Abstract
Highly active antiretroviral (ARV) therapy (HAART) for chronic suppression of HIV replication has revolutionized the treatment of HIV/AIDS. HAART is no panacea; treatments must be maintained for life. Although great progress has been made in ARV therapy, HIV continues to replicate in anatomical and intracellular sites where ARV drugs have restricted access. Nanotechnology has been considered a platform to circumvent some of the challenges in HIV/AIDS treatment. Dispersion polymerization was used to fabricate two types (PMM and ECA) of polymeric nanoparticles, and each was successfully loaded with four ARV drugs (zidovudine, lamivudine, nevirapine, and raltegravir), followed by physicochemical characterization: scanning electron microscope, particle size, zeta potential, drug loading, and in vitro availability. These nanoparticles efficiently inhibited HIV-1 infection in CEM T cells and peripheral blood mononuclear cells; they hold promise for the treatment of HIV/AIDS. The ARV-loaded nanoparticles with polyethylene glycol on the corona may facilitate tethering ligands for targeting specific receptors expressed on the cells of HIV reservoirs.
Keywords: nanoparticles, dispersion polymerization, ARV drugs, GALT, CEM T cells, PBMCs
Introduction
Highly active antiretroviral (ARV) therapy (HAART) for chronic suppression of HIV replication has revolutionized the treatment of HIV/AIDS: many patients are now in their second decade of treatment, with levels of plasma HIV RNA below the limits of detection of clinical assays.1 However, HAART is no panacea. Current treatments must be maintained for life. Although great progress has been made in ARV therapy, HIV continues to replicate in anatomical and intracellular sites where ARV drugs have restricted access.2 Thus, there is a great need for continued efforts in seeking innovative approaches for the treatment of HIV/AIDS. Cellular and anatomical reservoirs of HIV-1 represent major impediments to eradication.3 Cellular reservoirs include latently infected CD4+ T cells, macrophages, and follicular dendritic cells. The main anatomical reservoir sites for HIV include the lymphoid organs (particularly the gut-associated lymphoid tissue [GALT]) and the brain.3 They are the sites of persistent HIV replication. HIV is active in the GALT throughout the period of clinical latency, although minimal viral burden is observed in the blood.4–6
Viral replication occurs predominantly in the peripheral lymphoid organs, and the GALT is an important site of active viral replication, possibly reflecting high levels of T-cell activation.3,7–10 The maintenance of viral reservoirs revolves around the GALT of HIV-infected individuals despite long-term viral suppression.2,11 The belief is that the ongoing low level of replication in the GALT may replenish the cellular reservoir.12 Given the restricted access of HAART to anatomical reservoirs, nanotechnology has been considered a platform that can be brought to bear to circumvent some of the challenges in HIV/AIDS treatment. Nanomedicine platforms that deliver ARV agents to HIV reservoirs (drug polymer conjugates, dendrimers, micelles, liposomes, solid nanoparticles, nanosuspensions, polymeric nanoparticles, and cell-mediated nanoparticles) have been discussed recently.2 Polymeric nanoparticles have higher drug-loading capacity and the potential to target HIV reservoirs by manipulation of their surface characteristics.13–15 Polymeric nanoparticles are preferentially taken up by the M cells of Peyer’s patches of the GALT.16 Although M cells possess the capability to transcytose synthetic nanoparticles, the absorption occurs at very low levels. Thus, there is scope for the application of the concept of site-specific targeting of ARV drug-loaded nanoparticles to M cells to enhance the efficacy of orally administered ARV drugs.
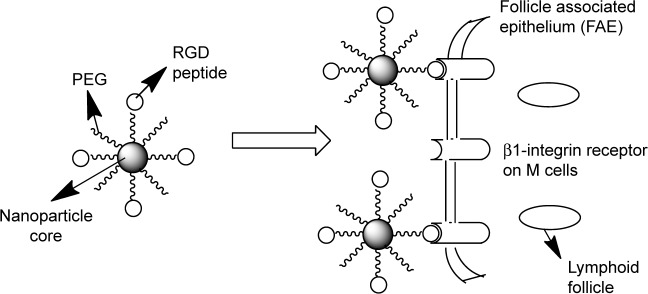
We believe that grafting an M-cell-targeting ligand to the nanoparticle surface could improve M-cell oral delivery strategies by influencing the endocytic mechanisms involved in their uptake by M cells.17–22 Given the report that the ongoing low level of replication in the GALT may replenish the latent reservoir,6,12 this series of projects will result in targeted ARV drugs-loaded nanoparticle delivery system to facilitate uptake and transcellular transport of the ARV drugs to minimize GALT reservoir responsible for the persistent ongoing replication of HIV-1 (ie, adapting drug delivery system properties to cell requirements), as shown in Scheme 1. We have used dispersion polymerization (in situ polymerization) technique previously for the fabrication of nanoparticles for the delivery of bioactive agents.23–26 In situ polymerization of monomers, including cross-linkers in the fabrication of polymeric nanoparticles, allows one-pot synthesis of nanoparticles with many advantages.26 All the advantages of in situ polymerization derive from the possibility of simultaneous encapsulation of bioactive agents during polymerization and, by copolymerization, the possibility of adding surface functionalities in one-batch process without further modifications compared to nanoparticle fabrication by dispersion of preformed polymers.23–27 Our in situ polymerization technique involves the use of a macromonomer or a monomer that forms the core of the nanoparticles, a cross-linker that cross-links the core, a redox initiator system, and a hydrophilic macromonomer (polyethylene glycol [PEG]), which confers the stealth property to the nanoparticles and which also serves as a stabilizer. We have developed polymerizable macromonomers bearing a site-specific ligand (GRGDS peptide) for the receptor expressed uniquely on the apical surface of M cells (cell surface β1 integrin) using low molecular weight (MW) polydisperse polyethylene glycol methacrylates MAv = 400 (n = 7) and 2000 (n = 44; MA-PEGs) capable of forming nanoparticles with a hydrophobic core and a hydrophilic crown or corona exposed to the aqueous medium.28
Scheme 1.
Surface-modified nanoparticles capable of crossing the intestinal epithelium via β1-integrin receptors on M cells.
Here, we fabricated polymeric nanoparticles that were loaded with four different ARV drugs: zidovudine (nucleoside reverse transcriptase inhibitor), lamivudine (nucleoside reverse transcriptase inhibitor), nevirapine (nonnucleoside reverse transcriptase inhibitor), and raltegravir (HIV integrase strand transfer inhibitor). We tested encapsulation efficiency and release of ARV drugs and the effect of ARV drugs-loaded nanoparticles on T cells and macrophages during one-round HIV-1 infection.
