Abstract
Objective
Ubiquitin-specific protease 7 (USP7) is a common target of herpesviruses and is important in the DNA damage response, which is also upregulated in several cancers, including prostate, colon, liver, and lung cancers. However, less is known about its expression in ovarian cancer tissues. The role of USP7 in epithelial ovarian cancer (EOC) has not yet been investigated.
Materials and methods
We recruited 141 patients from Linyi People’s Hospital between June 1999 and June 2013, all pathologically diagnosed with primary EOC. Their clinical data were collected, and the expression of USP7 in the tumor tissues was determined using immunohistochemistry. The correlations between USP7 expression and the clinicopathological variables of patients with EOC were assessed using Spearman’s rank correlation test. Kaplan–Meier analysis and Cox regression analysis were used to identify the prognosis value of USP7. The function of USP7 in the EOC cells was also detected in vitro.
Results
Among the 141 cases, USP7 expression was high in 59 EOC samples (41.8%), and was significantly correlated with lymphatic invasion; USP7 can act as independent prognostic indicator for the overall survival (OS) of EOC, and its high expression was associated with poor OS rate. The RNA inteference and overexpression assays indicated that USP7 can positively regulate the ovarian cell vitality and invasion process.
Conclusion
Patients with EOC expressing high level of USP7 have worse OS compared with those with low USP7 expression. USP7 may be involved in the proliferation and invasion of EOC cells, and USP7 expression can serve as an independent predictor of EOC.
Keywords: epithelial ovarian cancer, USP7, lymph node metastasis, invasion, prognosis
Introduction
Ovarian cancer (OC) is the most lethal gynecological tumor, constituting the fifth common cause of cancer-related death in women.1–3 OC constitutes 90% of malignant ovarian tumors, while epithelial ovarian cancer (EOC) accounts for over 90% of primary OCs, which consists of several histologic subtypes, including serous, clear cell, endometrioid, and mucinous adenocarcinomas, and the high-grade serous carcinoma is the most common one.4 Only ~15% of patients are diagnosed with EOC at an early stage.5 The morbidity of EOC is increasing worldwide,6 and the high lethal rate of EOC mainly results from its early metastasis and silent symptoms in early stage.7 The later symptoms mainly include pelvic pain, bloating, and abdominal swelling, with lung, liver, and lymph nodes being the most frequently invaded organs.2 Nowadays, the primary methods used to screen for OC are manual pelvic examination, transvaginal ultrasound, and cancer antigen (CA)-125 detection, and all have limited specificity and sensitivity.8 Despite current advances in surgery and adjuvant chemotherapy, the clinical outcome of patients with OC remains uncertain, with the 5-year survival rate being ~30% for advanced-stage disease.9 The development of EOC is associated with a series of molecular signals, so the requirement to identify an effective biomarker for early diagnosis and prognosis of EOC has been growing in recent years.
The ubiquitin-mediated protein degradation plays important roles in controlling the abundance of intracellular proteins and is critical in maintaining the normal cellular process, including cell cycle and tumor suppression. Negative regulation of ubiquitin-mediated proteolysis can result in the development of various cancers.10 The removal of ubiquitin from ubiquitylated proteins is also important in determining the ubiquitylation state of various E3-ubiquitin ligase substrates, and the de-ubiquitylating enzymes (DUBs) have emerged as important regulators in controlling a wide variety of different cellular functions, including the response to DNA damage.11,12 De-ubiquitylating enzymes act significantly as the E3 ligases deregulating the reversible protein ubiquitylation process in the cancer development. Several DUBs have been identified in the DNA damage response, including ubiquitin-specific protease (USP) 1, USP7, and USP28.13–15 Among them, the USP7 (herpesvirus-associated USP), which was first discovered through its strong and specific interaction with the ICP0 protein of HSV-1, has received a lot of attention.16 USP7 can deubiquitinate various tumor suppressors (p53, PTEN, FOXO, and claspin), E3 ligases (MDM2/MDMX and viral protein ICP0), as well as chromatin-associated proteins (histone H2B, UHRF1, and Tip60); therefore, it regulates important cellular processes involved in the tumorigenesis.17–24 Consistent with its functions, USP7 is upregulated in many cancers, including prostate, colon, liver, and lung cancers.20,23,25
However, the expression of USP7 in EOC has not been investigated; there has been no study on the role of USP7 in the tumorigenesis, metastasis, and the prognostic significance of EOC. In this study, we detected the expression of USP7 in OC cells and tissues, and then analyzed the correlation between USP7 expression and the clinicopathologic parameters and overall survival (OS) rates. Furthermore, we identified USP7 as an independent prognostic factor by multivariate analysis. To explore the mechanism by which USP7 overexpression leads to unfavorable prognosis, we further performed in vitro experiments to identify the USP7’s function of increasing the viability and invading the OC cells.
