Abstract
Motivation: We addressed the problem of inferring gene regulatory network (GRN) from gene expression data of knockout (KO) experiments. This inference is known to be underdetermined and the GRN is not identifiable from data. Past studies have shown that suboptimal design of experiments (DOE) contributes significantly to the identifiability issue of biological networks, including GRNs. However, optimizing DOE has received much less attention than developing methods for GRN inference.
Results: We developed REDuction of UnCertain Edges (REDUCE) algorithm for finding the optimal gene KO experiment for inferring directed graphs (digraphs) of GRNs. REDUCE employed ensemble inference to define uncertain gene interactions that could not be verified by prior data. The optimal experiment corresponds to the maximum number of uncertain interactions that could be verified by the resulting data. For this purpose, we introduced the concept of edge separatoid which gave a list of nodes (genes) that upon their removal would allow the verification of a particular gene interaction. Finally, we proposed a procedure that iterates over performing KO experiments, ensemble update and optimal DOE. The case studies including the inference of Escherichia coli GRN and DREAM 4 100-gene GRNs, demonstrated the efficacy of the iterative GRN inference. In comparison to systematic KOs, REDUCE could provide much higher information return per gene KO experiment and consequently more accurate GRN estimates.
Conclusions: REDUCE represents an enabling tool for tackling the underdetermined GRN inference. Along with advances in gene deletion and automation technology, the iterative procedure brings an efficient and fully automated GRN inference closer to reality.
Availability and implementation: MATLAB and Python scripts of REDUCE are available on www.cabsel.ethz.ch/tools/REDUCE.
Contact: rudi.gunawan@chem.ethz.ch
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
Gene regulatory networks (GRNs) describe the regulatory interactions among genes in a cell, i.e. how the transcription of one gene is regulated by those of the others. Such networks are often described as a directed graph with nodes representing genes and edges representing gene regulations. Perturbations to the GRN in a cell, for example because of gene mutations, could lead to changes in the gene expression profile and correspondingly in the cellular phenotype. The understanding of gene–gene interactions and their pathological alterations could shed light on the mechanism underlying genetic diseases such as cancer. For this reason, the inference of GRNs has garnered much attention in systems biology. Further fueled by the ever-growing public databases of gene expression profiles, hundreds of algorithms have been developed for inferring GRNs from such data. These methods borrow and extend techniques from different fields, such as statistics, machine learning and systems model identification.
Despite the large number of available methods, the inference of GRN from gene expression data still remains unsolved to date. The crux of the problem is the underdetermined nature of this inference, leading to the lack of network identifiability or inferability (Szederkényi et al., 2011; Ud-Dean and Gunawan, 2014). This issue often leads to the existence of multiple equivalent solutions to the inference problem, i.e. there exist a family of network graphs that are consistent with the available expression data. We recently developed an ensemble inference algorithm called Transitive Reduction and Closure Ensemble (TRaCE) with the goal of generating an ensemble of digraphs that are consistent with a given gene knockout (KO) dataset (Ud-Dean and Gunawan, 2014). More specifically, TRaCE produces upper and lower bound digraphs of the ensemble where any network in the ensemble is a subgraph of the upper bound and a supergraph of the lower bound. Edges in the upper bound that are missing from the lower bound are deemed uncertain as these edges could not be verified by the provided data. The GRN is therefore inferable when there exists no uncertain edges (i.e. the upper and lower bounds meet). Using TRaCE, we demonstrated that systematic KO experiments such as performing single-gene KOs (SKOs) and double-gene KOs (DKOs) is a strongly suboptimal strategy for inferring GRN (Ud-Dean and Gunawan, 2014).
The attention given to designing experiments for GRN inference pales in comparison to developing inference methods. Only a handful of strategies having been proposed previously. For example, Ideker et al. (2000) proposed an optimization of perturbation experiments for a Boolean model of acyclic GRNs using minimum set cover. Tegnér et al. (2003) formulated a heuristic strategy of ranking gene perturbations where genes with weaker differential expression or those associated with more uncertain interactions are more likely selected for KOs. Meanwhile, Spieth et al. (2004) employed an evolutionary strategy to create an ensemble of S-system models of GRN. The design of experiments (DOE) involved performing virtual gene KOs using the model ensemble and choosing the most informative KO experiment. On the other hand, Steinke et al. (2007) developed a DOE strategy based on Bayesian linear regression with a sparse prior distribution of the GRN, where experiments were selected according to the possible information gain. More recently, Lang et al. (2014) proposed a DOE strategy for cellular reaction networks based on selecting a set of measurements, using which low confidence reactions are isolated in disjoint subnetworks or modules. Finally, Birget et al. (2012) used a graph theory concept called node cut-sets or vertex separators, to formulate a systematic procedure for inferring GRN digraphs. Briefly, the strategy involves systematically knocking out the vertex separators of gene pairs with an indirect regulation, i.e. gene pair i and j where gene i regulates gene j through other gene(s). The authors showed theoretically that the inference of acyclic GRN with n genes would require gene KO experiments, while those with cycles would need experiments.
In this work, we developed an algorithm called REDuction of UnCertain Edges (REDUCE) for selecting the optimal gene KO experiment based on an ensemble of GRNs, particularly using the upper and lower bounds of the ensemble. REDUCE was formulated as a constrained optimization problem to maximize the number of uncertain edges that could potentially be verified. We introduced the concept of edge separatoid, similar to vertex separators, as the basis to count the number of possible edge verification associated with a given gene KO combination. Finally, we proposed an iterative procedure for the GRN inference, in which the upper and lower bounds of the ensemble are continually updated during iterations of wet-lab KO experiments and dry-lab optimal DOE using REDUCE. As a proof of concept, we applied the iterative procedure to infer the GRN of E.coli under ideal conditions. We further demonstrated the efficacy of REDUCE using benchmark gene expression simulator GeneNetWeaver (GNW) (Schaffter et al., 2011) in the inference of five 100-gene gold standard networks from DREAM 4 in silico network inference challenge (Schaffter et al., 2011). We compared the performance of the iterative inference with performing systematic KO experiments.