Materials and Methods
Materials
Zidovudine, lamivudine, and nevirapine (Sigma-Aldrich), raltegravir (APAC Pharmaceutical, LLC), hydroxylamine hydrochloride (99%; Sigma-Aldrich), pyridine (99+%; Sigma-Aldrich), diethyl ether (99.8%; Sigma-Aldrich), ethyl alcohol (denatured; Sigma-Aldrich), methyl alcohol (99.93%; Sigma-Aldrich), acetonitrile (99.93+%; Sigma-Aldrich), and hydrochloric acid (37%; Fisher Scientific) were used as received. Methacryloyl chloride (97%; Sigma-Aldrich) and methyl methacrylate (99%; Sigma-Aldrich) were vacuum distilled prior to use. Poly(ethylene glycol)n monomethyl ether monomethacrylate (PEG-MA; n = 1000; Polysciences Inc.), benzoyl peroxide (BP; 97%; Sigma-Aldrich), and N-phenyldiethanolamine (NPDEA; 97%; Sigma-Aldrich) were also used without further purification. ε-Caprolactone (Acros) was dried over molecular sieves (5 Å) for 48 hours and distilled under negative pressure before use. 2-Hydroxyethyl methacrylate (HEMA; 97%; Sigma-Aldrich) was dried over molecular sieves (4 Å) for 24 hours and distilled under negative pressure before use. Toluene (99%; Acros) and pyridine (≥99%; Sigma-Aldrich) were refluxed over calcium hydride for one hour and distilled prior to use. Stannous octoate (SnOct; 95%; Sigma-Aldrich) was used as received. Phosphorus pentoxide (97%; Sigma-Aldrich) was also used as received.
Cells and media
CD4+ T cells (CEM) were obtained from the NIH AIDS Research and Reference Reagent Program. Peripheral blood mononuclear cells (PBMCs) were purchased from Astra Biologics. Cells were cultured at 37°C in a 5% CO2 atmosphere in RPMI 1640 medium (Invitrogen) containing 10% fetal bovine serum and 1% antibiotic solution (penicillin and streptomycin; Invitrogen). PBMCs were stimulated with 2.5 μg/mL phytohemagglutinin for 24 hours and then activated for another 24 hours with 10 units/mL interleukin 2 before the infection with vesicular stomatitis virus g-glycoprotein (VSVg)-pseudotyped HIV-1 virus expressing luciferase (HIV-1 Luc).
Methods
Synthesis of hydrolyzable cross-linker (N,O-dimethylacryloylhydroxylamine)
The cross-linker, N,O-dimethacryloylhydroxylamine, was synthesized in our laboratory, as described previously in the literature.23–26,29,30 It involved simultaneous N,O-acylation of hydroxylamine; it was characterized by proton nuclear magnetic resonance (1H NMR) analysis and melting point determination.
Synthesis and characterization of methacrylate-terminated poly-ε-caprolactone macromonomer
The synthesis was done as reported previously.26 ε-Caprolactone (13.14 g), HEMA (4.0 g), and SnOct (three drops) were placed in a 50-mL round-bottom flask equipped with a magnetic stir bar. The reaction was kept under vacuum for 10 minutes, and polymerization was carried out in an inert atmosphere by flushing the flask with nitrogen gas for 24 hours on a silicone oil bath kept at 128°C. The resulting macromonomer was dissolved in dichloromethane and filtered. The macromonomer was precipitated out of this solution using an excess amount of hexane and dry ice. Then, the material was filtered and dried in a vacuum oven at room temperature under reduced pressure over phosphorus pentoxide for 24 hours. The FT-IR and 1H NMR spectra of the synthesized macromonomer were consistent with the expected structure.26,31,32 The number average MW (Mn) was determined by 1H NMR and gel permeation chromatography (GPC), while the weight average MW and polydispersity index of the macromonomer were determined by GPC using a Waters 2690 GPC system equipped with a Waters 2410 differential refractive index detector.24–26 Polystyrene standards were used for calibration and tetrahydrofuran was used as the mobile phase.
Synthesis of nanoparticles
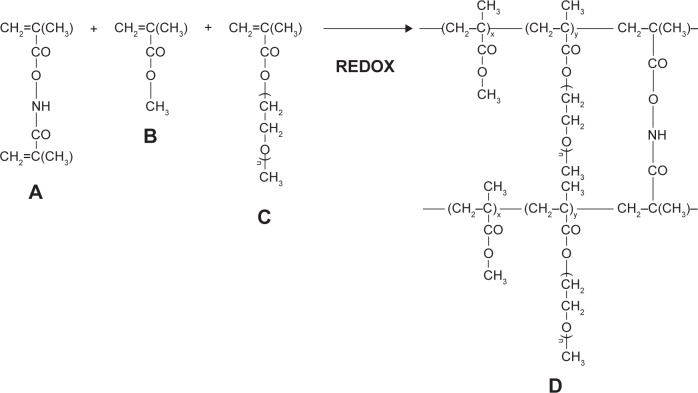
Methyl methacrylate-based nanoparticles (polymethyl meth-acrylate [PMM], Scheme 2)
Scheme 2.
Synthesis and structure of stealth hydrolyzable crosslinked PEG-PMMA (PMM) nanoparticles (A = N,O-dimethacryloylhydroxylamine (crosslinker); B = methyl methacrylate (MMA); C = poly(ethylene glycol)n monomethyl ether monomethacrylate (PEG-MA; n = 1000); D = crosslinked polymer).
8 mmol of methyl methacrylate, 0.1 mmol of PEG–methyl methacrylate (PEG-MMA) macromonomer, and 0.162 mmol of the cross-linker (N,O-dimethacryloylhydroxylamine) were dissolved in a binary solvent system ethanol:water (10 mL). Ten milligrams of each of the drugs (zidovudine, lamivudine, nevirapine, and raltegravir) were added. Then, the components of the redox initiator system (0.021 mmol of NPDEA and 0.021 mmol BP) were injected into the reaction mixture at 10 minutes and 20 minutes, respectively, through a rubber closure under continuous flushing with nitrogen gas and with continuous stirring. The overall polymerization time was 24 hours. After the polymerization, the resulting product was recovered by centrifugation and lyophilized.
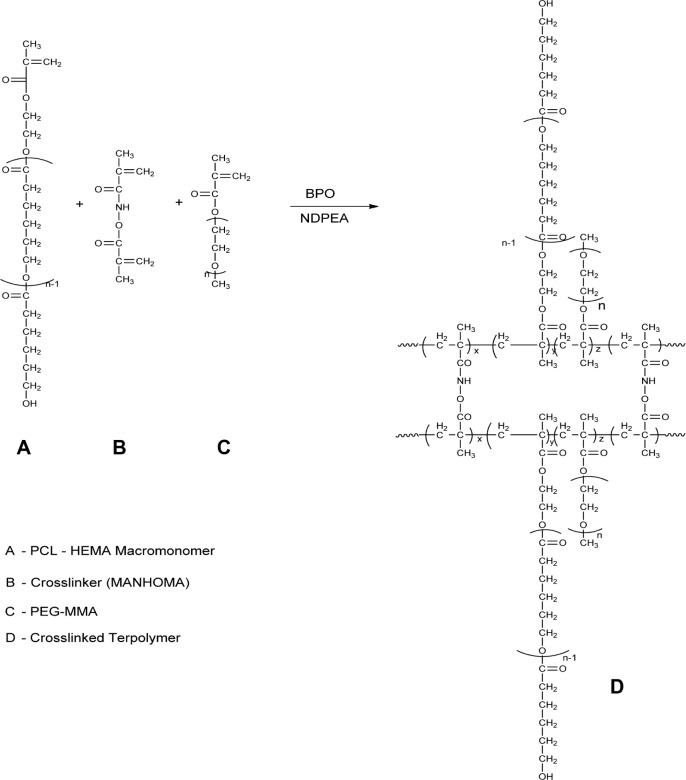
ε-Caprolactone-based nanoparticles (ECA; Scheme 3)
Scheme 3.