Materials and methods
Patients and samples
One hundred and forty-one patients who were diagnosed with EOC and underwent surgery at Linyi People’s Hospital were recruited between June 1999 and June 2013. All diagnoses were based on morphological criteria, immunohistochemical staining, and clinical findings. None of the patients had any prior history of chemoradiation, radiation, or hormonal therapy before the surgery. The histopathologic subtypes of the 141 EOC included 121 serous cystadenocarcinomas, 16 mucinous cystadenocarcinomas, and four endometrioid adenocarcinomas. The histopathologic grading of the 141 adenocarcinoma patients revealed that 39 patients had G1 (stage I), 58 patients had G2, and 44 patients had G3 adenocarcinomas. Tumor-nodes-metastais (TNM) staging was determined using the International Federation of Gynecology and Obstetrics (FIGO) staging system. According to the FIGO system, 17 cases had stage I, 29 had stage II, 71 had stage III, and 24 had stage IV. Among the 141 patients with adenocarcinomas, 77 patients had regional lymph nodes metastasis, while 64 had no metastasis. Survival information of 141 patients with ovarian adenocarcinomas was obtained through letters and phone calls. The postoperative follow-up period continued until the date of death or September 2013. The study was approved by the Ethics Committee of Linyi People’s Hospital. Signed informed consent forms were obtained from all subjects who participated in the study. The Code of Ethics of the World Medical Association (Declaration of Helsinki) was followed.
Cell culture and transfection
The human OC cell lines OVCAR3 and SKOV3 were obtained from the Cell Bank of the Chinese Academy of Sciences, Shanghai, People’s Republic of China. Ethical permission for the use of human cell lines was approved by the Ethics Committee of Linyi People’s Hospital. The ovarian benign cell line Hs893 was obtained from the American Type Culture Collection (Manassas, VA, USA). The cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% (v/v) fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin, under 5% CO2 in a humidified incubator at 37°C.
pcDNA3.1-USP7 was purchased from Addgene (Cambridge, MA, USA). USP7 knockdown was accomplished by oligo-small interfering RNA (siRNA) transfection. Approximately 2×105 cells/well were seeded in six-well plates and cultured in serum-free RPMI 1640 medium to 40% confluence by the time of transfection. Knockdown and overexpression of USP7 were realized by Lipofectamine 2000 reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer instructions. After 4 hours of incubation, the cells were cultured in RPMI 1640 medium with 10% FBS. The USP7-targeting siRNA and the negative control siRNA were designed and purchased from Shanghai Gene Pharma Co., Ltd. (Shanghai, People’s Republic of China). The siRNA sequences used in this study were as follows: USP7 siRNA: 5′-ACCCUUGGACAAUAUUCCUdTdT-3′26 and negative control RNA (ncRNA): 5′-UUCUCCGAACGUGU CACGUdTdT-3′.
Cell viability assay
The OVCAR3 and SKOV3 cells were seeded at 1×104 cells/well in 96-well plates 24 hours after transfection. To test the cell viability, PrestoBlue Cell Viability Reagent (Thermo Fisher Scientific) was used according to the manual, and we chose the time points as 8, 12, 24, 48, and 72 hours after seeding. The fluorescence was read at a wavelength of 560 and 590 nm using the microplate reader. Higher fluorescence represents greater total metabolic activity.
Immunohistochemistry
Briefly, paraffin-embedded specimens were cut into 3–4 μm serial sections; the sections were then deparaffinized and incubated with peroxidase solution (3% H2O2) in the dark for 10 minutes, followed by incubation at room temperature for 30 minutes with nonimmune animal serum. The sections were then incubated with USP7 primary antibody (ab4080; Abcam, Cambridge, UK) at 4°C overnight, followed by second antibody for 30 minutes, and washed with phosphate-buffered saline. Solution A was added to the sections for 30 minutes followed by 3,3′-diaminobenzidine staining and hematoxylin counter-staining. The slides were then dehydrated with alcohol, soaked in xylene for 3×5 minutes, and finally mounted with neutral balsam. A 5% FBS was used in place of primary antibodies as a negative control.
Semiquantitative analysis of immunohistochemistry results
The immunohistochemical results were evaluated by two pathologists from our hospital and followed the criterions of double-blind trials. Twelve visual fields were observed in each slide by a light microscope at a high power (×400). The staining intensity was scored (score 0, negative staining; score 1, pale yellow; score 2, dark yellow; score 3, brown), and the percentage of positive cells was also observed (score 0, <25% positive cells/field; score 1, 25%–50% positive cells/field; score 2, 50%–75% positive cells/field; and score 3, >75% positive cells/field). Based on the product of the two scores, the USP7 staining results were classified into low expression (<4) and high expression (≥4).
Western blotting
Total cell extracts were prepared with NP-40 Buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate) and protease inhibitor cocktails (1:1,000). Approximately 40 μg of each protein sample was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose. Membranes were blocked with 5% skim milk in Tris-buffered saline with 1% Tween and then incubated with antibodies against USP7 (ab4080 from Abcam; 1:2,000 dilution) and actin (sc-1616 from Santa Cruz Biotechnology, Dallas, TX, USA; 1:2,000 dilution). After washing, blots were probed with goat anti-rabbit peroxidase (Santa Cruz Biotechnology; 1:5,000 dilution) and developed using chemiluminescence reagents (Santa Cruz Biotechnology).
Matrigel invasion assay
The ability of tumor invasion was evaluated by Matrigel transwell assay with 8 μm pore Matrigel-coated transwells (BD Biosciences, San Jose, CA, USA). Briefly, after transfection for 24 hours, ~2×105 cells were passaged into the Matrigel invasion chambers and cultured for 12 hours. Then, cells were fixed with 4% paraformaldehyde for 30 minutes and stained with 0.05% gentian violet for 1 hour. Cells on the upper surface were carefully removed with a swab, and the cell number on the lower surface was counted from ten random fields. The cell number of the control group was set as a baseline, and fold change was calculated by the ratio to the control group. Each subclone was seeded in triplicate, and the analyzed data were from three different experiments. Statistical significance was evaluated by Student’s t-test.
