NIMA-related kinases have emerged as key regulators of ciliogenesis, and human mutations in several Nrks are associated with cilia-related disorders. This study examines CNK4, a Chlamydomonas Nrk, which regulates axonemal microtubule stability and intraflagellar transport to control flagellar maintenance and length.
Abstract
NIMA-related kinases (Nrks or Neks) have emerged as key regulators of ciliogenesis. In human, mutations in Nek1 and Nek8 cause cilia-related disorders. The ciliary functions of Nrks are mostly revealed by genetic studies; however, the underlying mechanisms are not well understood. Here we show that a Chlamydomonas Nrk, CNK4, regulates ciliary stability and length. CNK4 is localized to the basal body region and the flagella. The cnk4-null mutant exhibited long flagella, with formation of flagellar bulges. The flagella gradually became curled at the bulge formation site, leading to flagellar loss. Electron microscopy shows that the curled flagella involved curling and degeneration of axonemal microtubules. cnk4 mutation resulted in flagellar increases of IFT trains, as well as its accumulation at the flagellar bulges. IFT speeds were not affected, however, IFT trains frequently stalled, leading to reduced IFT frequencies. These data are consistent with a model in which CNK4 regulates microtubule dynamics and IFT to control flagellar stability and length.
INTRODUCTION
Eukaryotic flagella and cilia are motile and sensory organelles. Impairment of proper ciliogenesis or ciliary signaling is associated with a class of diseases and developmental disorders termed ciliopathies (Badano et al., 2005; Brown and Witman, 2014). Ciliary assembly and maintenance require intraflagellar transport (IFT), which delivers ciliary precursors to the assembly site at the ciliary tip and returns turnover products to the ciliary base within the cell body (Rosenbaum and Witman, 2002; Scholey, 2003). Anterograde and retrograde transport of IFT are driven by kinesin-2 and cytoplasmic dynein 1b, respectively. IFT motors carry large protein complexes called IFT complexes, consisting of complexes A and B, that serve as cargo adaptors for ciliary components. IFT complexes appear as small particles that are bridged with each other to form so-called IFT trains (Pigino et al., 2009). Defects in IFT may block or impair ciliogenesis (Pedersen and Rosenbaum, 2008).
In differentiated ciliated cells, cilia keep their structural integrity and length to perform ciliary functions (Ishikawa and Marshall, 2011). Recent evidence indicates that the ciliary length is controlled by a length-sensing system (Rosenbaum, 2003; Chan and Marshall, 2012; Cao et al., 2013). The length-generated signaling is predicted affect several cellular processes, including transcription, IFT, dynamics, and stability of axonemal microtubules (Ishikawa and Marshall, 2011; Chan and Marshall, 2012). Various protein kinases play key roles in ciliary length control (Cao et al., 2009; Lefebvre, 2009; Ishikawa and Marshall, 2011). The balance between the rates of axonemal assembly and disassembly mediated by IFT and protein turnover, respectively, was suggested to determine the final ciliary length (Marshall and Rosenbaum, 2001).
NIMA-related kinases (Nrks or Neks) are a group of kinases that share closer homology with NIMA than any other class of kinases and have been identified in organisms ranging from unicellular eukaryotes to multicellular organisms (O’Connell et al., 2003; Fry et al., 2012). Eleven genes in the human genome encode Nek1 to Nek11. In Chlamydomonas, 12 Nrks have been identified, including FA2 and CNK1-11 (Parker et al., 2007; Lin et al., 2015). Nrks play essential roles in mitotic progression, DNA repair, and microtubule dynamics. They are also involved in ciliogenesis, as demonstrated in Chlamydomonas, Tetrahymena, Trypanosome, mice, and humans (Fry et al., 2012; Meirelles et al., 2014; Kim et al., 2015).
In Chlamydomonas, CNK2 suppresses the dynamics of axonemal microtubules to control flagellar length (Hilton et al., 2013). CNK11 inhibits flagellar length only in mutants defective in flagellar substructures, although the underlying mechanism is unknown (Lin et al., 2015). Human mutations in Nek1 cause short-rib polydactyly syndrome, type Majewski, characterized by malformation of the brain, polydactyly, and kidney cyst, consistent with ciliary defects (Thiel et al., 2011). Depletion of Nek1 in kat2J mouse mesenchymal fibroblasts or in cell cultures reduces the number of ciliated cells. Surprisingly, a portion of remaining cilia are long and branched (Shalom et al., 2008; Wang et al., 2014).
We characterized CNK4 in Chlamydomonas. cnk4-null mutation induces long flagella that are gradually destabilized, resulting in final flagellar loss.
RESULTS
A Chlamydomonas flagellar mutant defective in CNK4
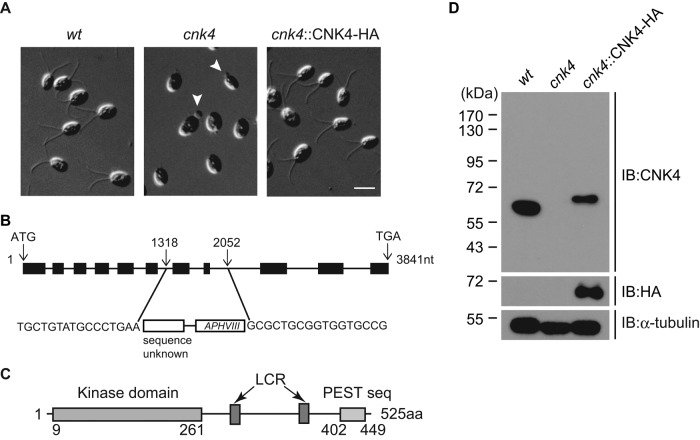
We used insertional mutagenesis to identify genes required for ciliogenesis in Chlamydomonas. A DNA fragment harboring the paromomycin resistance gene AphVIII was used for transformation (Sizova et al., 2001; Meng et al., 2014). The mutations can be caused by random insertion and/or deletion of flanking genomic DNA (Galvan et al., 2007; Jinkerson and Jonikas, 2015). Wild-type cells have two equal-length flagella of ∼12 μm. One mutant has no flagella or only flagellar buds, indicating ciliogenesis defects (Figure 1A). The flanking sequence of the insert in the mutant was cloned by restriction enzyme site–directed amplification (RESDA) PCR (Gonzalez-Ballester et al., 2005; Meng et al., 2014) and sequenced. The disrupted gene was identified as CNK4, encoding a protein kinase of 525 amino acids (aa). Further analysis showed that the CNK4 nucleotide sequence 1318–2052 was deleted and replaced by the AphVIII gene fragment and an unknown sequence, resulting in removal of exons 7 and 8 (Figure 1B).
FIGURE 1:
cnk4 is defective in ciliogenesis. (A) Wild-type (wt), cnk4, and cnk4::CNK4-HA cells were imaged by DIC microscopy. Note that cnk4 cells are aflagellated or have flagellar buds (arrowheads). Bar, 10 μm. (B) Schematic illustration of the CNK4 gene, showing the replacement of exons 7 and 8 by foreign DNA fragment during random DNA insertion. The insertion site was identified by PCR and DNA sequencing. (C) The domain structure of CNK4. Numbers are amino acid positions. LCR, low-complexity region. (D) Western blot of whole-cell lysates from indicated strains with antibodies against CNK4, HA, and α-tubulin. No CNK4 was detected in the cnk4 mutant.
CNK4 has an N-terminal kinase domain, a PEST sequence at the C-terminus, and two low-complexity regions predicted by SMART and EMBOSS websites (Figure 1C). A BLAST search of CNK4 kinase domain in the human proteome showed that CNK4 has closer sequence identity to NEK1 (Supplemental Table S1). However, CNK4 has only 525 aa, whereas NEK1 has 1258 aa, and does not have the coil-coil domain predicted in Nek1 (Fry et al., 2012). Immunoblot analysis showed that cnk4 does not express CNK4 (Figure 1D), indicating that cnk4 is a null mutant. Transformation of cnk4 with hemagglutinin (HA)-tagged CNK4 restored CNK4 expression and recovered the wild-type (wt) flagellar phenotype (Figure 1, A and D). Because >20% of transformants rescued the mutant phenotype from at least three independent transformations, this rescue was caused by incorporation of the transgene and not by disruption of some other genes. Therefore CNK4 is the causal gene in the cnk4 mutant.
