Abstract
Mouse skin mesenchymal stem cells (msMSCs) are dermis CD105+CD90+CD73+CD29+CD34− mesodermal precursors which, after in vitro induction, undergo chondro, adipo and osteogenesis. Extensive metabolic reconfiguration has been found to occur during differentiation, and the bioenergetic status of a cell is known to be dependent on the quality and abundance of the mitochondrial population, which may be regulated by fusion and fission. However, little is known regarding the impact of mitochondrial dynamics on the differentiation process. We addressed this knowledge gap by isolating MSCs from Swiss female mice, inducing these cells to differentiate into osteo, chondro and adipocytes and measuring changes in mass, morphology, dynamics and bioenergetics. Mitochondrial biogenesis was increased in adipogenesis, as evaluated through confocal microscopy, citrate synthase activity and mtDNA content. The early steps of adipo and osteogenesis involved mitochondrial elongation, as well as increased expression of mitochondrial fusion proteins Mfn1 and 2. Chondrogenesis involved a fragmented mitochondrial phenotype, increased expression of fission proteins Drp1, Fis1 and 2 and enhanced mitophagy. These events were accompanied by profound bioenergetic alterations during the commitment period. Moreover, knockdown of Mfn2 in adipo and osteogenesis and the overexpression of a dominant negative form of Drp1 during chondrogenesis resulted in a loss of differentiation ability. Overall, we find that mitochondrial morphology and its regulating processes of fission/fusion are modulated early on during commitment, leading to alterations in the bioenergetic profile that are important for differentiation. We thus propose a central role for mitochondrial dynamics in the maintenance/commitment of mesenchymal stem cells.
Keywords: Stem cells, mitochondrial dynamics, adipogenesis, osteogenesis, chondrogenesis, bioenergetics
Introduction
Mesenchymal stem cells (MSCs) are found in virtually all post-natal organs and tissues and display the ability to differentiate in vitro and in vivo into bone, cartilage and adipose tissues, among others [1, 2]. Mouse skin mesenchymal stem cells (msMSCs) are CD105+CD90+CD73+CD29+CD34− mesodermal precursors located in the dermis that, after induction, display classic differentiation potential in vitro [3, 4]. Recent studies suggest that extensive metabolic reconfiguration occurs during differentiation. Indeed, metabolic pathways may be controlled by the same signals that control cell proliferation/differentiation [5] and important metabolic changes may occur not only when a proliferating cell terminally differentiates but also during gradual changes in the differentiation state. For example, the expression of genes involved in oxidative metabolism is associated with more differentiated cells during mesodermal differentiation from immature somites to muscle progenitors in zebrafish [6].
Differentiation is clearly an energy-demanding process, and thus would be expected to affect and be affected by energy metabolism. The bioenergetic status of a cell is dependent on the overall quality and relative abundance of the mitochondrial population, a highly coordinated characteristic. Mitochondrial abundance is determined by the processes of mitochondrial degradation and biogenesis, which includes de novo synthesis of organelle components, formation of mitochondrial membranes and division of pre-existing mitochondria [7]. In addition, a complex machinery regulates mitochondrial morphology, dynamics and quality through the fusion and fission of these organelles. While fusion of healthy populations ensures content exchange and maintains the quality of the organellar population [8], sub-populations of low functioning mitochondria destined for turnover are segregated through fission. These defective organelles are then removed through a specialized type of autophagy called mitophagy [9, 10]. This method of mitochondrial surveillance is useful because it allows changes in mitochondrial parameters such as membrane potentials and oxidative phosphorylation activity to be placed in the dynamic context of cell function [11, 12].
Although an increasing body of knowledge has established that mitochondrial dynamics are important for regulating cellular energy metabolism, to date little is known regarding the impact of mitochondrial dynamics on the differentiation process [13]. In order to examine if mitochondrial network reorganization plays a role during the process of MSC commitment to differentiation, we studied changes in bioenergetics and mitochondrial morphology in murine dermis-derived mesenchymal stem cells differentiated into adipo, chondro and osteocytes.
Material and methods
Isolation and characterization of msMSCs
Cells were obtained as described in [3, 4]. Briefly, subcutaneous fat was removed from the skin with a scalpel, and the whole skin was placed dermis-down in trypsin (Gibco) at 37°C for 30 min. After scraping the epidermis away, single cell suspensions were obtained from the dermis through mechanical dissociation. The cells were then filtered with strainers (70 mm followed by 40 mm) and plated in colony forming unit densities (10 cells/cm2). After achieving a first 80% confluence round (passage 1), full characterization was performed using CD105, CD90, CD73, CD29 and CD34 as markers. Experiments were conducted using an Attune Cytometer (Life Technologies) and at least 50,000 events in duplicate were recorded. Negative controls were used and comprised of the same class IgG isotype controls. Compensation and analysis were performed using FlowJo X (TriStar Inc). All experiments were conducted in agreement with the National Institutes of Health guidelines for humane treatment of animals and were reviewed and approved by the local Animal Care and Use Committee (Permit number: 2013/17).
Differentiation induction
Mesenchymal cells were induced to undergo adipogenesis with high glucose DMEM (Gibco, Rockville, MD), 10% fetal calf serum (FCS), 1 μM dexamethasone (Sigma, USA), 0.5 mM IBMX (Sigma, USA), 10 μg/uL insulin (Invitrogen, USA) and 100 μM indomethacin (Sigma, USA). Chondrogenesis was promoted using DMEM-F12 (Gibco, Rockville, MD) 3:1, 0.5 μg/mL insulin (Invitrogen, USA), 50 μM ascorbic acid (Sigma, USA), 10 ng/mL TGFβ-1 (R&D Systems, USA) and 1 mM buffered sodium pyruvate (Sigma, USA). Osteogenesis was induced using αMEM (Gibco, Rockville, MD), 10% FCS, 0.1 μM dexamethasone (Sigma, USA), 2 mM ascorbic acid (Sigma, USA), and 10 mM buffered glycerol 2-phosphate (Sigma, USA). Differentiation was induced for 7 and 14 days for validation experiments and from 30 min to 96 h for commitment phase experiments, as stated.
q-RT-PCR
RNA was purified using Trizol (Invitrogen) and checked for quality using 260/230 nm and 260/280 nm scores added to denaturating gel detection of ribossomal bands. Equivalent amounts of RNA were reverse-transcribed using a Super-Script III cDNA Synthesis Kit (Invitrogen). cDNAs were normalized to equal amounts using primers against β-actin, HMBS and HPRT. The Genorm algorithm was used to create a normalization factor [14]. cDNAs were mixed with the indicated primers and Power SYBR Green PCR Master Mix (Applied Biosystems). qRT-PCR was performed using an Applied Biosystems 7300 Real-Time PCR system. qRT-PCR primer sequences were obtained from the Harvard database PrimerBank and had their efficiency (90% minimum) and concentration standardized [15]. For primer sequences see Supplementary Table 1.
Differentiation induction and histochemical detection
Osteogenic differentiation was detected using the Alizarin Red S staining protocol, adipogenic differentiation with Oil Red O and chondrogenesis with Toluidine Blue staining, as described elsewhere [16].
Citrate synthase activity
Undifferentiated and differentiated msMSCs were homogenized in lysis buffer (50 mM sodium phosphate, pH 7.4, 10% glycerol, 1% octylphenol ethoxylate, 10 mM sodium orthovanadate, 10 mM sodium fluoride and 10 mM sodium pyrophosphate, supplemented with Sigma protease inhibitor mixture). After 30 min over ice, the lysates were centrifuged (13,000 g, 20 min, 4°C), and the resulting supernatants were collected. Total protein (20 μg) was incubated for 5 min at 37°C in 20 mM Tris-HCl, pH 8.0, 0.42 mM acetyl-coenzyme A, and 0.1 mM 5,5′-dithiobis (2-nitrobenzoic acid). The reaction was initiated by the addition of 0.5 mM oxaloacetate and the reduction of 5′, 5′-dithiobis (2-nitrobenzoic acid) by citrate synthase was measured spectrophotometrically at 412 nm for 5 min (extinction coefficient = 13.6 mM−1 cm−1). Activities are expressed as nmol citrate min−1 mg−1 protein.