2 Methods
2.1 Definitions
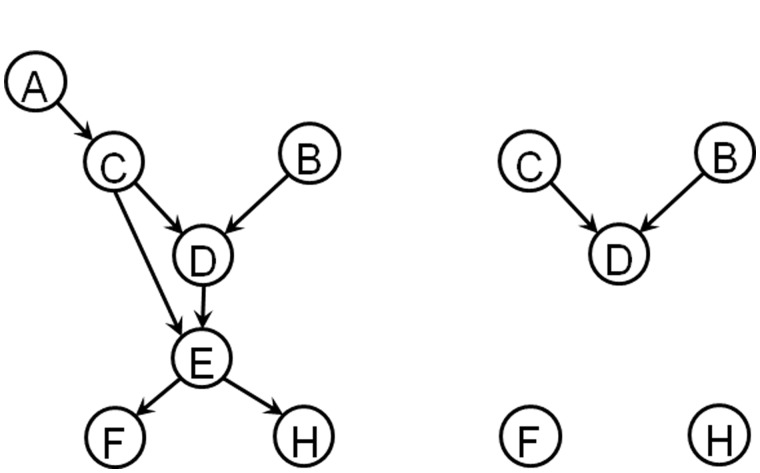
In this section, we review several basic concepts of graph theory which will be used in the development of the DOE algorithm. A graph G is defined by the pair (V(G),E(G)), where V(G) denotes the set of vertices (nodes) and denotes the set of edges. The number of vertices n(G) and edges m(G) are called the order and size of the graph, respectively. In a digraph, an edge is defined by an ordered pair of vertices (i,j) denoting the edge direction, from vertex i pointing to vertex j. In this case, vertex i is a parent of vertex j, and correspondingly vertex j is a child of vertex i. Here, we consider a digraph model of GRN where the edges are unsigned and unweighted. In such a digraph, the nodes represent genes while the edges describe the gene regulatory interactions. The directed edge (i,j) indicates that gene i regulates gene j. In the following, the graph denotes the digraph of the GRN of interest, which is also referred to as wild-type GRN. Meanwhile, the digraph corresponding to knocking out or deleting a set of genes is denoted by Figure 1a shows an example of a GRN digraph with 7 genes (n(G) = 7) and 7 gene interactions (m(G) = 7). Here, genes B and C are parents of gene D, and genes D and E are children of gene C. Figure 1b further shows the digraph corresponding to knocking out genes A and E from the GRN in Figure 1a.
Fig. 1.
(a) Example of a digraph of GRN . (b) The GRN resulting from knocking out genes A and E from
A directed path in a digraph is a sequence of vertices where a directed edge exists from one vertex to the next. For example, in Figure 1a the vertex sequence ACDEF gives a directed path from vertex A to vertex F. A vertex j is said to be accessible from a vertex i when there exists a directed path from vertex i to vertex j. In this regard, vertex i is an ancestor of vertex j, and vertex j is correspondingly a descendant of vertex i. The accessibility matrix of a digraph G, denoted by Acc(G), is an n(G) × n(G) matrix with Acci,j = 1 when vertex j is accessible from vertex i, and Acci,j = 0 otherwise. In Figure 1a, gene F is accessible from gene A. Furthermore, genes A, B, C, D and E are the ancestors of gene H, while genes D, E, F and H are the descendants of gene B.
2.2 TRaCE
To address the underdetermined nature of GRN inference, we previously developed TRaCE with the goal of identifying the ensemble of digraphs that are consistent with steady-state expression data from gene KO experiments. TRaCE is based on the assumption that gene KOs cause steady-state differential expressions among genes that are downstream or accessible from the deleted genes (i.e. all genes that are directly or indirectly regulated by the deleted genes). Thus, in the first step of TRaCE, we construct the perturbation graphs corresponding to the accessibility matrices of and all appropriate from gene KO data. For example, given the differential gene expression of SKO experiments, one could construct the accessibility matrix of . Here, the accessibility matrix represents the largest network (in size) that agrees with the data and is an estimate of the transitive closure of (Aho et al., 1972). However, many digraphs can share the same transitive closure. This ambiguity illustrates the underdetermined nature of the GRN inference problem. For directed acyclic graphs (DAGs), one can identify the smallest digraph among those sharing the same transitive closure by applying a transitive reduction algorithm (Aho et al., 1972). In the second step, TRaCE applies a modified transitive reduction called Condensation, Transitive Reduction and Expansion (ConTREx) to the accessibility matrices. ConTREx involves the condensation of a perturbation graph into a DAG of the strong components (strongly connected components), the transitive reduction of this DAG and the expansion of the resulting reduced graph where edges associated with cycles are removed (see Ud-Dean and Gunawan, 2014 for more details). In the last step of TRaCE, the accessibility matrices and their reductions are combined to produce the upper and lower bound digraphs of the ensemble, denoted by GU and GL, respectively. Edges in every member digraph of the ensemble are a subset of those in the upper bound and a superset of those in the lower bound. If desired, the ensemble of digraphs can be constructed from GU and GL. Edges in GU that do not appear in GL, defined by the set EU = {(i, j): (i, j) ∈GU, (i, j)∉GL}, are referred to as uncertain edges since their existence could not be verified by the available data. The number of uncertain edges (i.e. the cardinality of EU or N(EU)) gives a measure of the uncertainty in a particular GRN inference problem.
