.
Fig 8. T1ρ and LGE MRI in patients.
Subjects with A, PVCs, B, chronic myocardial infarction, and C-D, HCM. Spatial correspondence between increased T1ρ and LGE MRI is indicated with white arrows. LGE images were obtained in end-diastole and T1ρ in end-systole.
Discussion
The motivation for this work was to develop a robust, breath-held T1ρ mapping sequence for assessment of myocardial disease in humans, determine potential sources of T1ρ mapping error associated with pulse sequence parameters, and measure the accuracy and reproducibility of T1ρ relaxation times in normal human subjects. In cardiac parametric mapping, pulse sequence parameters may introduce bias or reduce measurement precision. It is essential to account for these possible errors in order to detect disease with high sensitivity or minimize the number of individuals recruited for research studies. Several elements of this work have been presented [25,26].
Previous CMR T1ρ mapping studies [6,27] were reported using T1ρ-prepared bSSFP in humans. In [6], relaxation times in normal myocardium were lower than reported here, 54±6.0 msec compared to 64.5±2.1 msec. Although spin lock amplitude was higher (B1 = 750 Hz), these differences may be accounted for by reduced flip angle (50°), increased TE and TR, and multishot instead of single shot readout. In [27], T1ρ relaxation times were very different (42.2±1.6) which might be attributed to a lower readout flip angle (15°) and lower spin lock amplitude [340 Hz]. In both previous reports, motion and heart rate correction were not discussed, but could reduce T1ρ reproducibility. As was observed in Fig 7, significant bias was introduced without heart rate correction and the bias was affected by whichever TSL was acquired first.
There was moderate qualitative correspondence between T1ρ and LGE lesions. Specifically, for the patient with ischemic heart disease, T1ρ MRI was consistent with non-transmural scar at a mid-ventricular imaging slice. The reduced wall thickness indicated a very large apical aneurysm and this was supported by LGE hyperenhancement. The spatial distribution of the hyperintense region was consistent with scar, since the lesion appeared predominantly in the mid-myocardium. For HCM patients, there was asymmetric septal wall thickening of the anterosuperior right ventricular insertion zone and this corresponded to elevated T1ρ and LGE lesions. In a fourth patient with non-ischemic cardiomyopathy, there was elevated midwall T1ρ corresponding to a similar LGE lesion pattern. There were several features that were inconsistent, such as the apparent LGE scar size in the ischemic patient or relative lesion position in HCM patients. Myocardial or patient motion may have contributed to incomplete overlap. In this study, LGE scans were performed 10–20 minutes after pre-contrast T1ρ scans and LGE was performed in diastole and T1ρ in systole. Other studies have also reported moderate correspondence between LGE and T1ρ. In van Oorschot et al., double-blinded overlap of LGE and T1ρ in 21 ischemic patients was 72% and unblinded was 74% [6]. Inconsistent visual scoring was attributed to low T1ρ contrast-to-noise ratio and inadequate radiologist training to discriminate scar on T1ρ vis-à-vis LGE. It is unclear if HR correction, image alignment, and single-shot imaging may have improved overlap, although the use of a longer T1 recovery may have reduced the need for HR correction. An alternative interpretation is that inconsistent visual overlap indicates true differences between Gd-based contrast agent (GBCA) and native methods. Such differences may arise due to GBCA perfusion kinetics, fibrosis, water content, or other unknown mechanisms, although we can only speculate from phantom studies and animal experiments. If verified, the two methods would appear to be complementary and together improve stratification or clinical diagnosis. The low number of subjects and absence of causal data linking fibrosis to relaxation time changes was inadequate to support this finding and should be a subject of future investigation.
We observed that single-shot bSSFP imaging at high flip angle (70°) had high accuracy compared to the standard, partially because bSSFP has lower perturbation of the transverse relaxation compared to spoiled gradient echo sequences, even at very high flip angles [28]. The accuracy was somewhat higher than was previously observed for cartilage T1ρ MRI using similar parameters [29], primarily because the cardiac sequence included a ramp-up period, minimizing perturbation of the PSF. As shown in Fig 2, this period helped stabilize the transient magnetization prior to spatial encoding. Nevertheless, the measured relaxation times are sensitive to the overall readout properties. While we identified potential sources of error due to PSF variability, a detailed further analysis is needed to assess the effect of spatial encoding parameters on the width of the voxel PSF. A number of factors potentially affect this measurement including linear or centric spatial encoding, readout flip angle, echo spacing, and echo train length. To ensure accurate and reproducible T1ρ measurements, it was desirable to change the field-of-view instead of parameters that would affect the number segments and readout duration. The bias for all flip angles and segment sizes was low. The range of T1ρ relaxation times was 10 msec in the normal subject group, but does not account for potential effects of age or gender in these populations and may impact the detectability of diffuse disease.
