Muscle hypertrophy is used as a physiological model to study how global transcription is affected by changes in cell size and DNA content. Myonuclear transcription is sensitive to both mechanical load and DNA content, with smaller myofibers unexpectedly having the highest level of transcription during hypertrophy.
Abstract
Myofibers increase size and DNA content in response to a hypertrophic stimulus, thus providing a physiological model with which to study how these factors affect global transcription. Using 5-ethynyl uridine (EU) to metabolically label nascent RNA, we measured a sevenfold increase in myofiber transcription during early hypertrophy before a change in cell size and DNA content. The typical increase in myofiber DNA content observed at the later stage of hypertrophy was associated with a significant decrease in the percentage of EU-positive myonuclei; however, when DNA content was held constant by preventing myonuclear accretion via satellite cell depletion, both the number of transcriptionally active myonuclei and the amount of RNA generated by each myonucleus increased. During late hypertrophy, transcription did not scale with cell size, as smaller myofibers (<1000 μm2) demonstrated the highest transcriptional activity. Finally, transcription was primarily responsible for changes in the expression of genes known to regulate myofiber size. These findings show that resident myonuclei possess a significant reserve capacity to up-regulate transcription during hypertrophy and that myofiber transcription is responsive to DNA content but uncoupled from cell size during hypertrophy.
INTRODUCTION
Global transcription is tightly coupled with cell size (Zhurinsky et al., 2010; Kempe et al., 2015; Padovan-Merhar et al., 2015). By adjusting the level of transcription, protein and RNA content are kept proportionate to cell size, ensuring optimal conditions for biochemical reactions (Zhurinsky et al., 2010; Marguerat and Bahler, 2012). Unlike RNA and protein, DNA content does not appear to scale with cell size (Schmidt and Schibler, 1995). For example, hepatocytes and neurons can become exceptionally large while still harboring two copies of the genome; however, transcriptional output per unit of DNA is increased to maintain mRNA concentration levels in such large cells (Sato et al., 1994; Miettinen et al., 2014). Consistent with the idea that transcriptional output is sensitive to cell size and DNA content, Padovan-Merhar et al. (2015) reported that cells are able to decrease transcriptional burst frequency or increase transcription burst magnitude to accommodate an increase in gene dosage or cytosolic volume, respectively.
Skeletal muscle cells (myofibers) are unique among the cells of the body in that each myofiber is a large syncytium containing hundreds of nuclei. In response to a hypertrophic stimulus, such as mechanical load, myofibers are able to significantly increase both size and total RNA content (von Walden et al., 2012). The increase in RNA content during myofiber hypertrophy is believed to be primarily driven by transcription, but it is unknown how such an increase in global transcription is coordinated among myonuclei. Does transcriptional output increase proportionally across all myonuclei, or is a subset of myonuclei transcriptionally active? Under resting conditions, transcription of specific mRNAs has been shown to occur in a pulsatile manner within a subset of myonuclei, but whether this behavior reflects global transcriptional output remains to be determined (Newlands et al., 1998).
It is well established that hypertrophic growth induced by mechanical overload results in myonuclear accretion via satellite cell fusion (McCarthy et al., 2011), yet it is unknown how transcriptional output is affected by such an increase in myofiber DNA content. One hypothesis proposes that myonuclear accretion is necessary during hypertrophy to increase the transcriptional capacity of the cell; however, myonuclear accretion does not appear to be absolutely required to support myofiber growth despite the observation that DNA content does limit cell size (Blaauw et al., 2009; McCarthy et al., 2011; Lee et al., 2012). We reported that total RNA content and myofiber growth persist in the absence of myonuclear accretion, although we did not investigate how this affected global transcription (McCarthy et al., 2011).
In the present study, we used skeletal muscle hypertrophy as a model system to determine how changes in DNA content and cell size affect nascent transcription. Unlike previous studies, skeletal muscle hypertrophy provides a nonpathological, in vivo model to study how these factors affect global transcription (Zhurinsky et al., 2010; Padovan-Merhar et al., 2015). Using 5-ethynyl uridine (EU) to label newly synthesized RNA, we tested the hypotheses that during muscle hypertrophy, nascent transcription 1) scales with myofiber size and 2) would be lower per myonucleus with increased DNA content. The results of the study demonstrate for the first time that during skeletal muscle hypertrophy, myofibers are almost entirely responsible for the significant increase in transcriptional output and that, like other cell types, global transcription of myofibers was sensitive to a change in DNA content; however, transcriptional output of myofibers did not scale with cell size, such that small myofibers had the highest level of transcription. Although the underlying mechanism for this uncoupling awaits further investigation, it may reflect the higher oxidative metabolism of small fibers, thereby providing a greater metabolic capacity to better support increased transcription.
RESULTS
Myogenic cells are the predominant source of nascent transcription during muscle hypertrophy
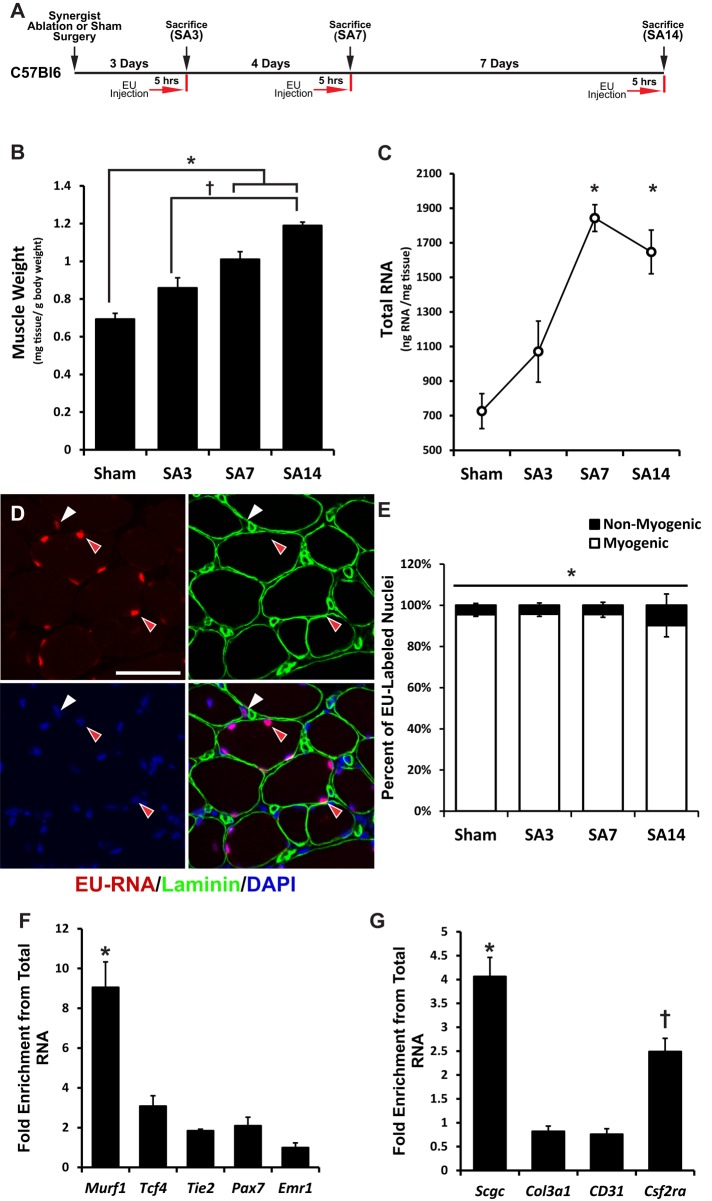
Synergist ablation surgery was used to place mechanical overload on skeletal muscle to induce hypertrophy, and global transcription was assessed via incorporation of a uridine analogue (EU; Figure 1A). Mechanical overload of the plantaris muscle induced by synergist ablation (SA) resulted in a progressive increase in muscle mass (Figure 1B) and myofiber cross-sectional area (Supplemental Figure S1), which was accompanied by a significant, ∼2.5-fold increase in total RNA per unit of tissue by day 7 (Figure 1C). Newly synthesized RNA that had incorporated the EU nucleotide (EU-RNA) was visualized on muscle cross-sections, in combination with a laminin antibody to define the basement membrane surrounding each myofiber. After a 5-h pulse, EU-RNA was only detectable within nuclei, which allowed for quantification of the percentage of skeletal muscle nuclei that were stained positive for EU-RNA (Figure 1D). Regardless of the time point, the vast majority (92–96%) of EU-positive nuclei were located within the laminin border, thus belonging to myogenic cells, that is, both myofiber and satellite cells (Figure 1E). Double labeling of EU-RNA and the satellite cell–specific transcription factor Pax7 revealed minimal colocalization (Supplemental Figure S2, related to Figure 1, C and D), indicating that myonuclei are the predominate source of newly synthesized RNA in response to mechanical overload.
