Abstract
The developing mouse retina is a tractable model for studying neurogenesis and differentiation. Although transgenic Cre mouse lines exist to mediate conditional genetic manipulations in developing mouse retinas, none of them act specifically in early developing rods. For conditional genetic manipulations of developing retinas, we created a Nrl-Cre mouse line in which the Nrl promoter drives expression of Cre in rod precursors. Our results show that Nrl-Cre expression is specific to the retina where it drives rod-specific recombination with a temporal pattern similar to endogenous Nrl expression during retinal development. This Nrl-Cre transgene does not negatively impact retinal structure and function. Taken together, our data suggest that the Nrl-Cre mouse line is a valuable tool to drive Cre-mediated recombination specifically in developing rods.
Keywords: differentiation, neuronal development, transgenic, retina-specific Cre
INTRODUCTION
Differentiation of rods, the predominant photoreceptor in the mouse retina, occurs over a long period during which rod precursor cells undergo a series of changes in gene expression, cellular structure and function (Brzezinski and Reh 2015). This process is controlled by an intrinsic genetic program as well as extrinsic signaling pathways (Brzezinski and Reh 2015, Cepko 2014). Mutations in genes that are a part of this genetic program lead to developmental defects and visual impairment in human patients (Swaroop 2010).
One of these genes is the neural retina leucine zipper protein (NRL), a key retinal transcription factor that belongs to the MAF family of transcription factors (Swaroop 1992). Nrl is highly expressed in the retina where it is specific to developing and mature rods (Liu 1996, Swain 2001). Nrl is essential for rod differentiation, and loss of Nrl results in rod precursors developing into cone-like photoreceptors (Mears 2001, Oh 2007). NRL interacts with other transcription factors including CRX to induce the expression of rod genes (Mitton 2000). The promoter of Nrl has been used previously to drive expression of an EGFP transgene specifically in rods and their precursors (Akimoto 2006).
There is a critical need to understand the intrinsic program and extrinsic signaling pathways that influence rod development and differentiation. Research on these regulatory mechanisms requires a genetic tool that allows conditional genetic manipulations in developing rods. Rho-Cre (Li 2005) and Cone-Cre (Le 2004) mice are currently available and are specific to rods or cones, respectively. However these Cre lines turn Cre on late in already differentiated photoreceptors. Thus, they are not suitable for investigating the function of genes whose products act in photoreceptor precursor cells during development and are critical for photoreceptor cell fate specification or differentiation. Alternatively, Crx-Cre and Crx-CreERT2 lines were created to specifically inactivate floxed genes from developing photoreceptors (Muranishi 2011, Nishida 2003, Prasov and Glaser 2012). Although early reports indicated that recombination was photoreceptor specific (Nishida 2003), these Crx-Cre transgenic and BAC transgenic mouse lines had Cre activity in all retinal cell types (Prasov and Glaser 2012). As a result, no reliable mouse Cre line is available that expresses Cre specifically in developing photoreceptors. Thus, generating a Cre line allowing for recombination during the specification of rod photoreceptors is important for the vision research community. We therefore created a transgenic mouse line expressing Cre under the control of the Nrl promoter. We report that this Nrl-Cre transgenic line expresses Cre in a spatiotemporal pattern similar to endogenous Nrl in developing and mature mouse retinas without affecting overall retinal integrity and maintenance. This Nrl-Cre transgenic line can effectively mediate loxP site recombination specifically in developing rods. Thus, Nrl-Cre is a valuable resource for investigating early regulatory factors important for cell fate specification of rods as opposed to cones, or for developing proper rod structure and function in the mammalian retina.