Western blots
Cell lysate proteins were diluted in Laemmli sample buffer (100 mM Tris-HCl, 2% w/v SDS, 10% v/v glycerol, 0.1% w/v bromophenol blue) containing 100 mM dithiothreitol. After heating at 90°C for 5 min, proteins were separated by SDS-PAGE and transferred onto nitrocellulose membranes. Membranes were blocked with 5% bovine serum albumin and the detection of individual proteins was carried out by blotting with specific primary antibodies (see Supplementary Table 2). Near-infrared (NIR) detection using a secondary antibody linked to an infrared dye (LI-COR) was used and analyzed with LI-COR Odyssey software. Signals were quantified by densitometry using Image J (NIH software). The detected proteins were normalized to actin or, where stated, a reference from the same sample.
Seahorse oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) measurements
Oxygen consumption rates (OCR) were measured in MSCs using the XF24 Analyzer (extracellular flux, 24-well plate, Seahorse Bioscience), as previously described [17, 18]. An assay medium composed of 114 mM NaCl, 4.7 mM KCl, 1.2 mM KH2PO4, 1.16 mM MgSO4, 2.5 mM CaCl2, pH 7.2, and 2.8 mM glucose was used. The cells were seeded in an XF24 24-well culture microplate at 60,000 cells/well (0.32-cm2 growth area) in 500 μL of growth medium and incubated overnight at 37°C in a humidified atmosphere of 95% air and 5% CO2. Prior to the assay, the medium was removed and replaced by 500 μL of assay medium. The cells were preincubated for 1 h at 37°C in air. Respiration-driven ATP synthesis and proton leak-driven respiration were determined by the addition of oligomycin (4 μg/mL). After 3 measurement cycles, 5 μM of the uncoupler carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (CCCP) was added to determine maximal respiratory capacity. After a further 3 measurement cycles, 1 μM rotenone was added to block complex I in addition to 1 μM antimycin A to inhibit complex III, thereby ablating mitochondrial oxygen consumption.
Immunofluorescence
Cells were fixed in 4% w/v paraformaldehyde and subjected to immunofluorescence microscopy as previously described [4]. The antibodies and their dilutions are listed in Supplementary Table 2. Secondary antibodies conjugated to Alexa-488 and Alexa-647 (Molecular Probes) were used for imaging. Nuclei were stained using 4′,6-diamidino-2-phenylindole (DAPI). In the experiments monitoring mitophagy, nucleus counterstaining was performed using Hoescht 33342 and lysossome staining was performed with Lysotracker Red and mitochondria with Mitotracker Green (Molecular Probes), as recommended by the manufacturer. In order to evaluate mito/autophagic flux, cells stimulated to differentiate into chondrocytes for 48h were exposed to 5 mM 3-methyladenine (3-MA, Santa Cruz, USA) or 100 μM chloroquine (CQ, Molecular Probes, USA) for 8 h before imaging (the drug loading control groups were exposed to equal quantities of the solvent DMSO). Imaging was performed using a Zeiss LSM 510-Meta laser scanning confocal microscope. Figures were prepared using ImageJ.
Live cell imaging
The preparation of a DNA plasmid containing mitochondrial matrix-targeted photoactivatable GFP (PA-GFPmt) was reported previously in detail [19]. The plasmid was delivered to the cells (~80% confluent) by an adenovirus (with addition of polybrene) carrying the PAGFPmt sequence under a CMV-actin promoter and PAGFPmt was allowed to accumulate for 72 hours. Mitochondria were labeled with 15 nM TMRE (tetramethylrhodamine-ethyl-ester-perchlorate; Invitrogen, Eugene, OR) for 45 min prior to imaging to allow mitochondrial visualization. Photoconversion of PA-GFPmt to its active (fluorescent) form was achieved using a 2-photon laser (750 nm) to promote a 375 nm photon-equivalence at the focal plane. This allowed for selective activation of regions that have submicron thickness and are less than 0.5 μm2 (Zeiss LSM510). Using the multi-track scanning mode, red-emitting TMRE was excited with a 1 mW 543 nm helium/neon laser set at 0.3% and emission was recorded through a BP 650–710 nm filter. PA-GFPmt protein was excited using a 25 mW 488 nm argon laser set at 0.2 %. The non-photoactivated PAGFPmt protein molecules remained stable in preactivated form and the presence of preactivated PAGFPmt was detected using a fully-opened collection pinhole. Emission was recorded through a BP 500–550 nm filter.
Mfn2 knockdown and overexpression of a dominant negative form of Drp1
DNA fragments covering part of mouse Mfn2 cDNA were cloned in pSRP vectors in order to produce double-stranded RNAs as previously described [20]. Each pSRP vector was co-transfected in 293T cells with gag-pol-env constructs derived from Moloney leukemia virus, and the virus-rich supernatants were used to infect msMSCs. Drp1 Dominant Negative mutant (DN, K38A) overexpression was also achieved using an adenovirus-mediated system described elsewhere [21].
Statistical analysis
Data were analyzed using GraphPad Prism and Origin software. Figures represent averages ± SEM of 3–12 measurements and were compared using ANOVA and when suitable with Tukey as a post-test. Two-tailed p values under 0.05 were considered significant.
Results
Clonogenic isolation of CD105+CD90+CD73+CD29+CD34− MSCs and in vitro adipo, chondro and osteogenesis induction
After clonogeneic selection and expansion of murine dermis-derived MSCs, we performed a full flow cytometry-based characterization of these cells, validating the expression of several cluster of differentiation (CD) markers, as recommended by the International Society for Cell Therapy [22]. We found that the cells isolated were positive for CD105, CD90, CD73 and 29 and did not present CD34 as a membrane marker (Supplementary Fig 1 A–E). We also induced these cells to differentiate and validated the process through histochemistry and qPCR. After 14 days of exposure to the adipogenic differentiation media, the cells increased relative mRNA levels of Creb, a transcription factor that coordinates adipogenesis (Sup Fig 1F, p ≤ 0.001) and displayed lipid droplet vesicles in the cytoplasm, as seen through Oil Red O staining (Sup Fig 1I). When induced to undergo chondrogenic differentiation, these cells presented a very significant increase in the relative Col2a mRNA levels (Sup Fig 1H, p ≤ 0.001) and a proteo/glycosaminoglycan matrix deposition pattern, seen through toulidine staining in Sup Fig 1K. Finally, osteogenesis induction promoted upregulation of Osterix mRNA levels (Sup Fig 1G, p ≤ 0.05) as well as the deposition of the calcified matrix observed through Alizarin Red positive ECM deposition (Sup Fig 1J). We thus considered our MSC differentiation models valid, and focused our next experiments on alterations in mitochondrial mass and morphology during the early steps of commitment to differentiation, namely after 48 h of induction.
Mitochondrial biogenesis and fusion are induced during MSC commitment to adipogenesis and osteogenesis; fragmentation occurs during early chondrogenesis
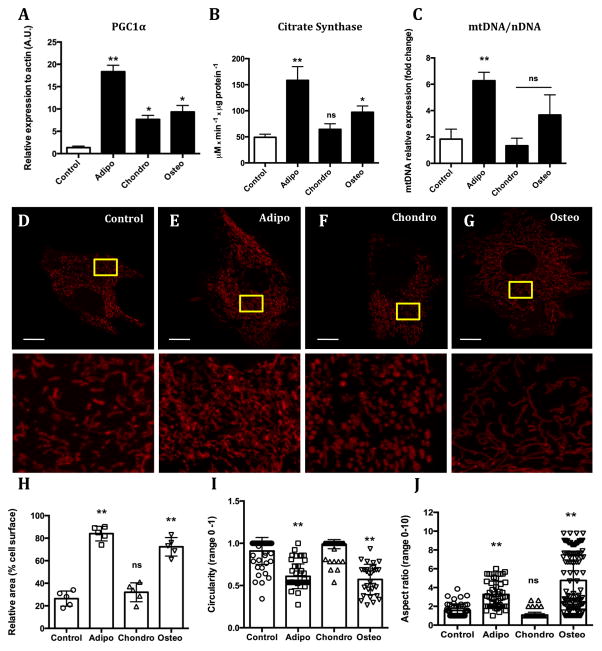
Because differentiation is an energy-demanding process, we hypothesized that changes in mitochondrial density could occur during commitment to differentiation. We thus measured markers of mitochondrial biogenesis and mass. The levels of PGC1α, a transcriptional co-activator that promotes mitochondrial biogenesis, were significantly increased in all forms of differentiation induced (Fig 1A). However, citrate synthase (Fig 1B) and mtDNA content (Fig 1C), markers for mitochondrial mass, were only increased during commitment toward osteo and, more strikingly, adipogenesis. In order to validate the suggestion that mitochondrial mass is increased during adipo and osteo, but not chondrogenesis, we imaged the cells in the presence of the mitochondrially-accumulated dye TMRE. Our results confirmed that relative to undifferentiated cells, mitochondrial area was increased in commitment toward adipo and osteogenesis, but not chondrogenesis (Fig 1D–G show representative images; Fig 1H depicts the quantification).