2.3 DOE by REDUCE
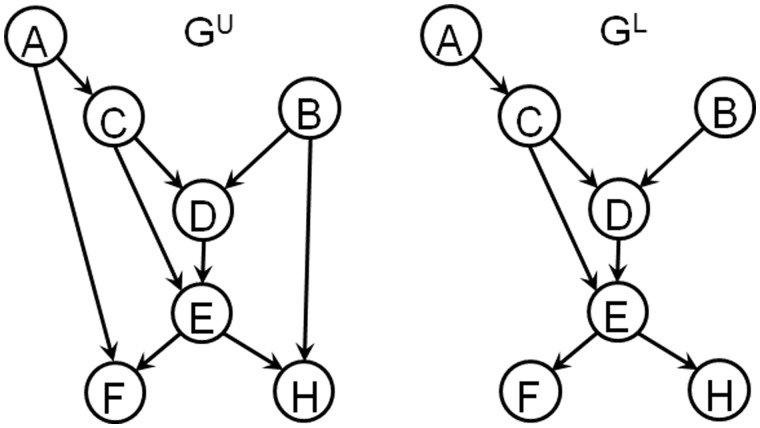
The premise behind REDUCE is to identify the optimal set of genes whose KO or deletion would enable the verification of the highest number of uncertain edges. As inputs, REDUCE requires the upper and lower bounds of the ensemble, such as those generated by TRaCE. We will illustrate the main concept of REDUCE using the following example. Consider the upper and lower bound digraphs shown in Figure 2. Here, there are two uncertain edges (A, F) and (B, H) since these edges appear in GU but are missing from GL. To confirm the uncertain edge (A, F), we consider knocking out (disconnecting) all indirect paths from gene A to gene F in GU. Removing any one of the genes in the set {C, E} or both genes would accomplish this task. When there exists no indirect path from gene A to gene F, the verification of the edge (A, F) becomes simple. For example, if perturbing gene A leads to a differential expression of gene F in the background of gene C KO, then we can confirm the existence of (A, F). Otherwise, the edge (A, F) does not exist. Similarly, for the edge (B, H), knocking out one of the genes in the set {D, E} or both genes would remove all indirect paths from B to H in GU. In this exercise, the optimal KO experiment would therefore be to knock out gene E as this would simultaneously disconnect the indirect paths from A to F and from B to H. Note that we still need to perturb or knock out gene A and B individually in the background of gene E deletion to verify the uncertain edges.
Fig. 2.
Example of upper and lower bounds of a GRN
In the earlier illustration, we call the sets of genes {C}, {E} and {C, E} as the edge separatoids of (A, F). More precisely, we define an edge separatoid of (i, j)∈EU as the set of nodes whose removal would disconnect all indirect paths from node i to node j in GU. There could be more than one (edge) separatoid for an uncertain edge as demonstrated in the previous example. In addition, two or more uncertain edges could share the same separatoid as in the case of gene E with respect to the uncertain edges (A, F) and (B, H). In REDUCE, we consider the following separatoids for each uncertain edge (i, j)∈EU:
S1(i, j) = children of i in GU ∩ ancestors of j in GU
S2(i, j) = descendants of i in GU ∩ parents of j in GU
S3(i, j) = descendants of i in GU ∩ ancestors of j in GU
For the example in Figure 2, we have S1(A,F) = {C}, S2(A,F) = {E} and S3(A,F) = {C,D,E}. The separatoids S1(i, j) and S2(i, j) are both subsets of S3(i, j), and S3(i, j) is the largest separatoid of (i, j). For the optimization of gene KO, we further define the following sets of separatoids:
S1 = {S1(i, j): (i, j) ∈EU}
S2 = {S2(i, j): (i, j) ∈EU}
S3 = {S3(i, j): (i, j) ∈EU}
The three separatoids above do not, by any means, represent the complete set of separatoids for an uncertain edge (i, j). Rather, they are selected because of the ease in computing and storing them. One could also consider the grandchildrens of i or grandparents of j and so on. However, delineating all possible separatoids would constitute finding the longest path between two nodes in a graph, a problem which is known to be NP-hard (non-deterministic polynomial-time hard) (Bjorklund et al., 2004). In addition, the memory requirement would become prohibitively large as the separatoids are computed a priori and stored in memory during KO optimization.
As mentioned earlier, REDUCE involves finding the optimal combination of nodes whose removal would enable the verification of the highest number of uncertain edges. Given the aforementioned sets of edge separatoids S1, S2 or S3, we solve the following optimization problem
where ET,q(VKO) = {(i, j): (i, j) ∈ EU, Sq(i, j) ⊆ VKO, i, j ∉ VKO} and N(ET,q(VKO)) is the cardinality of ET,q(VKO). In this optimization, ET,q(VKO) represents the set of uncertain edges that could potentially be verified by according to the set of separatoids Sq (q = 1, 2, 3). One can impose constraints in the earlier optimization, such as to exclude essential genes or combinations of genes whose KOs are lethal, and to limit the number of KO genes (i.e. the cardinality of VKO or N(VKO)). In the implementation of REDUCE, the optimization is carried out for each Sq (q = 1, 2, 3) separately using a modified genetic algorithm (GA) (Holland, 1992), the maximum of which is selected after the completion of the GA optimizations (see pseudo-code in Supplementary Information).