Statistical analysis
Statistical analysis was performed using SPSS software (standard version 22.0; SPSS, Chicago, IL, USA). The correlation between USP7 expression and the clinicopathological variables of the patients was assessed using Spearman’s rank correlation test. Univariate survival analysis was assessed using the Kaplan–Meier method and compared by the log-rank test. The Cox proportional hazard regression analysis was employed for multivariate survival analysis. During the assays, clinicopathologic parameters were grouped as follows: age: ≤60 vs >60 years; histological grade: G1–G2 vs G3; FIGO stage: I–II vs III–IV; and CA-125: ≤900 vs >900 U/mL. A two-sided P-value of <0.05 was considered statistically significant.
Results
Patient characteristics
After exclusion of the unsuitable cases (ie, patients accepted the preoperative chemotherapy, pathological samples were unrepresentative, etc), 141 cases were included in this study, aged from 46 to 75 years (median 54.0 years). The distribution of FIGO stages was 17 patients with stage I (12.1%), 29 patients with stage II (20.5%), 71 patients with stage III (50.4%), and 24 patients with stage IV (17.0%). Table 1 summarizes the characteristics of this population.
Table 1.
Basic clinicopathologic parameters of patients and the correlation with USP7 expression
| Variables | Cases (n=141) | USP7 expression
|
P-value# | |
|---|---|---|---|---|
| Low (n=82) | High (n=59) | |||
| Age (years) | 0.795 | |||
| ≤60 | 103 | 61 | 42 (40.8%) | |
| >60 | 38 | 21 | 17 (44.7%) | |
| FIGO stage | 0.929a | |||
| I–II | 46 | 27 | 19 (41.3%) | |
| III–IV | 95 | 55 | 40 (42.1%) | |
| Histology type | 0.250 | |||
| Serous | 121 | 68 | 53 (43.8%) | |
| Non-serous | 20 | 14 | 6 (30.0%) | |
| Pathological grade | 0.881b | |||
| G1–G2 | 97 | 56 | 41 (42.3%) | |
| G3 | 44 | 26 | 18 (40.9%) | |
| Lymph node metastasis | 0.048* | |||
| No | 64 | 43 | 21 (32.8%) | |
| Yes | 77 | 39 | 38 (49.4%) | |
| Preoperative CA-125 (U/mL) | 0.858 | |||
| ≤900 | 73 | 42 | 31 (42.5%) | |
| >900 | 68 | 40 | 28 (41.2%) | |
Notes:
Spearman’s rank correlation test.
Statistically significant.
III + IV vs I + II.
G2 + G3 vs G1.
Abbreviations: USP7, ubiquitin-specific protease 7; FIGO, International Federation of Gynecology and Obstetrics; CA, cancer antigen.
USP7 expression in EOC tissues and its association with clinicopathologic features
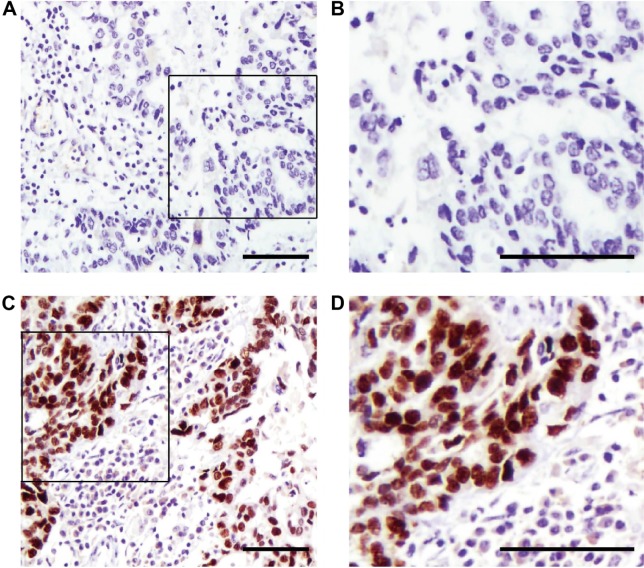
Immunohistochemistry (IHC) showed that USP7-positive (Figure 1) reactions were mainly localized in the nucleus of the EOC. According to the IHC score criteria described in the “Materials and methods” section, the USP7 expression was divided into higher expression and lower expression. USP7 was highly expressed in 59 patients (41.8%). The associations between the expression of USP7 and variable clinicopathologic features are summarized in Table 1. Of the 77 cases with lymph node metastasis, USP7 was positively expressed in 38 cases (49.4%), significantly higher than that in the cases with no lymph node metastasis (21 positive in 64 cases, 32.8%), showing high consistency between the USP7 expression and lymph node metastasis (P=0.048). This suggests that USP7 may play an essential role in the process of lymph node metastasis. No statistical correlations were observed between USP7 expression and age (P=0.795), FIGO stage (P=0.929), histology type (P=0.250), pathological grade (P=0.881), or preoperative CA-125 level (P=0.858).
Figure 1.
Representative figures of lower (A and B) and higher (C and D) USP7 immunohistochemistry staining in clinical samples of EOC (scale bar: 100 μm).