Figure 1. Mitochondrial mass, morphology and dynamics change during early commitment toward adipo, chondro and osteogenesis.
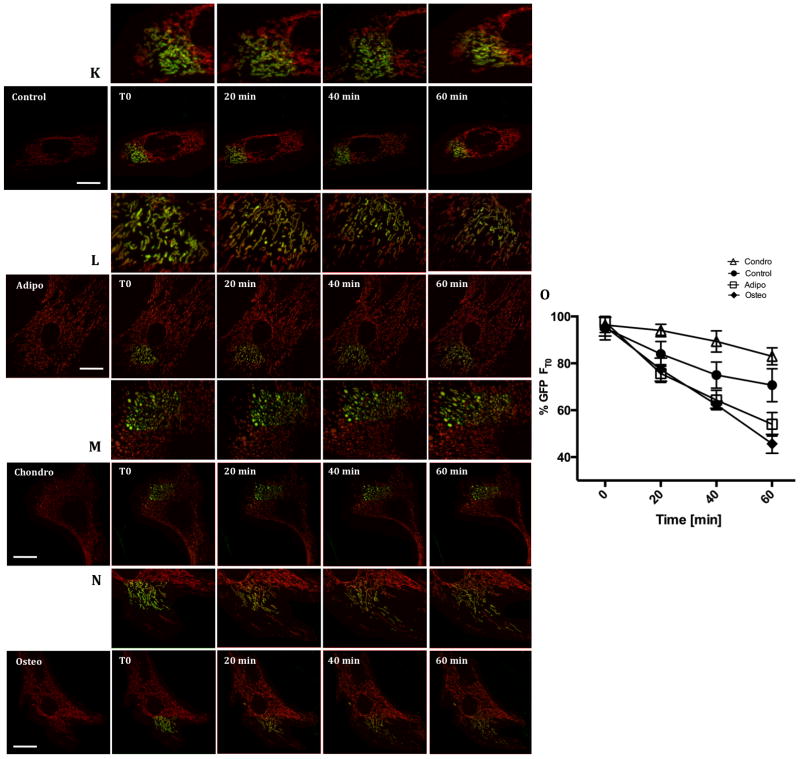
MSCs were stimulated to differentiate into adipo, chondro and osteocytes. After 48 h (early commitment) mitochondrial network content was evaluated through a series of biogenesis markers, namely (A) PGC1α protein expression, (B) Citrate synthase activity and (C) mtDNA/nDNA. TMRE-loaded cells were imaged (bar = 5 μm) (D) Control, (E) Adipogenesis, (F) Chondrogenesis and (G) Osteogenesis. Using these images, the percentage of area occupied by mitochondria per cell (H) was estimated, as well as Circularity (I) and the Form factor index (J). A photoactivatable GFP was targeted to mitochondria and its dilution in a selected region of interest was followed for 1 hour as seen in panels (K) Control, (L) Adipogenesis, (M) Chonndrogenesis and (N) Osteogenesis; the quantification of the GFP signal dilution is displayed in (O).* p ≤ 0.05, ** p ≤ 0.001, ns = not significant. Bar = 15 μm.
Mitochondrial staining with TMRE also unveiled striking changes in mitochondrial morphology between the differentiation processes. Indeed, mitochondria were more rounded during commitment toward chondrogenesis (Fig 1F; quantified in Fig 1I) and more elongated during commitment toward adipo and osteogenesis (Fig 1E and G; quantified in Fig 1J).
Changes in mitochondrial morphology often reflect differences in mitochondrial dynamics and the exchange and movement of mitochondrial content over time throughout the cell. Furthermore, enhanced mitochondrial dynamics has been correlated with optimized respiratory activity in many distinct cellular models [23]. We thus analyzed mitochondrial dynamics during commitment toward differentiation by visualizing and quantifying mitochondrial fusion using an assay based on the dilution rate of mitochondrially-targeted, photoactivable, GFP (mito-PAGFP) [24]. The assay is based on following the dilution of the localized GFP signal obtained after photoactivation throughout the TMRE-stained mitochondrial network over time, a process dependent on mitochondrial fusion. Fig 1K–N shows representative images of GFP dilution, which is quantified in Fig 1O. We observed that undifferentiated cells present an intermediate level of mitochondrial fusion made evident by the moderate dilution of the matrix-associated green fluorescence. Commitment toward adipogenesis and osteogenesis involved much higher rates of GFP dilution, reflecting active mitochondrial dynamics and exchange of matrix material. On the other hand, in chondrogenesis GFP fluorescence dilution was more limited, indicating that the fragmented mitochondrial phenotype observed reflects not only increased fission, but also decreased fusion and exchange of matrix content (i.e. decreased mitochondrial dynamics).
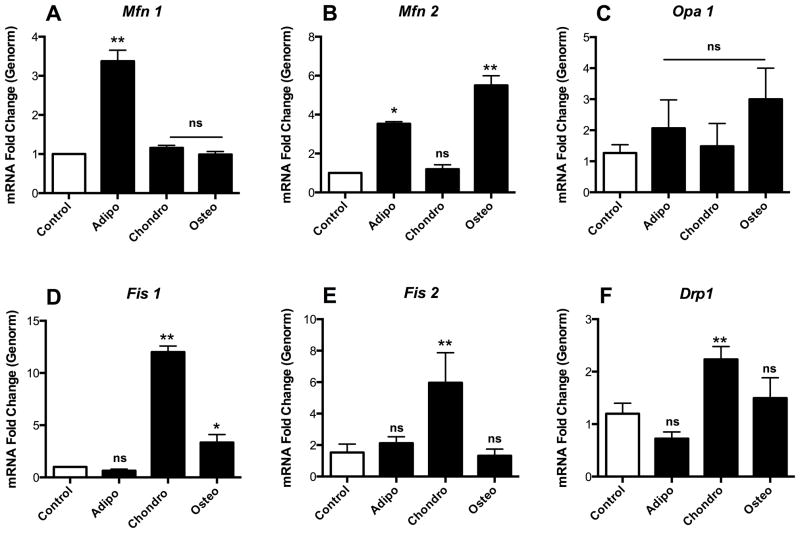
Changes in mitochondrial morphology and dynamics are mediated by fission (Fis1 and 2, Drp-1) and fusion (Mfn1 and 2, Opa-1) regulators, which were evaluated by q-RT-PCR during commitment to differentiation. Early adipogenesis presented high Mfn1 levels (Fig 2A, p ≤ 0.001), a minor increase in Mfn2 (Fig 2B, p ≤ 0.05) and unchanged opa1 levels (Fig 2C), accompanied by low levels of mitochondrial fission intermediates (Fig 2D, E and F, not significant when compared to undifferentiated cells). During osteogenesis, the increased fusion observed in Fig 1 correlated with high levels of Mfn2 (Fig 2B, p ≤ 0.001), accompanied by a more discreet increase in Fis1 expression (Fig 2D, p ≤ 0.05). In chondrogenesis, the levels of fission effectors Fis1, Fis2 and Drp1 were upregulated (Fig 2D–F, p ≤ 0.001), a result compatible with the fragmented phenotype observed in Fig 1. Overall, these experiments show that there are overt differences in the regulation of mitochondrial morphology and dynamics during the commitment phase of differentiation.
Figure 2. Mitochondrial fusion and fission mediator expression is altered during early commitment.
The relative mRNA levels of fusion markers Mfn1 (A), Mfn2 (B) and Opa1 (C) as well as fission mediators Fis1 (D) and Fis2 (E) and Drp1 (F) were measured. * p ≤ 0.05, ** p ≤ 0.001, ns = not significant.