CNK4 is a flagellar protein
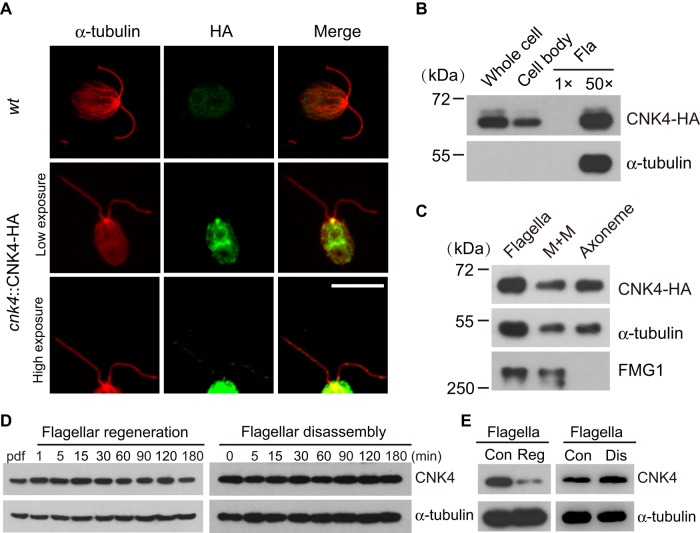
Nrks with ciliary functions are usually located at the ciliary structures. Chlamydomonas FA2 and CNK2 are localized to proximal flagella and along the axoneme, respectively (Mahjoub et al., 2004; Bradley and Quarmby, 2005). Both Nek1 and Nek2 localize to the basal body, whereas Nek8 is located at the proximal cilium (Mahjoub et al., 2005; Shalom et al., 2008; Sohara et al., 2008; Spalluto et al., 2012; Kim et al., 2015). To determine the cellular location of CNK4, we immunostained the rescued strain expressing CNK4-HA with antibodies against HA and α-tubulin with wt cells as control (Figure 2A). CNK4 is localized to several regions within the cell body, which includes prominent staining at the basal body. High exposure of the images also revealed punctate staining of CNK4 in the flagella. The presence of CNK4 in the flagella was further verified by immunoblotting of isolated flagella (Figure 2B). To pinpoint the location of CNK4 in the flagellar compartment, we further fractionated isolated flagella into membrane matrix (M+M) and axonemal fractions, followed by immunoblotting. The membrane protein FMG1 (Bloodgood and Salomonsky, 1994) and α-tubulin were used as controls. CNK4 is localized to both membrane matrix and axonemal fractions (Figure 2C).
FIGURE 2:
CNK4 is a flagellar protein. (A) CNK4 is localized to flagella and basal body. wt and cnk4::CNK4-HA cells were immunostained with anti-HA and anti–α-tubulin antibodies, respectively. High-exposure images show the flagellar location of CNK4. Bar, 10 μm. (B) cnk4::CNK4-HA cells were fractionated into cell body and flagella, followed by immunoblotting. Here 1×Fla (flagella) represents equal proportion of flagella to the cell body. (C) Isolated flagella of cnk4::CNK-HA cells were fractionated into membrane matrix (M+M) and axonemal fractions, followed by immunoblotting. FMG1 is a membrane protein. (D) wt cells were allowed to regenerate flagella after deflagellation (left) or treated with sodium pyrophosphate (NaPPi) to induce flagellar disassembly (right), followed by immunoblotting. (E) Flagella were isolated during flagellar regeneration after deflagellation or flagellar disassembly induced by NaPPi, followed by immunoblotting. Con, steady-state flagella; Dis, disassembling flagella; Reg, regenerating flagella.
The cellular location of CNK4 and the mutant flagellar phenotype implicate CNK4 in ciliogenesis. Next we examined CNK4 protein expression during flagellar assembly or disassembly. Cells were allowed to undergo flagellar assembly after deflagellation or treated with sodium pyrophosphate to induce flagellar disassembly. The expression level of CNK4 during both processes showed no apparent changes, as examined by immunoblotting of whole-cell extracts (Figure 2D). Nor did we find molecular shift of CNK4, which is usually associated with protein phosphorylation. Examination of isolated flagella by immunoblotting showed that CNK4 decreased during flagellar regeneration and slightly increased during flagellar disassembly, indicating that CNK4 is likely involved in flagellar assembly and disassembly (Figure 2E).
cnk4 fails to maintain flagella by forming bulges and flagellar balloons
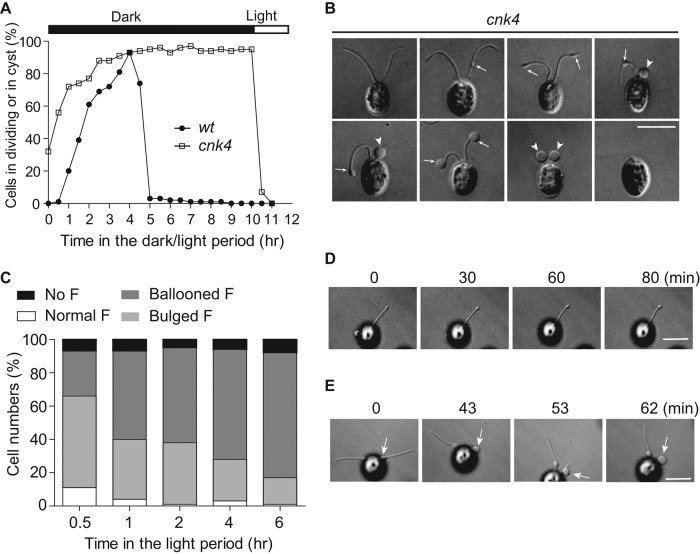
Under light/dark synchronization growth conditions, Chlamydomonas undergoes several rounds of continuous division within the mother cell wall in the dark period (Cross and Umen, 2015). The daughter cells with fully assembled flagella are then released. Cells that are unable to form flagella fail to be released from the cell wall, leading to formation of palmelloids or cysts (Kubo et al., 2009; Wood et al., 2013). Most cnk4 cells were aflagellate but did not form palmelloids (Figure 1A). Thus we examined whether cnk4 could form flagella after cell division (Figure 3A). wt cells divided soon after entering the dark period, and the flagellated daughter cells were completely released after 5 h. In contrast, cnk4 cells had an early onset of cell division, and the daughter cells were only released upon onset of the light period. This phenotype is also caused by CNK4 mutation, as CNK4-rescued strains did not show this phenotype. Treatment of cnk4 cells with light at 4 h in the dark period induced cell release, indicating that these cells did not require longer for completion of cell division but instead that cell release requires light.
FIGURE 3:
cnk4 loses flagellar maintenance by forming flagellar bulges and balloons. (A) Cells were synchronized by using light/dark cycles under 5% CO2. cnk4 cells exhibited early onset of cell division, and the daughter cells were released upon lighting, whereas wt cells released daughter cells after completion of cell division. (B) DIC images of cnk4 cells show distinct flagellar phenotypes. The flagellar bulges and balloons are marked as arrows and arrowheads, respectively. Bar, 10 μm. (C) Quantification of different types of flagella after cells were released in the light period. F, flagella. (D, E) Movie stills showing flagellar bulge formation (D) and flagellar curling at the bulge formation site (E). Arrows mark the bulges. Note that the distal flagellum to the bulge is retracted into the bulge. Bars, 10 μm.
cnk4 cells released from the mother cell wall were indeed flagellated. However, these cells had striking flagellar phenotypes (Figure 3B). Instead of having two normal flagella, some cells were aflagellate or had ballooned flagella, whereas others exhibited bulges along the axoneme or at the flagellar tips. Because most cnk4 cells were aflagellate in the late light period, these different shapes of flagella might represent different stages of flagella en route to flagellar loss. We then followed flagellar changes after cells were released from the mother cell wall. Indeed, bulged or partially curled flagella became completely curled, forming flagellar balloons as time progressed (Figure 3C).