Synthesis and structure of stealth crosslinked poly-ε-Caprolactone-HEMA Nanoparticle (ECA).
0.24 mmol of poly-caprolactone-HEMA macromonomer and 0.5 mmol of PEG-MMA, with 0.0209 mmol of cross-linker, were dispersed in acetone:water (10 mL). 0.3325 mmol of BP and 0.3325 mmol of NDPEA (as co-initiators) were injected through the rubber closure under continuous flushing with nitrogen gas and with continuous stirring at 400 rpm. Ten milligrams of each of the drugs (zidovudine, lamivudine, nevirapine, and raltegravir) were added. The overall polymerization time was 24 hours. The nanoparticles were recovered by centrifugation and then lyophilized.
Characterization of the nanoparticles
Surface morphology
The morphology of the nanoparticles was investigated using scanning electron microscope (SEM; JSM-5300 SEM; JEOL). The freeze-dried particles were fixed on a double-sided sticky carbon tape. Then, they were sputter coated with gold in a Denton Vacuum Inc. Desk II instrument (45 mA) and examined in the JSM-5300 SEM instrument at 15 kV.
Particle size analysis and zeta potential
Particle size and size distribution and zeta potential measurements of the nanoparticles were performed by photon correlation spectroscopy using Zetasizer® 3000 (Malvern Instruments). The measurement was performed at 25°C. The dried nanoparticles were suspended in filtered distilled water (3 mL) by vortexing for one minute on a touch mixer (Model 231; Fisher Scientific) and then sonicated for three minutes using a Sonics Vibracell™ Amplitude sonicator (Sonics & Materials, Inc.) before measurement.
Drug loading and in vitro drug availability
Preparation of calibration curves for zidovudine, lamivudine, nevirapine, and raltegravir
Hewlett Packard 1100 Series, a high-performance liquid chromatography (HPLC) system, was used. Separation was done at 35°C on Eclipse Plus C18 column (4.6 mm × 150 mm) from Agilent Technologies. The mobile phase comprised solution A (0.01 M KH2PO4) and B (acetonitrile). The injection volume was 20 μL. The analysis was done at gradient mode with a flow rate of 1 mL/minute; the ratio A/B changed from 90/10 to 20/80 gradually in five minutes and readily reequilibrated to 90/10. Total analysis time was eight minutes. The stock solution of raltegravir, nevirapine, zidovudine, and lamivudine (1 mg/mL) was prepared by dissolving 5 mg of each drug in 5 mL of methanol. Then, the stock solution was diluted with phosphate-buffered saline (PBS) for the preparation of the working solutions as follows: 0.025, 0.05, 0.1, 0.25, 0.5, 1.0, 2.5, 5.0, 10.0, 25.0, 50.0, and 100.0 μg/mL. Ultraviolet–visible detection was performed using an HP photodiode array detector. Blank samples showed no interfering substances eluting at the retention time of raltegravir (retention time, 5.7 minutes; detected at 310 nm), nevirapine (retention time, 4.6 minutes; detected at 285 nm), lamivudine (retention time, 2.7 minutes; detected at 267 nm), and zidovudine (retention time, 3.7 minutes; detected at 267 nm). Furthermore, linear calibration curves (peak areas versus drug concentrations) were established for raltegravir, nevirapine, zidovudine, and lamivudine over 0.05–100.0 μg/mL concentration range (r2 > 0.999).
In vitro availability studies
Drugs (raltegravir, nevi-rapine, zidovudine, and lamivudine)-loaded nanoparticles (known weight) were suspended in PBS (pH 7.4; IS 0.16), placed in a dialysis bag (membrane tubing with MW cutoff of 12,000–14,000) in centrifuge tubes containing known volume of PBS (pH 7.4; IS 0.16), and rotated at 360° using Labquake Shaker maintained at 37°C as reported previously.23–26 At specific time intervals, an aliquot of the release medium was taken and replaced with a fresh medium (maintained at 37°C) each time to keep the volume constant. Then, the aliquot release medium was filtered through a 0.45-μm syringe filter, and the amount of drugs (raltegravir, nevirapine, zidovudine, and lamivudine) released into the medium was quantified using a validated HPLC method (quantitation was carried out according to the standard curves prepared for the drugs).
Determination of drugs-loading capacity of the nanoparticles
The determination was done as reported previously.26 The HPLC analysis was the same as used for the in vitro availability studies of the drugs-loaded nanoparticles described earlier. The nanoparticles (ANP) were incubated in the release medium for more than 35 days till all the loaded drugs were released (each sample in triplicate). Then, quantitation of each drug (raltegravir, nevirapine, zidovudine, and lamivudine) was carried out using the standard curves. Encapsulation property of the nanoparticles was expressed as the loading capacity (amount of drug encapsulated expressed as a percentage of the nanoparticles’ weight), using the following equation:
| (1) |
VSVg-pseudotyped HIV-1 Luc
The 293T cells were cotransfected with HIV-1 proviral vector pNL4-3.Luc.R−E− and VSVg-expressing vector (both obtained from the NIH AIDS Research and Reference Reagent Program). At 48–72 hours post transfection, the media containing HIV-1 virions were collected, filter-sterilized, and kept frozen. HIV-1 Luc was added to CEM T cells or PBMCs. After 48 hours, the cells were washed in PBS and lysed with Steady Lite Luciferase buffer (PerkinElmer). Light emission was analyzed in Luminoskan (PerkinElmer). To adjust for the cell number, the samples were counted with trypan blue as described in the following section.
Cell viability assays
Cells were cultured in 96-well plates at 37°C and incubated with nanoparticles at the indicated concentrations for 24 hours. Cell viability was determined using trypan blue-based assay. The cells were supplemented with 0.2% trypan blue, transferred to a plastic disposable counting chamber, and counted on Cellometer Automatic Cell Counter (Nexcelom Bioscience).
Quantitative PCR for viral DNA analysis
PBMCs were infected with HIV-1 Luc and treated with nanoparticles for six hours. Total DNA was extracted from 4 × 106 cells using lysis buffer (10 mM Tris-HCl pH 8, 10 mM EDTA, 5 mM NaCl, and 200 μg/mL proteinase K). The cells were lysed for 20–30 minutes at room temperature, and proteinase K was inactivated by heating to 95°C for five minutes. For the real-time PCR analysis, 100 ng DNA was amplified in Roche LightCycler 480 II (Roche Diagnostics) and SYBR Green Master mix (Roche Diagnostics). PCR was carried out with initial preincubation for five minutes at 45°C and then for three minutes at 95°C followed by 45 cycles of denaturation at 95°C for 15 seconds, annealing and extension at 60°C for 45 seconds, and final extension at 72°C for 10 seconds. Quantification of an early LTR was carried out using β-globin DNA as a normalization standard. Primer sequences for early LTR were forward-GGCTAACTAGGGAACCCACTG and reverse-CTGCTAGAGATTTTCCACACTGAC and for β-globin were forward-CAACCTCAAACAGACACCATGG and reverse-TCCACGTTCACCTTGCCC (for primer information).33 Mean Cp values for early LTR and β-globin were determined, and AACT method was used to calculate the relative expression levels. Unpaired t-test was used to test for statistical significance.