FIGURE 1:
Mechanical overload results in a progressive increase in plantaris muscle mass and RNA content derived primarily from myogenic cells. Animals were subjected to synergist ablation (SA) for 3, 7, or 14 d and injected with 2 mg of EU 5 h before being killed. This paradigm enriched for nascent RNA (EU-RNA) labeling within myogenic cells. (A) Schematic for labeling of nascent RNA after mechanical overload. (B) Plantaris muscle mass normalized to the body weight of the animal. *Significant difference between SA7 and SA14 compared with Sham. †Significant difference between SA14 relative to SA3. (C) RNA abundance within the muscle tissue normalized to tissue weight. *Significant difference compared with Sham. (D) Red arrows indicate myogenic nuclei identified by their location within the laminin stain surrounding the myofiber. White arrow indicates a nonmyogenic nucleus outside the laminin stain of the myofiber. Scale bar, 50 μm. (E) Quantification of the percentage of either myogenic or nonmyogenic nuclei that stain positive for EU-RNA during mechanical overload of skeletal muscle. *Significantly greater percentage of EU-RNA–positive nuclei in myogenic cells than in nonmyogenic cells. (F) Enrichment for short-lived (∼4–6 h) cell type–specific markers. Murf1 (myofiber) was significantly enriched relative to Tcf4 (fibroblast cell), Tie2 (endothelial cell), Pax7 (satellite cell), and Emr1 (macrophage cell). *Significantly greater enrichment than with other cell-specific mRNAs. (G) Enrichment for intermediate-lived (12–18 h) cell type–specific markers. Scgc (myofiber) was significantly enriched relative to Col3a1 (fibroblast cell), CD31 (endothelial cell), and Csf2ra (macrophage cell). *Significantly greater enrichment than with all other markers. †Significant enrichment relative to Col3a1 and CD31. For all analyses, p < 0.05 and n = 5–7 at each time point. All data are presented as mean ± SEM.
Nascent transcription is highly enriched for muscle-specific transcripts
The limited amount of EU-RNA labeling observed in nonmuscle cells suggests that the rate of transcription was significantly less than in myofibers, and, as a result, the intensity of labeling in nonmuscle cell nuclei was below the level required for fluorescent detection. Therefore, in a complementary approach to the microscopic analysis, we affinity purified the labeled nascent RNA and performed quantitative PCR (qPCR) for cell type–specific transcripts and compared the expression profile to the profile obtained from the total RNA pool. If a cell type–specific transcript is enriched in the nascent RNA pool, independent of differences in transcript half-life, this would suggest that a greater proportion of the nascent RNA is being produced by that cell type. We focused on the major cells types that comprise skeletal muscle, including myofibers, satellite cells, fibroblasts, endothelial cells, and macrophages. The EU labeling microscopy data showed that the proportion of labeling between myogenic and nonmyogenic cells did not change during hypertrophic growth; therefore we focused qPCR analysis on sham-control muscle samples. To control for the potential effect of transcript half-life, we selected cell type–specific transcripts that had comparable half-lives and characterized them as either short lived or intermediate lived. As would be expected, the nascent RNA pool was enriched for short-lived transcripts. The only cell type–specific mRNA that was significantly enriched was Murf1, the myofiber-specific transcript (Figure 1F), which remained enriched across the hypertrophic time course (Supplemental Figure S3, A–C, related to Figure 1F). In agreement with the short-lived transcripts, the intermediate-lived, myofiber-specific transcript Sgcg was significantly enriched (Figure 1G). The intermediate-lived, macrophage-specific transcript Csf2ra also showed significant enrichment compared with Col3a1 and CD31, which are fibroblast and endothelial markers, respectively (Figure 1G).
EU-RNA has been readily detected in nonmyogenic cell types in other tissues, using the same pulse duration (Supplemental Figure S4; Jao and Salic, 2008). Thus it seems unlikely that nonmyogenic cells are not taking up the EU analogue. Instead, the inability to detect EU-labeled RNA in nonmyogenic cells suggests that the level of transcription of nascent RNA was significantly lower under resting conditions and during periods of hypertrophic growth. Collectively both microscopic and gene expression analyses demonstrate that metabolic labeling of nascent RNA in skeletal muscle was enriched for RNA produced within the myofiber. Thus we used this enrichment to assess how nascent transcription was affected by changes in DNA content and myofiber size in response to a hypertrophic stimulus.
Increased nascent transcription in individual myonuclei during hypertrophy
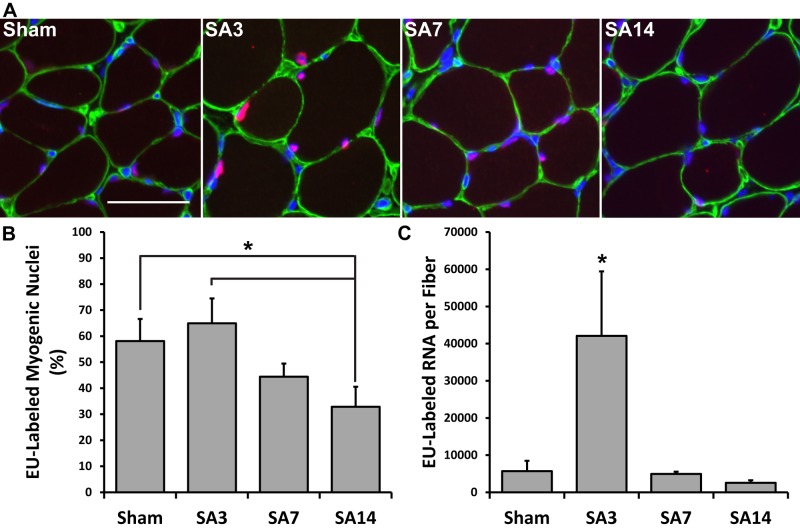
To determine whether the increase in total RNA content observed with hypertrophy is due to an increase in the percentage of transcriptionally active myonuclei, we quantified the number of EU-positive myonuclei relative to total myonuclei during mechanical overload–induced hypertrophy (Figure 2A). In comparison to sham (58%), there was no significant difference at SA3 and SA7, with ∼65 and 45% of myonuclei labeled, respectively; however, at SA14, the percentage of EU-positive myonuclei decreased by 43%, with only 32% of myonuclei being positive for EU-RNA (Figure 2B). Given that the increase in total RNA content observed at SA7 did not appear to be caused by an increase in the number of active myonuclei, we next quantified transcriptional output by determining the amount of EU-RNA at each time point using integrated fluorescence density. There was a significant, 7.4-fold increase in the amount of EU-RNA at SA3 compared with sham, with no difference at SA7 and SA14 (Figure 2C).
FIGURE 2:
Increased transcriptional demand during mechanical overload is met by elevated transcription within each myonucleus. EU-RNA within myonuclei was quantified on muscle cross-sections at various time points after mechanical overload. (A) Representative images of EU-RNA labeling during mechanical overload. Scale bar, 50 μm. (B) Quantification of the percentage of myonuclei that stained positive for EU-RNA during mechanical overload. *Significantly higher percentage of EU-RNA–positive nuclei at Sham and SA3 time points relative to SA14 (p < 0.05 and n = 5 or 6 mice per time point). (C) Quantification of the integrated density of EU-RNA labeling per myofiber during mechanical overload. *Significantly higher integrated density of EU-RNA at the SA3 time point relative to all other time points (p < 0.05 and n = 5 or 6 mice per time point). Data are presented as mean ± SEM.