The bulges were formed randomly along the flagellar length, and their formation was uncoordinated in the two flagella (Figure 3B). Bulge formation is likely spontaneous after flagella are assembled. Live imaging captured the formation of bulges at the flagellar tip, which took ∼30 min to be clearly visible and continued to enlarge (Figure 3D and Supplemental Video S1). Of interest, we also observed that the bulge formed in the middle of flagella could migrate toward the flagellar tip (Supplemental Video S2). Large flagellar bulges and flagellar balloons were likely formed by retraction of the flagellar axoneme into the bulge formation site (Figure 3E and Supplemental Video S3). Thus the flagella gradually form bulges, which induce flagellar curling or retraction and lead to formation of ballooned flagella, which are eventually detached from the cell body.
cnk4 loses flagellar length control
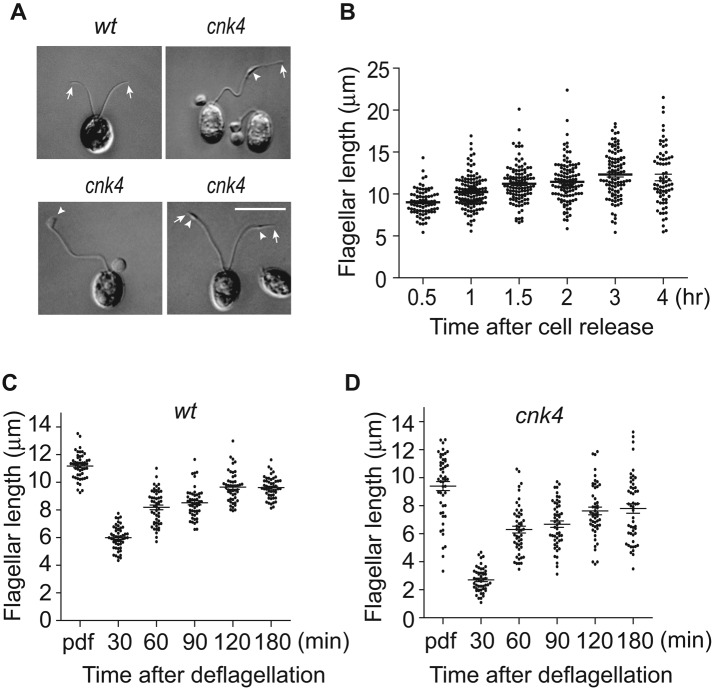
cnk4 cells exhibit flagella with bulges or flagellar balloons. Surprisingly, we also observed extremely long flagella (Figure 4A). These long flagella were also associated with bulges formed randomly along the flagellar length. Next we monitored changes of flagellar length after cell release from the mother cell wall. Cells that did not form flagella or had ballooned flagella were excluded for measurement. The average flagellar length increased over time (Figure 4B). However, some cells grew extremely long flagella, >20 μm, whereas the flagellar length of wt cells rarely exceeds 15 μm (Berman et al., 2003). Approximately 17% of flagella at 4 h after cell release are >15 μm in length. These data indicate that cnk4 loses not only flagellar maintenance but also flagellar length control. Next we examined flagellar regeneration. Consistent with previous work wt cells showed rapid flagellar growth, followed by decreased rate of assembly toward steady-state length (Figure 4C; Rosenbaum et al., 1969). In contrast, although cnk4 was able to assemble flagella after deflagellation, the initial flagellar assembly rate was reduced (Figure 4D). These data demonstrate that flagellar assembly in cnk4 is dysregulated.
FIGURE 4:
cnk4 loses flagellar length control. (A) DIC images of wt and cnk4 cells. cnk4 cells form extremely long flagella with bulge formation (arrowheads). Arrows mark the flagellar tips. Bar, 10 μm. (B) Flagellar length increase after cells are released from the mother cell wall of cnk4 cells. Cells were fixed at different times after cell release in the light period and imaged by DIC microscopy, followed by flagellar length measurement. Normal-looking flagella and flagella with bulges but not ballooned flagella were analyzed. (C, D) Flagellar regeneration of wt (C) and cnk4 (D) cells. Cells at 1 h in the light were deflagellated by pH shock to allow flagellar regeneration, followed by flagellar length measurement. Ballooned flagella were excluded from measurement. Note that cnk4 has a reduced rate of assembly before 30 min after deflagellation.
Accumulation of amorphous materials at flagellar bulges and destabilization of the flagellar axoneme
The formation of flagellar bulges is an early event leading to loss of flagellar maintenance. Electron microscopy revealed that cnk4 flagellar substructures, including outer doublet microtubules, central pair apparatus, dynein arms, and radial spokes, appeared normal (Figure 5A). However, the flagellar bulges contained amorphous materials accumulated between the flagellar membrane and the outer doublet microtubules. The axonemes of the ballooned flagella were curled within the flagellar membrane, and some of the outer doublet microtubules were disorganized or degenerated (Figure 5B).
FIGURE 5:
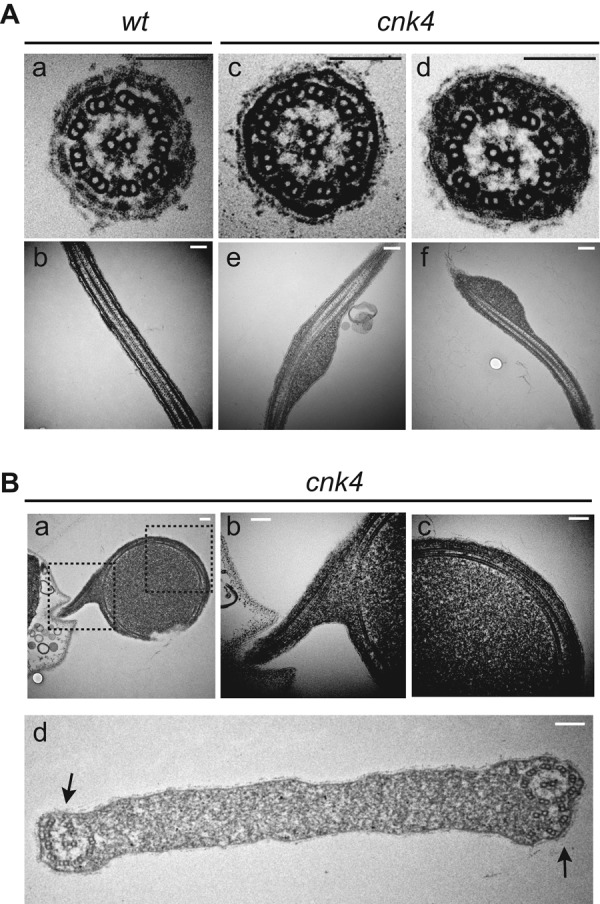
Electron microscopic analysis of cnk4 flagella. (A) Cross- and longitudinal sections of wt (a, b) and cnk4 (c–f) flagella. Note that cnk4 flagella accumulate amorphous materials between the membrane and the axoneme (d–f), whereas its substructures appear normal (d). Bars, 150 nm. (B) Flagellar balloon cross-sectioned to show curled axoneme (a–d) and degenerated axonemal microtubules (d, arrows). Dashed box images in (a) are enlarged and shown in (b) and (c), respectively. Bars, 150 nm.
Accumulation of IFT proteins along the flagella, particularly at the bulges in cnk4 flagella
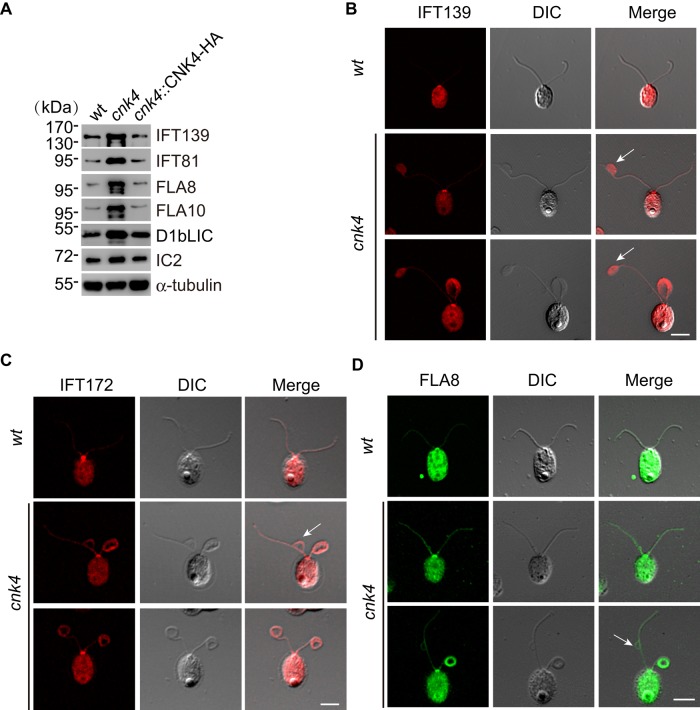
Defects in IFT regulation or IFT components are associated with ciliary length control and accumulation of materials within the flagellum (Pedersen and Rosenbaum, 2008). Temperature-sensitive mutants fla15 and fla17, which are defective in IFT144 and IFT139, respectively, form flagellar bulges randomly along the length of the flagellum (Piperno et al., 1998; Iomini et al., 2009). Null mutation of MAK induces long cilia with accumulation of IFT particles (Omori et al., 2010). Therefore we examined whether cnk4 mutation affects IFT. Examination of isolated flagella of cnk4 revealed increases of IFT-A component IFT139, IFT-B component IFT81, anterograde IFT motor subunits FLA10 and FLA8, and retrograde motor subunit D1bLIC (Figure 6A). Because the overall protein level of IFT in whole cells of cnk4 did not change (Supplemental Figure S1), these results indicate that cnk4 mutation results in increased IFT proteins in the flagellum.