Commitment toward adipogenesis and osteogenesis involves enhanced respiration; chondrogenesis commitment decreases respiration
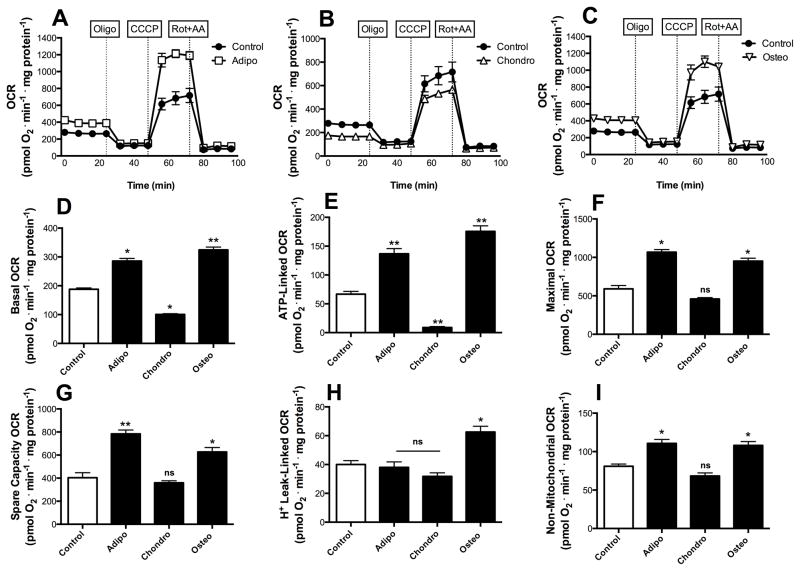
Changes in mitochondrial morphology lead to changes in mitochondrial function in many cellular models [23]. Thus, we investigated if early commitment to differentiation was associated with altered bioenergetic profiles by measuring real-time O2 consumption (OCR) in cell cultures (Fig 3A–C shows typical time scans, while panels D–H show quantitative comparisons derived from these scans). We found that the levels of basal respiration (which occurs in the absence of mitochondrial inhibitors and represents the sum of all mitochondrial O2-consuming processes) were increased both in early adipocytes and osteocytes (Fig 3D; p ≤ 0.001 for osteo and p ≤ 0.05 for adipo). This demonstrates that adipo and osteogenesis involve enhanced respiratory phenotypes during the early commitment phase. On the other hand, we found that the opposite occurs in early chondrogenesis, where significantly decreased levels of basal respiration were found, indicating that the fragmented mitochondrial network observed in Fig 1 is accompanied by lower respiratory function than that seen in undifferentiated cells (Fig 3D, p ≤ 0.05).
Figure 3. Early differentiation involves fate-specific bioenergetic shifts.
Real time whole-cell respiration was obtained in each of the three differentiation fates, namely adipogenesis (A), chondrogenesis (B) and osteogenesis (C). Oligomycin, CCCP and rotenone plus antymycin were added where indicated. Basal oxygen consumption rates (D), ATP-production dependent OCR (E), Maximal (F), Spare respiratory capacity (G), H+ Leak-linked OCR (H) and Non-mitochondrial respiration (I) were calculated from data such as those shown in panels A–C. * p ≤ 0.05, ** p ≤ 0.001, ns = not significant.
The injection of oligomycin, an inhibitor of the mitochondrial ATP synthase, leads to a decrease in basal respiration that is proportional to mitochondrial activity used to generate ATP (Fig 3A–C). Interestingly, in addition to displaying low levels of basal respiration, cells undergoing chondrogenesis presented very low ATP-linked OCR (Fig 3E, p ≤ 0.05). Osteo-and adipogenesis-induced cells displayed higher ATP production-dependent oxygen consumption (Fig 3E, p ≤ 0.001). The further addition of CCCP, a proton ionophore which uncouples respiration from oxidative phosphorylation, allows for the measurement of maximal respiratory rates (Fig 3A–C). Maximal respiration was substantially increased in both adipocytes and osteocytes, and remained unchanged in chondrocytes (Fig 3F). The reserve respiratory capacity (or spare respiratory capacity), calculated by subtracting the CCCP-stimulated from the basal OCR, was increased both in osteogenesis and adipogenesis (Fig 3G p ≤ 0.001). The proton leak, or the post-oligomycin injection OCR subtracted from non-mitochondrial respiration, associated with the natural proton leak across the mitochondrial inner membrane independent of ATP synthase activity, was significantly increased only in osteogenesis (Fig 3H). Finally, the extent of non-mitochondrial oxygen-consuming processes was estimated by inhibiting the respiratory chain with antimycin A plus rotenone (Fig 3A–C). Non-mitochondrial respiration was increased in both adipo and osteogenesis (Fig 3I), but accounted for only a small fraction of cellular OCR.
Overall, these results show that increased mitochondrial mass, a fused morphology and enhanced dynamics in adipo and osteogenesis are accompanied by a more respiratory phenotype. On the other hand, mitochondrial fragmentation during chondrogenesis is accompanied by dampened respiratory activity.
Chondrogenesis commitment involves LC3-dependent mitophagy
Mitochondrial fragmentation and decreased respiration could potentially indicate that these organelles are being targeted for degradation [9]. In order to observe if this was the case during early chondrogenesis, we performed a kinetic analysis of the autophagic flux during the commitment phase. Twenty-four hours after induction, we found that levels of lipidated LC3-II were significantly higher (more than 4 fold, p ≤ 0.05, Supplementary Fig 2A). These levels were sustained and even increased over longer periods (more than 6 fold at 48 h, p ≤ 0.001). The levels of Beclin were also increased (more than 3 fold at 9 h, p ≤ 0.001). Moreover, loading the cells with Mitotracker Green and Lysotracker Red uncovered a large amount of chondrogenic-stimulated cells in which mitochondria co-localized with lysosomes, a result not seen in undifferentiated cells (Supplementary Fig 2B). To further characterize chondrogenesis-induced mitophagy in MSC cells undergoing commitment to differentiation, we assessed the autophagic flux using two inhibitors, chloroquine and 3-methyladenine (3-MA) (Supplementary Fig 2D–F). Chloroquine inhibits autophagy by inhibiting the acidification of the lysosome, while 3-MA inhibits the formation of functional pre-autophagosomes by inhibiting the PI3K-mediated nucleation step of the autophagic process. We observed an accumulation of lysossomes with mitochondrial content (mitophagic puncta) in chloroquine treated cultures after 48 h of chondrogenic induction (a typical image is shown in Supplementary Fig 2D and a quantification of 100 cells is shown in Supplementary Fig 2F), a strong indicator that mitophagy is involved in early chondrogenesis. These results indicate that chondrogenesis involves a reduction in mitochondrial activity, through fragmentation and mitophagy.
Mfn2 knockdown abrogates the bioenergetic changes in adipo and osteogenesis commitment
Our results until this point show strong differences in mitochondrial shape, dynamics and activity in different forms of differentiation. The fact that these changes occur during the early commitment phase suggests they may play an active role in the differentiation process itself, but this hypothesis remains to be demonstrated. Furthermore, although increased mitochondrial fusion and dynamics are often associated with enhanced respiratory rates, our results to this point do not prove that the changes in respiratory activity are caused by the morphological and dynamic mitochondrial alterations observed. In order to establish cause and consequence relationships, we knocked down Mfn2 (Supplementary Fig 3A) and overexpressed a truncated form of Drp1 (Supplementary Fig 3B) to evaluate if the bioenergetic alterations associated with commitment toward differentiation were dependent on mitochondrial morphological changes.
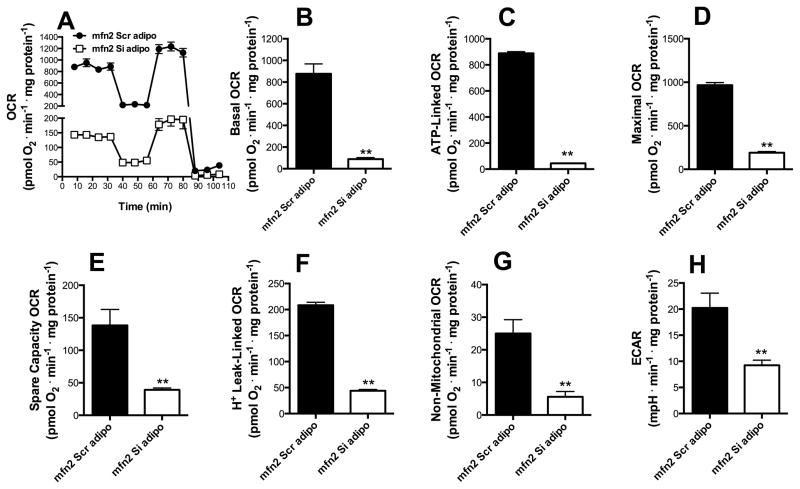
Adipogenesis, which involves increased mitochondrial fusion (Figs 1–2), was initiated in Mfn2 knockdown cells (mfn2Si) and respiratory patterns were compared to cells transfected with a scrambled sequence (mfn2scr, Fig 4). The knockdown of Mfn2 promoted a striking decrease in respiratory activity after 48 h of the induction of differentiation, involving basal (Fig 4B), ATP-linked (Fig 4C), maximal (Fig 4D), spare respiratory capacity (Fig 4E) and H+-leak-linked respiratory activity (Fig 4F). Most surprisingly, mfn2 knockdown decreased non-mitochondrial OCR (Fig 4G); the decreased respiration observed in these cells was not compensated by an increase in glycolysis, as suggested by the lower levels of extracellular acidification rates in mfn2Si MSCs (ECAR), observed in Figure 4H, p ≤ 0.001. This demonstrates that, in adipogenesis, the metabolic reprograming observed is dependent on mitochondrial fusion.