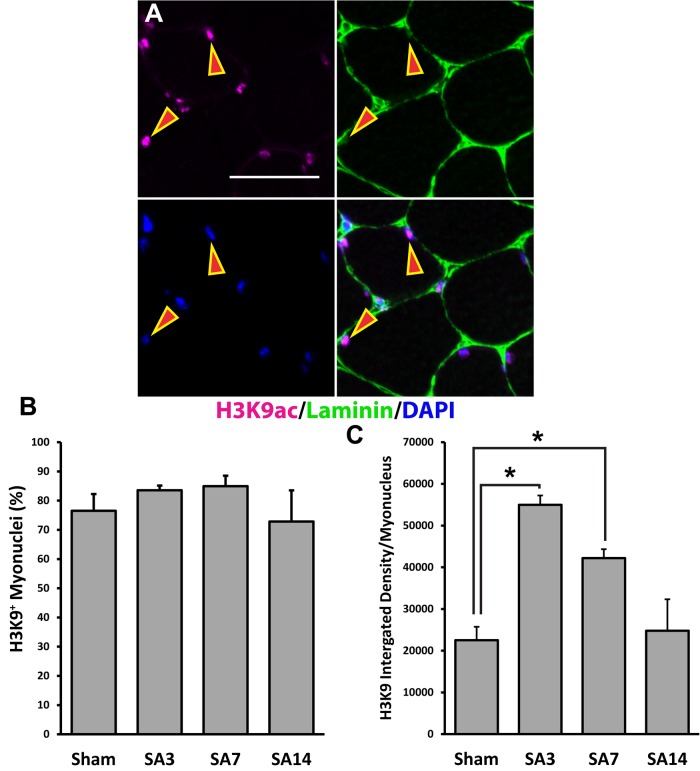
To determine whether the increase in EU-RNA at SA3 was the result of increased gene transcription in myogenic nuclei, we quantified the amount of H3K9ac, a histone modification indicative of active gene transcription (Figure 3A and Supplemental Figure S5) (Karmodiya et al., 2012; Wang et al., 2008). The percentage of myonuclei immunoreactive for H3K9ac did not change across the time course (Figure 3B); however, the intensity of H3K9ac immunofluorescence was significantly increased at SA3 and SA7, reflecting a global increase in promoter activation (Figure 3C). Because histone acetylation is short lived (Barth and Imhof, 2010), these results indicate the increased transcription observed at SA3 was due to higher transcriptional activity per myonucleus rather than more active myonuclei.
FIGURE 3:
Increased global transcription is associated with increased H3K9ac labeling intensity in myonuclei. H3K9ac staining within myonuclei was quantified on muscle cross-sections at various time points after mechanical overload. Arrows indicate H3K9ac-positive myonuclei. (A) Representative images for H3K9ac staining in skeletal muscle cross-sections. Scale bar, 50 μm. (B) Quantification of the percentage of myonuclei that stained positive for H3K9ac during mechanical overload. There was no significant change in the percentage of H3K9ac staining (p < 0.05 and n = 3 or 4 mice per time point). (C) Quantification of the integrated density of H3K9ac labeling per myonucleus during mechanical overload. *Significantly higher integrated density of H3K9ac labeling at the SA3 and SA7 time points relative to Sham (p < 0.05 and n = 3 or 4 mice per time point). Data are presented as mean ± SEM.
Decreased nascent transcription with increased DNA content
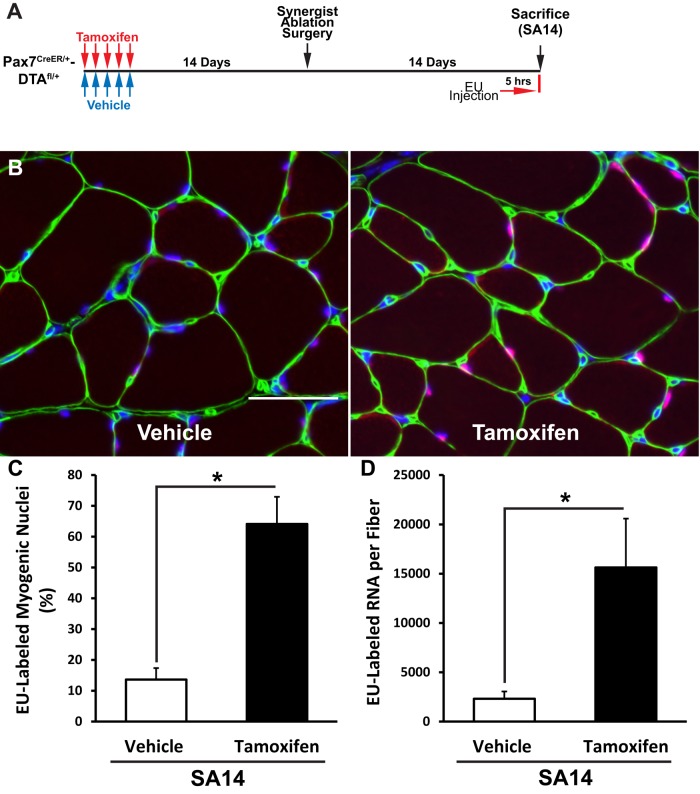
We reported that the synergist ablation model of hypertrophy induced a significant increase in the number of myonuclei per myofiber as the result of satellite cell fusion (McCarthy et al., 2011). Thus perhaps the reduced percentage of EU-labeled myonuclei observed at SA14 was the result of an increase in myofiber DNA content at this time point. To assess the effect of DNA content on transcription, we prevented myonuclear accretion by the conditional depletion of satellite cells using the previously described Pax7-DTA mouse strain (McCarthy et al., 2011; schematic in Figure 4A). After tamoxifen administration, the satellite cell pool was depleted by 90% (Supplemental Figure S6, A and B, related to Figure 4), resulting in these myofibers having 32% fewer myonuclei at SA14 than with vehicle-treated control muscle (Supplemental Figure S6C, related to Figure 4). The absence of nuclear fusion in satellite cell–depleted muscle resulted in a significantly greater percentage of myonuclei staining positive for EU-RNA at SA14 than with vehicle-treated muscle, such that it was comparable to the percentage of EU-labeled myonuclei observed at SA3 in wild-type mice (compare Figure 4, B and C, to Figure 2B). The amount of EU-RNA per myofiber was also significantly higher in tamoxifen-treated muscle at SA14 than with vehicle-treated muscle (Figure 4D). Thus resident myonuclei were sensitive to myofiber DNA content and able to maintain elevated levels of nascent transcription in the absence of myonuclear accretion to presumably meet the transcriptional demand required for hypertrophy.
FIGURE 4:
Myonuclei modulate transcriptional output to accommodate nuclear accretion. Nuclear accretion was inhibited by the specific and conditional depletion of satellite cells using the Pax7-DTA mouse strain. (A) Schematic. (B) Representative images of EU-RNA intensity with (vehicle) or without (tamoxifen) myonuclear accretion. (C) Increased relative number of EU-RNA–positive myonuclei in the absence of myonuclear accretion (tamoxifen). (D) Increased EU-RNA per myofiber in the absence of myonuclear accretion. *Significant difference between vehicle- and tamoxifen-treated animals (p < 0.05 and n = 6 or 7 mice). Data are presented as mean ± SEM.