Abbreviations: USP7, ubiquitin-specific protease 7; EOC, epithelial ovarian cancer.
OS in patients with EOC
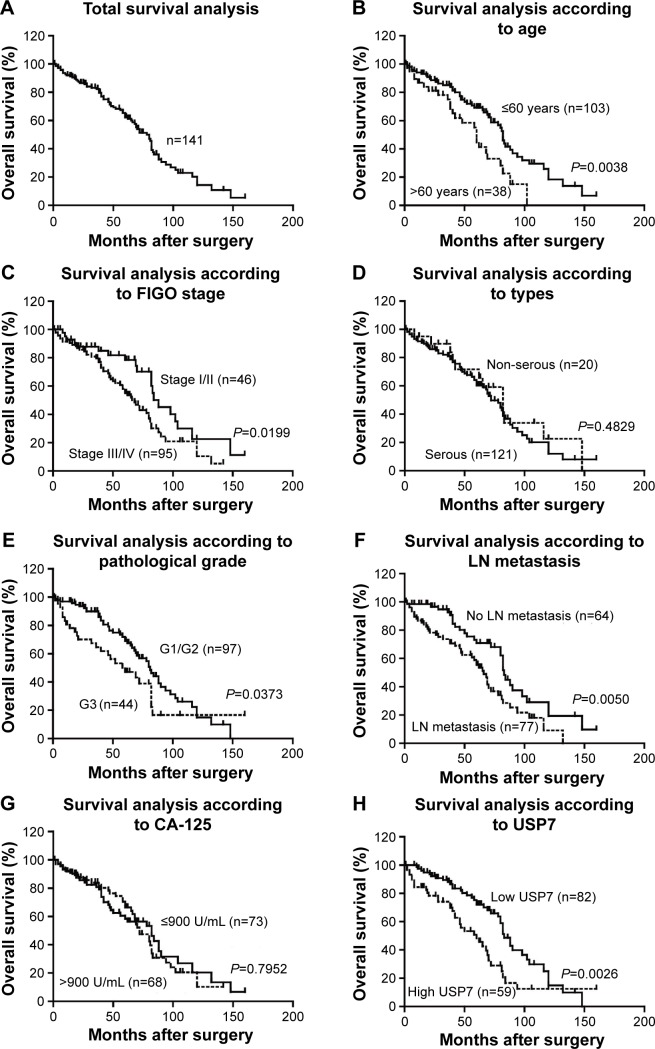
Survival information of 141 patients with EOC was obtained through letters and phone calls. The 5-year OS of the total patients was 62.34%, and the median survival time was 78 months. The Kaplan–Meier univariate survival analysis revealed that the age (P=0.004), FIGO stage (P=0.020), histological grade (P=0.037), lymph node metastasis (P=0.005), and USP7 expression (P=0.003) were significantly associated with the OS time in the EOC patients. Patients with low USP7 expression demonstrated longer OS compared with those with high USP7 expression (mean, 83 versus 60 months; P=0.003; Figure 2 and Table 2). The 5- and 10-year OS rates of the USP7-negative group were 72.01% and 14.95%, respectively, while those of the USP7-positive group were only 48.27% and 12.46%, respectively.
Figure 2.
Correlation between clinicopathologic parameters, USP7 expression, and prognosis in validation cohort.
Notes: Survival curve of the total patients was estimated (A). Survival analysis suggests that older age (B), advanced FIGO stage (C), higher pathological grade (E), positive lymph node metastasis (F), and high USP7 expression level (H) were significantly associated with poorer prognosis, while histology types (D) and CA-125 level (G) showed no statistically significant association.
Abbreviations: USP7, ubiquitin-specific protease 7; FIGO, International Federation of Gynecology and Obstetrics; LN, lymph node; CA, cancer antigen.
Table 2.
Kaplan–Meier survival analysis (log-rank test) according to USP7 expression in patients with EOC
| Variables | Cases (n=141) |
Overall survival time (months)
|
P-value# | |
|---|---|---|---|---|
| Mean | Median | |||
| Age (years) | 0.004* | |||
| ≤60 | 103 | 83.07 | 82 | |
| >60 | 38 | 57.09 | 60 | |
| FIGO stage | 0.020* | |||
| I–II | 46 | 91.68 | 88 | |
| III–IV | 95 | 68.96 | 68 | |
| Histology type | 0.483 | |||
| Serous | 121 | 74.90 | 72 | |
| Non-serous | 20 | 84.09 | 82 | |
| Pathological grade | 0.037* | |||
| G1–G2 | 97 | 81.80 | 81 | |
| G3 | 44 | 64.89 | 60 | |
| Lymph node metastasis | 0.005* | |||
| No | 64 | 89.45 | 83 | |
| Yes | 77 | 64.48 | 66 | |
| Preoperative CA-125 (U/mL) | 0.795 | |||
| ≤900 | 73 | 78.20 | 82 | |
| >900 | 68 | 74.89 | 72 | |
| USP7 expression | 0.003* | |||
| Low expression | 82 | 85.75 | 83 | |
| High expression | 59 | 62.34 | 60 | |
Notes:
Log-rank correlation test.
Statistically significant.
Abbreviations: USP7, ubiquitin-specific protease 7; EOC, epithelial ovarian cancer; FIGO, International Federation of Gynecology and Obstetrics; CA, cancer antigen.