RESULTS AND DISCUSSION
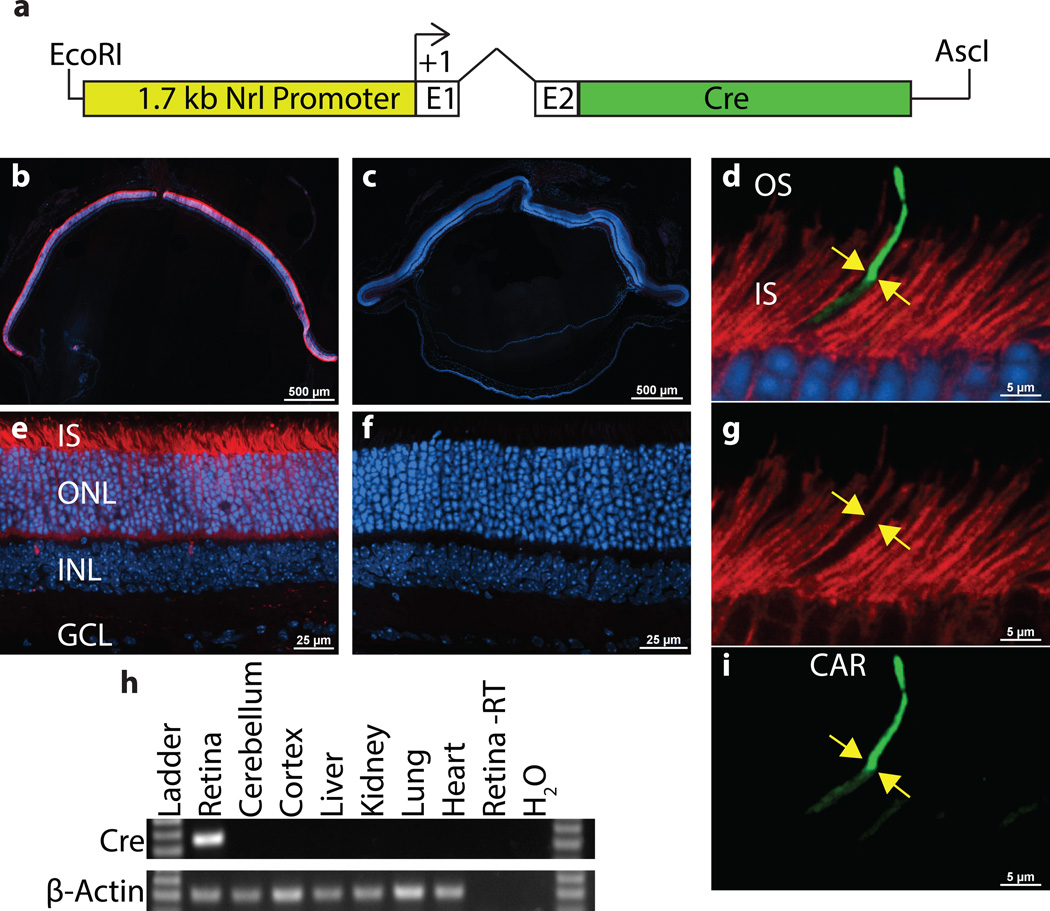
To conditionally knock out genes during early photoreceptor development, we generated a Nrl-Cre transgenic mouse line in which the 1.7 kb Nrl promoter drives Cre expression in rods and their precursors (Fig. 1a). To determine the cell type specificity of Cre-mediated recombination, Nrl-Cre mice were crossed to Ai9 Cre Reporter mice (Madisen 2010). The Ai9 Cre Reporter mouse uses a ubiquitously expressed reporter to drive expression of the TdTomato fluorophore. This expression is dependent on Cre mediated recombination because a STOP codon flanked by LoxP sites is located upsteam of the TdTomato coding sequence. As a result, TdTomato fluorescence will only be seen in cells where Cre-mediated recombination occurs, and cells without Cre activity will lack TdTomato fluorescence. As expected, Nrl-Cre drove Cre recombination and TdTomato fluorescence specifically in the photoreceptor layer of young adult mice (Fig. 1b,e). This expression was uniform across the retina and appears to be complete (Fig. 1b). To confirm that Cre-mediated recombination did not occur in cones, we immunostained Nrl-Cre; Ai9 retinal cross-sections for Cone-Arrestin (CAR), which specifically marks cone outer segments (Zhu 2002). As expected, tdTomato fluorescence was absent from cone photoreceptors labeled with CAR in Nrl-Cre; Ai9 retinal cross-sections (Fig. 1d,g,i), demonstrating that the Cre activity is restricted to rod photoreceptors. To survey the expression pattern of Cre across different tissues, reverse transcriptase polymerase chain reaction (RT-PCR) was performed on total RNA purified from retina, cerebellum, cortex, liver, kidney, lung and heart of 1 month old Nrl-Cre mice. In agreement with the retina-specific expression pattern of endogenous Nrl, RT-PCR amplification of a single Cre band was exclusively detected in retinal RNA, demonstrating retinal specificity of the Nrl-Cre transgenic line (Fig. 1h).