Results and Discussion
The data obtained from the synthesis of the cross-linker (N,O-dimethacryloylhydroxylamine) [m.p.: 55–56°C; 1H NMR (400 MHz, CDCl3): (C8H11NO3) δ = 2.04 (s, 3H), 2.08 (s, 3H) 5.54, 5.8 (s, 2H), 5.9, 6.38 (s, 2H), 9.25 (s, 1H)] were in agreement with previously obtained data in the literature and in our laboratory.23–26,29,30 Controlled hydrolysis of the cross-linker in the core of the nanoparticles will facilitate controlled release of the encapsulated ARV drugs. FT-IR and 1H NMR spectra of the poly(ε-caprolactone) macromonomer, the average Mn determined by 1H NMR, and the average MW determined by GPC together with the polydispersity index are consistent with the expected structure.26,31,32 The polycaprolactone macromonomer was employed in the dispersion polymerization for the fabrication of ARV drugs-loaded nanoparticles (ECA nanoparticles).
Two types of nanoparticles were synthesized using dispersion polymerization technique: stealth methyl methacrylate-based (PMM; Scheme 2) and stealth ε-caprolactone-based (ECA; Scheme 3) nanoparticles. Methyl methacrylate monomer (Scheme 2) or poly-ε-caprolactone macromonomer (Scheme 3), PEG-MA (n = 1,000), N,O-dimethacryloylhydroxylamine as the cross-linking agent, and BP/NPDEA (BP/NPDEA as redox initiator system) were used for in situ nanoparticle synthesis by free radical dispersion polymerization. PEG macro-monomer was used both as a co-monomer and a steric stabilizer and it formed the corona of the nanoparticles.
Morphological evaluation of the nanoparticles
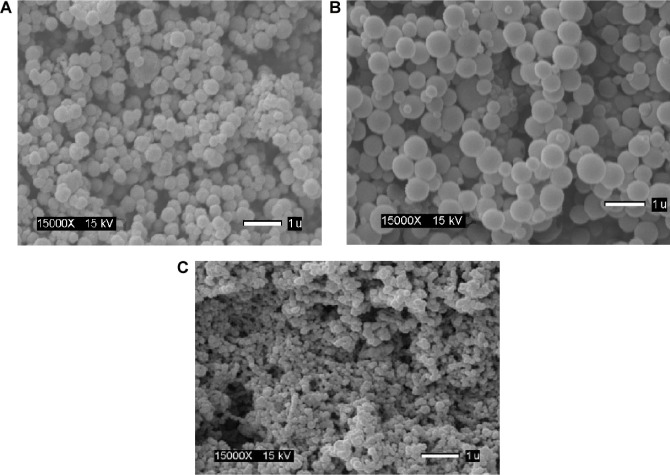
Nanoparticle fabrication by dispersion polymerization was confirmed by SEM for the two types of nanoparticles (stealth methyl methacrylate based [Scheme 2] and stealth ε-caprolactone based [Scheme 3]). Scanning electron micrographs of ARV drugs-loaded hydrolyzable cross-linked PEG–poly-ε-caprolactone nanoparticles (Fig. 1A and 1B) and ARV drugs-loaded hydrolyzable cross-linked PEG–polymethyl methacrylate nanoparticles (Fig. 1C) show the formation of smooth spherical nanoparticles.
Figure 1.
Scanning electron micrographs of ARV drugs-loaded hydrolyzable crosslinked PEG-poly caprolactone (ECA) nanoparticles (A and B) and ARV drugs-loaded hydrolyzable crosslinked PEG-poly methylmethacrylate (PMM) nanoparticles (C).
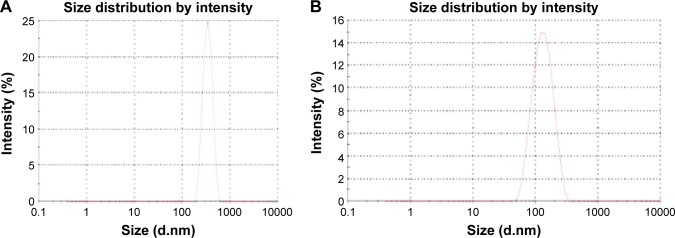
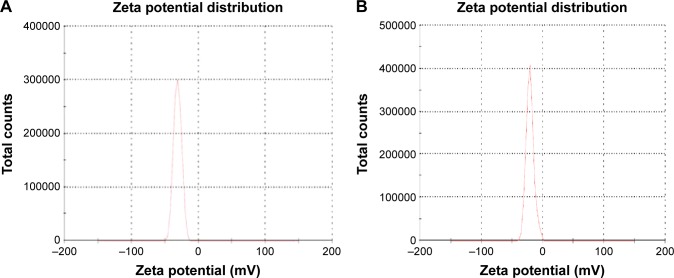
The average particle size for cross-linked PEG–polycaprolactone nanoparticles (Fig. 2A) is 321 ± 36 nm with 0.344 ± 0.023 PDInanoparticle, while the average particle size for cross-linked PEG-PMM nanoparticles (Fig. 2B) is 137.9 ± 1.3 nm with 0.280 ± 0.0.021 PDInanoparticle. Thus, the nanoparticles obtained with poly-ε-caprolactone as the hydrophobic core are bigger than the nanoparticles obtained with PMM as the hydrophobic core. The particle size distribution is given by the PDInanoparticle (a PDInanoparticle value of < 0.1 indicates a homogenous monodisperse formulation, while a PDInanoparticle of >0.3 indicates polydispersity with large variations in particle size).24 The PDInanoparticle values obtained in this work indicate that PEG-PMM nanoparticles are more homogenous than PEG–poly-ε-caprolactone nanoparticles. The zeta potential on the surface of PEG–polycaprolactone-based nanoparticles is more negative (indicating greater tendency for colloidal stability: −31.4 ± −3.6 mV) than the zeta potential on the surface of PEG-PMM-based nanoparticles (−19.2 ± −2.8 mV). It has been reported that particle size plays an important role in determining drug release behavior of drug-loaded nanoparticles and their fate after in vivo administration.34 Furthermore, the zeta potential results obtained in this work show predominantly negative values. Following administration, nanoparticles with a positive zeta potential pose a threat of causing transient embolism and rapid clearance compared to negatively charged particles. Thus, the pegylated poly-ε-caprolactone-based nanoparticles and pegylated PMM-based nanoparticles fabricated by dispersion polymerization are very promising: capable of carrying the drug load to its site of action.35
Figure 2.
Typical particle size distribution for PEG-polycarpolactone (A) and PEG-polymethymethacrylate (B). ARV drugs-loaded hydrolyzable crosslinked nanoparticles.