Figure 4. Mfn2 is necessary for the metabolic switch observed during adipogenesis.
mfn2Scr (black) and mfn2Si (white) MSCs were exposed to adipogenesis media for 48 h (commitment time). Real time whole-cell respiration traces were obtained (A). Basal oxygen consumption rates (B), ATP-production dependent OCR (C), Maximal (D), Spare respiratory capacity (E), H+ Leak-linked OCR (F) and Non-mitochondrial respiration (G) were obtained from traces such as those in Panel A. Extracellular acidification rates (ECAR) (H) were measured by extracellular flux analysis as described in the Methods section. * p ≤ 0.05, ** p ≤ 0.001, ns = not significant.
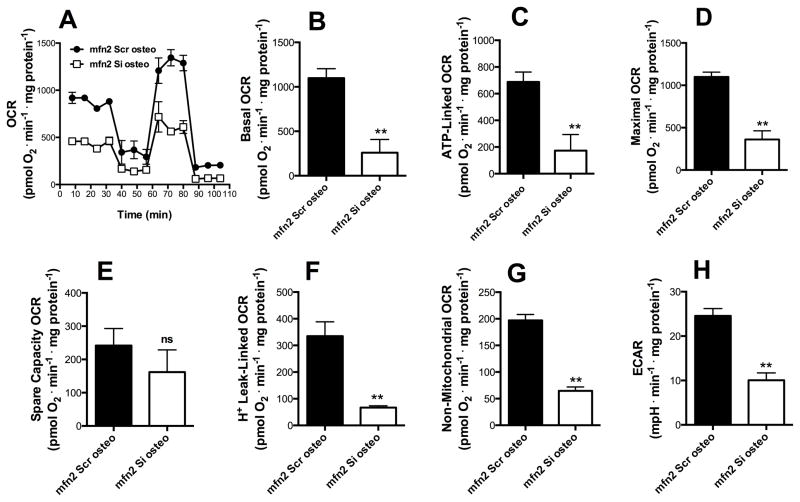
The same experiments were conducted during the commitment phase of osteogenesis, with similar results (Fig 5). All components of the respiratory profile (except for spare respiratory capacity, Fig 5E) were decreased in mfn2Si cells induced for 48 h with osteogenic induction media (Fig 5B, C, D, F and G p ≤ 0.001). Thus, the incapacity to modulate the mitochondrial network towards a fused state deeply impacts upon metabolic alterations observed during osteogenesis commitment.
Figure 5. Mfn2 is necessary for the metabolic switch observed during osteogenesis.
mfn2Scr (black) and mfn2Si (white) MSCs were exposed to osteogenesis media for 48 h (commitment time). Real time whole-cell respiration traces were obtained (A). Basal oxygen consumption rates (B), ATP-production dependent OCR (C), Maximal (D), Spare respiratory capacity (E), H+ Leak-linked OCR (F) and Non-mitochondrial respiration (G) were obtained from traces such as those in Panel A. Extracellular acidification rates (ECAR) (H) were measured by extracellular flux analysis as described in the Methods section. * p ≤ 0.05, ** p ≤ 0.001, ns = not significant.
Metabolic alterations during chondrogenesis are partially abrogated by dominant negative Drp1
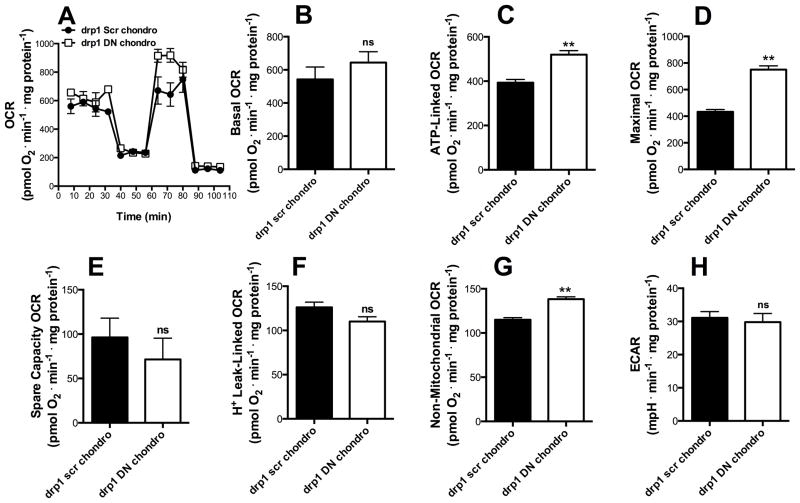
The transient overexpression of a dominant negative form of Drp1 impairs mitochondrial fission, leading to a fused/hyperfused mitochondrial network [25]. In order to evaluate if this inability to acquire a fragmented phenotype impacts upon the dampened bioenergetic activity observed in chondrogenesis (Fig 3), we induced drp1scr and drp1DN MSCs to differentiate into chondrocytes (Fig 6A) and analyzed their metabolic profile after 48 h. We found that basal (Fig 6B) OCRs present a non-significant increase in drp1DN MSCs relative to drp1scr cells. On the other hand, ATP-linked and maximal respiration in cells undergoing chondrogenesis were increased by drp1DN expression (Fig 6C–D, p ≤ 0.001). Spare respiratory capacity and the proton leak were unaltered (Fig 6E–F), while non-mitochondrial respiration also increased (Fig 6G) and the acidification rate was unchanged (Fig 6H). Overall, we find that a truncated version of the DRP1 protein abrogates at least partially the decreased respiratory capacity seen in cells during the chondrogenic commitment phase.
Figure 6. Drp1 mediates respiratory loss observed during chondrogenesis.
drp1Scr (black) and drp1DN (white) MSCs were exposed to chondrogenesis media for 48 h (commitment time). Real time whole-cell respiration traces were obtained (A). Basal oxygen consumption rates (B), ATP-production dependent OCR (C), Maximal (D), Spare respiratory capacity (E), H+ Leak-linked OCR (F) and Non-mitochondrial respiration (G) were obtained from traces such as those in Panel A. Extracellular acidification rates (ECAR) (H) were measured by extracellular flux analysis as described in the Methods section. * p ≤ 0.05, ** p ≤ 0.001, ns = not significant.
Mitochondrial network remodeling is critical for MSC differentiation into adipo, chondro and osteocytes
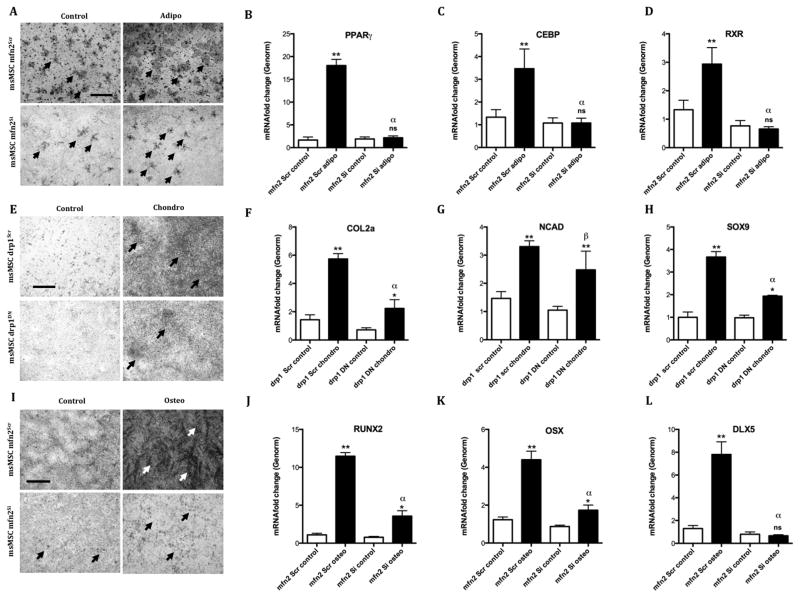
Since we found that the metabolic alterations associated with adipo, chondro and osteogenesis were dependent on Mfn2 and Drp1, we asked ourselves if the differentiation proficiency would be altered under the same conditions. We thus induced drp1Scr and drp1DN MSCs to undergo chondrogenesis and mfn2Scr and mfn2Si to undergo adipogenesis and osteogensis and evaluated their capacity to differentiate after 7 days. Oil red analysis of mfn2Scr and mfn2Si MSCs revealed a clear decrease in the number of both spontaneous (Control) and induced (Adipo) lipid-droplet containing immature adipocytes (Fig 7A), with preserved cellular survival. Moreover, increased levels of PPARγ (Fig 7B, p ≤ 0.001 relative to control media), Creb (Fig 7C, p ≤ 0.001) and Rxr (Fig 7D, p ≤ 0.001) observed in the scrambled-construct transduced cells after adipogenesis induction are completely absent in silenced cells (p ≤ 0.001 comparing scrambled and silenced cells that were induced to differentiate). This indicates that adipogenesis is dependent on the reorganization of the mitochondrial network, increasing mass and fusion and resulting in enhanced respiratory rates.