Nascent transcription is uncoupled from cell size during hypertrophy
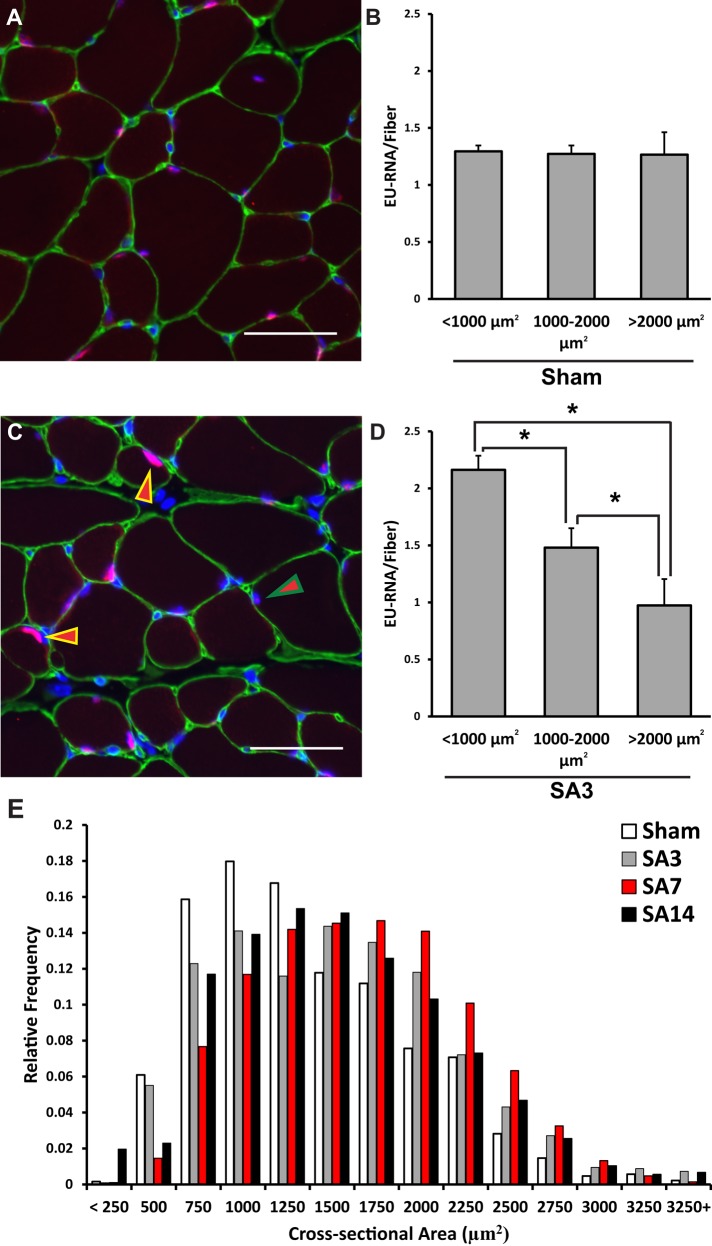
Given that global transcription is tightly coupled to cell size, we sought to determine the relationship between myofiber size and nascent transcription. We focused our analysis on SA3 samples because it was the only time point that showed a significant increase in EU-RNA per myofiber (Figure 2, A and C). Under sham conditions, there was no significant difference in the amount of EU-RNA produced in myofibers of different sizes (Figure 5, A and B). Although nascent transcription was increased in all myofibers at SA3 relative to sham, the smallest myofibers (<1000 μm2) showed the highest amount of EU-RNA (Figure 5, C and D). Furthermore, a greater proportion of myonuclei in these small fibers stained positive for EU-RNA than with sham (Supplemental Figure S7, A and B, related to Figure 5). However, by SA7 and SA14, there was a rightward shift toward a greater proportion of larger fibers containing an EU-RNA label (Supplemental Figure S7, C and D), demonstrating a similar distribution to the sham condition. The rightward shift at SA14 was also observed in the Pax7-DTA strain (Supplemental Figure S8, A and B), indicating that nuclear accretion was not responsible for the higher transcription in larger fibers at SA14. Fibers were binned based on their size to determine how fiber cross-sectional area changed during overload-induced hypertrophy (Figure 5E). Based on the relative frequency of fiber cross-sectional area, it appeared that the most pronounced growth occurred in small myofibers (<1000 μm2), with a large decrease in the frequency of these fibers by SA7. These findings show that, unlike other cell types, transcriptional output is uncoupled from cell size in myofibers during hypertrophic growth.
FIGURE 5:
Increased global transcription preferentially occurs in small myofibers during hypertrophic growth. Myofibers were binned based on their cross-sectional area to determine the effect of cell size on EU-RNA integrated density levels. (A) Representative image of EU-RNA labeling intensity under Sham conditions. Scale bar, 50 μm. (B) No significant difference in EU-RNA per fiber with across cell size under sham conditions. (C) Representative image for increased EU-RNA labeling in small myofibers at SA3. Yellow/red arrowhead indicates a nucleus in a cell <1000 μm2, and green/red arrow indicates a nucleus in a cell >2000 μm2. Note the difference in EU-RNA staining intensity in the fibers <1000 μm2. Scale bar, 50 μm. (D) Progressive decrease in EU-RNA integrated density with increasing cell size. *Significant difference between cell sizes (n = 5 for each cell size bin). (E) Mechanical overload results in a rightward shift in the relative frequency of large fibers (>2000 μm2) at SA7 (n = 5 or 6 mice per time point). All data are presented as mean ± SEM.
Labeling of nascent RNA reveals potential posttranscriptional regulation of gene expression
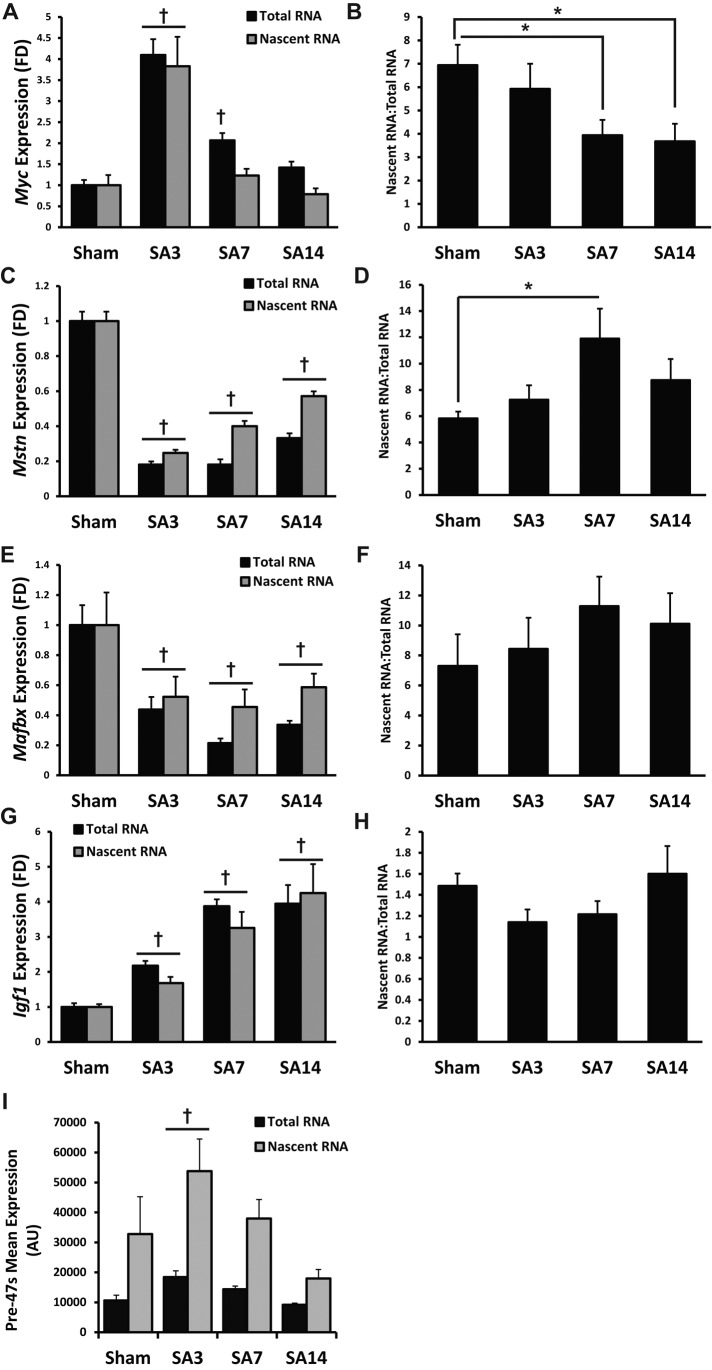
The in vivo imaging results showed that nascent transcription was dramatically up-regulated during the early stage of muscle hypertrophy. We wanted to take advantage of the myofiber enrichment of nascent RNA to quantify the expression of genes known to have a role in regulating global transcription and, secondarily, determine whether changes in gene expression were the result of transcription and/or mRNA stability. Two well-characterized pathways that are known to regulate muscle hypertrophy are the mammalian target of rapamycin complex 1 (mTORC1) and Wnt/β-catenin pathways (Armstrong et al., 2006; Goodman et al., 2011). Downstream targets of these pathways have been shown to influence global transcription, as well as ribosomal DNA transcription (Nader et al., 2005; Iadevaia et al., 2012; Lin et al., 2012). Taking a candidate approach, we selected genes known to be regulated by these pathways and responsive (either up- or down-regulated) to mechanical overload–induced muscle hypertrophy; we measured by qPCR the total RNA and nascent RNA expression levels of cMyc, Mstn, Mafbx, and Igf1 to determine whether the change in expression was regulated at the level of transcription and/or mRNA stability.