In our analysis, we identified major sources of measurement error in cardiovascular T1ρ MRI arising from pulse sequence parameter selection. In particular, Bloch equation simulations showed that proper choice of pulse sequence parameters is essential to reduce bias and reproducibly measure T1ρ relaxation times in the heart. Motion was a major contributing factor to error in cardiovascular T1ρ mapping. Our initial experience using breath-held multi-shot T1ρ imaging was highly unsatisfactory due to the presence of image artifacts, which caused inconsistency in phase encoded k-space data. Single-shot imaging and OF motion correction was found to improve the quality of the T1ρ map (Figs 4–6)
We introduced a method for motion correction of cardiovascular T1ρ MRI images using optical flow, which was validated for T1ρ mapping using data collected from phantoms and human subjects. The varying contrast in T1ρ images could potentially compromise robust alignment of images. The OF method was robust to contrast change and applied regularization to produce piecewise smooth deformation fields and physiologically relevant deformations. The computation time to estimate and apply deformation fields was ~2 sec and the computation time for linear T1ρ fitting and R2 map creation was ~0.1 sec, so registered and parametric maps could be produced in clinical workflow. In addition, it was not necessary to manually draw reference contours and, instead, the registration was performed between the reference and moving images as a global optimization.
We observed that the moving images would not be well corrected if they were significantly different from the chosen reference image. These observations were confirmed in basal or apical slices where longitudinal cardiac motion partially shifted myocardium out of the image frame, such as in Fig 5, or in the presence of spin lock artifacts in individual frames. Methods to identify a more consistent reference frame for registration would reduce these artifacts. For instance, an optimal reference image or template could be generated from the most consistent or prevalent features of the acquired images. Images with highly inconsistent longitudinal motion would therefore not be used as the reference. Synthetic T1ρ image estimates, using the approach of Xue, et al, would be a useful approach for eliminating this type of motion inconsistency [12].
We assessed the impact of the fitting models in T1ρ mapping using 3 models of T1ρ relaxation time maps. The rationale for testing three fitting models for T1ρ mapping was that the choice of the fitting models may impact the accuracy and precision of T2 mapping techniques; for a more detailed discussion see [23] and the references therein. We did not observe significant differences in T1ρ relaxation times between the 3 fitting model types in normal myocardium (Fig 7C). Nevertheless, we note that a much more detailed analysis would need to be performed for a systematic study of the effects of the different fitting models in T1ρ mapping.
There is considerable interest to characterize myocardial edema and fibrosis in ischemic patients and those with non-ischemic cardiomyopathy. Qualitative visual comparison of the injured myocardium in Fig 8 depicts small variations in the measured LGE and T1ρ signals. These differences may reflect differences in the LGE and T1ρ contrast generating mechanisms. Variations in extracellular volume (ECV) fraction, upstream coronary perfusion or tracer kinetics may alter contrast agent distribution in tissue and LGE signal intensity. T1ρ is sensitive to interactions between tissue water 1H and the macromolecular environment. Increased myocardial fibrosis associated with extracellular matrix expansion may reduce water 1H rotational correlation times and increase T1ρ. In addition, T1ρ imaging has shown increased signal intensity in areas of edema [2,5] and thus has the potential to assess therapies that limit myocardial injury during or after an acute MI. Recent reports have observed very close correspondence between the size of injury detected by LGE and T1ρ MRI. A study by Wang et al. [27] showed strong spatial correspondence of injury determined by T1ρ maps and LGE cardiac magnetic resonance using quantitative thresholding in HCM patients. In this group, myocardial disarray and replacement fibrosis may contribute to increased T1ρ relaxation.
The putative advantage of T1ρ to T2 mapping, is that T1ρ limits low frequency contributions to the measured relaxation rate, increasing the dynamic range of transverse relaxation times in normal and diseased myocardial tissue. This property was observed in ex vivo myocardial tissue and scar at several B1 amplitudes [3] and would be expected for acute injuries as well. Preliminary investigations [26] have nonetheless suggested that acute ischemia may result in more complicated and time-dependent T1ρ patterns, which may depend on reperfusion time, ischemia-reperfusion injury, extent of coronary ischemia, and hemorrhage. Animal models and prospective studies of T1ρ in humans will be crucial to address these issues.