USP7 as an independent prognostic factor in EOC
In addition, when the aforementioned parameters were included in the Cox regression multivariate analysis, the results indicated that USP7 can act as an independent prognostic factor (hazard ratio [HR] =1.733, confidence interval [CI] =1.099–2.860, P=0.019). Moreover, older age (HR =1.862, CI =1.108–3.132, P=0.019), advanced FIGO stages (HR =1.753, CI =1.028–2.987, P=0.039), and lymph node metastasis (HR =1.895, CI =1.162–3.090, P=0.010) were all negatively correlated with OS. This suggests that age, FIGO stage, lymph node metastasis, and USP7 expression are all risk factors and have a strong impact on prognosis of EOC (Table 3).
Table 3.
Cox multivariate analysis of the clinicopathological parameters for overall survival
| Variables | Hazard ratio | 95% confidence interval | P-value# |
|---|---|---|---|
| Age | 1.862 | 1.108–3.132 | 0.019* |
| FIGO stage | 1.753 | 1.028–2.987 | 0.039* |
| Pathological grade | 1.437 | 0.872–2.366 | 0.154 |
| Lymph node metastasis | 1.895 | 1.162–3.090 | 0.010* |
| USP7 expression | 1.733 | 1.099–2.860 | 0.019* |
Notes:
Cox proportional hazards regression.
Statistically significant.
Abbreviations: FIGO, International Federation of Gynecology and Obstetrics; USP7, ubiquitin-specific protease 7.
USP7 could promote the viability and invasion of OC cells
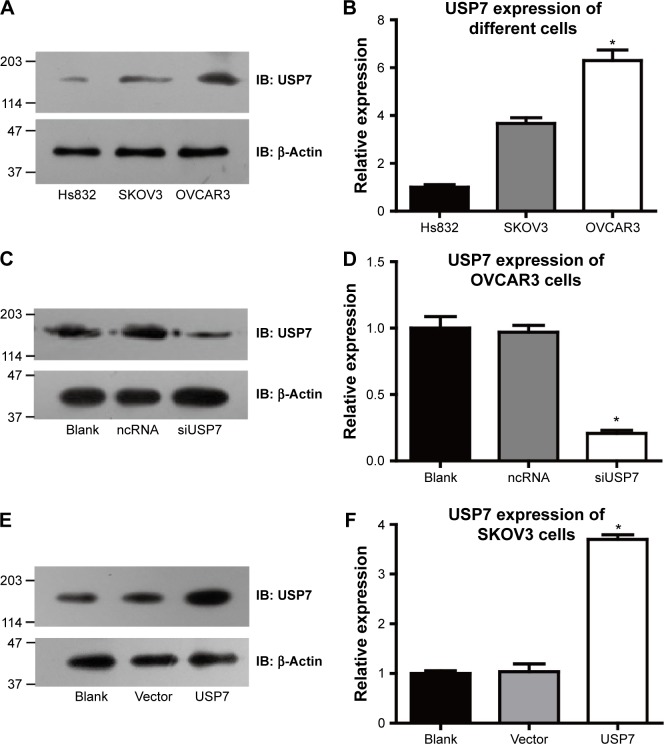
We detected the expression levels of USP7 in various OC cell lines to further confirm the results concluded from clinical data. Hs832 cells have a relatively low expression of USP7 compared with OVCAR3 and SKOV3 cell lines, indicating that the USP7 may be associated with the malignant characteristics of cancer cells. Even for the malignant cell lines, the USP7 showed different expression levels with higher expression in OVCAR3 cells than that in SKOV3 cells (Figure 3A and B). So we chose OVCAR3 to knock down USP7, while SKOV3 as the USP7 overexpression model. Successful knockdown by siRNA and overexpression were verified by Western Blot (Figure 3C–F).
Figure 3.
USP7 expression in different ovarian cell lines.
Notes: USP7 showed different expression patterns among ovarian benign cell line Hs832 and malignant cell lines OVCAR3 and SKOV3 (A and B). The expression level of USP7 was detected using immunoblotting after 48 hours after transfection with USP7 siRNA in OVCAR3 cells (C and D) and overexpression of USP7 in SKOV3 cells (E and F). *Statistically significant.
Abbreviations: USP7, ubiquitin-specific protease 7; ncRNA, negative control siRNA; siUSP7, siRNA of USP7; si, small interfering.