cMyc was significantly more abundant in both the total and nascent RNA fractions (∼4.1- and ∼3.8-fold, respectively) at SA3, indicating that transcription was the primary mechanism driving this increase (Figure 6, A and B). cMyc mRNA remained significantly elevated at SA7 as the result of increased mRNA stability, given that nascent RNA returned to baseline; cMyc mRNA at SA14 was no different from sham, although mRNA stability remained significantly higher (Figure 6, A and B). Mstn expression was significantly decreased in both RNA fractions after mechanical overload, which initially (SA3) was caused by a decrease in transcription that at a later time point (SA7) was the result of a decrease in mRNA stability (Figure 6, C and D). Similarly, Mafbx mRNA was significantly decreased during mechanical overload as the result of a decrease in transcription with no change in transcript stability (Figure 6, E and F). Finally, Igf1 mRNA significantly increased in response to mechanical overload, and this was due to an increase in transcription with no apparent change in mRNA stability (Figure 6, G and H).
FIGURE 6:
Nascent RNA labeling reveals altered stability of mRNAs involved in regulating global transcription. Nascent RNA was affinity purified from total RNA, and both fractions were used to determine changes in transcript levels and transcript stability in response to mechanical overload. A decrease in the nascent RNA:total RNA indicates increased transcript stability, whereas an increase in the nascent RNA:total RNA indicates decreased transcript stability. (A, B) cMyc. (A) cMyc shows increased abundance during mechanical overload. †Significant increase within the respective RNA fraction relative to Sham conditions. (B) cMyc demonstrates increased transcript stability at SA7 and SA14 relative to Sham. *Significant difference from Sham. (C, D) Mstn. (C) Mstn shows decreased expression during mechanical overload. †Significant decrease within the respective RNA fraction relative to Sham conditions. (D) Mstn demonstrates decreased transcript stability at SA7 relative to Sham. *Significant difference from Sham. (E, F) Mafbx. (E) Mafbx shows decreased expression during mechanical overload. †Significant decrease within the respective RNA fraction relative to Sham conditions. (F) No significant change to Mafbx stability during mechanical overload. (G) Igf1 expression after mechanical overload. †Significant increase within the respective RNA fraction relative to Sham conditions (p < 0.05 and n = 5 mice per time point). (H) No significant change to Igf1 stability with mechanical overload. (I) Nascent RNA is enriched for rRNA pre-47S, which increased at SA3 in response to mechanical overload. †Significant increase within the respective RNA fraction relative to Sham conditions (p < 0.05 and n = 5 mice per time point for all genes). All data are presented as mean ± SEM.
Given that the vast majority of cellular RNA is rRNA, it is likely that the significant increase in EU-RNA observed at SA3 was rRNA. Consistent with the microscopic analysis, pre-47S expression was significantly higher at SA3 relative to all other time points (Figure 6I). This finding indicated that ribosome biogenesis was up-regulated at SA3, providing the basis for the significant increase in total RNA content observed at SA7 and SA14 (Figure 1B).
DISCUSSION
Skeletal muscle myofibers comprise a multinucleated syncytium capable of readily increasing cellular volume and DNA content in response to a hypertrophic stimulus. The purpose of this study was to determine how these factors influence nascent transcription during hypertrophy, which was induced by mechanical overload. The major findings of the study are as follows: 1) myofibers are overwhelmingly the most transcriptionally active cell type in skeletal muscle at rest and during hypertrophy, 2) in response to mechanical overload, resident myonuclei demonstrate the intrinsic capacity to significantly up-regulate transcription by greater than sevenfold, 3) when myofiber DNA content is held constant during hypertrophy, transcription remains significantly elevated for a prolonged period of time in resident myonuclei, and 4) in contrast to other cell types, in mechanically overloaded muscle, transcription is highest in the smallest myofibers.
Whether myofibers partition global transcriptional output equally across all myonuclei within the syncytium or instead restrict transcription to a subset of nuclei was unknown. Further, it was not known how the partitioning of transcriptional output might be altered in response to a hypertrophic stimulus and what influence cell size and DNA content may have on this process. A previous study showed that myonuclear transcription of a few select genes occurred in pulses and that the proportion of active myonuclei changed during postnatal growth (Newlands et al., 1998). Taking a global approach, we show that nascent transcription is heterogeneous across myonuclei at rest, and, in response to mechanical overload, transcription is significantly up-regulated but only at the initial stage (SA3) of hypertrophy. This up-regulation of transcription is the result of individual myonuclei increasing their transcriptional output rather than simply increasing the proportion of transcriptionally active myonuclei. Furthermore, this increase in transcription cannot be explained by an initial larger myonuclear domain, as smaller fibers actually have a smaller myonuclear domain (van der Meer et al., 2011a, b). Although the mechanisms regulating the increase in myonuclear transcription remain to be determined, increases in cytoplasmic volume were reported to increase the amplitude of transcriptional burst (Padovan-Merhar et al., 2015); however, we did not observe a significant increase in myofiber size until SA7. This finding indicates that myofibers are able to dramatically increase global transcription, independent of detectable changes in cell size. Like protein synthesis, it may be that the rate of transcription within the myofiber is up-regulated in response to an increase in mechanical load (Goodman et al., 2012).
A unique property of muscle cells is the ability to increase their DNA content during hypertrophy via the fusion of satellite cells (McCarthy et al., 2011). This characteristic has historically been viewed as necessary during hypertrophy to maintain the myonuclear domain size, thereby ensuring the transcription output of each myonucleus is sufficient to meet the global transcriptional demand required to support hypertrophic growth. Contrary to this idea, we showed previously that myofibers can undergo robust growth when DNA content is held constant after the depletion of satellite cells (McCarthy et al., 2011). The results of the present study suggest that muscle is able to compensate for the loss of satellite cells by maintaining a higher level of transcription in the existing myonuclei. How myonuclei are able to sustain an elevated level of transcription with a relatively lower DNA content remains to be determined, but one possibility is increased transcriptional pulse frequency. Padovan-Merhar et al. (2015) reported that transcriptional pulse frequency changed according to genomic content, such that higher DNA content caused a decrease in pulse frequency.