Following the simple illustration earlier, the verification of uncertain edges involves obtaining gene expression data from the following experiments:
deletion of genes in the set
perturbation of KO of each gene i from the set I* in the background deletion, where
I* = {i: (i, j) ∈ }
Using the data from the earlier experiments, we perform a two-sample t-test to determine if the perturbation of gene i leads to a differential expression of gene j in the background of KO for each edge (i, j) ∈ Based on the t-test, we update the ensemble bounds as follows:
if the null hypothesis is rejected, then we add (i, j) to the lower bound GL;
otherwise, we remove (i, j) from the upper bound GU.
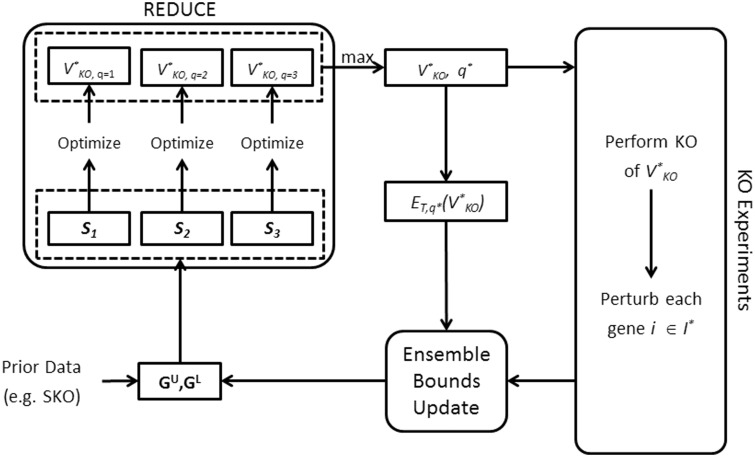
One can repeat applying REDUCE, carrying out KO experiments and updating the ensemble bounds until the upper and lower bounds of the ensemble converge or until the distance between these bounds does not reduce further or until a given number or budget of experiments is reached. As outlined in Figure 3, the inference of GRN can therefore be carried out iteratively. The total number of KO experiments is thus given by the summation between the number of iterations and the cumulative number of elements of I*s. In our experience, when the GRNs contain cycles, the sets of edge separatoids defined earlier may become sensitive to errors, particularly to false negatives (FNs) in the upper bound GU. In the implementation of the iterative procedure, we employed the following sets for REDUCE:
S1(i, j) = children of i in ∩ ancestors of j in
S2(i, j) = descendants of i in ∩ parents of j in
S3(i, j) = descendants of i in ∩ ancestors of j in
where denotes the upper bound of the ensemble in the k-th iteration and is the initial upper bound.
Fig. 3.
Iterative procedure for GRN inference. The procedure starts with an initial GU and GL, for example from the outputs of TRaCE. REDUCE uses the ensemble bounds to find the optimal set of gene KOs V*KO for the subsequent experiments. The resulting gene expression data are then used to update the ensemble bounds. The procedure is repeated until convergence
In practice, multiplex assay such as in RNA sequencing plays an important role in cost- and time-saving by processing a large number of samples simultaneously. If desired, the iterative inference procedure can be adapted for multiplexing. Briefly, the modified procedure involves running REDUCE sequentially without bound updates until the uncertain edges are exhausted or until no feasible solution can be found or until a desired number of KO experiments is produced (Supplementary Information). In each iteration, we thus obtain a ranked list of instead of a single optimal , with non-increasing whose analyses can be parallelized using multiplex assay.
2.4 Comparison to other DOE strategies
A direct comparison between REDUCE and several existing DOE algorithms for GRN inference in the case studies is complicated by (i) differences in the modeling framework used to represent GRNs (e.g. Boolean acyclic graph (Ideker et al., 2000) and ordinary differential equations (Spieth et al., 2004)), (ii) the types or the parameterizations of network perturbations (e.g. using network input functions (Steinke et al., 2007)) and (iii) ambiguity in the published procedure (e.g. in the generation of ensemble of solutions to the linear regression problem in the method by Tegnér et al. (2003)). For these reasons, in the next section we compared REDUCE to systematic gene KO procedures, including the complete set of DKOs, DKOs from ancestor–descendant (AD) pairing and the DOE proposed by Birget et al. (2012) which used a similar graph theoretic concept to edge separatoids, namely vertex separators. In DKOs from AD pairing, we knock out a gene pair i and j, if gene j is accessible from gene i and if there exist at least one gene k (k ∉ {i, j}) which is accessible from both i and j. When the KO of genes i and j leads to a differential expression of gene k with respect to the KO of only gene j, then we add the edge (i, k) to the lower bound. Otherwise, we remove the edge (i, k) from the upper bound.
3 Results
In this section, we demonstrate the performance of REDUCE by applying the iterative procedure in Figure 3 to three case studies, involving 100-gene random scale-free networks under ideal conditions, E.coli GRN under ideal conditions and five 100-gene gold standard GRNs from DREAM 4 challenge using GNW data (Schaffter et al., 2011). As the starting point, we assume that the complete set of SKO experiments have been performed. We used TRaCE to construct the initial GU and GL from the expression data from SKO. Note that in this case GU,0 is the accessibility matrix of . For the 100-gene DREAM 4 GRNs, we employed GNW for the data generation using the same settings as in the challenge. In the implementation of REDUCE, we used GA with a population size of 100 and a maximum generation of 50, which we found to give a good balance between finding globally optimal solution and reducing computational cost. All other GA optimization parameters were set to the default values (see Supplementary Information).