Figure 1.
Nrl-Cre is expressed specifically in rods. The 1.7 kb Nrl promoter upstream of Nrl exons 1 (E1) and 2 (E2) with a 724bp intron was used to drive Cre expression (a). tdTomato fluorescence (red) in Nrl-Cre; Ai9 (b,e) and Cre negative, Ai9 (c,f) frozen retinal cross-sections at 1 month of age are shown. Immunostaining of 1 month old Nrl-Cre; Ai9 retinas with Cone Arrestin (CAR, green) shows that tdTomato is not expressed in CAR+ cones (yellow arrows in d,g,i). Reverse Transcriptase-PCR (RT-PCR) analysis of Cre expression in total RNA from the indicated tissues of 1 month old Nrl-Cre mice (h). β-Actin served as a control for RNA quality. A sample without reverse transcriptase (Retina -RT) serves as a negative control. Cre expression was only detected in the retina. ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer; OS, outer segment; IS, inner segment.
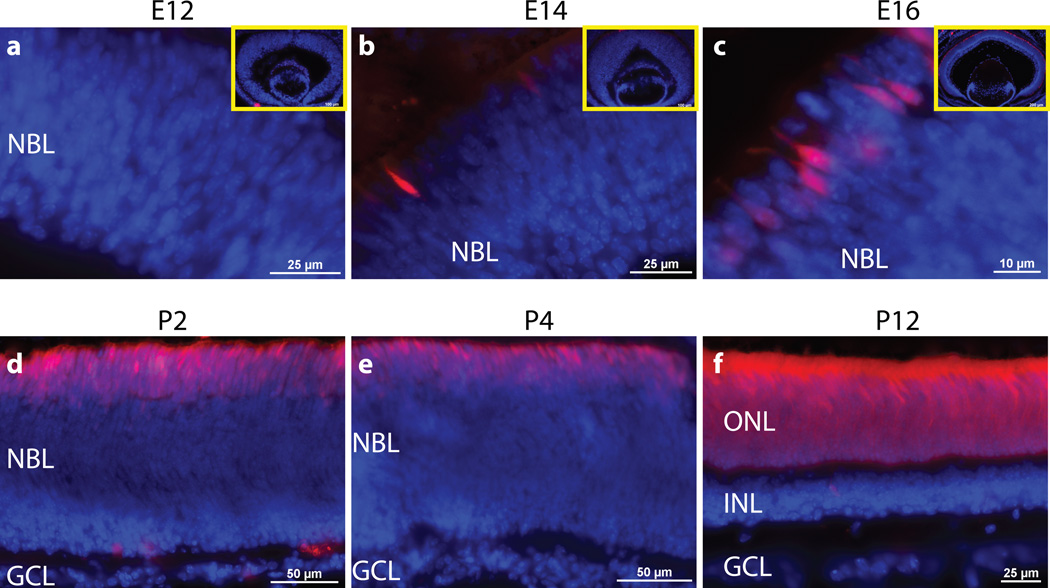
To determine the time course of Cre-mediated recombination, Nrl-Cre; Ai9 retinas were collected at several time points throughout development (Fig. 2). At embryonic day 12 (E12), Nrl-Cre; Ai9 did not display any detectable expression of tdTomato (Fig. 2a). At E14, a few tdTomato-positive cells appeared on the outer part of the neuroblast layer (Fig. 2b). Subsequently, the number of tdTomato+ cells gradually increased up to postnatal day 12 (P12), by which time all rods in the photoreceptor layer expressed tdTomato (Fig. 2c–f). This recombination time course is consistent with the temporal expression pattern of endogenous Nrl in developing rods (Akimoto 2006). In contrast, Cre-mediated recombination in Rho-Cre and Cone-Cre mice starts later than P5 in differentiated rods or cones and is incomplete until P12 (Hennig 2013, Le 2004, Li 2005).
Figure 2.