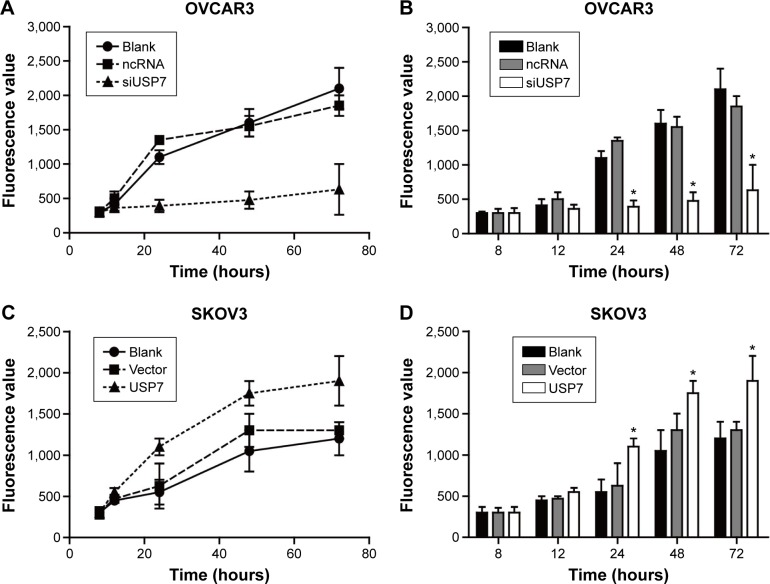
The viability of cell lines was then detected by Presto-Blue Cell Viability Reagent. We used OVCAR3 to compare the differences among USP7 siRNA-transfected cells, ncRNA-transfected cells, and untreated controls (Figure 4A and B). USP7 siRNA-transfected OVCAR3 cells showed reduced cell viability by 60%–70% compared with the ncRNA-transfected OVCAR3 cells and the untreated control after they were seeded to the 96-well plate for 72 hours. Consequently, when we overexpressed the USP7 in the SKOV3 cells, the cell viability was 1.3-fold higher than the control cells (Figure 4C and D). These findings demonstrated that USP7 can positively regulate the OC cell viability.
Figure 4.
USP7 could increase the viability of malignant ovarian cell lines.
Notes: Cell viability was detected by the PrestoBlue Cell Viability Reagent at different time points. The siRNA-transfected OVCAR3 cells showed decreased cell viability compared with blank control and the ncRNA-OVCAR3 (A and B), while the overexpression of UPS7 can significantly increase the cell viability (C and D). *Statistically significant.
Abbreviations: USP7, ubiquitin-specific protease 7; ncRNA, negative control siRNA; siUSP7, siRNA of USP7; si, small interfering.
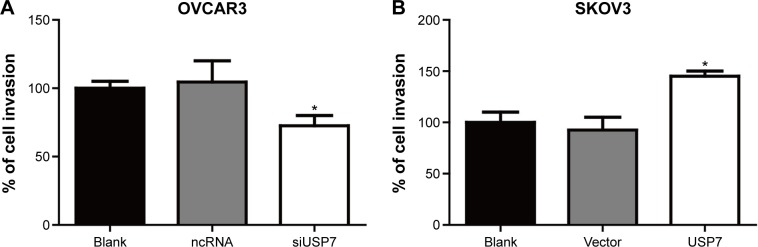
Through clinical investigation, we detected that USP7 was significantly associated with positive lymph node metastasis, which indicated that USP7 may promote EOC metastasis. Therefore, we performed the transwell invasion assay to explore the exact role of USP7 in ovarian cell invasion. According to our detection, knockdown of USP7 could decrease the invasion of OVCAR3 cells by 25%, and overexpression of USP7 could remarkably increase the invasion of SKOV3 cells by ~50%, suggesting the critical role of USP7 in EOC invasion (Figure 5A and B).
Figure 5.
USP7 could promote the invasion of malignant ovarian cells.
Notes: Invasion of OVCAR3 and SKOV3 cells was detected by transwell method after the USP7 was knocked down or overexpressed. Knockdown of USP7 could reduce the invasion of the OVCAR3 cells to ~25% (A), and the overexpression of USP7 in SKOV3 cells could significantly promote the invasion ability by ~50% compared with control cells (B). The statistical significance was calculated by Student’s t-test (comparing the tested group and control group at the same time, such as the siUSP7 vs ncRNA, and the vector vs USP7 overexpression). *Statistically significant.
Abbreviations: USP7, ubiquitin-specific protease 7; ncRNA, negative control siRNA; siUSP7, siRNA of USP7; si, small interfering.
Discussion
EOC is associated with a high incidence of disease recurrence and mortality.9 The utility of new biomarkers could promote a more consistent method to identify the proliferation potential of the EOC and a better understanding of its biological behavior. Many of the molecules and mechanisms mediating the prognosis of EOC have been characterized in recent years, and significant progress has been made.27 This study mainly investigated the expression of USP7 in EOC. Our aim was to investigate whether changes in expression of USP7 could influence the behavior or clinical outcomes of EOC.
USP7 was initially identified through its interaction with the ICP0 protein of herpes simplex virus.28 Subsequently, USP7 was also shown to interact with the EBNA1 protein from Epstein–Barr virus, vIRF4 and LANA proteins from Kaposi’s sarcoma herpesvirus, UL35 from cytomegalovirus, and E1B-55K from adenovirus.29–32 Targeted by various viral proteins, USP7 can promote cell survival and viral infection, living up to its original name of herpesvirus-associated USP. However, USP7 is also a cellular protein that can catalyze the deubiquitination of many cellular proteins and promote the protein stabilization, and thus involved in DNA damage and immune responses, neural stem cell maintenance, and tumor suppression.23,33–38 Its involvement in multiple signaling pathways including oncogenesis makes it emerge as a new potential target for cancer therapy. Recently, Pfoh et al have made great progress in the crystal structure of USP7 ubiquitin-like domains, promoting an advanced understanding of its regulation mechanisms.39 Up to now, USP7 has been proved to be upregulated in many cancers, including prostate, bladder, colon, liver, and lung cancers. However, the expression and functions of USP7 in EOC have not been reported.