Drug-loading capacity and in vitro availability of ARV drugs from nanoparticles
Drug-loading capacity was determined by a validated HPLC method as described previously and found to be 6.44 ± 0.13% w/w (raltegravir), 2.13 ± 0.25% w/w (lamivudine), 0.88 ± 0.16% w/w (zidovudine), and 4.15 ± 0.39% w/w (nevirapine) for PEG-PMM-based nanoparticles. For PEG–polycaprolactone-based nanoparticles, the results are as follows: 6.79 ± 0.8% w/w (raltegravir), 1.02 ± 0.12% w/w (lamivudine), 2.11 ± 0.38% w/w (zidovudine), and 5.13 ± 0.58% w/w (nevirapine). While the drug-loading capacity values for each of the drug are better than the values obtained for paclitaxel loaded in PEG–poly(L-lactide)25 and docetaxel loaded in PEG–poly(ε-caprolactone) nanoparticles26 (prepared by dispersion polymerization), which are 0.25% and 0.8%, respectively, effort will be made to improve the loading capacity.
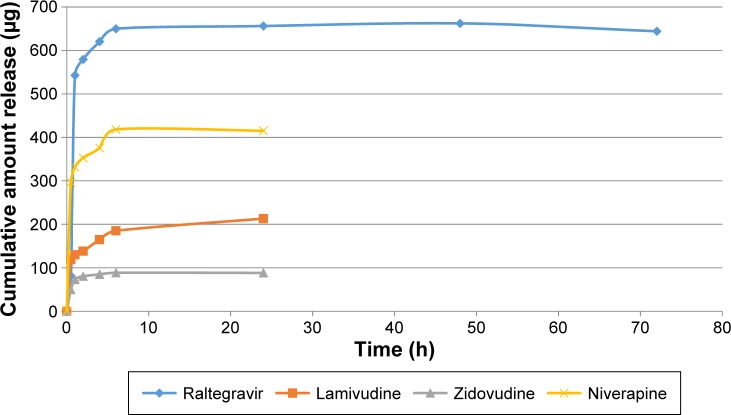
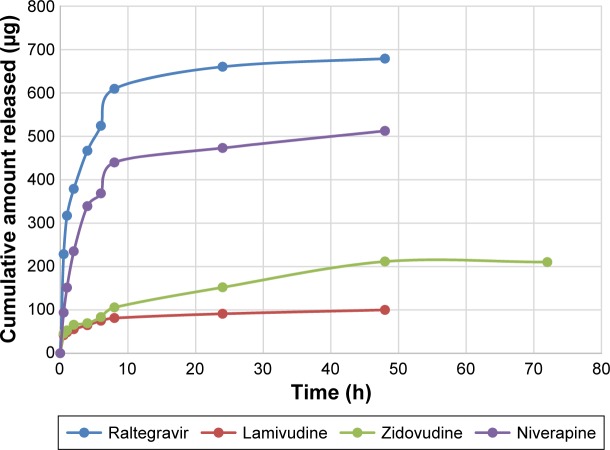
Figures 4 and 5 show the in vitro release isotherms of the ARV drugs from PEG-PMM-based and PEG–polycaprolactone-based nanoparticles, respectively. The release is biphasic with an initial rapid release followed by a slower rate of release. Drug-loading capacity and in vitro availability of the drugs from the nanoparticles appear to be independent of the type of nanoparticles (PEG-PMM-based or PEG–polycaprolactone-based nanoparticles). It appears that the physicochemical properties of the drugs control drug loading and drug release. Raltegravir used in this work (MW: 443.42 g/mol) exhibits poor aqueous solubility (ie, <1 mg/mL); it is very slightly insoluble according to the classification of Amidon and his group,36 whereas the potassium salt of raltegravir is significantly more soluble in water.37 As shown in Figures 4 and 5, raltegravir-loaded nanoparticles, with the highest drug-loading capacity, released the highest amount of drug, followed by nevirapine (MW: 266.31 g/mol), lamivudine (MW: 229.26 g/mol), and zidovudine (MW: 267.4 g/mol). Based on the data from NIH AIDS Research and Reagent Program (Reagent Information, September 2011),38 the rank order of their aqueous solubilities is as follows: nevirapine (0.1 mg/mL: very slightly insoluble in water) > zidovudine (20 mg/mL: sparingly soluble) > lamivudine (70 mg/mL: soluble). Thus, the more hydrophobic ARV drugs have a greater affinity for the hydrophobic cores of the nanoparticles; they exhibit high drug-loading capacity and high drug release.
Figure 4.
Cumulative amount released versus time for ARV drugs-loaded PEG-polymethylmethacrylate based nanoparticles (Raltegravir: ε = 8.15–31.28 μg; Niverapine: ε = 7.5–36.84 μg; Lamivudine: ε = 1.20–3.59 μg; and Zidovudine: ε = 2.06–16.74 μg).
Figure 5.
Cumulative amount released versus time for ARV drugs-loaded PEG-polycaprolactone based nanoparticles (Raltegravir: ε = 6.31–30.2 μg); Niverapine: ε = 6.7–57 μg; Lamivudine: ε = 0.14–4.71 μg; and Zidovudine: ε = 0.56–37.46 μg).
HIV-1 inhibition experiments
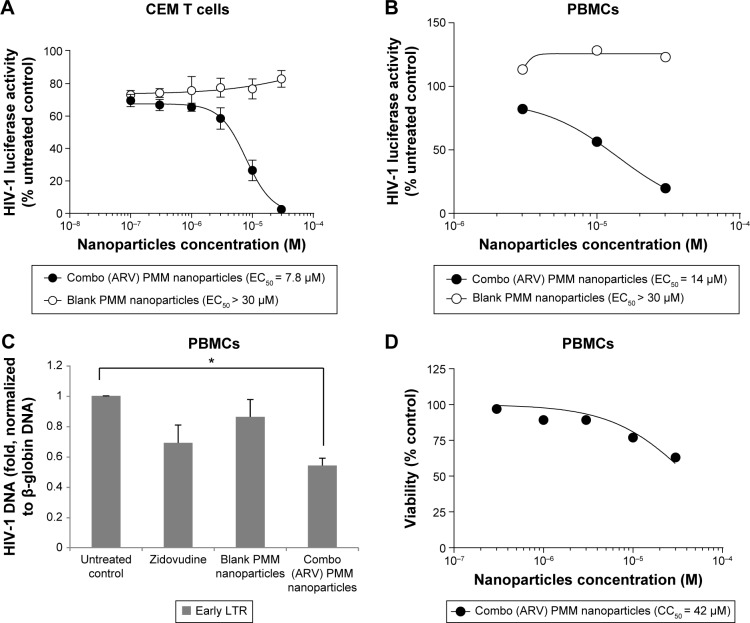
ARV drugs-loaded stealth PMM-based nanoparticles (containing raltegravir, lamivudine, zidovudine, and nevirapine) inhibit one-round HIV-1 infection in cultured T cells and primary PBMCs (Fig. 6). The ARV-loaded PMM-based nanoparticles inhibited HIV-1 infection in CEM T cells by more than 90% with low micromolar EC50 (Fig. 6A). We next tested the effect of the PMM nanoparticles on one-round HIV-1 infection of PBMCs. The ARV-loaded but not blank nanoparticles markedly inhibited HIV-1 replication with EC50 = 14 μM (Fig. 6B). To confirm that the mechanism of HIV-1 inhibition by the ARV-loaded PMM nanoparticles was due to the inhibition of HIV-1 reverse transcription (RT), we analyzed early RT by quantifying HIV-1 DNA.33 The established HIV-1 inhibitor, zidovudine, inhibited RT (Fig. 6C). The ARV-loaded PMM nanoparticles (10 μM) inhibited RT, while blank nanoparticles had little effect (Fig. 6C), suggesting that HIV-1 inhibition was due to the reduction of RT. To access the toxicity of PMM-based nanoparticles, PBMCs were treated with different concentrations of PMM-based nanoparticles and a trypan blue exclusion assay was performed to assess cell’s viability. The assay showed that the ARV-loaded PMM-based nanoparticles were toxic at higher concentration (CC50 = 42 μM; Fig. 6D).