Figure 7. MSC differentiation is dependent on mitochondrial fusion and fission.
MSCs were transduced with adenovirus harboring siRNA Mfn2 or carrying a superexpression vector for a dominant negative form of drp1. Adipogenic differentiation proficiency of mfn2Scr versus mfn2Si was evaluated through Oil Red O lipid storage vacuole histochemistry (A) and validated through qPCR for specific markers (B) PPARγ (B) CREB and (C) RXR. Chondrogenic differentiation proficiency of drp1KD cells was evaluated in comparison to drp1Scr cells through the presence of proteoglycan rich ECM stained with toluidine blue (E) and validated through qPCR for specific markers (F) Col2a, N-cadherin (Ncad) (G) and Sox9 (H). Osteogenic differentiation proficiency of mfn2Scr versus mfn2Si was evaluated through Alizarin Red calcified ECM histochemistry (I) and validated through qPCR for specific markers (J) Runx2 (K) Osterix (Osx) and (L) Dlx-5. * p ≤ 0.05, ** p ≤ 0.001, ns = not significant. Bar = 100 μm.
In drp1DN MSCs induced to undergo chondrogenesis, less significant proteo/glycosaminoglycan-rich extracellular matrix formation was observed by staining with toluidine blue (Fig 7E). Collagen 2a expression increases promoted by differentiation induction (Fig 7F) were significantly decreased in the dominant negative cells (p ≤ 0.001, induced scramble versus DN cells). The same pattern was observed in Sox9 mRNA relative levels (Fig 7H), but not in Ncad expression, that was equally increased in both scramble and DN cells (Fig 7G), leading to the conclusion that chondrogenesis was partially affected by the inability to promote mitochondrial fragmentation.
Osteogenesis-induced mfn2Scr and mfn2Si MSCs stained with Alizarin Red displayed a very significant decrease in the area and amount of calcified matrix deposited when differentiation was induced (Fig 7I). Even more strikingly, the relative mRNA levels of several mediators of the osteoblast differentiation that were upregulated in mfn2scr cells exposed to the induction media were decreased in mfn2Si cells (Runx2, Fig 7J and Osterix, Fig 7K and Dlx-5 Fig 7L p ≤ 0.001). This demonstrates that osteogenesis heavily relies on the capacity of MSCs to reorganize their mitochondrial network into hyperfused units.
Discussion
Stem and progenitor cells divide and produce different cell types in a process that requires extensive functional rearrangements. Indeed, metabolic demands vary widely in different cell types, and therefore the activity of pathways that import and allocate nutrients is expected to change during differentiation [5]. Nonetheless, few studies to date have addressed how these metabolic changes happen and whether they are determinant during cell differentiation itself, and not only after achieving the fully differentiated state. The controlled regulation of mitochondrial abundance through biogenesis or degradation is critical for proper cell function, maintaining mitochondrial abundance finely tuned to the energetic needs of the cell [13]. Moreover, bioenergetic adaptation has been shown to involve the remodeling of mitochondrial architecture in many models [23], but the the relationship between stem cell differentiation and mitochondrial dynamics remains uninvestigated. Here, we studied how mitochondrial abundance, morphology and dynamics affect bioenergetic function and differentiation during murine MSC commitment to adipo, chondro and osteogenesis. We found that an extensive remodeling of mitochondrial architecture occurs and is necessary for differentiation, demonstrating that the energetic landscape of differentiation directly impacts upon mitochondrial network organization in a cell-fate dependent manner.
Accumulating evidence suggests that mitochondria are vital for metabolic homeostasis in white adipocytes due to their involvement in adipogenesis, fatty acid synthesis and esterification, branched-chain amino acid catabolism and lipolysis [26]. We found that during early stages of commitment to adipogenesis MSCs undergo mitochondrial biogenesis and network reorganization involving enhanced fusion. This leads to a metabolic switch toward a more oxidative profile that is abolished when these cells are incapable of undergoing mitochondrial fusion due to the knockdown of Mfn2. More importantly, in the absence of Mfn2, the potentiality of these cells is impaired and they lose their capacity to be addressed to undergo adipogenesis almost completely. The importance of Mfn2 for adipogenesis has already been suggested in a 3T3-L1 fat cell differentiation model, where the expression of this protein increases with the accumulation of lipid droplets [27]. Supporting our findings in MSCs, Wilson-Fritch et al. [28] reported an increase in the concentration of numerous mitochondrial proteins during 3T3-L1 cells adipogenic differentiation. Zhang et al [29] also described the occurrence of mitochondrial biogenesis and increased oxygen consumption during adipogenic differentiation, and found that reducing mitochondrial respiration by hypoxia or by inhibition of the mitochondrial electron transport chain significantly suppresses adipogenic differentiation, a different approach that supports our finding that mitochondrial remodeling leading to respiratory enhancement is essential for this differentiation process. The molecular mechanisms underlying mitochondrial biogenesis during adipogenesis are not fully characterized, but involve PGC1α [26], that we found to be very significantly upregulated very early during the commitment phase.
A metabolic shift towards a more oxidative phenotype also occurs in other models. Muscle precursor differentiation is associated with the translation of oxidative metabolism components [6]. The same oxidative shift was found in P19 neural differentiation [30], embryonic stem cell cardiac differentiation [31], human embryonic stem cell mesendoderm commitment [32] and spontaneous differentiation of human embryonic stem cells [33]. In an elegant manuscript, Chen and collaborators found that, upon osteogenic induction, human MSCs present increases in mitochondrial DNA content, respiratory enzyme protein subunits, oxygen consumption rates and intracellular ATP content, suggesting a coordinated regulation of mitochondrial biogenesis during osteogenic differentiation [34]. In our model we were unable to detect significantly increased levels of mtDNA copy numbers during osteogenesis, but several other parameters that indicate that mitochondrial mass is increased, including the measurement of the mitochondrial area, citrate synthase activity and maximal respiratory capacity. This enhanced respiratory capacity is strongly dependent on extensive mitochondrial fusion observed during commitment toward osteogenesis, as indicated by abrogation of these effects by Mfn2 knockdown. Importantly, osteogenic differentiation has been previously found to be prevented by hypoxia, in which an oxidative shift is impaired [35]. We found that the inability to organize mitochondria into a hyperfused network due to Mfn2 knockdown results in loss of their osteoblast potentiality. Supporting our findings, the loss of Mfn2, but not Mfn1, causes severe defects in the placental trophoblast giant cell layer during embryonic development, failure in dendritic outgrowth, spine formation, and Purkinje cell survival and leads to skeletal muscle atrophy characterized by mitochondrial dysfunction, mtDNA depletion and mutations [36].