FIGURE 6:
Increase of IFT proteins in the cnk4 flagella. (A) Isolated flagella from wt, cnk4, and cnk4::CNK4-HA were analyzed by immunoblotting with antibodies against IFT proteins (IFT139, IFT-A; IFT81, IFT-B) and IFT motor subunits (FLA8, FLA10, and D1bLIC). IC2 and α-tubulin were used as controls. (B–D) wt and cnk4 cells were immunostained with antibodies against IFT139 (B), IFT172 (C), and FLA8 (D) antibodies. cnk4 flagella exhibit an overall increase of IFT proteins along the flagellar length compared with wt flagella. Flagellar bulges (arrows) are filled with IFT139, and IFT172 and FLA8 attach to the flagellar membrane. Bars, 10 μm.
Next we investigated where the IFT proteins were increased in the flagella. We used antibodies against representative IFT proteins IFT139, IFT172, and FLA8 for immunostaining. In wt cells, IFT proteins showed prominent staining at the basal body, with faint staining along the length of flagella, which is consistent with previous work (Figure 6, B–D; Cole et al., 1998). In contrast, cnk4 flagella accumulated more IFT proteins along the length of flagella. In particular, IFT proteins heavily accumulated in the bulge region, (Figure 6B, arrow). In some bulges, IFT proteins were associated with a bulged flagellar membrane (Figure 6, C and D), indicating that IFT trains likely fall off the axonemal microtubules.
cnk4 is defective in IFT frequency, with frequent stalling of IFT
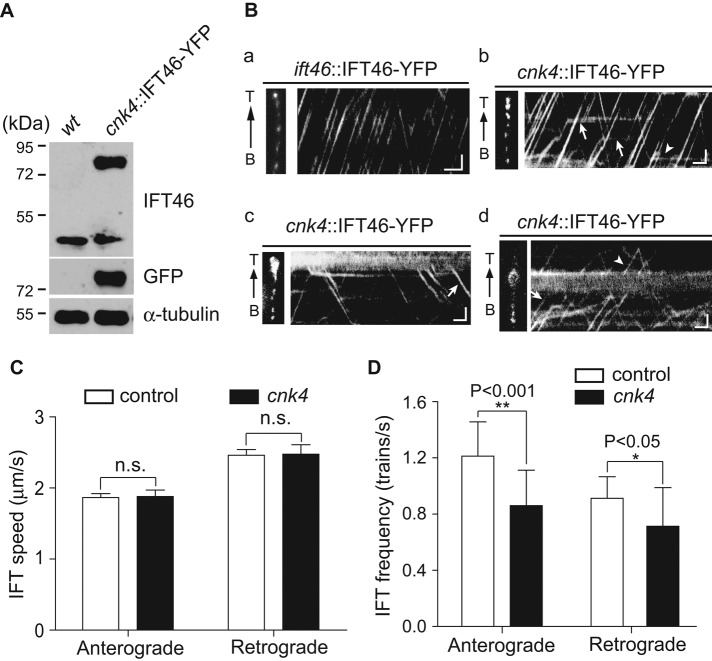
To gain further insight into the effect of cnk4 mutation on IFT regulation, we used total internal reflection fluorescence (TIRF) microscopy to observe IFT. IFT46-YFP was expressed in cnk4 cells (Figure 7A). An ift46 mutant strain rescued by expression of IFT46-YFP was used as a control. In control cells, IFT trains moved smoothly in both anterograde and retrograde directions (Figure 7Ba and Supplemental Video S4). In contrast, in cnk4 cells, IFT frequently stalled in both anterograde and retrograde directions (Figure 7Bb and Supplemental Video S5). At the bulged flagellar tip, some IFT trains underwent proper turnaround, whereas others were trapped, and trapped IFT trains might spontaneously initiate retrograde movement (Figure 7Bc and Supplemental Video S6). At the bulges formed in the middle of the flagellum, whereas some IFT trains passed the bulges freely, others were trapped (Figure 7Bd and Supplemental Video S7).
FIGURE 7:
cnk4 flagella exhibit normal IFT speed but reduced IFT frequency, with frequent IFT stalling. (A) Expression of yellow fluorescent protein (YFP)–tagged IFT46 in cnk4. Cells were analyzed by immunoblotting with antibodies against IFT46, GFP, and α-tubulin. (B) Kymographs of IFT46-YFP in the flagella of ift46::IFT46-YFP (a) and cnk4::IFT46-YFP (b–d) cells. Left, images of recorded flagella. (b) Kymograph from apparently normal flagellum. The arrows and arrowhead indicate stalled IFT in anterograde and retrograde directions, respectively. (c) Kymograph highlighting IFT turnaround at the bulged flagellar tip. Arrow, spontaneously initiated retrograde IFT. (d) Kymograph showing IFT at the flagellar bulge in the middle of the flagellum. Arrow, trapped IFT; arrowhead, IFT passing though the bulge. B, flagellar base; T, flagellar tip. Bars, 1 μm and 1 s. (C) The velocities of anterograde and retrograde IFT are not affected by cnk4 mutation. n.s., not significant (p > 0.05). (D) IFT frequencies of both directions in cnk4 flagella are reduced compared with those of wild-type flagella.
Using apparently normal flagella of cnk4 cells, we analyzed IFT speed and frequency. The anterograde and retrograde speeds of IFT of cnk4 cells were similar to those of controls (Figure 7C). The velocities of anterograde IFT for control and cnk4 flagella were 1.87 ± 0.06 (64 tracks) and 1.88 ± 0.09 μm/s (202 tracks), respectively, and those of retrograde IFT were 2.46 ± 0.08 (72 tracks) and 2.48 ± 0.13 μm/s (100 tracks), respectively. As expected, anterograde and retrograde IFT frequencies were significantly reduced compared with those of the control, which is consistent with frequent stalling or derailing of IFT trains (Figure 7D). The anterograde frequencies for control and cnk4 flagella were 1.21 ± 0.24 (10 flagella) and 0.86 ± 0.25 trains/s (64 flagella), respectively, and those for retrograde IFT were 0.91 ± 0.15 (10 flagella) and 0.72 ± 0.27 trains/s (65 flagella), respectively. The retrograde frequency reported here for the wild-type strain is less than that reported by using differential interference contrast (DIC) microscopy. For example, Dentler (2005) reported 1.3–1.5 trains/s. This might reflect different experimental conditions and the methods used.
DISCUSSION
We identified CNK4, an Nrk in Chlamydomonas, as a key regulator of ciliogenesis. cnk4-null mutation induces formation of long flagella with axonemal instability, leading to eventual flagellar loss, indicating that CNK4 plays an important role in ciliary length and stability.