In conclusion, highly robust T1ρ imaging methods are necessary for parametric mapping of myocardial disease with high accuracy and reproducibility in humans. The methods shown here significantly reduce measurement error of T1ρ relaxation times associated with pulse sequence parameters, magnetic field heterogeneity, and myocardial and respiratory motion and will be essential in the assessment of myocardial disease.
Supporting Information
(DOCX)
(DOCX)
Data Availability
Due to ethical restrictions imposed by the University of Pennsylvania Institutional Review Board, patient and human subject data are available upon request from the corresponding author (witschey@mail.med.upenn.edu). All other data are within the paper.
Funding Statement
This work was supported by the National Institutes of Health NHLBI R00-HL108157 and the Mr. and Mrs. Thomas B. McCabe Fund. The project described was supported by the National Center for Research Resources, Grant UL1RR024134, and is now at the National Center for Advancing Translational Sciences, Grant UL1TR000003. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This work was also supported in part by the Institute for Translational Medicine and Therapeutics’ (ITMAT) Transdisciplinary Program in Translational Medicine and Therapeutics (http://www.itmat.upenn.edu/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Witschey WRT, Zsido G a, Koomalsingh K, Kondo N, Minakawa M, Shuto T, et al. In vivo chronic myocardial infarction characterization by spin locked cardiovascular magnetic resonance. J Cardiovasc Magn Reson. 2012;14(1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Musthafa H-SN, Dragneva G, Lottonen L, Merentie M, Petrov L, Heikura T, et al. Longitudinal rotating frame relaxation time measurements in infarcted mouse myocardium in vivo. Magn Reson Med. 2013;69(5):1389–95. 10.1002/mrm.24382 [DOI] [PubMed] [Google Scholar]
- 3.Witschey WR, Pilla JJ, Ferrari G, Koomalsingh K, Haris M, Hinmon R, et al. Rotating frame spin lattice relaxation in a swine model of chronic, left ventricular myocardial infarction. Magn Reson Med. 2010;64(5):1453–60. 10.1002/mrm.22543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huber S, Muthupillai R, Lambert B, Pereyra M, Napoli A, Flamm SD. Tissue characterization of myocardial infarction using T1rho: influence of contrast dose and time of imaging after contrast administration. J Magn Reson Imaging. 2006;24(5):1040–6. [DOI] [PubMed] [Google Scholar]
- 5.Muthupillai R, Flamm SD, Wilson JM, Pettigrew RI, Dixon WT. Acute myocardial infarction: tissue characterization with T1rho-weighted MR imaging—initial experience. Radiology. 2004;232(2):606–10. [DOI] [PubMed] [Google Scholar]
- 6.van Oorschot JW, El Aidi H, Jansen of Lorkeers SJ, Gho JM, Froeling M, Visser F, et al. Endogenous assessment of chronic myocardial infarction with T1ρ-mapping in patients. J Cardiovasc Magn Reson. 2014;16:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han Y, Liimatainen T, Gorman R, Witschey W. Assessing myocardial disease using T1rho MRI. Curr Cardiovasc Imaging Rep. 2014;7:65–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixon WT, Oshinski JN, Trudeau JD, Arnold BC, Pettigrew RI. Myocardial suppression in vivo by spin locking with composite pulses. Magn Reson Med. 1996;36:90–4. [DOI] [PubMed] [Google Scholar]
- 9.Witschey WRT, Borthakur A, Elliott M, Mellon E, Niyogi S, Wallman DJ, et al. Artifacts in T1 rho-weighted imaging: compensation for B(1) and B(0) field imperfections. J Magn Reson. 2007;186(1):75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Witschey WR, Borthakur A, Elliott MA, Mellon E, Niyogi S, Wang C, et al. Compensation for spin-lock artifacts using an off-resonance rotary echo in T1rhooff-weighted imaging. Magn Reson Med. 2007;57(1):2–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen W, Takahashi A, Han E. Quantitative T1rho imaging using phase cycling for B0 and B1 field inhomogeneity compensation. Magn Reson Imaging. 2011;29(5):608–19. 10.1016/j.mri.2011.02.002 [DOI] [PubMed] [Google Scholar]