Figure 6.
Inhibition of HIV-1 one round infection by PMM nanoparticles. (A) Inhibition of HIV-1 in CEM T cells by ARV combination PMM nanoparticles. CEM T cells were seeded in 96 wells plates, infected with VSVG HIV-Luc and allowed to incubate overnight. The following day, the cells were treated with different concentrations of combination anti-retroviral nanoparticles containing lamivudine, zidovudine, nevirapine, and raltegravir. After incubating overnight, a luciferase activity was measured. The half maximal effective concentration (EC50) was determined with GraphPad Prizm 6 Software. (B) Inhibition of HIV-1 in peripheral blood mononuclear cells (PBMCs) by ARV combination PMM nanoparticles. Activated PBMCs were seeded in 96 wells plates, infected with VSVG HIV-Luc and allowed to incubate overnight. The following day, the cells were treated with different concentrations of combination anti-retroviral nanoparticles containing lamivudine, zidovudine, nevirapine, and raltegravir. After incubating overnight, a luciferase activity was measured. Results were analyzed in Prism. (C) Inhibition of HIV-1 reverse transcriptase (RT) in PBMCs. Activated PBMCs were infected with HIV-1 Luc and then treated with DMSO, zidovudine, blank PMM nanoparticles or combo (ARV) PMM nanoparticles for 6 hours. DNA was extracted and analyzed by real-time PCR on Roche 480 II using primers for early HIV-1 LTR and β-globin gene as a reference. The asterisk indicates p = 0.005. Unpaired t-test was used to determine statistical significance. (D) Effect of ARV combination PMM nanoparticles on PBMCs viability. PBMCs were seeded in 96 well plates, treated with combination anti-retroviral nanoparticles containing lamivudine, zidovudine, nevirapine, and raltegravir and incubated overnight. Cell viability was determined with trypan blue exclusion assay. The half maximal cytotoxicity concentration (CC50) was determined with GraphPad Prizm 6 Software.
HIV-1 inhibition by ARV drugs-loaded stealth polycaprolactone (ECA) nanoparticles
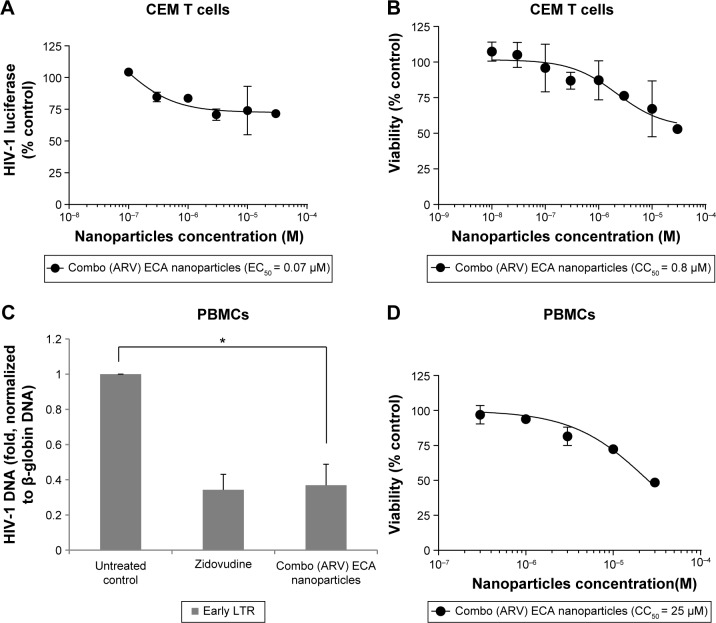
We tested ARV-loaded ECA nanoparticles for HIV-1 inhibition. In CEM T cells infected with HIV-1 Luc, the combo (ARV) ECA-based nanoparticles achieved only ~25% inhibition with EC50 = 0.07 μM (Fig. 7A). This effect was not due to toxicity as nanoparticles were not toxic at this concentration (CC50 = 0.8 μM; Fig. 7B). Data were not available for blank nanoparticles as we could not disperse them well enough in culture media, possibly due to a low level of PEG on nanoparticle surface. The ARV-loaded ECA nanoparticles (10 μM) inhibited HIV-1 RT in the infected PBMCs similar to zidovudine (Fig. 7C). This was not due to toxicity as the ARV-loaded PMM-based nanoparticles were toxic at higher concentrations (CC50 = 25 μM; Fig. 7D).
Figure 7.
Inhibition of HIV-1 one round infection by ECA nanoparticles. (A) Inhibition of HIV-1 in CEM T cells by ARV combination ECA nanoparticles. CEM T cells were seeded in 96 wells plates, infected with VSVG HIV-Luc and allowed to incubate overnight. The following day, the cells were treated with different concentrations of combination anti-retroviral nanoparticles containing lamivudine, zidovudine, nevirapine, and raltegravir. After incubating overnight, a luciferase activity was measured. EC50 was determined in GraphPad Prizm 6. (B) Effect of ARV combination ECA nanoparticles on cell viability in CEM cells. CEM cells were seeded in 96 well plates, treated with combo (ARV) ECA nanoparticles for 6 hours and incubated overnight. Cell viability was determined with trypan blue exclusion assay. CC50 was determined with GraphPad Prizm 6 Software. (C) Inhibition of HIV-1 RT in PBMCs by ECA nanoparticles. Activated PBMCs were infected with HIV-1 Luc and treated with combo (ARV) ECA nanoparticles or zidovudine for 6 hours or left untreated. DNA was extracted and analyzed by real-time PCR on Roche 480 II using primers for early HIV-1 LTR and β-globin gene as a reference. The asterisk indicates p = 0.01. Unpaired t-test was used to determine statistical significance. (D) Effect of ARV combination ECA nanoparticles on cell viability in PBMCs. PBMCs were seeded in 96 well plates and treated with combo (ARV) ECA nanoparticles for 6 hours and then incubated overnight. Cell viability was determined with trypan blue exclusion assay. CC50 was determined with GraphPad Prizm 6 Software.
Taken together, our results indicate that HIV-1 inhibition can be achieved by PMM- and ECA-based nanoparticles fabricated by dispersion polymerization and loaded with ARV drugs. As expected, one-round HIV-1 replication was inhibited at the level of RT. These nanoparticles, fabricated by dispersion polymerization, are very promising, as shown in our current data and as exemplified by our recent report on the nanoparticle packaging of HIV-1-activating small molecule, SMAPP1, which was not stable in mouse serum; its activity was preserved and the stability was improved when encapsulated in similar nanoparticles.39 We also have preliminary data showing successful nanoparticle packaging of HIV-1 transcription inhibitor, 1E7-03 (Nekhai and Akala, unpublished observation).