A somewhat surprising finding of the present study was the apparent disconnect between cell size and global transcription during hypertrophy, with smaller myofibers showing the highest level of nascent transcription. A possible explanation for this finding may involve the metabolic phenotype of small myofibers, which is quite different from that of the larger myofibers (Goodman et al., 2012; Fry et al., 2014). Small fibers in the mouse plantaris muscle are classified as type IIa and have a high oxidative metabolic capacity due to a high mitochondrial content. Given that global transcription is highly dependent on cellular ATP levels and mitochondrial mass (das Neves et al., 2010), type IIa myofibers may have a greater biosynthetic activity than larger, more glycolytic myofibers. This notion is supported by the work of Goodman et al. (2012), who showed that the rate of protein synthesis was highest in small, type IIa myofibers during hypertrophy, with these fibers also undergoing the greatest hypertrophy. It is likely that the disconnect between cell size and transcription that we observed is not a unique property of skeletal muscle but instead reflects a difference between cells under homeostatic conditions and cells undergoing hypertrophy. Alternatively, it may be that multinucleated myofibers regulate global transcription differently than mononucleated cells during hypertrophic growth.
In addition to investigating transcription dynamics, the labeling of nascent RNA also provides the ability to determine whether changes in gene expression are regulated at the level of transcription and/or mRNA stability. We focused on genes that have a known role in regulating global transcription (cMyc) or skeletal muscle size (Mstn, Murf1, and Igf1) and are responsive to changes in mechanical load (Adams and Haddad, 1996; Yamaguchi et al., 2006; Chaillou et al., 2014). In general, early (SA3) changes in mRNA abundance, regardless of whether an mRNA was increased or decreased, were primarily driven by transcription, whereas at later time points (SA7 and SA14), changes in mRNA stability made a greater contribution to mRNA abundance. This finding, together with the fact that nascent transcription was highly myofiber specific, provides the foundation for a more comprehensive study aimed at developing regulatory networks based on shared mechanisms of regulation.
Together the findings of this study provide the first evidence describing the transcriptional response of myonuclei to a hypertrophic stimulus. In particular, the evidence presented shows that, similar to other cell types, the transcriptional output of resident myonuclei is sensitive to changes in myofiber DNA content and that, unexpectedly, during hypertrophy the level of transcription does not scale with cell size, suggesting that small myofibers require an unusually high transcriptional output. Moreover, the results of this study demonstrate that skeletal muscle hypertrophy provides a powerful physiological model with which to better understand how changes in DNA content and cell size affect global transcription.
MATERIALS AND METHODS
Mice
All animal procedures were conducted in accordance with institutional guidelines for the care and use of laboratory animals as approved by the Institutional Animal Care and Use Committee of the University of Kentucky. For initial time course experiments, male and female C57BL/6J mice 4–6 mo of age (Jackson Laboratory, Bar Harbor, ME) were housed in a temperature- and humidity-controlled room and maintained on a 14:10 h light:dark cycle with food and water ad libitum. For myonuclear accretion experiments, male and female Pax7-DTA mice were used as previously described (McCarthy et al., 2011; Fry et al., 2014; Lee et al., 2015). Briefly, these mice allow for the inducible and specific depletion of the primary stem cell (satellite cell) in adult skeletal muscle after administration of tamoxifen. At 4 mo of age, Pax7-DTA mice were administered by intraperitoneal injection either vehicle (15% ethanol in sunflower seed oil) or tamoxifen (2 mg/d) for 5 consecutive days, followed by a 2-wk washout period before synergist ablation surgery.
The mice were subjected to bilateral synergist ablation surgery to induce hypertrophy of the plantaris muscle as previously described in detail (McCarthy and Esser, 2007). Briefly, after anesthetization with a mixture of 95% oxygen and 5% isoflurane gas, the soleus and the majority of the gastrocnemius muscles were surgically excised, with particular attention made to ensure that the neural and vascular supply remained intact and undamaged for the remaining plantaris muscle. Sham surgery controls involved similar procedures without gastrocnemius and soleus muscle excision. After recovery from surgery, mice were anesthetized at the designated time point by an intraperitoneal injection of ketamine (100 mg/kg) and xylazine (10 mg/kg), and plantaris muscles were excised and weighed. Tissue used for RNA was flash frozen in liquid nitrogen and stored at −80°C until further use. Tissue used for microscopy was pinned to a cork block at resting length, covered with a thin layer of Tissue Tek optimum cutting temperature (OCT) compound (Sakura Finetek, Torrance, CA), and then quickly frozen in liquid nitrogen–cooled isopentane and stored at −80°C until sectioning. For time-course experiments, plantaris muscle was collected at 3, 7, and 14 d after the surgery (SA3, SA7, and SA14, respectively; n = 5 per time point). Control plantaris muscle (n = 5) was collected from mice subjected to a sham synergist ablation surgery at each of the time points (Sham). For nuclear accretion experiments, plantaris was collected 14 d after surgery (n = 6). After collection of the plantaris muscle, the mice were killed by cervical dislocation under anesthesia.
Nascent RNA labeling
For nascent RNA visualization and expression experiments, mice were given an intraperitoneal injection of 2 mg of EU (Jena Biosciences, Jena, Germany) suspended in sterile phosphate-buffered saline 5 h before being killed. Five hours was chosen after it was determined this pulse period allows for maximal visual detection of nascent RNA within the nucleus in both sham and overloaded skeletal muscle (Supplemental Figure S9). EU is a uridine analogue that is incorporated specifically into newly synthesized RNA (Jao and Salic, 2008). Each EU molecule had been modified to contain an alkyne group, which can then be used for detection with molecules containing an azide group via copper-mediated “click” chemistry.
Histochemistry
Frozen tissue was sectioned (7 μm), air dried for ∼20 min, and then immediately fixed in 4% paraformaldehyde. For EU-RNA detection, sections were incubated in a solution containing Tris base (100 mM), copper(II) sulfate (4 mM), biotin-conjugated azide (100 μm; Jena Biosciences), and ascorbic acid (100 mM) for 30 min at room temperature and washed, and this procedure was repeated. Sections were then incubated in Texas red–streptavidin (1:150; Vector Labs, Burlingame, CA) for 60 min at room temperature. Sections were postfixed in 4% paraformaldehyde (PFA), blocked in 1% bovine serum albumin (BSA) for 1 h, and then incubated with an anti-laminin antibody (1:100; Sigma-Aldrich, St. Louis, MO) overnight at 4°C. Sections were washed, and laminin immunoreactivity was visualized with Alexa Fluor 488 goat anti-rabbit secondary (1:500; Invitrogen, Carlsbad, CA) and counterstained with 4′,6-diamidino-2-phenylindole (DAPI). For histine 3 lysine 9 acetylation (H3K9ac), sections were air dried for 1 h, fixed in 4% PFA, blocked using 1% BSA, and incubated in primary antibody (1:1000; ab10812; Abcam, Cambridge, MA) overnight 4°C using a 1:500 dilution in 1% BSA. H3K9ac was visualized using Alexa Fluor 594 chicken anti-rabbit secondary (1:500; Invitrogen). Sections were washed and reacted with laminin antibody and DAPI as described.
For Pax7 detection, sections were fixed in 4% PFA, followed by epitope retrieval using sodium citrate (10 mM, pH 6.5) at 92°C for 20 min. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide in phosphate-buffered saline for 7 min, followed by an additional blocking step with Mouse-on-Mouse Blocking Reagent (Vector Laboratories). Incubation with Pax7 antibody (1:100; Developmental Studies Hybridoma Bank, Iowa City, IA) was followed by incubation with the biotin-conjugated secondary antibody (1:1000; Jackson ImmunoResearch, West Grove, PA), and the signal was amplified using streptavidin–horseradish peroxidase included within a tyramide signal amplification kit (Invitrogen). Tyramide signal amplification–Alexa Fluor 488 or 594 was used to visualize antibody binding.
