Abstract
Recent efforts to design a synthetic extracellular matrix for cell culture, engineering, and therapies greatly contributed to addressing biological roles of types and spatial organization of cell adhesion ligands. It is often suggested that ligand-matrix bond strength is another path to regulate cell adhesion and activities; however tools are lacking. To this end, this study demonstrates that a hydrogel coupled by integrin-binding deoxyribonucleic acid (DNA) tethers with pre-defined rupture forces can modulate cell adhesion, differentiation, and secretion activities due to the changes in the number and, likely, force of cells adhered to a gel. The rupture force of DNA tethers was tuned by altering spatial arrangement of matrix-binding biotin groups. The DNA tethers were immobilized on a hydrogel of alginate grafted with biotin using avidin. Mesenchymal stem cells showed enhanced adhesion, neural differentiation, and paracrine secretion when cultured on the gel coupled with DNA tethers with higher rupture forces. Such innovative cell-matrix interface engineering would be broadly useful to a series of materials used for fundamental and applied studies on biological cells.
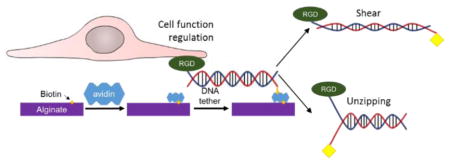
Graphical Abstract
Integrin-binding peptides and proteins have been extensively employed to regulate adhesion of biological cells to materials of interests used as a synthetic extracellular matrix (ECM).1 These cell adhesion ligands are chemically conjugated to or physically associated with molecular building blocks of the synthetic ECM. Previous studies have demonstrated that cell adhesion and functionality could be controlled with the type, density, and spatial organization of cell adhesion ligands.2–4 For instance, the affinity and density of the cell adhesion ligands on a matrix can regulate differentiation levels of various precursor, progenitor, and stem cells.5 The distance between the ligands at the nanometer scale can also change the morphology, focal adhesion formation, and differentiation of cells.6 Additionally, the ligand density can direct cells adhered to a soft matrix to behave like those adhered to a rigid material.4, 7
Furthermore, linkage of the cell adhesion ligands to a matrix may mediate phenotypic activities of cells. For example, the absence of chemical linkages between cell adhesion ligands and a matrix significantly reduces the cell adhesion force and, subsequently, the extent of cell spreading and frequency of cell division.8 Therefore, it has been suggested that tailoring strength of the cell-matrix linkage may further mediate cellular activities in a refined manner.9 Recently, it was demonstrated that deoxyribonucleic acid (DNA) with pre-defined rupture forces can be used as a tension gauge at the molecular level by inserting it between cell adhesion peptides and a matrix. Then, cells can rupture or retain DNA tethers, termed tension gauge tethers (TGTs) depending on the magnitude of the force that cells exert.10
We therefore hypothesized that the strength of a molecular linker that connects a cell adhesion ligand to a material would play important roles in regulating cellular function. To examine this hypothesis, DNA tether coupled with biotin and cell adhesion peptide containing Arg-Gly-Asp (RGD) sequence was used as a model linker that tunes the strength of RGD peptide-matrix bonds. The biotin group was connected to different sites in the DNA tether in order to regulate the rupture force. The resulting DNA tether was linked to hydrogels of alginate conjugated with biotins via specific biotin-avidin bonds. Bone marrow-derived mesenchymal stem cells (MSCs) were cultured on the hydrogels. Then, the role of DNA tether was evaluated by analyzing adhesion, neural differentiation, and proangiogenic factor secretion of MSCs. Taken together, the results of this study would greatly serve to advance the ability to regulate cellular activities by engineering cell-matrix interface.