- 12.Xue H, Shah S, Greiser A, Guetter C, Littmann A, Jolly MP, et al. Motion correction for myocardial T1 mapping using image registration with synthetic image estimation. Magn Reson Med. 2012;67:1644–55. 10.1002/mrm.23153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue H, Greiser A, Zuehlsdorff S, Jolly MP, Guehring J, Arai AE, et al. Phase-sensitive inversion recovery for myocardial T1 mapping with motion correction and parametric fitting. Magn Reson Med. 2013;69:1408–20. 10.1002/mrm.24385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roujol S, Foppa M, Weingärtner S, Manning WJ, Nezafat R. Adaptive registration of varying contrast-weighted images for improved tissue characterization (ARCTIC): Application to T1 mapping. Magn Reson Med. 2014;00:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.von Knobelsdorff-Brenkenhoff F, Prothmann M, Dieringer M a, Wassmuth R, Greiser A, Schwenke C, et al. Myocardial T1 and T2 mapping at 3 T: reference values, influencing factors and implications. J Cardiovasc Magn Reson. 2013;15(1):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang DJJ, Bi X, Avants BB, Meng T, Zuehlsdorff S, Detre JA. Estimation of Perfusion and Arterial Transit Time in Myocardium Using Free-Breathing Myocardial Arterial Spin Labeling With Navigator-Echo. 2010;1295:1289–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murase K, Tanki N. Numerical solutions to the time-dependent Bloch equations revisited. Magn Reson Imag. 2011;29(1):126–31. [DOI] [PubMed] [Google Scholar]
- 18.Horn BK, Schunck BG. Determining Optical Flow Pearson JJ, editor. Artif Intell. International Society for Optics and Photonics; 1981. November;17:185–203. [Google Scholar]
- 19.Bruce D. Lucas TK. An Iterative Image Registration Technique with an Application to Stereo Vision. In: IJCAI’81 Proceedings of the 7th international joint conference on Artificial intelligence.
- 20.Liu C. Beyond Pixels : Exploring New Representations and Applications for Motion Analysis. 2009. [Google Scholar]
- 21.Brox T, Bruhn A, Papenberg N, Weickert J. High accuracy optical flow estimation based on a theory for warping. European Conference on Computer Vision; 2004;25–36.
- 22.Bruhn A, Weickert J., Schnorr C. Lucas / Kanade Meets Horn / Schunck : Combining Local and Global Optic Flow Methods. 2005;61(3):211–31. [Google Scholar]
- 23.Akçakaya M, Basha T a., Weingärtner S, Roujol S, Berg S, Nezafat R. Improved quantitative myocardial T2 mapping: Impact of the fitting model. Magn Reson Med. 2014; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fleiss JL. The Design and Analysis of Clinical Experiments. Hoboken, NJ, USA: John Wiley & Sons, Inc.; 1999. [Google Scholar]
- 25.Han JQ, Han Y, Witschey WR. Accuracy and reproducibility of T1rho mapping sequences. J Cardiovasc Magn Reson. BioMed Central Ltd; 2015;17(Suppl 1):P22. [Google Scholar]
- 26.Han Q, Han Y, Gorman RC, Witschey WR. The influence of static and RF field heterogeneity on T1rho cardiovascular MRI. J Cardiovasc Magn Reson. 2014;16(Suppl 1):P70. [Google Scholar]
- 27.Wang C, Zheng J, Sun J, Wang Y, Xia R, Yin Q, et al. Endogenous contrast T1rho cardiac magnetic resonance for myocardial fibrosis in hypertrophic cardiomyopathy patients. J Cardiol. 2015. May;S0914-5087(15):00090–8. [DOI] [PubMed] [Google Scholar]
- 28.Scheffler K, Hennig J. T1 quantification with inversion recovery TrueFISP. Magn Reson Med. John Wiley & Sons, Inc.; 2001;45(4):720–3. [DOI] [PubMed] [Google Scholar]
- 29.Witschey WR, Borthakur A, Elliott MA, Fenty M, Sochor MA, Wang C, et al. T1rho-prepared balanced gradient echo for rapid 3D T1rho MRI. J Magn Reson Imag. 2008;28(3):744–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
Data Availability Statement
Due to ethical restrictions imposed by the University of Pennsylvania Institutional Review Board, patient and human subject data are available upon request from the corresponding author (witschey@mail.med.upenn.edu). All other data are within the paper.