This study is the first to explore the USP7 expression in EOC. By IHC and clinicopathological parameters analysis, we detected the expression of USP7 in 141 samples of EOC, and identified that the higher USP7 expression rate was 41.84%. In addition, we found that high USP7 expression in EOC tissues was closely correlated with the lymph node metastasis using Spearman’s rank correlation test. Furthermore, the cumulative 5-year OS rates of patients with high USP7 expression were lower than those with low USP7 expression, indicating that patients with high USP7 expression have a poorer prognosis. With Kaplan–Meier univariate and Cox multivariate analysis, we demonstrated that USP7 expression could be identified as an independent prognostic factor for EOC.
Since we detected that the higher expression of USP7 was significantly associated with lymphatic invasion, we presumed that USP7 may promote the invasive ability of OC cells, and we verified our hypothesis through the in vitro study. To illuminate the underlying mechanisms by which how USP7 affects the invasion process, we may need more fundamental experiments, such as coimmunoprecipitation, microarray, confocal immunofluorescence, and in vivo experiments. However, our study was mainly focused on the clinical significance of USP7 in the prognosis of EOC. We hope our finding that USP7 can act as a prognostic biomarker in EOC could be helpful in the management of the disease.
Conclusion
Our findings demonstrated that patients with EOC expressing high level of USP7 have worse OS compared with those with low USP7 expression. USP7 may be involved in the proliferation and invasion of OC cells, and USP7 expression can be regarded as an independent predictor of EOC. Therefore, the USP7 could be a potential drug target, and further studies are required to reveal the role of USP7 in EOC.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Hensley ML. A step forward for two-step screening for ovarian cancer. Journal of Clinical Oncology: Official Journal of the American Society of Clinical Oncology. 2010;28(13):2128–2130. doi: 10.1200/JCO.2009.26.6346. [DOI] [PubMed] [Google Scholar]
- 2.Partridge EE, Barnes MN. Epithelial ovarian cancer: prevention, diagnosis, and treatment. CA: A Cancer Journal for Clinicians. 1999;49(5):297–320. doi: 10.3322/canjclin.49.5.297. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA: A Cancer Journal for Clinicians. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 4.Basu M, Mukhopadhyay S, Chatterjee U, Roy SS. FGF16 promotes invasive behavior of SKOV-3 ovarian cancer cells through activation of mitogen-activated protein kinase (MAPK) signaling pathway. The Journal of Biological Chemistry. 2014;289(3):1415–1428. doi: 10.1074/jbc.M113.535427. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Siegel R, Naishadham D, Jemal A. Cancer statistics for Hispanics/Latinos, 2012. CA: A Cancer Journal for Clinicians. 2012;62(5):283–298. doi: 10.3322/caac.21153. [DOI] [PubMed] [Google Scholar]
- 6.Lozano R, Naghavi M, Foreman K, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet (London, England) 2012;380(9859):2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goff BA, Mandel L, Muntz HG, Melancon CH. Ovarian carcinoma diagnosis. Cancer. 2000;89(10):2068–2075. doi: 10.1002/1097-0142(20001115)89:10<2068::aid-cncr6>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 8.Jelovac D, Armstrong DK. Recent progress in the diagnosis and treatment of ovarian cancer. CA: A Cancer Journal for Clinicians. 2011;61(3):183–203. doi: 10.3322/caac.20113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colombo PE, Fabbro M, Theillet C, Bibeau F, Rouanet P, Ray-Coquard I. Sensitivity and resistance to treatment in the primary management of epithelial ovarian cancer. Critical Reviews in Oncology/Hematology. 2014;89(2):207–216. doi: 10.1016/j.critrevonc.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 10.Hoeller D, Hecker CM, Dikic I. Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nature Reviews Cancer. 2006;6(10):776–788. doi: 10.1038/nrc1994. [DOI] [PubMed] [Google Scholar]
- 11.Jacq X, Kemp M, Martin NM, Jackson SP. Deubiquitylating enzymes and DNA damage response pathways. Cell Biochemistry and Biophysics. 2013;67(1):25–43. doi: 10.1007/s12013-013-9635-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nijman SM, Luna-Vargas MP, Velds A, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123(5):773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 13.Nijman SM, Huang TT, Dirac AM, et al. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Molecular Cell. 2005;17(3):331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 14.Morra F, Luise C, Merolla F, et al. FBXW7 and USP7 regulate CCDC6 turnover during the cell cycle and affect cancer drugs susceptibility in NSCLC. Oncotarget. 2015;6(14):12697–12709. doi: 10.18632/oncotarget.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang D, Zaugg K, Mak TW, Elledge SJ. A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell. 2006;126(3):529–542. doi: 10.1016/j.cell.2006.06.039. [DOI] [PubMed] [Google Scholar]
- 16.Meredith M, Orr A, Everett R. Herpes simplex virus type 1 immediate-early protein Vmw110 binds strongly and specifically to a 135-kDa cellular protein. Virology. 1994;200(2):457–469. doi: 10.1006/viro.1994.1209. [DOI] [PubMed] [Google Scholar]