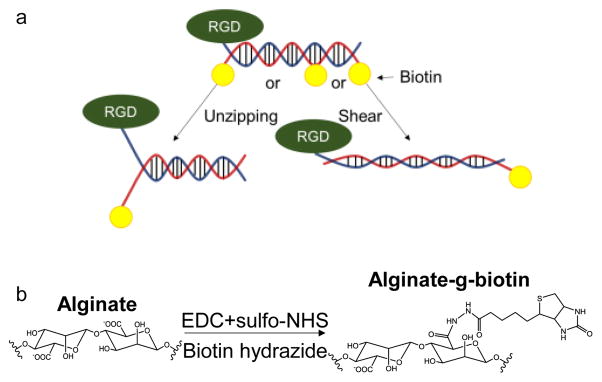
As previously reported, integrin-binding DNA tethers were prepared by annealing two single-stranded DNA (ssDNA) chains (Fig. 1a).10 One ssDNA chain was modified with thiol group at 3′ end and Cy5 at 5′ end, while the complementary DNA strand is modified with biotin at different sites. The thiolated strand of DNA tether was further modified to present RGD peptides via Michael addition reaction between amine ends (or lysine residue) of the peptides and the thiol group. The resulting RGD-ssDNA was annealed with the complementary ssDNA conjugated with biotin at different locations by incubating two molecules in the annealing buffer. A previous study demonstrated that the tension tolerance value (Ttol) of the TGT in the unzipping mode, where biotin is attached to the complementary strand opposite to the RGD peptide, is about 12 pN regardless of the length of DNA tether.11 On the other hand, Ttol of TGTs in shear mode or intermediate mode, where the other end or internal locations of the complementary strand are modified with biotin, varies depending on the site of biotin labeling.12 In this study, we used DNA tethers with Ttol values of 12, 43, and 54 pN, which were estimated by the equation , where 2fc is the rupture force for one single bond, L is the number of DNA base pairs, where Q is the spring constant of the backbone, and R is the spring constant of the bond between base pairs. Empirical data measured using magnetic tweezers showed that fcp = 3.9 pN and X−1 = 6.8.12
Figure 1. Synthesis and characterization of RGD-DNA tether and alginate-g-biotin.
a. Scheme of different force geometry of DNA tethers. b, Reaction scheme to synthesize alginate-g-biotin.
Separately, alginate molecules, a molecular building block of a hydrogel, were chemically coupled with biotin groups using N-hydroxy-succinimide/ethyldimethyl-aminopropyl-carbodiimide (NHS/EDC) chemistry (Fig. 1b). The molar ratio between biotin and alginate units was kept constant at 20%. The conjugation of biotin to alginate was confirmed with peaks in the 1H nuclear magnetic resonance (NMR) spectra. (Fig. S1).
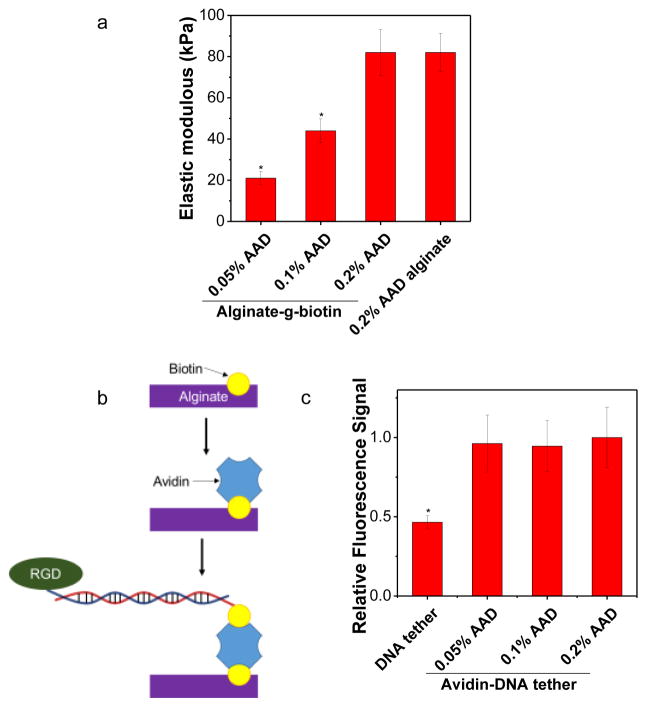
The DNA tether coupled with RGD peptides, termed RGD-DNA tether, was immobilized to alginate conjugated with biotin, termed alginate-g-biotin. The hydrogel was formed by cross-linking alginate-g-biotin molecules with adipic acid dihydrazide (AAD) in aqueous media using NHS/EDC chemistry. By varying the molar ratio between the AAD cross-linker and uronic acids of the alginate-g-biotin from 0.05 to 0.2, the elastic modulus of alginate-g-biotin gel was tuned from 21 to 82 kPa (Fig. 2a). The conjugation of biotin minimally affected gel elastic modulus compared to unconjugated alginate gels (Fig. 2a).
Figure 2. Surface conjugation of RGD-DNA tethers to the alginate-g-biotin hydrogel.
a, Elastic modulus of alginate-g-biotin hydrogels controlled with different molar ratio between the AAD cross-linker and alginate-g-biotin. b. Schematics of the process to immobilize RGD-DNA tether to the alginate-g-biotin gel. c, Fluorescence from the DNA tether coupled to the gel due to biotin-avidin bonds, independent of the molar ratio between the AAD cross-linker and alginate-g-biotin. “DNA tether” represents a condition at which RGD-DNA tether was incubated with the gel in the absence of avidin. The error bars represent the standard deviation of three measurements. * indicates statistical significance of the difference compared to any other group (*p < 0.05).