Image acquisition and quantification
Images were acquired using a Zeiss upright microscope (AxioImager M1; Carl Zeiss, Oberkochen, Germany) at 20× magnification, and analysis was carried out using the AxioVision Rel 4.8 software (Zeiss). During image acquisition of EU-RNA and H3K9ac, careful consideration was taken to select an exposure that fell within the linear portion of the dynamic range of the camera to avoid overexposure. EU-RNA– and H3K9ac-positive staining was determined by thresholding a lower-density cutoff, which was then maintained for all images analyzed. An integrated density value for each EU-RNA– and H3K9ac-positive area was calculated using the formula
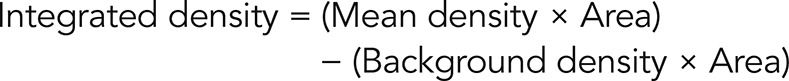 |
Densities across different exposure times were normalized, taking into account the dark pixel intensity (Pang et al., 2012). EU-RNA– and H3K9ac-positive nuclei were considered to be from myogenic cells when a DAPI-positive nucleus fell inside the laminin stain of the myofiber or nonmyogenic when the nucleus fell outside the laminin border. Nuclei that did not contain an EU-RNA– or H3K9ac-positive area were manually counted. Based on the results of the EU-RNA experiments, H3K9ac quantification was restricted to myogenic cells. For each animal, an average of 786 ± 37 and 609 ± 48 fibers were analyzed per cross-sectional area for C57BL6/J and Pax7-DTA mouse strains, respectively.
Cell-size global transcription analysis
Myofiber cross-sectional area was quantified on the same images used for EU-RNA quantification. Cross-sectional area was determined using automated segmentation of the myofiber by selecting an intensity threshold below that of the laminin fluorescent stain. For each animal, an average of 509 ± 36 fibers per cross-sectional area were analyzed. A relative fiber frequency based on cross-sectional area was generated for each animal and averaged for each time point. Based on the inherent variability between the cross-sectional area on male and female mice, the cross-sectional area of male mice was normalized using a correction factor (0.779) derived from a large cohort of control mice (n = 24 female and n = 24 male) from previous studies (McCarthy et al., 2011; Fry et al., 2014) and unpublished data.
To determine the influence of cell size on transcriptional output, data from the cross-sectional and EU-RNA analyses were aligned; only myofibers that contained an EU-RNA–positive area were included in the analysis. If a single myofiber contained multiple EU-RNA labels (i.e., multiple nuclei), the data were summed to give a total amount of EU-RNA for each fiber. About 200 myofibers were quantified per animal, with each EU-RNA quantity normalized to the geometric mean of all EU-RNA quantities. This was performed to facilitate comparisons across animals with different overall labeling intensities, since the primary objective was to determine how variation in cell size affected EU-RNA levels within each animal.
Nascent RNA affinity purification and cDNA synthesis
Total RNA was prepared from plantaris muscle using TRIzol reagent (Invitrogen) according to the manufacturer’s directions. RNA samples were treated with TURBO DNase (Ambion, Austin, TX) to remove genomic DNA contamination. Total RNA concentration and purity were assessed by measuring the optical density (230, 260, and 280 nm) with a Nanodrop 1000 Spectrophotometer (ThermoFisher Scientific, Wilmington, DE). Nascent RNA was affinity purified from 4.5 μg of total RNA using the commercially available Click-iT Nascent RNA Capture Kit (Life Technologies, Carlsbad, CA). cDNA was generated on 500 ng of total RNA and all EU affinity-purified RNA using the SuperScript VILO cDNA Synthesis Kit (Life Technologies).
Cell type–specific transcript analysis
qPCR was performed using cDNA generated from both total and nascent RNA. To minimize the effect of transcript half-life on the enrichment analysis, cell type–specific transcripts were selected based on a narrow range of half-lives (Sharova et al., 2009). Short-lived transcripts had a half-life of 4-6.5 h, whereas intermediate-lived transcripts had a half-life of 16–18 h. Short-lived, cell type–specific transcripts included muscle RING-finger protein-1 (Murf1 or Trim63; myofiber), paired box 7 (Pax7; satellite cell), epidermal growth factor-like module–containing mucin-like hormone receptor–like 1 (Emr1 or F4/80; macrophage), transcription factor 4 (Tcf4; fibroblast), and endothelial-specific receptor tyrosine kinase (Tek or Tie2; endothelial cell). Intermediate-lived (16 to 18-h half-life) cell type–specific transcripts included γ-sarcoglycan (Scgc; myofiber), colony-stimulating factor 2 receptor, α, low-affinity (granulocyte-macrophage) (Csf2ra; macrophage), collagen, type III, α,1 (Col3a1; fibroblast) and platelet/endothelial cell adhesion molecule 1 (CD31 or Pecam1; endothelial cell). The limited number and low expression of satellite cell–specific genes precluded detection of an intermediate-lived transcript for satellite cells. Cycle threshold (Ct) was determined using KiCqStart qPCR ReadyMix (KCQS07; Sigma-Aldrich) using 5 μl of diluted cDNA (1/20 dilution from stock cDNA mixture). qPCR was performed using an ABI 7900HT Fast RT-PCR system (Invitrogen), and Ct was determined using ABI 7900HT Sequence Detection Systems software, version 2.3. Absolute quantification was achieved by exponential conversion of the Ct using the qPCR efficiency, which was estimated from standard curves obtained by serial dilutions (one-log range) of a pooled sample for each RT set. Relative quantification for each transcript was determined using the relative standard curve method, in which absolute values are generated for both the transcript of interest and four stable transcripts (Rn7sk, Gapdh, Tmx4, and Capzb) known to show little change in abundance in the synergist ablation model of skeletal muscle hypertrophy (Chaillou et al., 2013). The geometric mean of the four stable transcripts was used to normalize the data (Vandesompele et al., 2002). Nascent RNA enrichment for each mRNA was determined by calculating its fold difference between nascent RNA and total RNA. Primers for all transcripts are listed in Supplemental Table S1.
Transcript stability assessment
Transcript stability was assessed by qPCR for myelocytomatosis oncogene (cMyc), myostatin (Mstn), F-box protein 32 (Mafbx, Fbxo32, atrogin-1), and insulin-like growth factor 1 (Igf1). Transcript stability was calculated as the ratio between the amounts of nascent RNA and total RNA (nascent RNA:total RNA). All comparisons were made relative to the sham condition ratio. A decrease in the nascent RNA:total RNA ratio indicated that less mRNA was actively being transcribed relative to the total RNA fraction, which was interpreted as an increase in transcript stability. Conversely, an increase in the nascent RNA:total RNA ratio indicated that more mRNA was actively being transcribed relative to the total RNA fraction, which was interpreted as a decrease in transcript stability.
Statistical analyses
Data were analyzed using either a one-way or two-way analysis of variance, followed by Tukey’s post hoc analysis to discriminate which means differed. In cases of nonnormal distribution, data were log transformed. For all analyses, significance was set at p < 0.05. Data are presented as mean ± SEM.
Supplementary Material
Acknowledgments
We thank Chris Fry for helping with tissue collection and Sarah White for statistical advice. We also thank Sami Michaelis for helping with image analysis. This work was supported by National Institutes of Health Grant AR060701 to J.J.M. and C.A.P.
Abbreviations used:
- EU
5-ethynyl uridine
- SA
synergist ablation.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-08-0585) on January 13, 2016.