Developmental time course of Nrl-Cre activity. Microscopy of tdTomato fluorescence (red) with DAPI costain (blue) in Nrl-Cre; Ai9 frozen retinal cross-sections at embryonic day 12 (E12) (a), E14 (b), E16 (c), P2 (d), P4 (e) and P12 (f). Insets are of entire embryonic retinal cross-section (a–c). NBL, neuroblastic layer.
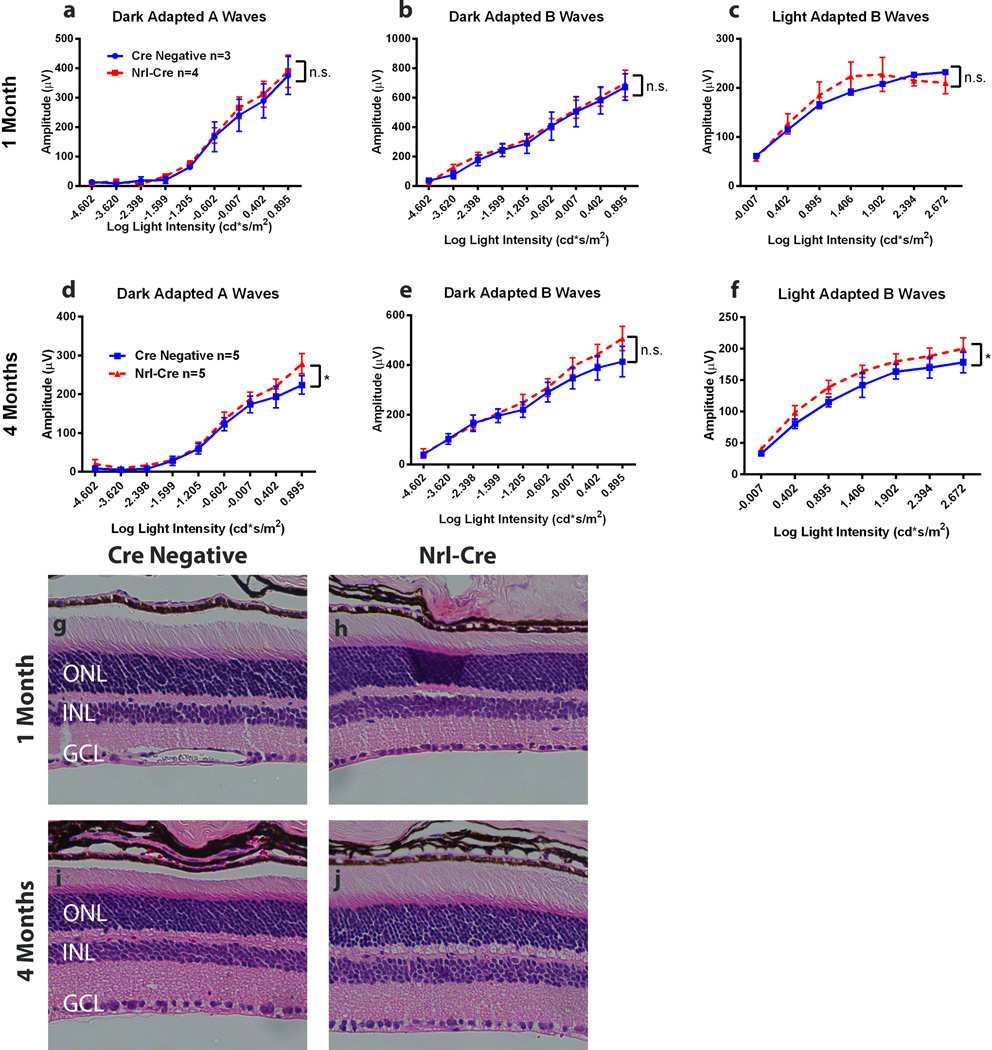
To rule out undesired side effects of Cre expression on retina integrity, we first examined retinal function by in vivo electroretinograms (ERGs) (Fig. 3a–f). At 1 month of age, the ERG responses from both dark-adapted (Fig. 3a, b) and light-adapted (Fig. 3c) Nrl-Cre mice were no different from Cre negative littermate controls, suggesting these mice developed normal rod and cone function (Fig. 3a–c). By 4 month of age, Nrl-Cre mice maintained rod and cone function compared to the controls: They actually showed slight but significant increases in dark-adapted A wave amplitudes (Fig. 3d). However, dark-adapted B wave amplitudes were not significantly different from controls (Fig. 3e). The light-adapted B wave amplitudes were also slightly increased (Fig. 3f). These ERG results suggest the Nrl-Cre mice develop normal light responses from rods and cones, and the neuronal signal is transmitted normally from photoreceptors to the inner retina. Nrl-Cre mice also maintain the normal function at least up to 4 months of age.