Nrks have been increasingly recognized as critical regulators of cilia (Fry et al., 2012). Four Nrks in mammalian cells have been implicated in ciliary function. Depletion of Nek1 results in reduced ciliation, as well as long and branched cilia (Shalom et al., 2008; Wang et al., 2014). Nek2 suppresses ciliogenesis at G2/M to ensure ciliary disassembly (Kim et al., 2015). Down-regulation of Nek4 leads to a decrease in ciliary assembly (Coene et al., 2011). nek8 missense mutation in jck mice leads to long cilia (Smith et al., 2006; Sohara et al., 2008). However, complete depletion of Nek8 does not affect ciliogenesis (Manning et al., 2013), indicating that the nek8 missense mutation is likely a gain-of-function mutation. In Chlamydomonas, FA2 is required for flagellar autonomy, whereas CNK2 suppresses axonemal microtubule dynamics to control ciliary length (Finst et al., 1998; Hilton et al., 2013). CNK11 regulates flagellar length only in mutants defective in flagellar substructures (Lin et al., 2015). In Tetrahymena, overexpression of Nrks induces ciliary shortening (Wloga et al., 2006), whereas overexpression of NRKC in Trypanosome inhibits ciliary formation (Pradel et al., 2006). These data demonstrate that Nrks are involved in different aspects of ciliary regulation, although the molecular mechanisms are mostly unknown.
Ciliary length control by CNK4
In steady-state or differentiated cells, ciliary length is kept at a constant length, which is required for optimal signaling capacity or proper motility of cilia (Ishikawa and Marshall, 2011). How ciliary length is controlled is not completely understood. The initial discovery of a long flagellar mutant lf4 that is defective in a mitogen-activated protein (MAP) kinase suggested the possible existence of a length-sensing system (Berman et al., 2003; Rosenbaum, 2003). Subsequent studies validated this notion. Chlamydomonas Aurora-like kinase and FLA8/KIF3B change their phosphorylation states in response to alteration of flagellar length (Luo et al., 2011; Cao et al., 2013; Liang et al., 2014). In addition, flagellar length is associated with inverse scaling of IFT train size, and accumulation of IFT trains at the basal body is also dependent on length (Engel et al., 2009; Ludington et al., 2013). Ciliary length sensing is expected to trigger a signaling cascade to balance the assembly and disassembly of axonemal microtubules to achieve final ciliary length (Marshall and Rosenbaum, 2001). Multiple kinases are implicated in ciliary length control, which include LF2, LF4, LF5, GSK3, and CNK2 in Chlamydomonas (Berman et al., 2003; Wilson and Lefebvre, 2004; Tam et al., 2007, 2013; Hilton et al., 2013) and intestinal cell kinase (ICK), male germ cell–associated kinase (MAK), Nek1, and Nek8 in mammalian cells (Smith et al., 2006; Sohara et al., 2008; Omori et al., 2010; Yang et al., 2013; Chaya et al., 2014; Moon et al., 2014). The molecular mechanisms of these kinases underlying ciliary length regulation are largely unknown. Signaling mediated by protein phosphorylation should influence processes involved in axonemal assembly such as IFT and microtubule dynamics to control ciliary length.
Alteration of IFT is expected to tilt the balance between assembly and disassembly, leading to changes of ciliary length. Temperature-sensitive mutant fla10, which is defective in IFT motor KIF3A/FLA10, shortens its flagella at restrictive temperature (Lux and Dutcher, 1991; Walther et al., 1994; Kozminski et al., 1995). Long flagellar mutant lf4 appears to increase IFT particles at the flagellar tip (Berman et al., 2003). In mammalian cells, depletion of protein kinase ICK or MAK induces formation of long cilia with accumulation of IFT proteins (Omori et al., 2010; Yang et al., 2013; Chaya et al., 2014). The increase of IFT trains in cnk4 flagella might function similarly to promote ciliary length.
Modulation of microtubule dynamics controls the incorporation and turnover of tubulins of axonemal microtubules at the ciliary tip. Microtubule depolymerases CrKinesin13 and KIF19A are implicated in ciliary length control (Niwa et al., 2012; Wang et al., 2013). Reducing polyglutamylation of axonemal microtubules increases ciliary length (Kubo et al., 2015). CNK2 suppresses microtubule dynamics, and its mutation induces flagellar elongation of lf4 (Hilton et al., 2013). Given that cnk4 mutation induces axonemal instability, CNK4 might also act on microtubule dynamics to regulate ciliary length. Future identification of the targets of CNK4 will pinpoint the mechanism of CNK4 in ciliary length control.
IFT control by CNK4
IFT is under tight control. In Chlamydomonas, CDPK1 controls IFT ciliary entry and turnaround at the ciliary tip by regulating the phosphorylation states of the anterograde motor subunit FLA8/KIF3B (Liang et al., 2014). ICK phosphorylates KIF3A, and loss of ICK leads to accumulation of IFT proteins at the tip of primary cilia (Chaya et al., 2014; Moon et al., 2014). Accumulation of IFT proteins in the middle of cilia is also observed in the null mutant of MAK, although the mechanism is unknown (Omori et al., 2010). Loss of DFY-5, a MAP kinase in C. elegans, prevents undocking of kinesin-II at the distal end of the middle cilia segment (Burghoorn et al., 2007).
cnk4 mutation leads to overall increase of IFT trains in the flagellum. We found that the frequency of IFT rather than IFT speed is affected by cnk4 mutation. Frequent stalling of IFT is consistent with an overall increase of IFT proteins, although we could not exclude the possibility that CNK4 also regulates the IFT injection rate to bring about an increase of IFT particles (Ludington et al., 2013). Frequent IFT stalling might be due to dysregulation of IFT motors, IFT particles, axonemal microtubules, and the connection between IFT trains with the ciliary membrane. We observed that IFT trains that are detached from the axonemal microtubules still associate with the flagellar membrane, indicating that this linkage is not affected. The anterograde and retrograde motors have divergent sequences. Given that IFT stalling occurs in both directions, IFT motors might not be directly affected by CNK4. The fact that both IFT-A and IFT-B components accumulate in cnk4-mutant flagella suggests that the target of CNK4 is not individual components of IFT particles. Gene mutations of IFT-B components either block ciliary assembly or lead to formation of short flagella, whereas IFT particles rarely accumulate (Pazour et al., 2000; Brazelton et al., 2001; Hou et al., 2007). In contrast, defects in IFT-A components result only in IFT-B accumulation (Piperno et al., 1998; Iomini et al., 2009). One possibility is that CNK4 affects axonemal microtubules to affect IFT. Posttranslational modifications of microtubules are implicated in regulation of motor velocity, processivity and microtubule stability (Gaertig and Wloga, 2008; Schneider et al., 2008; Konno et al., 2012; Sirajuddin et al., 2014).
CNK4, ciliary curling, and axonemal instability
cnk4 cells form flagellar bulges, and the flagella gradually become curled at the bulge formation site, leading to eventual flagellar loss. How bulge formation leads to flagellar curling is not clear. Chlamydomonas fla15 and fla17 mutants form flagellar bulges due to accumulation of IFT proteins but their flagella do not curl (Piperno et al., 1998; Iomini et al., 2009). Thus simple accumulation of IFT proteins may not cause flagellar curling. The curling of the flagellar axoneme might involve both instability of the axoneme and loss of control of axonemal dyneins that control axonemal microtubule sliding (Witman, 1992).
MATERIALS AND METHODS
Strains and culture conditions
Chlamydomonas reinhardtii strain 21gr (mt+; CC-1690) is available from the Chlamydomonas Resource Center (University of Minnesota, St. Paul, MN). ift46::IFT46-YFP was kindly provided by Kaiyao Huang (Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China). Cells were cultured on 1.5% agar plates or in liquid M medium at 23°C with aeration under a 14:10-h light/dark cycle. In experiments involving cell cycle progression, cells were synchronized in 14:10-h light/dark cycles with aeration of 5% CO2 in a Light Incubator (Percival AL-36, Percival, Perry, IA; Hu et al., 2015).
Insertional mutagenesis, gene cloning, and transformation
Insertional mutants were generated by transformation with electroporation of AphVIII fragments (Sizova et al., 2001; Liang and Pan, 2013). The genomic sequences flanking the AphVIII insertion site were identified by RESDA PCR and sequencing (Gonzalez-Ballester et al., 2005; Meng et al., 2014). To make a construct for expressing CNK4-HA in cnk4, the CNK4 gene with an ∼1.2-kb fragment upstream of the start codon was cloned by PCR, and a 3×HA tag sequence followed by a Rubisco terminator was cloned from plasmid pKL-3xHA (kindly provided by Karl F. Lechtreck, University of Georgia, Athens, GA; Lechtreck et al., 2009). The resulting construct was cloned into a modified vector pHyg3 harboring a hygromycin B resistance gene (Berthold et al., 2002). The final construct was linearized with AhdI-HF and transformed into the cnk4 mutant using electroporation.