3.1 Comparison to the DOE by Birget et al.
We first compare REDUCE to the KO design procedure by Birget et al. using vertex separators. We applied the DOEs under the ideal scenario where we can accurately detect differential expressions of genes that are accessible from the deleted genes. In this case data noise does not play any role in the inferability of GRN. REDUCE has several key advantages over this strategy. First, REDUCE allows constraints of practical significance in the gene KO optimization, such as excluding essential genes whose KO would leave the cell inviable. In addition, prior knowledge and expression data could be easily taken into account in the ensemble bounds input for REDUCE (see Section 4). Meanwhile, the systematic KO procedure of Birget et al. was developed specifically for GRN digraphs without cycles. The procedure also did not consider any constraints nor allow incorporation of prior data.
As shown in Table 1, for 10 randomly generated 100-gene scale-free acyclic GRNs (Albert and Barabási, 2002), the DOE of Birget et al. prescribed significantly more KO experiments than the iterative inference using REDUCE. Furthermore, the KO experiments from the systematic procedure involved a very high number of genes (up to 50 genes). Using our iterative network inference procedure, we could consistently reach the true GRNs using fewer KO experiments, while limiting the number of KO genes in a given experiment (up to 10 genes). Because of the clear advantages of REDUCE over the DOE of Birget et al., in the remaining case studies we will compare REDUCE only to strategies using DKOs.
Table 1.
Inference of random 100-gene scale-free acyclic GRNs under ideal conditions
| Network | Number of KO experiments: iterative DOE (at most 10 genes in VKO) | Number of KO experiments: Birget et al. (2012) | Maximum number of genes in KO experiments: Birget et al. (2012) |
|---|---|---|---|
| 1 | 678 | 1320 | 35 |
| 2 | 626 | 1884 | 28 |
| 3 | 644 | 1292 | 36 |
| 4 | 666 | 1574 | 37 |
| 5 | 744 | 2120 | 46 |
| 6 | 738 | 2222 | 26 |
| 7 | 760 | 2138 | 50 |
| 8 | 747 | 2766 | 44 |
| 9 | 666 | 2234 | 24 |
| 10 | 670 | 1830 | 39 |
The inference was carried out until completion, leading to the true GRN in all cases.
3.2 Inference of E.coli GRN under ideal condition
As a proof of the applicability of REDUCE to large realistic GRNs, in this case study, we used the E.coli GRN reported in GNW with 1565 nodes and 3758 edges (Schaffter et al., 2011). Here, we again considered data under ideal conditions. Assuming that we started with data from the complete SKO experiments, we constructed the accessibility matrix of and its reduction by ConTREx and set these as the initial ensemble bounds GU,0 and GL,0, respectively. We constrained REDUCE such that the optimal solution excludes essential genes, whose KOs are detrimental to E.coli viability (Kato and Hashimoto, 2007). We implemented two versions of the iterative procedure. The first implementation used a fixed maximum number of 10 genes in VKO (N(VKO) ≤ 10). In the second implementation, we started with a maximum of one gene in VKO and gradually increased this limit when the GA optimization could not find any feasible solution (i.e. when the remaining separatoids involved more genes than the prescribed limit). The ensemble bounds update followed a modified procedure as outlined in Supplementary Information.
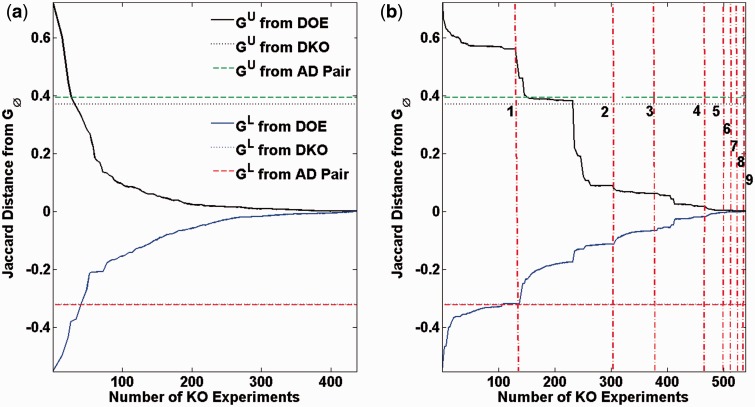
Figure 4 shows the Jaccard distances between GU and and between GL and . The Jaccard distance between two digraphs G1 and G2 is a measure of the difference between the sets of edges in G1 and G2, defined by (Levandowsky and Winter, 1971):
A Jaccard distance of 1 means that there exist no common edge in G1 and G2, while a Jaccard distance of 0 means that G1 and G2 have the same sets of edges. When limiting the number of genes in the VKO to 10, the iterative procedure converged to the true in 166 iterations with a total of 437 KO experiments, as shown in Figure 4a. We could also obtain the true GRN by gradually increasing the limit of genes in VKO (Fig. 4b), but not surprisingly, this implementation required more iterations and more KO experiments (247 iterations and 539 KO experiments). Meanwhile, GU and GL from the complete set of DKO experiments with a total of ∼1.22 million KO experiments did not meet (see dotted lines in Fig. 4). The DKOs from AD pairing produced ensemble bounds GU and GL with Jaccard distances similar to those using the complete DKOs (see dashed lines in Fig. 4) but using much fewer experiments (700 KO experiments). This case study thus demonstrated that the iterative network inference using REDUCE could provide much more informative experiments than systematic designs using DKOs.
Fig. 4.
Iterative inference of E.coli GRN under ideal conditions. The plots show the Jaccard distances of and , and the negative Jaccard distances of and . The number of genes in VKO is limited to 10 genes in (a), and increased from 1 to 9 genes (demarcated by vertical lines) in (b)
3.3 Inference of 100-gene DREAM 4 challenge GRNs
In this case study, we applied the iterative procedure to infer five 100-gene gold standard networks in the DREAM 4 in silico network inference challenge (Prill et al., 2011; Schaffter et al., 2011). For each KO experiment, we simulated 10 replicates of steady-state gene expression data using GNW (Schaffter et al., 2011). GNW employed two types of biological noise: inherent stochastic noise associated with gene transcription process and additive measurement noise. The intrinsic stochastic noise was simulated using stochastic differential equations with independent Gaussian white-noise, while log-normal measurement noise was added to the simulated expression data (Stolovitzky et al., 2005).