Figure 3.
Nrl-Cre mice have no detectable functional or morphological defects. Whole animal electroretinograms (ERGs) of Nrl-Cre mice and Cre negative littermate controls were performed at 1 Month (a–c) and 4 Months of age (d–f). Responses of Nrl-Cre mice were no different from Cre negative littermate controls for all physiological tests at 1 month of age. 4 month old Nrl-Cre mice showed marginal but significant increases in dark-adapted A wave amplitudes (d) and light-adapted B wave amplitudes (f) responses but no individual light intensities were significantly increased. Dark-adapted B wave amplitudes at 4 months were not significantly different. Nrl-Cre (h,j) and Cre Negative littermate control (g, i) retinal cross-sections were stained with hematoxylin and eosin (H&E) at 1 month (g,h) and 4 months (i,j). Two-way ANOVA with Sidak’s multiple comparisons. p<0.05 CI: 95% Error bars = SEM.
Next, we examined retinal morphology by hematoxylin and eosin (H&E) staining of retinal cross-sections. At 1 and 4 months of age, Nrl-Cre retinas were the same thickness as Cre negative littermate controls and had normal retinal lamination (Fig. 3g–j). These results are consistent with normal electroretinograms, suggesting that the Nrl-Cre transgene does not cause defects in retinal function or structure at least up to four months. This non-toxic nature of the Nrl-Cre transgene allows faithful assessment of accumulative effects of conditional manipulations on retinal structure and function after retinal development is complete.
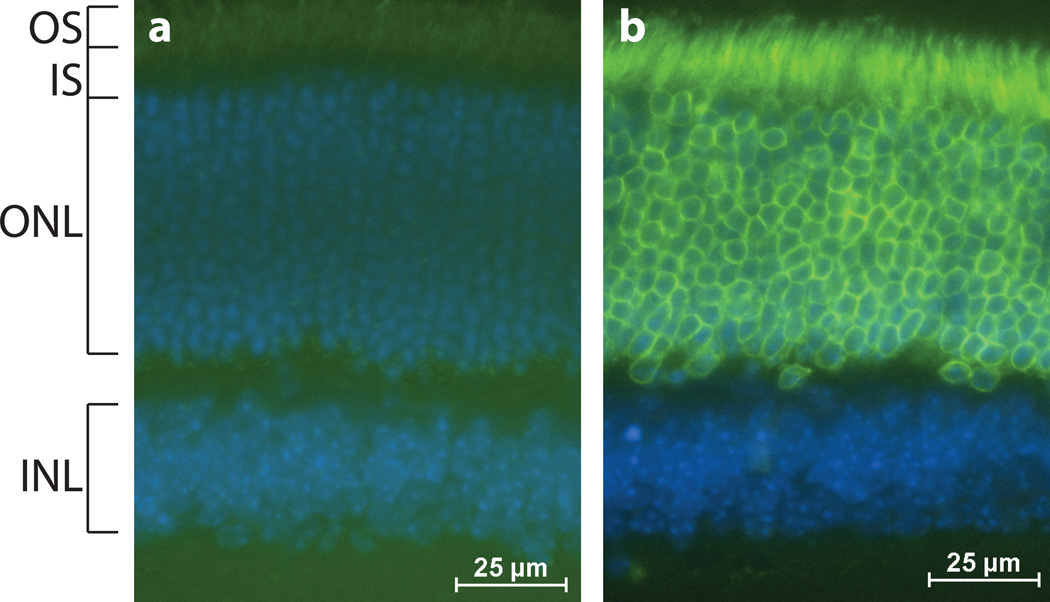
To confirm that Nrl-Cre could effectively recombine floxed alleles other than a Cre reporter, we crossed the Nrl-Cre mouse to the CAG-LacZ/EGFP-KASH2 mouse (Razafsky and Hodzic 2014). The latter conditionally expresses an EGFP-tagged version of the KASH domain of Nesprin2, a transmembrane polypeptide that is sufficient to mediate nuclear envelope localization. As shown in Figure 4, EGFP-KASH2 was efficiently expressed in the outer nuclear layer of Nrl-Cre; EGFP-KASH2 retinas where it exclusively decorated the nuclear envelope of rod photoreceptor nuclei. As expected, non-recombinant littermates did not display any expression of EGFP-KASH2, thereby demonstrating Cre-dependent EGFP-KASH2 expression in rods (Fig. 4a,b).