Cell fractionation, flagellar regeneration, flagellar shortening, flagellar length measurement, and DIC cell images
Cells were deflagellated by pH shock, followed by isolation of cell bodies and flagella or allowed for flagellar regeneration. Flagellar shortening was induced by adding 20 mM sodium pyrophosphate. Flagellar fractionation was carried out by adding 0.5% NP-40, followed by centrifugation. Cells for flagellar length measurement or examination of flagellar phenotypes were fixed by 1% glutaraldehyde and imaged by a Zeiss DIC microscope (Zeiss, Shanghai, China). The detailed procedures are essentially as described previously (Meng et al., 2014).
Primary antibodies
We used the following primary antibodies: rat monoclonal anti-HA (1:50 for immunofluorescence [IF] and 1:1000 for immunoblotting [IB]; clone 3F10; Roche); mouse monoclonal anti–α-tubulin (1:200 for IF and 1:2500 or 1:10,000 for IB; Sigma-Aldrich, Shanghai, China); mouse monoclonal anti-IC2 (1:20,000 for IB; Sigma-Aldrich); mouse monoclonal anti–green fluorescent protein (GFP; 1:2000 for IB; Abmart, Shanghai, China); mouse monoclonal anti-IFT139 (1:50 for IF and 1:10,000 for IB); mouse monoclonal anti-IFT172 (1:50 for IF); mouse monoclonal anti-IFT81 (1:1000 for IB); rabbit polyclonal anti-D1bLIC (1:250 for IB); rabbit polyclonal anti-FLA10 (1:1000 for IB), rabbit polyclonal anti-FLA8 (1:100 for IF and 1:2000 for IB; Liang and Pan, 2013); and rabbit polyclonal anti-CNK4 (1:2000 for IB).
IFT antibodies were kindly provided by Dennis Diener and Joel Rosenbaum (Yale University, New Haven, CT). Polyclonal D1bLIC antibody was made in rabbit using the 55- to 427–amino acid protein fragment (YingJi, Shanghai, China). Rabbit polyclonal anti-CNK4 antibody was made with the two peptides RPKTPQGDMNAPPEVAGC (amino acids 297–313) and CLAQQMEELAVDDGEDED (amino acids 383–399) and affinity purified (MBL, Beijing, China).
SDS–PAGE and immunoblotting
SDS–PAGE and immunoblotting analysis were essentially as described previously (Meng et al., 2014).
IF and electron microscopy
A previously published protocol was followed (Hu et al., 2015). The secondary antibodies used were Texas red–conjugated goat anti-mouse immunoglobulin G (IgG; 1:400) and Alexa Fluor 488–conjugated goat anti-rabbit IgG (1:400; Molecular Probes). The samples were viewed on a Zeiss LSM780 META Observer Z1 Confocal Laser Microscope. Images were acquired and processed by ZEN 2009 Light Edition (Zeiss) and Photoshop (Adobe). Analysis for electron microscopy followed published methods (Craige et al., 2010; Meng et al., 2014).
Live-cell imaging
To image flagellar bulge formation and flagellar curling, DIC images were captured at 1 frame per 30 s with a Zeiss microscope (Zeiss Axio) equipped with a charge-coupled device (CCD) camera (QuantEM 512SC; Photometrics) using a 100× objective. Images were exported and processed using Photoshop and ImageJ (National Institutes of Health, Bethesda, MD).
For TIRF microscopy, cells were immobilized on coverglasses with polylysine. Images were acquired at room temperature on a Nikon microscope (A1RSi) equipped with a 100×/numerical aperture 1.49 TIRF objective and a cooled electron-multiplying CCD camera. Images were analyzed with NIS-Elements software. Kymographs and videos were generated using ImageJ, and the IFT speed and frequency were calculated.
Supplementary Material
Acknowledgments
We thank Joel Rosenbaum, Dennis Diener, Robert Bloodgood, and Kaiyao Huang for provision of antibodies and strains and Wallace Marshall and Susan Dutcher for discussion during the course of this work. This work was supported by the National Basic Research Program of China (973 Program; 2012CB945003, 2013CB910700) and the National Natural Science Foundation of China (31330044; to J.P.).
Abbreviations used:
- CNK
Chlamydomonas MIMA–related kinase
- IFT
intraflagellar transport
- NaPPi
sodium pyrophosphate
- Nrks
NIMA-related kinases
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-10-0707) on January 13, 2016.
REFERENCES
- Badano JL, Teslovich TM, Katsanis N. The centrosome in human genetic disease. Nat Rev Genet. 2005;6:194–205. doi: 10.1038/nrg1557. [DOI] [PubMed] [Google Scholar]
- Berman SA, Wilson NF, Haas NA, Lefebvre PA. A novel MAP kinase regulates flagellar length in Chlamydomonas. Curr Biol. 2003;13:1145–1149. doi: 10.1016/s0960-9822(03)00415-9. [DOI] [PubMed] [Google Scholar]
- Berthold P, Schmitt R, Mages W. An engineered Streptomyces hygroscopicus aph 7” gene mediates dominant resistance against hygromycin B in Chlamydomonas reinhardtii. Protist. 2002;153:401–412. doi: 10.1078/14344610260450136. [DOI] [PubMed] [Google Scholar]
- Bloodgood RA, Salomonsky NL. The transmembrane signaling pathway involved in directed movements of Chlamydomonas flagellar membrane glycoproteins involves the dephosphorylation of a 60-kD phosphoprotein that binds to the major flagellar membrane glycoprotein. J Cell Biol. 1994;127:803–811. doi: 10.1083/jcb.127.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley BA, Quarmby LM. A NIMA-related kinase, Cnk2p, regulates both flagellar length and cell size in Chlamydomonas. J Cell Sci. 2005;118:3317–3326. doi: 10.1242/jcs.02455. [DOI] [PubMed] [Google Scholar]
- Brazelton WJ, Amundsen CD, Silflow CD, Lefebvre PA. The bld1 mutation identifies the Chlamydomonas osm-6 homolog as a gene required for flagellar assembly. Curr Biol. 2001;11:1591–1594. doi: 10.1016/s0960-9822(01)00485-7. [DOI] [PubMed] [Google Scholar]
- Brown J, Witman GB. Cilia and diseases. Bioscience. 2014;64:1126–1137. [Google Scholar]
- Burghoorn J, Dekkers MP, Rademakers S, de Jong T, Willemsen R, Jansen G. Mutation of the MAP kinase DYF-5 affects docking and undocking of kinesin-2 motors and reduces their speed in the cilia of Caenorhabditis elegans. Proc Natl Acad Sci USA. 2007;104:7157–7162. doi: 10.1073/pnas.0606974104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M, Li G, Pan J. Regulation of cilia assembly, disassembly, and length by protein phosphorylation. Methods Cell Biol. 2009;94:333–346. doi: 10.1016/S0091-679X(08)94017-6. [DOI] [PubMed] [Google Scholar]
- Cao M, Meng D, Wang L, Bei S, Snell WJ, Pan J. Activation loop phosphorylation of a protein kinase is a molecular marker of organelle size that dynamically reports flagellar length. Proc Natl Acad Sci USA. 2013;110:12337–12342. doi: 10.1073/pnas.1302364110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YH, Marshall WF. How cells know the size of their organelles. Science. 2012;337:1186–1189. doi: 10.1126/science.1223539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaya T, Omori Y, Kuwahara R, Furukawa T. ICK is essential for cell type-specific ciliogenesis and the regulation of ciliary transport. EMBO J. 2014;33:1227–1242. doi: 10.1002/embj.201488175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coene KL, Mans DA, Boldt K, Gloeckner CJ, van Reeuwijk J, Bolat E, Roosing S, Letteboer SJ, Peters TA, Cremers FP, et al. The ciliopathy-associated protein homologs RPGRIP1 and RPGRIP1L are linked to cilium integrity through interaction with Nek4 serine/threonine kinase. Hum Mol Genet. 2011;20:3592–3605. doi: 10.1093/hmg/ddr280. [DOI] [PubMed] [Google Scholar]
- Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craige B, Tsao CC, Diener DR, Hou Y, Lechtreck KF, Rosenbaum JL, Witman GB. CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. J Cell Biol. 2010;190:927–940. doi: 10.1083/jcb.201006105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross FR, Umen JG. The Chlamydomonas cell cycle. Plant J. 2015;82:370–392. doi: 10.1111/tpj.12795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentler W. Intraflagellar transport (IFT) during assembly and disassembly of Chlamydomonas flagella. J Cell Biol. 2005;170:649–659. doi: 10.1083/jcb.200412021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel BD, Ludington WB, Marshall WF. Intraflagellar transport particle size scales inversely with flagellar length: revisiting the balance-point length control model. J Cell Biol. 2009;187:81–89. doi: 10.1083/jcb.200812084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finst RJ, Kim PJ, Quarmby LM. Genetics of the deflagellation pathway in Chlamydomonas. Genetics. 1998;149:927–936. doi: 10.1093/genetics/149.2.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry AM, O’Regan L, Sabir SR, Bayliss R. Cell cycle regulation by the NEK family of protein kinases. J Cell Sci. 2012;125:4423–4433. doi: 10.1242/jcs.111195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertig J, Wloga D. Ciliary tubulin and its post-translational modifications. Curr Top Dev Biol. 2008;85:83–113. doi: 10.1016/S0070-2153(08)00804-1. [DOI] [PubMed] [Google Scholar]
- Galvan A, Gonzalez-Ballester D, Fernandez E. Insertional mutagenesis as a tool to study genes/functions in Chlamydomonas. Adv Exp Med Biol. 2007;616:77–89. doi: 10.1007/978-0-387-75532-8_7. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Ballester D, de Montaigu A, Galvan A, Fernandez E. Restriction enzyme site-directed amplification PCR: a tool to identify regions flanking a marker DNA. Anal Biochem. 2005;340:330–335. doi: 10.1016/j.ab.2005.01.031. [DOI] [PubMed] [Google Scholar]
- Hilton LK, Gunawardane K, Kim JW, Schwarz MC, Quarmby LM. The kinases LF4 and CNK2 control ciliary length by feedback regulation of assembly and disassembly rates. Curr Biol. 2013;23:2208–2214. doi: 10.1016/j.cub.2013.09.038. [DOI] [PubMed] [Google Scholar]
- Hou Y, Qin H, Follit JA, Pazour GJ, Rosenbaum JL, Witman GB. Functional analysis of an individual IFT protein: IFT46 is required for transport of outer dynein arms into flagella. J Cell Biol. 2007;176:653–665. doi: 10.1083/jcb.200608041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Liang Y, He W, Pan J. Cilia disassembly with two distinct phases of regulation. Cell Rep. 2015;10:1803–1810. doi: 10.1016/j.celrep.2015.02.044. [DOI] [PubMed] [Google Scholar]
- Iomini C, Li L, Esparza JM, Dutcher SK. Retrograde intraflagellar transport mutants identify complex A proteins with multiple genetic interactions in Chlamydomonas reinhardtii. Genetics. 2009;183:885–896. doi: 10.1534/genetics.109.101915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Marshall WF. Ciliogenesis: building the cell’s antenna. Nat Rev Mol Cell Biol. 2011;12:222–234. doi: 10.1038/nrm3085. [DOI] [PubMed] [Google Scholar]
- Jinkerson RE, Jonikas MC. Molecular techniques to interrogate and edit the Chlamydomonas nuclear genome. Plant J. 2015;82:393–412. doi: 10.1111/tpj.12801. [DOI] [PubMed] [Google Scholar]
- Kim S, Lee K, Choi JH, Ringstad N, Dynlacht BD. Nek2 activation of Kif24 ensures cilium disassembly during the cell cycle. Nat Commun. 2015;6:8087. doi: 10.1038/ncomms9087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno A, Setou M, Ikegami K. Ciliary and flagellar structure and function–their regulations by posttranslational modifications of axonemal tubulin. Int Rev Cell Mol Biol. 2012;294:133–170. doi: 10.1016/B978-0-12-394305-7.00003-3. [DOI] [PubMed] [Google Scholar]
- Kozminski KG, Beech PL, Rosenbaum JL. The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J Cell Biol. 1995;131:1517–1527. doi: 10.1083/jcb.131.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T, Hirono M, Aikawa T, Kamiya R, Witman GB. Reduced tubulin polyglutamylation suppresses flagellar shortness in Chlamydomonas. Mol Biol Cell. 2015;26:2810–2822. doi: 10.1091/mbc.E15-03-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T, Kaida S, Abe J, Saito T, Fukuzawa H, Matsuda Y. The Chlamydomonas hatching enzyme, sporangin, is expressed in specific phases of the cell cycle and is localized to the flagella of daughter cells within the sporangial cell wall. Plant Cell Physiol. 2009;50:572–583. doi: 10.1093/pcp/pcp016. [DOI] [PubMed] [Google Scholar]
- Lechtreck KF, Luro S, Awata J, Witman GB. HA-tagging of putative flagellar proteins in Chlamydomonas reinhardtii identifies a novel protein of intraflagellar transport complex B. Cell Motil Cytoskeleton. 2009;66:469–482. doi: 10.1002/cm.20369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre PA. Flagellar length control. In: Witman GB, editor. The Chlamydomonas Sourcebook: Cell Motilty and Behavior. Oxford, UK: Elsevier; 2009. pp. 115–129. [Google Scholar]
- Liang Y, Pan J. Regulation of flagellar biogenesis by a calcium dependent protein kinase in Chlamydomonas reinhardtii. PLoS One. 2013;8:e69902. doi: 10.1371/journal.pone.0069902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Pang Y, Wu Q, Hu Z, Han X, Xu Y, Deng H, Pan J. FLA8/KIF3B phosphorylation regulates kinesin-II interaction with IFT-B to control IFT entry and turnaround. Dev Cell. 2014;30:585–597. doi: 10.1016/j.devcel.2014.07.019. [DOI] [PubMed] [Google Scholar]
- Lin H, Zhang Z, Guo S, Chen F, Kessler JM, Wang YM, Dutcher SK. A NIMA-related kinase suppresses the flagellar instability associated with the loss of multiple axonemal structures. PLoS Genet. 2015;11:e1005508. doi: 10.1371/journal.pgen.1005508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludington WB, Wemmer KA, Lechtreck KF, Witman GB, Marshall WF. Avalanche-like behavior in ciliary import. Proc Natl Acad Sci USA. 2013;110:3925–3930. doi: 10.1073/pnas.1217354110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Cao M, Kan Y, Li G, Snell W, Pan J. The phosphorylation state of an aurora-like kinase marks the length of growing flagella in Chlamydomonas. Curr Biol. 2011;21:586–591. doi: 10.1016/j.cub.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lux FG, 3rd, Dutcher SK. Genetic interactions at the FLA10 locus: suppressors and synthetic phenotypes that affect the cell cycle and flagellar function in Chlamydomonas reinhardtii. Genetics. 1991;128:549–561. doi: 10.1093/genetics/128.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahjoub MR, Qasim Rasi M, Quarmby LM. A NIMA-related kinase, Fa2p, localizes to a novel site in the proximal cilia of Chlamydomonas and mouse kidney cells. Mol Biol Cell. 2004;15:5172–5186. doi: 10.1091/mbc.E04-07-0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahjoub MR, Trapp ML, Quarmby LM. NIMA-related kinases defective in murine models of polycystic kidney diseases localize to primary cilia and centrosomes. J Am Soc Nephrol. 2005;16:3485–3489. doi: 10.1681/ASN.2005080824. [DOI] [PubMed] [Google Scholar]
- Manning DK, Sergeev M, van Heesbeen RG, Wong MD, Oh JH, Liu Y, Henkelman RM, Drummond I, Shah JV, Beier DR. Loss of the ciliary kinase Nek8 causes left-right asymmetry defects. J Am Soc Nephrol. 2013;24:100–112. doi: 10.1681/ASN.2012050490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Rosenbaum JL. Intraflagellar transport balances continuous turnover of outer doublet microtubules: implications for flagellar length control. J Cell Biol. 2001;155:405–414. doi: 10.1083/jcb.200106141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meirelles GV, Perez AM, de Souza EE, Basei FL, Papa PF, Melo Hanchuk TD, Cardoso VB, Kobarg J. “Stop Ne(c)king around”: how interactomics contributes to functionally characterize Nek family kinases. World J Biol Chem. 2014;5:141–160. doi: 10.4331/wjbc.v5.i2.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng D, Cao M, Oda T, Pan J. The conserved ciliary protein Bug22 controls planar beating of Chlamydomonas flagella. J Cell Sci. 2014;127:281–287. doi: 10.1242/jcs.140723. [DOI] [PubMed] [Google Scholar]