Recently, a specialized type of autophagy that targets whole mitochondria and is known as mitophagy has been implicated in the regulation of quality control, number and subcellular distribution of these organelles during hematopoietic maintenance and differentiation [37]. Indeed, the idea that autophagy in general is crucial for quality control mechanisms that have direct impact on cellular homeostasis maintenance in stem cells is gaining wide acceptance, given the relatively long permanence of these cells in mammalian tissues [38]. In Drosophila, Drp1 ablation inactivates Notch and inhibits follicle cell differentiation in a manner dependent on mitochondrial fission [39]. More recently, Drp1 was shown to be required for proper neuronal differentiation [25], although, differently from our findings during chondrogenesis, mitochondrial function was unaffected during this process. We found that one of the early events associated with chondrogenesis commitment was the fragmentation of the mitochondrial network, possibly followed by its targeting toward mitophagy in a LC3 dependent-manner, corroborated by the analysis of the autophagic flux in the presence of 3-MA and chloroquine. The result is that early chondogenesis involves decreased basal and ATP-linked oxygen consumption rates. Indeed, mitophagy is triggered in a manner dependent on mitochondrial fission in many cell models [40]. Our results also indicate that Drp1 is functionally involved in chondrogenesis commitment, since this process is partially impaired by the incapacity to fragment the mitochondrial network through the overexpression of a dominant negative form of Drp1. Interestingly, since mitochondrial content does not seem to change during early chondrogenesis, despite increased PGC1α levels. This suggests a possible increase in biogenesis that is counter balanced by removal through mitophagy, leading to higher mitochondrial turnover. Moreover, the activation of mitophagy to selectively remove mitochondria during early chondrogenesis can reflect a developmentally programmed step towards maturation in a hypoxic environment occurring in vivo [41] that is conserved in vitro.
Finally, the cytoskeleton is known to play a large role in mechanotransduction. A growing number of studies show that it can contribute toward MSC differentiation, since chondrogenic differentiation is enhanced when MSCs have a spherical morphology associated with a dispersed actin cytoskeleton with few focal adhesions and altered microtubule patterning [42]. Cells allowed to attach onto micropatterned substrates, but prevented from spreading and flattening, present upregulated chondrogenic genes [43]. It is tempting to speculate that this dependency in cytoskeleton/cell shape that seems to regulate chondrogenesis is involved in the turnover of mitochondria, since the processes of fusion and fission rely heavily on the movement of these organelles along the cytoskeleton. This relationship remains to be elucidated.
Conclusion
In brief, mitochondrial morphology and dynamics and their regulating processes of fission and fusion are modulated early on during MSC commitment, leading to alterations in bioenergetic profiles during differentiation. The remodeling of the mitochondrial network is essential for the differentiation program, since these cells present delayed or completely abrogated capacity to differentiate when fusion or fission are abrogated. We thus propose that altered mitochondrial dynamics is not only a consequence, but rather a cause of commitment to differentiation in MSCs. Overall, our data show a central role for mitochondrial dynamics and resulting bioenergetic alterations in the maintenance and commitment of mesenchymal stem cells.
Supplementary Material
MSCs were isolated as described previously [3] and validated through their CD panel of positive (CD105, Panel A) (CD90, Panel B) (CD29, Panel c) (CD73 Panel D) and negative (CD34, Panel E) markers (black = IgG-stained cells, grey = antibody of interest). MSCs were also induced to differentiate and, after 7 days, the relative mRNA levels of (F) creb, (G) osterix and collagen 2a (H) were evaluated. Final validation of adipogenesis was obtained through Oil Red staining of lipid vesicles (I), calcified matrix stained with Alizarin Red for osteogenesis (J) and extracellular matrix rich in proteoglycans stained with toluidine blue for chondrogenesis (K). Control cells at the same time points can be observed in the inserts. * p ≤ 0.05, ** p ≤ 0.001, ns = not significant. Bar = 100 μm.
MSCs were induced to undergo chondrogenic differentiation and the early kinetics of macroautophagy markers total and lipidated LC3 (A) and Beclin (B) were measured by western blotting. Mitotracker Green and Lysotracker Red were used to stain mitochondria for lysosomes, respectively, and the co-localization pattern of mitochondria targeted for degradation was observed both in non-stimulated cells (left panel) and chondrogenic-fate induced cells (right panel) (C). Morevover, the analysis of autophagic flux through exposure to 3-MA and chloroquine was observed (D) and the number of mitophagic foci per cell was quantified in 100 cells (E, F). * p ≤ 0.05, ** p ≤ 0.001, ns = not significant. Bar = 10 μm.
Levels of Mfn2 protein were transiently downregulated using an adenovirus-mediated delivered system (A). Drp1 Dominant Negative mutant (DN, K38A) overexpression was achieved using an adenovirus-mediated system (B). * p ≤ 0.05, ** p ≤ 0.001, ns = not significant.
Antibodies used for Western Blot and Flow Cytometry
Acknowledgments
Grants acknowledgment: The authors would like to acknowledge the grants that supported this work namely FAPESP Temático (10/51906-1), Centro de Pesquisa e Desenvolvimento em Processos Redox em Biomedicina (CEPID Redoxoma 13/07937-8), FAPESP PD fellowship to MFF (13/04871-6), FAPESP IC fellowship to JP (14/17270-3), Núcleo de Apoio à Pesquisa em Processos Redox em Biomedicina (NAP Redoxoma). KT was supported by a National Science Foundation Graduate Research Fellowship under Grant No. DGE-0741448, and Levinsky Fellowship from Boston University School of Medicine. OS is funded by NIH grants R01 DK35914, R01 DK56690, and R01 DK074778.
The authors are in debt to Silvania M. P. Neves, Renata S. Fontes, Flavia M. P. Ong, and Maria de Fátima Rodrigues for excellent animal care. Eleni Ritou and Camille Caldeira were invaluable in their efficient lab management. The authors would also like to acknowledge the grants that supported this work namely FAPESP Temático (10/51906-1), FAPESP Redoxoma (13/07937-8), FAPESP PD fellowship to MFF (13/04871-6), FAPESP IC fellowship to JP (14/17270-3). We are in debt to Prof. Henning Ulrich for kindly letting us use the flow cytometer and Prof. Leticia Labriola for lending the LC3 antibody.
Footnotes
Disclosure of potential conflicts of interest
The authors declare no conflict of interest.
Author Contributions
MFF: Concept and design, Final approval of manuscript, Collection and/or assembly of data, Data analysis and interpretation, Manuscript writing.
KT and JP: Final approval of manuscript, Collection and/or assembly of data, Data analysis and interpretation.
OS: Final approval of manuscript, Financial support, Administrative support, Provision of study material, Manuscript writing.
AJK: Concept and design, Final approval of manuscript, Financial support, Administrative support, Provision of study material, Manuscript writing.