Figure 4.
Nrl-Cre recombines floxed genes in rods. Microscopy of 1 month old Cre negative; CAG-LacZ/EGFP-KASH2 (a) and Nrl-Cre;CAG-LacZ/EGFP-KASH2 (b) retinal cross-sections to visualize EGFP (green) and DAPI co-stain (blue). EGFP-KASH2 is specifically expressed in the photoreceptor layer of Nrl-Cre; CAG-LacZ/EGFP-KASH2 retinas where it exclusively decorates the nuclear envelope of rod photoreceptor nuclei.
Taken together, our results indicate that the Nrl-Cre transgenic mouse line is a valuable tool to drive Cre-mediated recombination specifically in rods on the expected developmental time course without detectable consequences to retinal structure and function. Of note is that a few sparse cells in the inner nuclear layer displayed Cre-mediated recombination (Fig. 1e). The physiological significance of that observation, if any, remains to be determined. In comparison to currently available Crx-Cre mice (Nishida 2003, Prasov and Glaser 2012), Nrl-Cre transgenic mice can specifically target floxed alleles in developing rods without targeting cones, providing a new genetic tool to study rod development and differentiation.
METHODS
Mice
Animal protocols used in this study adhered to the ethical and sensitive care and use of animals in research and were approved by the Washington University School of Medicine Animal Studies Committee [Animal Welfare Assurance Permit # A-3381-01, Protocols # 20130225 (Hodzic) and #20120246 (Chen)]. All mice were bred and maintained in Washington University School of Medicine barrier facilities. Nrl-Cre mice were generated by the Molecular Genetics Core of the Department of Ophthalmology and Visual Sciences, on the C57BL/6J background using a conventional transgenic protocol. The Nrlp-EGFP parent vector from Anand Swaroop included the 1.7kb Nrl promoter and Nrl exons 1 and 2 driving expression of EGFP. We replaced EGFP with the Cre coding sequence and linearized the transgene cassette using EcoRI and AscI restriction digestion (Fig. 1a) for pronuclear injection. One founder line was stably established after screening about 100 injected pronuclei. This male founder was bred to C57BL/6J mice from The Jackson Laboratory (#000664). Nrl-Cre mice were genotyped using CRE primers (5' GCATTACCGGTCGATGCAACGAGTGATGAG 3' and 5’GAGTGAACGAACCTGGTCGAAATCAGTGCG 3’). Ai9 Cre Reporter mice were from Jackson Laboratory (#007909) and transgenic CAG-LacZ/EGFP-KASH2 mice were previously described (Razafsky and Hodzic 2014). All mice used in experiments were heterozygous for the transgene. The Nrl-Cre strain will be made available to the research community upon acceptance of the manuscript.
Histology and Immunofluorescence
Mice were sacrificed via CO2 inhalation. Eyes were enucleated and dissected in phosphate buffered saline (PBS) to remove the cornea and lens. Eyecups were then fixed in 4% paraformaldehyde overnight and embedded in paraffin to slice 5 µm-thick retinal cross-sections on a Leica RM 2255 microtome. Hematoxylin and eosin staining was performed for histology. Slides were imaged on a Leica DB5500 microscope. For fluorescence and immunofluorescence microscopy, small incisions were performed through the cornea of whole enucleated eyes. Eyes were then rinsed in PBS before fixation in 4% PFA/PBS for 1 hour followed by overnight incubation in 30% sucrose and embedding in OCT compound. For direct fluorescence microscopy, cryosections (15µm) on Superfrost Plus slides were rinsed in PBS, permeabilized in 0.5% Triton X-100/10% donkey serum in PBS and counterstained with DAPI. Upon mounting, slices were imaged on an Eclipse Ti inverted microscope (Nikon, Melville, NY, USA) coupled to a CoolSnap HQ2 camera (Photometrics, Tucscon, AZ, USA) and controlled by NIS Elements software. For immunofluorescence microscopy, permeabilized cryosections were incubated with a cone-arrestin antibody (1:1000, Millipore) diluted in the permeabilization buffer for 1 hour at room temperature. After three washes in PBS, slices were incubated with donkey anti-rabbit Alexa-conjugated secondary antibodies (Invitrogen) for 1 hour in permeabilization buffer. After three washes, slices were counterstained with DAPI, mounted and imaged as described above.