- 17.Everett RD, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. The EMBO Journal. 1997;16(3):566–577. doi: 10.1093/emboj/16.3.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faesen AC, Dirac AM, Shanmugham A, Ovaa H, Perrakis A, Sixma TK. Mechanism of USP7/HAUSP activation by its C-terminal ubiquitin-like domain and allosteric regulation by GMP-synthetase. Molecular Cell. 2011;44(1):147–159. doi: 10.1016/j.molcel.2011.06.034. [DOI] [PubMed] [Google Scholar]
- 19.Gao Y, Kalkhoven E. TIPping the balance in adipogenesis: USP7-mediated stabilization of Tip60. Adipocyte. 2014;3(2):160–165. doi: 10.4161/adip.28307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hussain S, Zhang Y, Galardy PJ. DUBs and cancer: the role of deubiq-uitinating enzymes as oncogenes, non-oncogenes and tumor suppressors. Cell Cycle (Georgetown, Texas) 2009;8(11):1688–1697. doi: 10.4161/cc.8.11.8739. [DOI] [PubMed] [Google Scholar]
- 21.Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Molecular Cell. 2004;13(6):879–886. doi: 10.1016/s1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- 22.Nicholson B, Suresh Kumar KG. The multifaceted roles of USP7: new therapeutic opportunities. Cell Biochemistry and Biophysics. 2011;60(1–2):61–68. doi: 10.1007/s12013-011-9185-5. [DOI] [PubMed] [Google Scholar]
- 23.Song MS, Salmena L, Carracedo A, et al. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature. 2008;455(7214):813–817. doi: 10.1038/nature07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang ZM, Rothbart SB, Allison DF, et al. An allosteric interaction links USP7 to deubiquitination and chromatin targeting of UHRF1. Cell Reports. 2015;12(9):1400–1406. doi: 10.1016/j.celrep.2015.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen ST, Okada M, Nakato R, Izumi K, Bando M, Shirahige K. The deubiquitinating enzyme USP7 regulates androgen receptor activity by modulating its binding to chromatin. The Journal of Biological Chemistry. 2015;290(35):21713–21723. doi: 10.1074/jbc.M114.628255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He J, Zhu Q, Wani G, et al. Ubiquitin-specific protease 7 regulates nucleotide excision repair through deubiquitinating XPC protein and preventing XPC protein from undergoing ultraviolet light-induced and VCP/p97 protein-regulated proteolysis. The Journal of Biological Chemistry. 2014;289(39):27278–27289. doi: 10.1074/jbc.M114.589812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meredith M, Orr A, Elliott M, Everett R. Separation of sequence requirements for HSV-1 Vmw110 multimerisation and interaction with a 135-kDa cellular protein. Virology. 1995;209(1):174–187. doi: 10.1006/viro.1995.1241. [DOI] [PubMed] [Google Scholar]
- 29.Holowaty MN, Sheng Y, Nguyen T, Arrowsmith C, Frappier L. Protein interaction domains of the ubiquitin-specific protease, USP7/HAUSP. The Journal of Biological Chemistry. 2003;278(48):47753–47761. doi: 10.1074/jbc.M307200200. [DOI] [PubMed] [Google Scholar]
- 30.Jager W, Santag S, Weidner-Glunde M, et al. The ubiquitin-specific protease USP7 modulates the replication of Kaposi’s sarcoma-associated herpesvirus latent episomal DNA. Journal of Virology. 2012;86(12):6745–6757. doi: 10.1128/JVI.06840-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salsman J, Jagannathan M, Paladino P, et al. Proteomic profiling of the human cytomegalovirus UL35 gene products reveals a role for UL35 in the DNA repair response. Journal of Virology. 2012;86(2):806–820. doi: 10.1128/JVI.05442-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saridakis V, Sheng Y, Sarkari F, et al. Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization. Molecular Cell. 2005;18(1):25–36. doi: 10.1016/j.molcel.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 33.Du Z, Song J, Wang Y, et al. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Science Signaling. 2010;3(146):ra80. doi: 10.1126/scisignal.2001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang Z, Wu Q, Guryanova OA, et al. Deubiquitylase HAUSP stabilizes REST and promotes maintenance of neural progenitor cells. Nature Cell Biology. 2011;13(2):142–152. doi: 10.1038/ncb2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meulmeester E, Maurice MM, Boutell C, et al. Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced desta-bilization of Hdmx and Hdm2. Molecular Cell. 2005;18(5):565–576. doi: 10.1016/j.molcel.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 36.Oh YM, Yoo SJ, Seol JH. Deubiquitination of Chfr, a checkpoint protein, by USP7/HAUSP regulates its stability and activity. Biochemical and Biophysical Research Communications. 2007;357(3):615–619. doi: 10.1016/j.bbrc.2007.03.193. [DOI] [PubMed] [Google Scholar]
- 37.Schwertman P, Lagarou A, Dekkers DH, et al. UV-sensitive syndrome protein UVSSA recruits USP7 to regulate transcription-coupled repair. Nature Genetics. 2012;44(5):598–602. doi: 10.1038/ng.2230. [DOI] [PubMed] [Google Scholar]
- 38.van der Horst A, de Vries-Smits AM, Brenkman AB, et al. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nature Cell Biology. 2006;8(10):1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 39.Pfoh R, Lacdao IK, Georges AA, et al. Crystal structure of USP7 ubiquitin-like domains with an ICP0 peptide reveals a novel mechanism used by viral and cellular proteins to target USP7. PLoS Pathogens. 2015;11(6):e1004950. doi: 10.1371/journal.ppat.1004950. [DOI] [PMC free article] [PubMed] [Google Scholar]