- Moon H, Song J, Shin JO, Lee H, Kim HK, Eggenschwiller JT, Bok J, Ko HW. Intestinal cell kinase, a protein associated with endocrine-cerebro-osteodysplasia syndrome, is a key regulator of cilia length and Hedgehog signaling. Proc Natl Acad Sci USA. 2014;111:8541–8546. doi: 10.1073/pnas.1323161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa S, Nakajima K, Miki H, Minato Y, Wang D, Hirokawa N. KIF19A is a microtubule-depolymerizing kinesin for ciliary length control. Dev Cell. 2012;23:1167–1175. doi: 10.1016/j.devcel.2012.10.016. [DOI] [PubMed] [Google Scholar]
- O’Connell MJ, Krien MJ, Hunter T. Never say never. The NIMA-related protein kinases in mitotic control. Trends Cell Biol. 2003;13:221–228. doi: 10.1016/s0962-8924(03)00056-4. [DOI] [PubMed] [Google Scholar]
- Omori Y, Chaya T, Katoh K, Kajimura N, Sato S, Muraoka K, Ueno S, Koyasu T, Kondo M, Furukawa T. Negative regulation of ciliary length by ciliary male germ cell-associated kinase (Mak) is required for retinal photoreceptor survival. Proc Natl Acad Sci USA. 2010;107:22671–22676. doi: 10.1073/pnas.1009437108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JD, Bradley BA, Mooers AO, Quarmby LM. Phylogenetic analysis of the Neks reveals early diversification of ciliary-cell cycle kinases. PLoS One. 2007;2:e1076. doi: 10.1371/journal.pone.0001076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Dickert BL, Vucica Y, Seeley ES, Rosenbaum JL, Witman GB, Cole DG. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J Cell Biol. 2000;151:709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen LB, Rosenbaum JL. Intraflagellar transport (IFT) role in ciliary assembly, resorption and signalling. Curr Top Dev Biol. 2008;85:23–61. doi: 10.1016/S0070-2153(08)00802-8. [DOI] [PubMed] [Google Scholar]
- Pigino G, Geimer S, Lanzavecchia S, Paccagnini E, Cantele F, Diener DR, Rosenbaum JL, Lupetti P. Electron-tomographic analysis of intraflagellar transport particle trains in situ. J Cell Biol. 2009;187:135–148. doi: 10.1083/jcb.200905103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Siuda E, Henderson S, Segil M, Vaananen H, Sassaroli M. Distinct mutants of retrograde intraflagellar transport (IFT) share similar morphological and molecular defects. J Cell Biol. 1998;143:1591–1601. doi: 10.1083/jcb.143.6.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel LC, Bonhivers M, Landrein N, Robinson DR. NIMA-related kinase TbNRKC is involved in basal body separation in Trypanosoma brucei. J Cell Sci. 2006;119:1852–1863. doi: 10.1242/jcs.02900. [DOI] [PubMed] [Google Scholar]
- Rosenbaum J. Organelle size regulation: length matters. Curr Biol. 2003;13:R506–507. doi: 10.1016/s0960-9822(03)00440-8. [DOI] [PubMed] [Google Scholar]
- Rosenbaum JL, Moulder JE, Ringo DL. Flagellar elongation and shortening in Chlamydomonas. The use of cycloheximide and colchicine to study the synthesis and assembly of flagellar proteins. J Cell Biol. 1969;41:600–619. doi: 10.1083/jcb.41.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Schneider MJ, Ulland M, Sloboda RD. A protein methylation pathway in Chlamydomonas flagella is active during flagellar resorption. Mol Biol Cell. 2008;19:4319–4327. doi: 10.1091/mbc.E08-05-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey JM. Intraflagellar transport. Annu Rev Cell Dev Biol. 2003;19:423–443. doi: 10.1146/annurev.cellbio.19.111401.091318. [DOI] [PubMed] [Google Scholar]
- Shalom O, Shalva N, Altschuler Y, Motro B. The mammalian Nek1 kinase is involved in primary cilium formation. FEBS Lett. 2008;582:1465–1470. doi: 10.1016/j.febslet.2008.03.036. [DOI] [PubMed] [Google Scholar]
- Sirajuddin M, Rice LM, Vale RD. Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat Cell Biol. 2014;16:335–344. doi: 10.1038/ncb2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova I, Fuhrmann M, Hegemann P. A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene. 2001;277:221–229. doi: 10.1016/s0378-1119(01)00616-3. [DOI] [PubMed] [Google Scholar]
- Smith LA, Bukanov NO, Husson H, Russo RJ, Barry TC, Taylor AL, Beier DR, Ibraghimov-Beskrovnaya O. Development of polycystic kidney disease in juvenile cystic kidney mice: insights into pathogenesis, ciliary abnormalities, and common features with human disease. J Am Soc Nephrol. 2006;17:2821–2831. doi: 10.1681/ASN.2006020136. [DOI] [PubMed] [Google Scholar]
- Sohara E, Luo Y, Zhang J, Manning DK, Beier DR, Zhou J. Nek8 regulates the expression and localization of polycystin-1 and polycystin-2. J Am Soc Nephrol. 2008;19:469–476. doi: 10.1681/ASN.2006090985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalluto C, Wilson DI, Hearn T. Nek2 localises to the distal portion of the mother centriole/basal body and is required for timely cilium disassembly at the G2/M transition. Eur J Cell Biol. 2012;91:675–686. doi: 10.1016/j.ejcb.2012.03.009. [DOI] [PubMed] [Google Scholar]
- Tam LW, Ranum PT, Lefebvre PA. CDKL5 regulates flagellar length and localizes to the base of the flagella in chlamydomonas. Mol Biol Cell. 2013;24:588–600. doi: 10.1091/mbc.E12-10-0718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam LW, Wilson NF, Lefebvre PA. A CDK-related kinase regulates the length and assembly of flagella in Chlamydomonas. J Cell Biol. 2007;176:819–829. doi: 10.1083/jcb.200610022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel C, Kessler K, Giessl A, Dimmler A, Shalev SA, von der Haar S, Zenker M, Zahnleiter D, Stoss H, Beinder E, et al. NEK1 mutations cause short-rib polydactyly syndrome type majewski. Am J Hum Genet. 2011;88:106–114. doi: 10.1016/j.ajhg.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther Z, Vashishtha M, Hall JL. The Chlamydomonas FLA10 gene encodes a novel kinesin-homologous protein. J Cell Biol. 1994;126:175–188. doi: 10.1083/jcb.126.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Piao T, Cao M, Qin T, Huang L, Deng H, Mao T, Pan J. Flagellar regeneration requires cytoplasmic microtubule depolymerization and kinesin-13. J Cell Sci. 2013;126:1531–1540. doi: 10.1242/jcs.124255. [DOI] [PubMed] [Google Scholar]
- Wang W, Wu T, Kirschner MW. The master cell cycle regulator APC-Cdc20 regulates ciliary length and disassembly of the primary cilium. Elife. 2014;3:e03083. doi: 10.7554/eLife.03083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NF, Lefebvre PA. Regulation of flagellar assembly by glycogen synthase kinase 3 in Chlamydomonas reinhardtii. Eukaryot Cell. 2004;3:1307–1319. doi: 10.1128/EC.3.5.1307-1319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman GB. Axonemal dyneins. Curr Opin Cell Biol. 1992;4:74–79. doi: 10.1016/0955-0674(92)90061-g. [DOI] [PubMed] [Google Scholar]
- Wloga D, Camba A, Rogowski K, Manning G, Jerka-Dziadosz M, Gaertig J. Members of the NIMA-related kinase family promote disassembly of cilia by multiple mechanisms. Mol Biol Cell. 2006;17:2799–2810. doi: 10.1091/mbc.E05-05-0450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood CR, Huang K, Diener DR, Rosenbaum JL. The cilium secretes bioactive ectosomes. Curr Biol. 2013;23:906–911. doi: 10.1016/j.cub.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Roine N, Makela TP. CCRK depletion inhibits glioblastoma cell proliferation in a cilium-dependent manner. EMBO Rep. 2013;14:741–747. doi: 10.1038/embor.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.