As before, we started with initial data from the complete set of SKO experiments and constructed GU,0 and GL,0 using TRaCE. The differential expression analysis in TRaCE was performed using a procedure described previously (using zcutoff = 3 and zthreshold = 2) (Ud-Dean and Gunawan, 2014). We applied the iterative procedure using α = 0.01 for the two-sample t-test during the ensemble bounds update. Here, we gradually increased the maximum number of genes in VKO, starting from 1, and incremented this number by 1 when REDUCE could not find any feasible solution. For all five gold standard networks, the iterative procedure terminated in the convergence between GU and GL. The iterations for the inference of these networks involved at most three gene KOs (N(VKO) ≤2), except for Network 2 which required only two gene KOs (N(VKO) = 1). The accuracy of the resulting GRNs is summarized in Figure 5. For each gold standard network, we compared the iterative procedure to performing the complete set of DKO experiments and DKOs based on AD pairing.
Fig. 5.
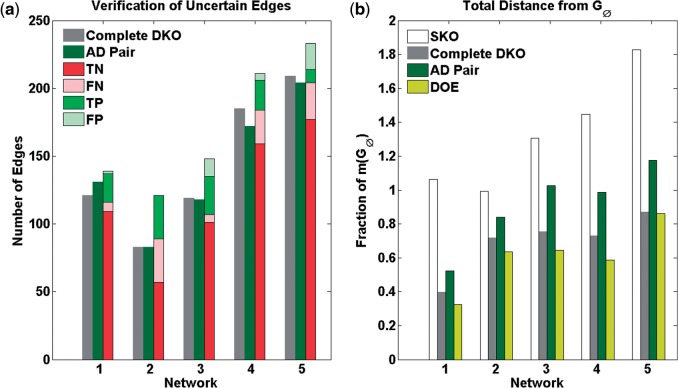
Comparison of REDUCE DOE and DKOs on DREAM 4 100-gene networks: (a) number of uncertain edge verifications and (b) total network distance (TD). (a) The number of uncertain edges verified by the iterative procedure using REDUCE, in comparison to the complete set of DKO experiments and to DKOs based on AD pairing. (TN, true negative; FN, false negative; TP, true positive and FP, false positive) (b) The TD among , and . TDs are reported as a fraction of the size (the number of edges) of
Under non-ideal scenario, we could not obtain the true GRN even when using the iterative procedure. For all gold standard networks, GU and GL from the iterative procedure converged to a GRN that was different from . Nevertheless, as shown in Figure 5a, the iterative procedure consistently led to the verification of more uncertain edges than DKO data (paired t-test P = 0.001 against complete DKOs and P = 0.003 against DKOs using AD pair). The fractions of false positives (FPs) and FNs among the verified edges did not significantly correlate with the number of uncertain edges (for FP: ρ = 0.46, P = 0.43; for FN: ρ = −0.20, P = 0.75). The numbers of experiments using the iterative procedure were between 30 and 110 times lower (Table 2) than the complete DKOs (4950 experiments) but were higher than the DKO experiments using AD pairing. Again, we did not notice significant correlations between the number of uncertain edges and the number of iterations as well as the number of KO experiments (for iterations: ρ = 0.75, P = 0.14; for KOs: ρ = 0.69, P = 0.20). Figure 5b shows the total distance (TD) among GU, GL and which is calculated as follows:
The TD gives a combined measure of uncertainty and accuracy of the ensemble with respect to the gold standard network. As shown in Figure 5b, for all five GRNs, the iterative inference procedure could provide lower TDs than the complete DKOs (paired t-test P = 0.009) and DKOs from AD pairs (P = 0.001).
Table 2.
Number of iterations and total KO experiments in the iterative inference of 100-gene DREAM 4 GRNs
| Network | Number of iterations using REDUCE | Number of KO experiments using REDUCEa | Number of KO experiments using AD pairinga |
|---|---|---|---|
| 1 | 34 | 48 | 41 |
| 2 | 21 | 22 | 22 |
| 3 | 55 | 106 | 57 |
| 4 | 80 | 120 | 64 |
| 5 | 54 | 77 | 57 |
anot including KOs involving single gene.
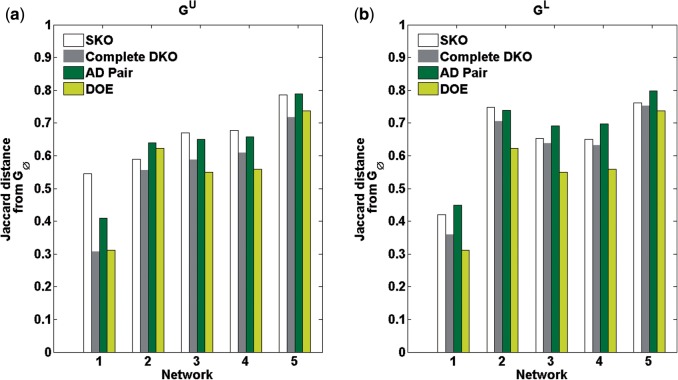
Additionally, we computed the Jaccard distances between GU and and between and GL, which are shown in Figure 6. The upper bounds GU from the proposed iterative inference had similar Jaccard distances to those from the complete DKOs (P = 0.48) despite using much fewer experiments. On the other hand, the optimal DOE consistently produced lower Jaccard distances for the lower bound GL than the complete DKOs (P = 0.005). In comparison to DKOs from AD pairs, the iterative procedure led to lower Jaccard distances for both GU (paired t-test P = 0.006) and GL (P = 0.001). We also implemented the modified iterative procedure for multiplex assay where we again gradually increased the cardinality of VKO. The resulting GRNs were of similar accuracy as those from the original procedure (see Supplementary Fig. S2). The use of multiplex assay expectedly led to fewer iterations ( ∼ 5-fold decrease), while the total number of KO experiments did not change appreciably (see Supplementary Table S1). Taken together, the results of the case studies demonstrated the power of REDUCE for inferring GRN and further suggested that systematic KOs of genes can be severely suboptimal for such a purpose.