RNA Purification and RT-PCR
Total RNA was purified from mouse tissues using the Perfect Pure RNA Tissue Kit (5Prime). RNA was quantified using NanoDrop ND-1000 (NanoDrop Technologies). 1 µg of total RNA was reverse transcribed into cDNA using the Transcriptor First Strand cDNA Synthesis Kit (Roche) and oligo-dT priming. cDNA was diluted 1:50 and polymerase chain reactions (PCR) were performed using Jumpstart RedTaq (Sigma-Aldrich) and Cre (5' ATCTGGCATTTCTGGGGATTGC 3' and 5' GCAACACCATTTTTTCTGACCCG 3') or β-Actin (5' CCAACTGGGACGACATGGAG 3' and 5' TGGTACGACCAGAGGCATACAG 3') primers with thirty amplification cycles. PCR products were run on 1.2% agarose gels and imaged with an AlphaImager 3400 (Alpha Innotech Corporation).
Electroretinograms
In vivo ERGs were performed on 1 month old and 4 month old Nrl-Cre mice and their Cre-negative littermate controls. Responses to light flashes were recorded using the UTAS-3000 Visual Electrodiagnostic System with EM for Windows (LKC Technologies, Inc.). Mice were dark-adapted overnight and anesthetized with 80 mg/kg ketamine and 15 mg/kg xylazine. Mice were set up under dim red light. The mouse body temperature was maintained at 37±0.5°C. Pupils were dilated with 1% atropine sulfate solution (Bausch & Lomb). 2.0mm platinum loop electrodes were placed on the corneas with 2.5% Hypromellose Demulcent Solution (GONAK, Akom Inc.). A ground electrode was placed under the skin of the mouse’s back and a reference electrode was placed under the skin of the mouse’s skull. Retinal activities were recorded in response to 10 µs light flashes of increasing intensity. Following dark-adapted tests, mice were light-adapted for 10 minutes with 29.2 cd/m2 and exposed to 10 µs light flashes of increasing intensity. Responses for several trials were recorded at each light intensity and averaged. The mean peak amplitudes of dark-adapted A waves and B waves and light-adapted B waves were graphed against log light intensity (cd*s/m2). Results for the two genotypes were compared by two-way ANOVA with Sidak’s multiple comparisons using Graphpad Prism software. p<0.05 CI: 95% Error bars = SEM.
Acknowledgments
The authors are grateful to Michael Casey (Mouse Genetics Core, Washington University), GuangYi Ling (Morphology and Imaging Core, Washington University), Anne Hennig (ERG Core, Washington University) and Mingyan Yang for technical assistance.
Funding: NIH grants EY017015 and EY012543 (to SC), EY022632 (to DH), and EY002687 (to WU-DOVS), and unrestricted funds from Research to Prevent Blindness (to WU-DOVS).
Footnotes
The authors have no conflict of interest to declare.