RREFERENCES
- Adams GR, Haddad F. The relationships among IGF-1, DNA content, and protein accumulation during skeletal muscle hypertrophy. J Appl Physiol. 1996;81:2509–2516. doi: 10.1152/jappl.1996.81.6.2509. [DOI] [PubMed] [Google Scholar]
- Armstrong DD, Wong VL, Esser KA. Expression of beta-catenin is necessary for physiological growth of adult skeletal muscle. Am J Physiol Cell Physiol. 2006;291:C185–C188. doi: 10.1152/ajpcell.00644.2005. [DOI] [PubMed] [Google Scholar]
- Barth TK, Imhof A. Fast signals and slow marks: the dynamics of histone modifications. Trends Biochem Sci. 2010;35:618–626. doi: 10.1016/j.tibs.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Blaauw B, Canato M, Agatea L, Toniolo L, Mammucari C, Masiero E, Abraham R, Sandri M, Schiaffino S, Reggiani C. Inducible activation of Akt increases skeletal muscle mass and force without satellite cell activation. FASEB J. 2009;23:3896–3905. doi: 10.1096/fj.09-131870. [DOI] [PubMed] [Google Scholar]
- Chaillou T, Kirby TJ, McCarthy JJ. Ribosome biogenesis: emerging evidence for a central role in the regulation of skeletal muscle mass. J Cell Physiol. 2014;229:1584–1594. doi: 10.1002/jcp.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaillou T, Lee JD, England JH, Esser KA, McCarthy JJ. Time course of gene expression during mouse skeletal muscle hypertrophy. J App Physiol. 2013;115:1065–1074. doi: 10.1152/japplphysiol.00611.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- das Neves RP, Jones NS, Andreu L, Gupta R, Enver T, Iborra FJ. Connecting variability in global transcription rate to mitochondrial variability. PLoS Biol. 2010;8:e1000560. doi: 10.1371/journal.pbio.1000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry CS, Lee JD, Jackson JR, Kirby TJ, Stasko SA, Liu H, Dupont-Versteegden EE, McCarthy JJ, Peterson CA. Regulation of the muscle fiber microenvironment by activated satellite cells during hypertrophy. FASEB J. 2014;28:1654–1665. doi: 10.1096/fj.13-239426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman CA, Frey JW, Mabrey DM, Jacobs BL, Lincoln HC, You JS, Hornberger TA. The role of skeletal muscle mTOR in the regulation of mechanical load-induced growth. J Physiol. 2011;589:5485–5501. doi: 10.1113/jphysiol.2011.218255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman CA, Kotecki JA, Jacobs BL, Hornberger TA. Muscle fiber type-dependent differences in the regulation of protein synthesis. PLoS One. 2012;7:e37890. doi: 10.1371/journal.pone.0037890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iadevaia V, Zhang Z, Jan E, Proud CG. mTOR signaling regulates the processing of pre-rRNA in human cells. Nucleic Acids Res. 2012;40:2527–2539. doi: 10.1093/nar/gkr1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao CY, Salic A. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc Natl Acad Sci USA. 2008;105:15779–15784. doi: 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmodiya K, Krebs AR, Oulad-Abdelghani M, Kimura H, Tora L. H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics. 2012;13:424. doi: 10.1186/1471-2164-13-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempe H, Schwabe A, Cremazy F, Verschure PJ, Bruggeman FJ. The volumes and transcript counts of single cells reveal concentration homeostasis and capture biological noise. Mol Biol Cell. 2015;26:797–804. doi: 10.1091/mbc.E14-08-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JD, Fry CS, Mula J, Kirby TJ, Jackson JR, Liu F, Yang L, Dupont-Versteegden EE, McCarthy JJ, Peterson CA. Aged muscle demonstrates fiber-type adaptations in response to mechanical overload, in the absence of myofiber hypertrophy, independent of satellite cell abundance. J Gerontol A Biol Sci Med Sci. 2015 doi: 10.1093/gerona/glv033. glv033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Huynh TV, Lee YS, Sebald SM, Wilcox-Adelman SA, Iwamori N, Lepper C, Matzuk MM, Fan CM. Role of satellite cells versus myofibers in muscle hypertrophy induced by inhibition of the myostatin/activin signaling pathway. Proc Natl Acad Sci USA. 2012;109:E2353–E2360. doi: 10.1073/pnas.1206410109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marguerat S, Bahler J. Coordinating genome expression with cell size. Trends Genet. 2012;28:560–565. doi: 10.1016/j.tig.2012.07.003. [DOI] [PubMed] [Google Scholar]
- McCarthy JJ, Esser KA. MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J Appl Physiol. 2007;102:306–313. doi: 10.1152/japplphysiol.00932.2006. [DOI] [PubMed] [Google Scholar]
- McCarthy JJ, Mula J, Miyazaki M, Erfani R, Garrison K, Farooqui AB, Srikuea R, Lawson BA, Grimes B, Keller C, et al. Effective fiber hypertrophy in satellite cell-depleted skeletal muscle. Development. 2011;138:3657–3666. doi: 10.1242/dev.068858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen TP, Pessa HK, Caldez MJ, Fuhrer T, Diril MK, Sauer U, Kaldis P, Bjorklund M. Identification of transcriptional and metabolic programs related to mammalian cell size. Curr Biol. 2014;24:598–608. doi: 10.1016/j.cub.2014.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nader GA, McLoughlin TJ, Esser KA. mTOR function in skeletal muscle hypertrophy: increased ribosomal RNA via cell cycle regulators. Am J Physiol Cell Physiol. 2005;289:C1457–C1465. doi: 10.1152/ajpcell.00165.2005. [DOI] [PubMed] [Google Scholar]
- Newlands S, Levitt LK, Robinson CS, Karpf AB, Hodgson VR, Wade RP, Hardeman EC. Transcription occurs in pulses in muscle fibers. Genes Dev. 1998;12:2748–2758. doi: 10.1101/gad.12.17.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padovan-Merhar O, Nair GP, Biaesch AG, Mayer A, Scarfone S, Foley SW, Wu AR, Churchman LS, Singh A, Raj A. Single mammalian cells compensate for differences in cellular volume and DNA copy number through independent global transcriptional mechanisms. Mol Cell. 2015;58:339–352. doi: 10.1016/j.molcel.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Z, Laplante NE, Filkins RJ. Dark pixel intensity determination and its applications in normalizing different exposure time and autofluorescence removal. J Microsc. 2012;246:1–10. doi: 10.1111/j.1365-2818.2011.03581.x. [DOI] [PubMed] [Google Scholar]
- Sato S, Burgess SB, McIlwain DL. Transcription and motoneuron size. J Neurochem. 1994;63:1609–1615. doi: 10.1046/j.1471-4159.1994.63051609.x. [DOI] [PubMed] [Google Scholar]
- Schmidt EE, Schibler U. Cell size regulation, a mechanism that controls cellular RNA accumulation: consequences on regulation of the ubiquitous transcription factors Oct1 and NF-Y and the liver-enriched transcription factor DBP. J Cell Biol. 1995;128:467–483. doi: 10.1083/jcb.128.4.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharova LV, Sharov AA, Nedorezov T, Piao Y, Shaik N, Ko MS. Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res. 2009;16:45–58. doi: 10.1093/dnares/dsn030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer SF, Jaspers RT, Jones DA, Degens H. Time-course of changes in the myonuclear domain during denervation in young-adult and old rat gastrocnemius muscle. Muscle Nerve. 2011a;43:212–222. doi: 10.1002/mus.21822. [DOI] [PubMed] [Google Scholar]
- van der Meer SF, Jaspers RT, Jones DA, Degens H. The time course of myonuclear accretion during hypertrophy in young adult and older rat plantaris muscle. Ann Anat. 2011b;193:56–63. doi: 10.1016/j.aanat.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Walden F, Casagrande V, Ostlund Farrants AK, Nader GA. Mechanical loading induces the expression of a Pol I regulon at the onset of skeletal muscle hypertrophy. Am J Physiol Cell Physiol. 2012;302:C1523–C1530. doi: 10.1152/ajpcell.00460.2011. [DOI] [PubMed] [Google Scholar]
- Wang L, Charroux B, Kerridge S, Tsai CC. Atrophin recruits HDAC1/2 and G9a to modify histone H3K9 and to determine cell fates. EMBO Rep. 2008;9:555–562. doi: 10.1038/embor.2008.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A, Fujikawa T, Shimada S, Kanbayashi I, Tateoka M, Soya H, Takeda H, Morita I, Matsubara K, Hirai T. Muscle IGF-I Ea, MGF, and myostatin mRNA expressions after compensatory overload in hypophysectomized rats. Pflugers Arch. 2006;453:203–210. doi: 10.1007/s00424-006-0127-9. [DOI] [PubMed] [Google Scholar]
- Zhurinsky J, Leonhard K, Watt S, Marguerat S, Bahler J, Nurse P. A coordinated global control over cellular transcription. Curr Biol. 2010;20:2010–2015. doi: 10.1016/j.cub.2010.10.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.