Fig. 6.
Comparison REDUCE DOE and DKOs on DREAM 4 100-gene networks: Jaccard distances. (a) Jaccard distance of GU and . (b) Jaccard distance of GL and
3.4 Runtime and computational complexity
The computational complexity of the optimization problem in REDUCE depend on the number of genes n; the number of uncertain edges (N(EU)); the GA parameters, specifically the population size (npop) and the number of generations (ngen), and the constraint on the maximum number of genes (Nmax) allowed in VKO. Specifically, the calculation of the separatoids prior to the GA optimization has a computational complexity that scales linearly with the number of gene n. Furthermore, the complexity of GA optimization scales linearly with the population size and the number of generations, while the computation of the objective function (see Supplementary Information) scales with . Meanwhile, the memory requirement of REDUCE is dominated by the storing of separatoids, which scales with . For the E.coli GRN example with 1565 genes and 11 411 initial uncertain edges, the GA optimization in REDUCE completed in around 90 s on a workstation with 3.33 GHz Intel Xeon W3680 Processor (6 cores), and used 1.4 GB of RAM.
4 Discussion
In this work, we developed a method called REDUCE for optimizing gene KO experiments for the purpose of inferring GRN digraphs. The method builds on the ensemble inference of GRNs using gene expression data (Ud-Dean and Gunawan, 2014). In particular, REDUCE uses the upper and lower bounds of an ensemble of GRNs to find the optimal set of gene KOs which would potentially reduce the most number of uncertain edges. We further proposed an iterative procedure which cycled over performing gene KO experiments, updating ensemble bounds and optimizing gene KO by REDUCE. As a proof of principle, we successfully applied the iterative procedure to infer the GRN of E.coli under ideal conditions (no data noise, infinite sensitivity). The iterative inference could converge to the true GRN, whereas performing all combinations of DKOs or DKOs based on AD pairing could not despite the larger number of experiments.
Using benchmark data generator and 100-gene gold standard networks of DREAM 4 challenge, the proposed iterative inference procedure could significantly outperform DKO experiments providing informative data, as judged by network distances from the true GRNs. In particular, the iterative procedure could converge to a unique digraph with a lower TD than the ensemble bounds from the complete set of DKO experiments, while using 1–2 orders of magnitude fewer experiments. For roughly the same number of experiments, DKO experiments based on AD pairing led to verifications of fewer uncertain edges and much larger TDs from the gold standard network.
We recommend implementing the iterative inference procedure using a gradual increase of KO genes, i.e. starting with N(VKO) = 1 and increasing N(VKO) when REDUCE could not find any feasible solution. Such a strategy is preferred because knocking out a large number of genes simultaneously could be detrimental to cell viability. As shown in Figure 4b, even with limiting the number of KO genes to no more than three (VKO with at most two genes), the iterative procedure could verify a large fraction (93%) of the uncertain edges in E.coli GRN. Also, in practice one would not necessarily want to carry out the iterations until completion (e.g. due to budget constraint). Here, REDUCE will still prove to be useful in maximizing the information return per KO experiment.
For the 100-gene networks in DREAM 4 challenge, the iterative procedure produced a unique digraph (i.e. GU and GL converge) but the inferred GRN deviated from the true GRN. Here, errors in the input upper and lower bounds contributed significantly (79%) to the total errors in the final GRNs (i.e. the majority of the errors were not due to REDUCE). FNs dominated over FPs (86% of errors were FN). We further noted that on average, 90% of the FNs were associated with fan-in motifs where a gene was regulated by two or more genes. The inference of such motif from steady-state gene KO data is fundamentally challenging because perturbing one of the regulators may not lead to any differential expression of the target gene due to compensatory effects. Meanwhile, the remaining errors were not associated with any particular network motifs. Since errors in the initial input to REDUCE were not included in the set of uncertain edges, the iterative inference could therefore not correct these errors.
The issue of FN above could be addressed by considering other types of data, for example time-series gene expression data and transcription factor binding sites. When considering time-series data, we ideally need (i) fast sampling to capture transient changes in the expression of target genes and (ii) slow compensation by other regulators. Meanwhile, if the binding sites of transcription factors are known (see for example FANTOM project Consortium, 2014), one could then construct a transcriptional regulatory network. Any edges in the transitive closure of this transcriptional network which do not appear in the input upper bound, are possible FNs and should be added to the upper bound.