Conclusion
We show here that four different ARV drugs (zidovudine [nucleoside reverse transcriptase inhibitor], lamivudine [nucleoside reverse transcriptase inhibitor], nevirapine [non-nucleoside reverse transcriptase inhibitor], and raltegravir [HIV integrase strand transfer inhibitor]) could be loaded into nanoparticles by dispersion polymerization and released in vitro. Furthermore, we show here that PMM and ECA nanoparticles loaded with the four antiretroviral drugs efficiently inhibited HIV-1 infection in CEM T cells and PBMCs. As expected, ARV-loaded nanoparticles inhibited HIV-1 RT. The ARV-loaded nanoparticles may help to improve the efficacy of ARV drugs. The ARV-loaded nanoparticles with PEG on the corona may facilitate tethering ligands for targeting specific receptors expressed on the cells of HIV reservoirs. Other advantages of the nanoparticles reported here are as follows: the fabrication of the nanoparticles is a one-pot process (simultaneous encapsulation of ARV drugs during polymerization) carried out at ambient temperature; it is possible to add surface functionalities in one-batch process without further modifications compared to nanoparticle fabrication by dispersion of preformed polymers; the process is surfactant free; and the ARV drugs combination is typical of clinical recommendations. We are optimizing these nanoparticles to target them to the gut and the brain. In conclusion, the ARV-loaded nanoparticles will be useful for future ARV therapy.
Figure 3.
Typical particle zeta potential distribution for PEG-polycarpolactone (A) and PEG-polymethymethacrylate (B). ARV drugs-loaded hydrolyzable crosslinked nanoparticles.
Footnotes
ACADEMIC EDITOR: Douglas MacPherson, Editor in Chief
PEER REVIEW: Four peer reviewers contributed to the peer review report. Reviewers’ reports totaled 987 words, excluding any confidential comments to the academic editor.
FUNDING: This work was supported by the District of Columbia Center for AIDS Research grant NIH/NIAID Grant # 5P30A1087714-02 (11-M56R CFDA # 93.855) and NIH Research Grants (1P50HL118006, 1R01HL125005, and 5G12MD007597). This work was carried out in facilities supported by NCRR/NIH Grants #1 C06 RR 020608-01 and #1 C06 RR 14469-01. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors would like to thank the NIH AIDS Research and Reference Reagent Program for pHEF-VSVG expression vector (courtesy of Dr. Lung-Ji Chang) and pNL4-3.Luc.R−E− (courtesy of Dr. Nathaniel Landau) and ARV drugs for comparison with those obtained commercially. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert single-blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived and designed the experiments: EOA, SN, and WAA. Analyzed the data: EOA, SN, WAA, OO, NK, KAS, OB, SA, and AG. Wrote the first draft of the manuscript: EOA, SN, WAA, OO, NK, KAS, OB, SA, and AG. Contributed to the writing of the manuscript: EOA, SN, WAA, OO, NK, KAS, OB, SA, and AG. Agreed with manuscript results and conclusions: EOA, SN, WAA, OO, NK, KAS, OB, SA, and AG. Jointly developed the structure and arguments for the paper: EOA, SN, WAA, OO, NK, KAS, OB, SA, and AG. Made critical revisions and approved the final version: EOA, SN, WAA, OO, NK, KAS, OB, SA, and AG. All the authors reviewed and approved the final manuscript.
REFERENCES
- 1.Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerant RJ. The challenge of finding a cure for HIV infection. Science. 2009;323:1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- 2.Edagwa BJ, Zhou T, McMillan JM, Liu X-M, Gendelman HE. Development of HIV reservoir targeted long acting nanoformulated antiretroviral therapies. Curr Med Chem. 2014;1(36):4186–4198. doi: 10.2174/0929867321666140826114135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tyagi M, Bukrinsky M. Human Immunodeficiency Virus (HIV) latency: the major hurdle in HIV eradication. Mol Med. 2012;18:1096–1108. doi: 10.2119/molmed.2012.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dembri A, Montisd M-J, Gantier JC, Chacon H, Ponchell G. Targeting of 3′–Azido 3′-deoxythymidine (AZT)-loaded poly(isohexylcyanoacrylate) nano-spheres to the gastrointestinal mucosa and associated lymphoid tissues. Pharm Res. 2001;18(4):467–473. doi: 10.1023/a:1011050209986. [DOI] [PubMed] [Google Scholar]
- 5.Schrager LK, D’Souza MP. Cellular and anatomical reservoirs of HIV-1 in patients receiving potent antiretroviral combination therapy. JAMA. 1998;280(1):67–71. doi: 10.1001/jama.280.1.67. [DOI] [PubMed] [Google Scholar]
- 6.Saksena NK, Haddad DN. Viral reservoirs an impediment to HAART: new strategies to eliminate HIV-1. Curr Drug Targets Infect Disord. 2003;3:179–206. doi: 10.2174/1568005033481187. [DOI] [PubMed] [Google Scholar]
- 7.Blankson JN, Persaud D, Siliciano RF. The challenge of viral reservoirs in HIV-1 infection. Annu Rev Med. 2002;53:557–593. doi: 10.1146/annurev.med.53.082901.104024. [DOI] [PubMed] [Google Scholar]
- 8.Pantaleo G, Graziosi C, Demarest JF. HIV infection is active and progressive in lymphoid tissue during the clinically latent stage of disease. Nature. 1993;362:355–358. doi: 10.1038/362355a0. [DOI] [PubMed] [Google Scholar]
- 9.Veazey RS, Demaria MA, Chalifoux LV. Gastrointestinal tract as a major site of CD4C T cell depletion and viral replication in SIV infection. Science. 1998;280:427–431. doi: 10.1126/science.280.5362.427. [DOI] [PubMed] [Google Scholar]
- 10.Gunthard HF, Havlir DV, Fiscus S. Residual Human Immunodeficiency Virus (HIV) Type 1 RNA and DNA in lymph nodes and HIV RNA in genital secretions and in cerebrospinal fluid after suppression of viremia for 2 years. J Infect Dis. 2001;183:1318–1327. doi: 10.1086/319864. [DOI] [PubMed] [Google Scholar]
- 11.Chun T-W, Nickle DC, Justement JS, et al. Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy. JID. 2008;197:714–720. doi: 10.1086/527324. [DOI] [PubMed] [Google Scholar]
- 12.Gandhi RT, Bosch RJ, Aga E, Xin-Ming Liu X-M, Gendelman HE. No evidence for decay of the latent reservoir in HIV-1–infected patients receiving intensive enfuvirtide-containing antiretroviral therapy. JID. 2010;201:293–296. doi: 10.1086/649569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khali NM, Carraro E, Cotica LF, Mainardes RM. Potential of polymeric nanoparticles in AIDS treatment and prevention. Expert Opin Drug Deliv. 2011;8(1):95–112. doi: 10.1517/17425247.2011.543673. [DOI] [PubMed] [Google Scholar]