The resulting gel was incubated with avidin, rinsed, and further incubated with DNA tethers (Fig. 2b). In the presence of avidin, alginate-g-biotin gel incubated with the Cy5-labeled DNA tether presented fluorescence emission upon excitation (Fig. 2c). The fluorescence emission intensity was independent of an elastic modulus of the gel, but the fluorescence emission was significantly lower on the gel incubated with Cy5-labeled DNA tether without avidin, although certain amount of DNA tether was non-specifically bound to the alginate-g-biotin hydrogel. (Fig. 2c).
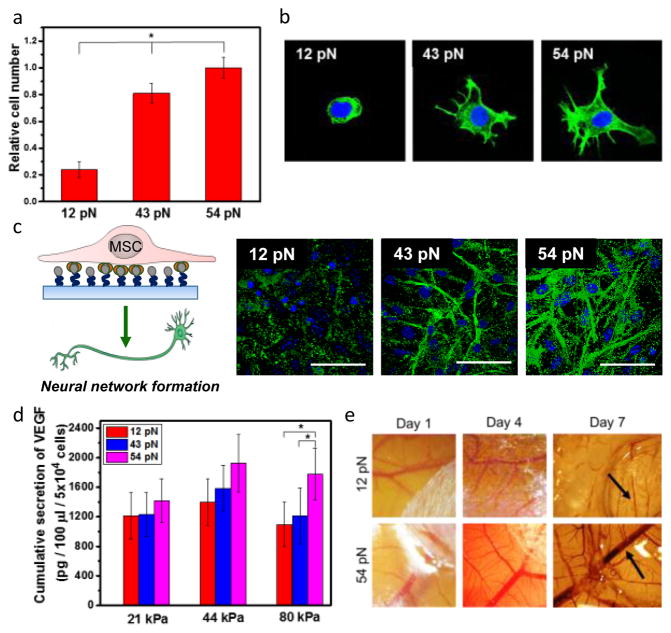
We studied of the effects of Ttol on cellular adhesion by plating MSCs on the gel coated with RGD-DNA tether. Previous studies demonstrated that alginate gel without RGD peptides does not support cellular adhesion.13 More cells adhered to the 44 kPa hydrogel modified with the RGD-DNA tether with higher Ttol values (Fig. 3a). Extent of cell spreading also increased with increasing Ttol from 12 to 54 pN (Fig. 3b).14, 15
Figure 3. Analysis of the role of Ttol of RGD-DNA tether on adhesion and phenotypic activities of MSCs.
a, Quantification of the number of MSCs adhered to a hydrogel modified with RGD-DNA tether with controlled Ttol. b, Fluorescence image of F-actin (green) of MSCs. c, Analysis of neural differentiation of MSCs. Differentiated neural cells were positively stained for MAP-2 (green). d, The total amount of VEGF secreted by MSCs cultured on alginate-g-biotin hydrogels with different elastic moduli modified with RGD-DNA tether with controlled Ttol. e, Microvascular networks of the CAM implanted with the MSC-hydrogel constructs. Arrows indicate blood vessels. The error bars represent the standard deviation of three measurements. * indicates statistical significance of the difference (*p < 0.05).
The hydrogel-TGT system was also used to examine whether the RGD-DNA tether can regulate neural differentiation level of MSCs at a given RGD density and elastic modulus. MSCs adhered to the hydrogel-TGT system were incubated in a media formulated to activate neural differentiation (Fig. 3c). Differentiated neural cells were marked by antibody to microtubule-associated protein 2 (MAP2). Interestingly, the ratio of MAP2-positive cells increased with increasing Ttol value in 44 kPa hydrogel-TGT system (Fig. 3c). On gels functionalized by the RGD-DNA tether with Ttol of 54 and 43 pN, 95% and 88% of the cells expressed MAP2, respectively (Fig. 3c). On the other hand, only 27% of the cells were MAP2 positive on hydrogel-TGT system with Ttol value of 12 pN (Fig. 3c).
We next tested if Ttol values can modulate cellular activities to secrete growth factors (Fig. 3d). Vascular endothelial growth factor (VEGF) is known to stimulate vascular sprouting from existing blood vessels, in a process known as angiogenesis.16 Thus, VEGF enhances outcomes of wound healing and tissue regeneration.17 Interestingly, the 82 kPa hydrogel-TGT system with Ttol value of 54 pN released 1.6-fold higher amount of VEGF than that with Ttol value of 12 pN (Fig. 3d). Additionally, the dependency of VEGF secretion level on Ttol value was mediated by an elastic modulus of the gel (Fig. 3d). In particular, the gel with an elastic modulus of 21 kPa abolished effects of Ttol values on cellular VEGF secretion level while increasing Ttol enhanced the secretion of VEGF in 44 and 82 kPa gels.