REFERENCES
- 1.Akimoto M, Cheng H, Zhu D, Brzezinski JA, Khanna R, Filippova E, Oh EC, Jing Y, Linares JL, Brooks M, Zareparsi S, Mears AJ, Hero A, Glaser T, Swaroop A. Targeting of GFP to newborn rods by Nrl promoter and temporal expression profiling of flow-sorted photoreceptors. Proc Natl Acad Sci U S A. 2006;103:3890–3895. doi: 10.1073/pnas.0508214103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brzezinski JA, Reh TA. Photoreceptor cell fate specification in vertebrates. Development. 2015;142:3263–3273. doi: 10.1242/dev.127043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cepko C. Intrinsically different retinal progenitor cells produce specific types of progeny. Nat Rev Neurosci. 2014;15:615–627. doi: 10.1038/nrn3767. [DOI] [PubMed] [Google Scholar]
- 4.Hennig AK, Peng GH, Chen S. Transcription coactivators p300 and CBP are necessary for photoreceptor-specific chromatin organization and gene expression. PloS one. 2013;8:e69721. doi: 10.1371/journal.pone.0069721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le YZ, Ash JD, Al-Ubaidi MR, Chen Y, Ma JX, Anderson RE. Targeted expression of Cre recombinase to cone photoreceptors in transgenic mice. Mol Vis. 2004;10:1011–1018. [PubMed] [Google Scholar]
- 6.Li S, Chen D, Sauve Y, McCandless J, Chen YJ, Chen CK. Rhodopsin-iCre transgenic mouse line for Cre-mediated rod-specific gene targeting. Genesis. 2005;41:73–80. doi: 10.1002/gene.20097. [DOI] [PubMed] [Google Scholar]
- 7.Liu Q, Ji X, Breitman ML, Hitchcock PF, Swaroop A. Expression of the bZIP transcription factor gene Nrl in the developing nervous system. Oncogene. 1996;12:207–211. [PubMed] [Google Scholar]
- 8.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mears AJ, Kondo M, Swain PK, Takada Y, Bush RA, Saunders TL, Sieving PA, Swaroop A. Nrl is required for rod photoreceptor development. Nat Genet. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- 10.Mitton KP, Swain PK, Chen S, Xu S, Zack DJ, Swaroop A. The leucine zipper of NRL interacts with the CRX homeodomain. A possible mechanism of transcriptional synergy in rhodopsin regulation. J Biol Chem. 2000;275:29794–29799. doi: 10.1074/jbc.M003658200. [DOI] [PubMed] [Google Scholar]
- 11.Muranishi Y, Terada K, Inoue T, Katoh K, Tsujii T, Sanuki R, Kurokawa D, Aizawa S, Tamaki Y, Furukawa T. An essential role for RAX homeoprotein and NOTCH-HES signaling in Otx2 expression in embryonic retinal photoreceptor cell fate determination. J Neurosci. 2011;31:16792–16807. doi: 10.1523/JNEUROSCI.3109-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishida A, Furukawa A, Koike C, Tano Y, Aizawa S, Matsuo I, Furukawa T. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat Neurosci. 2003;6:1255–1263. doi: 10.1038/nn1155. [DOI] [PubMed] [Google Scholar]
- 13.Oh EC, Khan N, Novelli E, Khanna H, Strettoi E, Swaroop A. Transformation of cone precursors to functional rod photoreceptors by bZIP transcription factor NRL. Proc Natl Acad Sci U S A. 2007;104:1679–1684. doi: 10.1073/pnas.0605934104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prasov L, Glaser T. Pushing the envelope of retinal ganglion cell genesis: context dependent function of Math5 (Atoh7) Dev Biol. 2012;368:214–230. doi: 10.1016/j.ydbio.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Razafsky D, Hodzic D. Temporal and tissue-specific disruption of LINC complexes in vivo. Genesis. 2014;52:359–365. doi: 10.1002/dvg.22755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swain PK, Hicks D, Mears AJ, Apel IJ, Smith JE, John SK, Hendrickson A, Milam AH, Swaroop A. Multiple phosphorylated isoforms of NRL are expressed in rod photoreceptors. J Biol Chem. 2001;276:36824–36830. doi: 10.1074/jbc.M105855200. [DOI] [PubMed] [Google Scholar]
- 17.Swaroop A, Kim D, Forrest D. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat Rev Neurosci. 2010;11:563–576. doi: 10.1038/nrn2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swaroop A, Xu JZ, Pawar H, Jackson A, Skolnick C, Agarwal N. A conserved retina-specific gene encodes a basic motif/leucine zipper domain. Proc Natl Acad Sci U S A. 1992;89:266–270. doi: 10.1073/pnas.89.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu X, Li A, Brown B, Weiss ER, Osawa S, Craft CM. Mouse cone arrestin expression pattern: light induced translocation in cone photoreceptors. Mol Vis. 2002;8:462–471. [PubMed] [Google Scholar]