- 14.Amiji MM, Vyas TK, Shah LK. Role of nanotechnology in HIV/AIDS treatment: potential to overcome the viral reservoir challenge. Discov Med. 2006;34:157–162. [PubMed] [Google Scholar]
- 15.Mamo T, Moseman EA, Kolishetti N, et al. Emerging nanotechnology approaches for HIV/AIDS treatment and prevention. Nanomedicine. 2010;5(2):269–285. doi: 10.2217/nnm.10.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akala EO. Strategies for transmembrane passage of polymer-based nano-structures. In: Pavel Broz., editor. Polymer-Based Nanostructures: Medical Applications. Cambridge, UK: Royal Society of Chemistry; 2010. pp. 16–80. (Royal Society of Chemistry Series 9: Series Editors). Harry Kroto, Paul O’Brien, and Harold Craighead. [Google Scholar]
- 17.Kernies S, Bogdanova A. Conversion by Peyer’s patch lymphocytes of human enterocytes into M cells that transport bacteria. Science. 1997;277(5328):949–952. doi: 10.1126/science.277.5328.949. [DOI] [PubMed] [Google Scholar]
- 18.Jepson MA, Simmons NL, O’Hagan DT, Hirst BH. Comparison of poly(DL-lactide-co-glycolide) and polystyrene microsphere targeting to intestinal M cells. J Drug Targeting. 1993;1:245–249. doi: 10.3109/10611869308996082. [DOI] [PubMed] [Google Scholar]
- 19.Eldridge JH, Meulbroek JA, Staas JK, Tice TR, Gilley RM. Vaccine-containing biodegradable microspheres specifically enter the gut-associated lymphoid tissue following oral administration and induce a disseminated mucosal immune response. Adv Exp Med Biol. 1989;251:191–202. doi: 10.1007/978-1-4757-2046-4_18. [DOI] [PubMed] [Google Scholar]
- 20.Eldridge JH. Controlled vaccine release in the gut-associated lymphoid tissues: I. Orally administered biodegradable microspheres target the Peyer’s patches. J Control Release. 1990;11:205–214. [Google Scholar]
- 21.Ermak TH, Dougherty EP, Bhagat HR, Kabok Z, Pappo J. Uptake and transport of copolymer biodegradable microspheres by Rabbit Peyer’s patch M cells. Cell Tissue Res. 1995;279:433–436. doi: 10.1007/BF00318501. [DOI] [PubMed] [Google Scholar]
- 22.Torche AM, Jouan H, Corre PL, et al. Ex vivo and in situ PLGA microspheres uptake by Pig Ileal Peyer’s patch segment. Int J Pharm. 2000;201:15–27. doi: 10.1016/s0378-5173(00)00364-1. [DOI] [PubMed] [Google Scholar]
- 23.Akala EO, Okunola O. Novel stealth degradable nanoparticles prepared by Dispersion Polymerization for the delivery of bioactive agents part I. Pharm Ind. 2013;75(7):1191–1196. [Google Scholar]; Pharm Ind. 2013;75(7):1346–1352. Part II. [Google Scholar]
- 24.Adesina SK, Wight SA, Akala EO. Optimization of the fabrication of novel stealth PLA-based nanoparticles by dispersion polymerization using D-optimal mixture design. Drug Dev Ind Pharm. 2014;40(11):1547–1556. doi: 10.3109/03639045.2013.838578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adesina SK, Holly A, Kramer-Marek G, Capala J, Akala EO. Polylactide based paclitaxel loaded nanoparticles fabricated by dispersion polymerization: characterization, evaluation in cancer cell lines, and preliminary biodistribution studies. J Pharm Sci. 2014;103(8):2546–2555. doi: 10.1002/jps.24061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogunwuyi O, Adesina SK, Akala EO. D-optimal mixture experimental design for stealth biodegradable crosslinked docetaxel-loaded poly-E-caprolactone nanoparticles fabricated by dispersion polymerization. Pharmazie. 2015;70:165–176. [PubMed] [Google Scholar]
- 27.Landfester K, Mailander V. Nanocapsules with specific targeting and release properties using miniemulsion polymerization. Expert Opin Drug Deliv. 2013;10(5):593–609. doi: 10.1517/17425247.2013.772976. [DOI] [PubMed] [Google Scholar]
- 28.Bolshakov O, Akala EO. MS-monitored conjugation of poly(ethylene glycol) monomethacrylate to RGD peptides. J Appl Polym Sci. 2014;131:40385–40395. doi: 10.1002/app.40385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yin W, Akala EO, Taylor R. Design of naltrexone-loaded hydrolyzable cross-linked nanoparticles. Int J Pharm. 2002;244(1–2):9–19. doi: 10.1016/s0378-5173(02)00297-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akala EO, Kopeckova P, Kopecek J. Novel pH-sensitive hydrogels with adjustable swelling kinetics. Biomaterials. 1998;19(11–12):1037–1047. doi: 10.1016/s0142-9612(98)00023-4. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y, Schulze M, Albertsson A-C. α-methacryloyl-ω-hydroxyl-poly(e-caprolactone) macromonomer: synthesis, characterization, and copolymerization. J Macromol Sci Pure Appl Chem. 1998;35(2):207–232. [Google Scholar]
- 32.Tortosa K, Miola C, Hamaide T. Synthesis of low molecular weight-hydroxy polycaprolactone macromonomers by coordinated anionic polymerization in protic conditions. J Appl Polymer Sci. 1997;65:2357–2372. [Google Scholar]
- 33.Vermeire J, Naessens E, Vanderstraeten H, et al. Quantification of reverse tran-scriptase activity by real-time PCR as a fast and accurate method for titration of HIV, lenti- and retroviral vectors. PLoS One. 2012;7:e50859. doi: 10.1371/journal.pone.0050859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin C, Wu H, Liu J, Bai L, Guo G. The effect of paclitaxel-loaded nanoparticles with radiation on hypoxic MCF-7 cells. J Clin Pharm Ther. 2007;32:41–44. doi: 10.1111/j.1365-2710.2007.00796.x. [DOI] [PubMed] [Google Scholar]
- 35.Li S, Huang L. Pharmacokinetics and biodistribution of nanoparticles. Mol Pharm. 2008;5:496–504. doi: 10.1021/mp800049w. [DOI] [PubMed] [Google Scholar]
- 36.Kasim NA, Whitehouse M, Ramachandran C, et al. Molecular properties of WHO essential drugs and provisional biopharmaceutical classification. Mol Pharm. 2004;1(1):85–96. doi: 10.1021/mp034006h. [DOI] [PubMed] [Google Scholar]
- 37.WO2013111100A1 Patent.
- 38.NIH AIDS Research and Reagent Program (Reagent Information) 2011 [Google Scholar]
- 39.Smith KA, Lin X, Bolshakov O, et al. Activation of HIV-1 with nanoparticle-packaged small-molecule protein phosphatase-1-taargeting compound. Sci Pharm. 2015;83:535–538. doi: 10.3797/scipharm.1502-01. [DOI] [PMC free article] [PubMed] [Google Scholar]