References
- 1.da Silva Meirelles L, Caplan AI, Nardi NB. In search of the in vivo identity of mesenchymal stem cells. Stem Cells. 2008;26:2287–2299. doi: 10.1634/stemcells.2007-1122. [DOI] [PubMed] [Google Scholar]
- 2.da Silva Meirelles L, Chagastelles PC, Nardi NB. Mesenchymal stem cells reside in virtually all post-natal organs and tissues. J Cell Sci. 2006;119:2204–2213. doi: 10.1242/jcs.02932. [DOI] [PubMed] [Google Scholar]
- 3.Halcsik E, Forni MF, Fujita A, et al. New insights in osteogenic differentiation revealed by mass spectrometric assessment of phosphorylated substrates in murine skin mesenchymal cells. BMC Cell Biology. 2013;14:47. doi: 10.1186/1471-2121-14-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forni M, Maia A, Ferreira A, et al. Simultaneous isolation of three stem cell populations from murine skin. PLoS One. doi: 10.1371/journal.pone.0140143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agathocleous M, Harris WA. Metabolism in physiological cell proliferation and differentiation. Trends Cell Biol. 2013;23:484–492. doi: 10.1016/j.tcb.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 6.Ozbudak EM, Tassy O, Pourquie O. Spatiotemporal compartmentalization of key physiological processes during muscle precursor differentiation. Proc Nat Acad Sci USA. 2010;107:4224–4229. doi: 10.1073/pnas.0909375107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim I, Lemasters JJ. Mitochondrial degradation by autophagy (mitophagy) in GFP-LC3 transgenic hepatocytes during nutrient deprivation. Am J Physiol Cell Physiol. 2011;300:C308–317. doi: 10.1152/ajpcell.00056.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michel S, Wanet A, De Pauw A, et al. Crosstalk between mitochondrial (dys)function and mitochondrial abundance. J Cell Physiol. 2012;227:2297–2310. doi: 10.1002/jcp.23021. [DOI] [PubMed] [Google Scholar]
- 9.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nature Reviews Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- 11.Nisoli E, Clementi E, Paolucci C, et al. Mitochondrial biogenesis in mammals: the role of endogenous nitric oxide. Science. 2003;299:896–899. doi: 10.1126/science.1079368. [DOI] [PubMed] [Google Scholar]
- 12.Cao W, Medvedev AV, Daniel KW, et al. beta-Adrenergic activation of p38 MAP kinase in adipocytes: cAMP induction of the uncoupling protein 1 (UCP1) gene requires p38 MAP kinase. J Biol Chem. 2001;276:27077–27082. doi: 10.1074/jbc.M101049200. [DOI] [PubMed] [Google Scholar]
- 13.Hill BG, Benavides GA, Lancaster JR, Jr, et al. Integration of cellular bioenergetics with mitochondrial quality control and autophagy. Biol Chem. 2012;393:1485–1512. doi: 10.1515/hsz-2012-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Spandidos A, Wang H, et al. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 2012;40:D1144–1149. doi: 10.1093/nar/gkr1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng YH, Xiong W, Su K, et al. Multilineage differentiation of human bone marrow mesenchymal stem cells and. Exp Ther Med. 2013;5:1576–1580. doi: 10.3892/etm.2013.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfiffer V, Prigione A. Assessing the bioenergetic profile of human pluripotent stem cells. Methods Mol Biol. 2015;1264:279–288. doi: 10.1007/978-1-4939-2257-4_25. [DOI] [PubMed] [Google Scholar]
- 18.Nicholls DG, Darley-Usmar VM, Wu M, et al. Bioenergetic profile experiment using C2C12 myoblast cells. J Vis Exp. 2010;6:2511. doi: 10.3791/2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wikstrom JD, Mahdaviani K, Liesa M, et al. Hormone-induced mitochondrial fission is utilized by brown adipocytes as an amplification pathway for energy expenditure. EMBO J. 2014;33:418–436. doi: 10.1002/embj.201385014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shibata M, Lu T, Furuya T, et al. Regulation of intracellular accumulation of mutant Huntingtin by Beclin 1. J Biol Chem. 2006;281:14474–14485. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- 21.Twig G, Elorza A, Molina AJ, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dominici M, Le Blanc K, Mueller I, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 23.Liesa M, Shirihai OS. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab. 2013;17:491–506. doi: 10.1016/j.cmet.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 25.Wang L, Ye X, Zhao Q, et al. Drp1 is dispensable for mitochondria biogenesis in induction to pluripotency but required for differentiation of embryonic stem cells. Stem Cells Dev. 2014;23:2422–2434. doi: 10.1089/scd.2014.0059. [DOI] [PubMed] [Google Scholar]
- 26.Boudina S, Graham TE. Mitochondrial function/dysfunction in white adipose tissue. Exp Physiol. 2014;99:1168–1178. doi: 10.1113/expphysiol.2014.081414. [DOI] [PubMed] [Google Scholar]
- 27.Ducluzeau PH, Priou M, Weitheimer M, et al. Dynamic regulation of mitochondrial network and oxidative functions during 3T3-L1 fat cell differentiation. J Physiol Bioch. 2011;67:285–296. doi: 10.1007/s13105-011-0074-6. [DOI] [PubMed] [Google Scholar]
- 28.Wilson-Fritch L, Burkart A, Bell G, et al. Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol Cell Biol. 2003;23:1085–1094. doi: 10.1128/MCB.23.3.1085-1094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Marsboom G, Toth PT, et al. Mitochondrial respiration regulates adipogenic differentiation of human mesenchymal stem cells. PLoS One. 2013;8:e77077. doi: 10.1371/journal.pone.0077077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watkins J, Basu S, Bogenhagen DF. A quantitative proteomic analysis of mitochondrial participation in p19 cell neuronal differentiation. J Proteome Res. 2008;7:328–338. doi: 10.1021/pr070300g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chung S, Arrell DK, Faustino RS, et al. Glycolytic network restructuring integral to the energetics of embryonic stem cell cardiac differentiation. J Mol Cell Cardiol. 2010;48:725–734. doi: 10.1016/j.yjmcc.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prowse AB, Chong F, Elliott DA, et al. Analysis of mitochondrial function and localisation during human embryonic stem cell differentiation in vitro. PLoS One. 2012;7:e52214. doi: 10.1371/journal.pone.0052214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cho YM, Kwon S, Pak YK, et al. Dynamic changes in mitochondrial biogenesis and antioxidant enzymes during the spontaneous differentiation of human embryonic stem cells. Biochem Biophys Res Comm. 2006;348:1472–1478. doi: 10.1016/j.bbrc.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 34.Chen CT, Shih YR, Kuo TK, et al. Coordinated changes of mitochondrial biogenesis and antioxidant enzymes during osteogenic differentiation of human mesenchymal stem cells. Stem Cells. 2008;26:960–968. doi: 10.1634/stemcells.2007-0509. [DOI] [PubMed] [Google Scholar]
- 35.Hsu SH, Chen CT, Wei YH. Inhibitory effects of hypoxia on metabolic switch and osteogenic differentiation of human mesenchymal stem cells. Stem Cells. 2013;31:2779–2788. doi: 10.1002/stem.1441. [DOI] [PubMed] [Google Scholar]
- 36.Kasahara A, Scorrano L. Mitochondria: from cell death executioners to regulators of cell differentiation. Trends Cell Biol. 2014;24:761–770. doi: 10.1016/j.tcb.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 37.Joshi A, Kundu M. Mitophagy in hematopoietic stem cells: the case for exploration. Autophagy. 2013;9:1737–1749. doi: 10.4161/auto.26681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guan JL, Simon AK, Prescott M, et al. Autophagy in stem cells. Autophagy. 2013;9:830–849. doi: 10.4161/auto.24132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitra K, Rikhy R, Lilly M, et al. DRP1-dependent mitochondrial fission initiates follicle cell differentiation during Drosophila oogenesis. J Cell Biol. 2012;197:487–497. doi: 10.1083/jcb.201110058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frank M, Duvezin-Caubet S, Koob S, et al. Mitophagy is triggered by mild oxidative stress in a mitochondrial fission dependent manner. Biochim Biophys Acta. 2012;1823:2297–2310. doi: 10.1016/j.bbamcr.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 41.Craft AM, Ahmed N, Rockel JS, et al. Specification of chondrocytes and cartilage tissues from embryonic stem cells. Development. 2013;140:2597–2610. doi: 10.1242/dev.087890. [DOI] [PubMed] [Google Scholar]
- 42.Mathieu PS, Loboa EG. Cytoskeletal and focal adhesion influences on mesenchymal stem cell shape, mechanical properties, and differentiation down osteogenic, adipogenic, and chondrogenic pathways. Tissue Eng B. 2012;18:436–444. doi: 10.1089/ten.teb.2012.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao L, McBeath R, Chen CS. Stem cell shape regulates a chondrogenic versus myogenic fate through Rac1 and N-cadherin. Stem Cells. 2010;28:564–572. doi: 10.1002/stem.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MSCs were isolated as described previously [3] and validated through their CD panel of positive (CD105, Panel A) (CD90, Panel B) (CD29, Panel c) (CD73 Panel D) and negative (CD34, Panel E) markers (black = IgG-stained cells, grey = antibody of interest). MSCs were also induced to differentiate and, after 7 days, the relative mRNA levels of (F) creb, (G) osterix and collagen 2a (H) were evaluated. Final validation of adipogenesis was obtained through Oil Red staining of lipid vesicles (I), calcified matrix stained with Alizarin Red for osteogenesis (J) and extracellular matrix rich in proteoglycans stained with toluidine blue for chondrogenesis (K). Control cells at the same time points can be observed in the inserts. * p ≤ 0.05, ** p ≤ 0.001, ns = not significant. Bar = 100 μm.
MSCs were induced to undergo chondrogenic differentiation and the early kinetics of macroautophagy markers total and lipidated LC3 (A) and Beclin (B) were measured by western blotting. Mitotracker Green and Lysotracker Red were used to stain mitochondria for lysosomes, respectively, and the co-localization pattern of mitochondria targeted for degradation was observed both in non-stimulated cells (left panel) and chondrogenic-fate induced cells (right panel) (C). Morevover, the analysis of autophagic flux through exposure to 3-MA and chloroquine was observed (D) and the number of mitophagic foci per cell was quantified in 100 cells (E, F). * p ≤ 0.05, ** p ≤ 0.001, ns = not significant. Bar = 10 μm.
Levels of Mfn2 protein were transiently downregulated using an adenovirus-mediated delivered system (A). Drp1 Dominant Negative mutant (DN, K38A) overexpression was achieved using an adenovirus-mediated system (B). * p ≤ 0.05, ** p ≤ 0.001, ns = not significant.
Antibodies used for Western Blot and Flow Cytometry