Beside the FN issue, a recent study demonstrated that the ordering of gene deletions aceA and pgi in E.coli could influence cell’s transcriptomic profiles (Gawand et al., 2015). In the analysis of the data from this study (Supplementary Data File), we found that the order of deleting the two genes affected the expression of 53 out of 4690 genes (at false discovery rate < 5%). Furthermore, gene aceA showed a differential expression upon deleting gene pgi, suggesting that aceA is accessible from pgi. However, pgi was not differentially expressed in the KO of aceA. Consequently, by analyzing the differential expression of DKO Δpgi and ΔaceA in the background of ΔaceA, we should be able to verify edges emanating from pgi to all genes that are accessible from both pgi and aceA. Among the verifiable edges satisfying the condition above (ignoring antisense transcripts), the transcriptomic discrepancy between deleting ΔaceA-then-Δpgi and Δpgi-then-ΔaceA did not cause any difference in the verification outcomes of these edges (0 out of 3). Nevertheless, the influence of the order of gene deletions demonstrated in the above study could complicate the GRN inference, which future DOEs and network inference algorithms would need to address.
The initial ensemble bounds for the iterative procedure in the case studies came from applying TRaCE to expression data of SKO experiments. If the transcription factor genes are known a priori, then TRaCE requires only the SKO data of each transcription factor gene to construct the initial upper and lower bounds of the GRN ensemble. Beside using TRaCE, one could also construct the initial upper and lower bounds of the ensemble from prior knowledge. When the members of the initial ensemble are known a priori, the upper and lower bounds could be constructed by taking the union and intersection of the members, respectively. For example, one could obtain the initial ensemble from the GRN predictions using different network inference algorithms, following the idea of wisdom of crowds (Marbach et al., 2012). When observational data (as opposed to KO data) are available, one could also construct a Markov equivalence class, for example using PC algorithm (Kalisch and Bühlmann, 2007; Maathuis et al., 2010). The Markov equivalence class represents the ensemble of DAGs encoding the same independence and conditional relationships that result from a Bayesian network learning using such data. Again, the upper and lower bound could be constructed by taking the union and intersection of the DAGs in this equivalence class.
Supplementary Material
Acknowledgement
The authors acknowledge Dr Lakshmi Narayanan Lakshmanan for his help in setting up REDUCE webpage.
Funding
This work was supported by grants from the Swiss National Science Foundation (grant number: 137614 and 157154)
Conflict of Interest: none declared.
References
- Aho A.V., et al. (1972) The transitive reduction of a directed graph. SIAM J. Comput., 1, 131–137. [Google Scholar]
- Albert R., Barabási A.-L. (2002) Statistical mechanics of complex networks. Rev. Mod. Phys., 74, 47–97. [Google Scholar]
- Birget J.C., et al. (2012) A theoretical approach to gene network identification. Information Theory Workshop (ITW), IEEE, New York, NY, pp. 432–436. [Google Scholar]
- Bjorklund A., et al. (2004) Approximating longest directed paths and cycles. In: Lecture Notes in Computer Science, Automata, Languages and Programming, Springer, Heidelberg, Germany: Vol 3142, pp. 222–233. [Google Scholar]
- Consortium T.F. (2014) A promoter-level mammalian expression atlas. Nature, 507, 462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawand P., et al. (2015) Sub-optimal phenotypes of double-knockout mutants of Escherichia coli depend on the order of gene deletions. Integr. Biol., 7, 930–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland J.H. (1992) Adaptation in Natural and Artificial Systems: An Introductory Analysis With Applications to Biology, Control, and Artificial Intelligence. MIT press, Cambridge, MA. [Google Scholar]
- Ideker T.E., et al. (2000) Discovery of regulatory interactions through perturbation: inference and experimental design. Pac. Symp. Biocomput., 5, 302–313. [DOI] [PubMed] [Google Scholar]
- Kalisch M., Bühlmann P. (2007) Estimating high-dimensional directed acyclic graphs with the PC-algorithm. J. Mach. Learn. Res., 8, 613–636. [Google Scholar]
- Kato J.I., Hashimoto M. (2007) Construction of consecutive deletions of the Escherichia coli chromosome. Mol. Syst. Biol., 3, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang M., et al. (2014) Cutting the wires: modularization of cellular networks for experimental design. Biophys. J., 106, 321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levandowsky M., Winter D. (1971) Distance between sets. Nature, 234, 34–35. [Google Scholar]
- Maathuis M.H., et al. (2010) Predicting causal effects in large-scale systems from observational data. Nat. Methods, 7, 247–248. [DOI] [PubMed] [Google Scholar]
- Marbach D., et al. (2012) Wisdom of crowds for robust gene network inference. Nat. Methods, 9, 796–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prill R.J., et al. (2011) Crowdsourcing network inference: the DREAM predictive signaling network challenge. Sci. Signal., 4, mr7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffter T., et al. (2011) GeneNetWeaver: in silico benchmark generation and performance profiling of network inference methods. Bioinformatics, 27, 2263–2270. [DOI] [PubMed] [Google Scholar]
- Spieth C., et al. (2004) Iteratively inferring gene regulatory networks with virtual knockout experiments. In: Applications of Evolutionary Computing. Springer, Heidelberg, Germany, pp. 104–112. [Google Scholar]
- Steinke F., et al. (2007) Experimental design for efficient identification of gene regulatory networks using sparse Bayesian models. BMC Syst. Biol., 1, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolovitzky G., et al. (2005) Statistical analysis of MPSS measurements: application to the study of LPS-activated macrophage gene expression. Proc. Natl. Acad. Sci. USA, 102, 1402–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szederkényi G., et al. (2011) Inference of complex biological networks: distinguishability issues and optimization-based solutions. BMC Syst. Biol. 5, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegnér J., et al. (2003) Reverse engineering gene networks: Integrating genetic perturbations with dynamical modeling. Proc. Natl. Acad. Sci. USA, 100, 5944–5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ud-Dean S.M.M., Gunawan R. (2014) Ensemble inference and inferability of gene regulatory networks. PLoS One, 9, e103812. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.