The MSC-adhered gel was further implanted on chicken chorioallantoic membrane in order to assess the capability of the RGD-DNA tether to promote vascularization in vivo. The 82 kPa gel immobilized with the RGD-DNA tether with Ttol of 54 pN led to a significant increase of vascular density at the site of implantation as compared with the gel coupled with the RGD-DNA tether with Ttol of 12 pN (Fig. 3e).
Taken together, this study demonstrates that the ability of cell-laden hydrogels to form neural networks and release proangiogenic factor can be modulated by coupling integrin-binding RGD-DNA tether with predefined Ttol values to the gel. We propose that such dependency of cell-laden gel function on Ttol values is mainly attributed to the number of cells adhered to the gel as experimentally confirmed. As expected, cells plated on a gel functionalized by the RGD-DNA with a lower Ttol value, in particular 12 pN, would readily separate ssDNA and fail to adhere to the gel. Conversely, the RGD-DNA with higher Ttol values may resist cell-induced separation, because the ultimate strength is higher than cell-exerting forces. Furthermore, the stronger RGD-DNA tether may be advantageous in activating cellular signaling involved in differentiation and secretion activities, because a small increase of Ttol from 43 to 54 pN elevated cellular differentiation and secretion levels despite the small difference of the number of cells adhered to the gel. These results implicate that RGD-DNA tether may partially modulate cellular traction forces by controlling cell spreading and further cell signaling pathways, independent of gel mechanics and cell adhesion ligand type and density. To the best of our knowledge, this is the first work to demonstrate a capability to regulate cell differentiation and growth factor secretion with ligand-matrix bond strength.
More interestingly, effects of tether strength on cellular secretion activities are mediated by gel stiffness. This result implicates combinatorial effects of matrix stiffness and interfacial mechanics on cellular signaling. This perspective will be more thoroughly studied in near future studies via both theoretical modeling and experimental measurements.
In conclusion, a hydrogel engineered with RGD-DNA tethers of pre-defined strength allows us to regulate adhesion and phenotypic activities of biological cells by modulating both number and, likely, traction force of cells adhered to the gel matrix. We propose that the results of this study would be advantageous in assembling cell-laden gel implants that should remain structurally stable at the site of implantation while providing cells an environment appropriate to stimulate cellular activities desirable for therapies. This method would also allow us to orthogonally regulate cellular activities with bulk gel properties and cell-gel interface. Overall, the results of this study to engineer cell-hydrogel interface mechanics present a new perspective of material design used for culture and therapies of a wide array of stem and progenitor cells.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation (CBET-140349 & STC-EBICS Grant CBET-0939511 to H.K. and PHY-1430124 to T.H.), partially National Institute of Health (1R01 HL109192 to H.K.), National Research Foundation of Korea (NRF) funded by the Korea Ministry of Science, ICT and Future Planning (Engineering Research Center 2014R1A5A1009799 to J. L.), and Basic Science Research Program through NRF funded by the Ministry of Education, Science and Technology (2015R1A6A3A03015834 to J. P.).
Notes and references
- 1.Lutolf MP, Hubbell JA. Nat Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 2.Harbers GM, Healy KE. J Biomed Mater Res A. 2005;75a:855–869. doi: 10.1002/jbm.a.30482. [DOI] [PubMed] [Google Scholar]
- 3.Maheshwari G, Brown G, Lauffenburger DA, Wells A, Griffith LG. J Cell Sci. 2000;113:1677–1686. doi: 10.1242/jcs.113.10.1677. [DOI] [PubMed] [Google Scholar]
- 4.Roein-Peikar M, Xu Q, Wang X, Ha T. Phys Rev X. 2015 doi: 10.1016/j.bpj.2015.10.029. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kilian KA, Mrksich M. Angewandte Chemie-International Edition. 2012;51:4891–4895. doi: 10.1002/anie.201108746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frith JE, Mills RJ, Cooper-White JJ. J Cell Sci. 2012;125:317–327. doi: 10.1242/jcs.087916. [DOI] [PubMed] [Google Scholar]
- 7.Trappmann B, Gautrot JE, Connelly JT, Strange DGT, Li Y, Oyen ML, Stuart MAC, Boehm H, Li BJ, Vogel V, Spatz JP, Watt FM, Huck WTS. Nat Mater. 2012;11:642–649. doi: 10.1038/nmat3339. [DOI] [PubMed] [Google Scholar]
- 8.Cha C, Jeong JH, Tang X, Zill AT, Prakash YS, Zimmerman SC, Saif TA, Kong H. Bioconjug Chem. 2011;22:2377–2382. doi: 10.1021/bc200339s. [DOI] [PubMed] [Google Scholar]
- 9.Huebsch N, Arany PR, Mao AS, Shvartsman D, Ali OA, Bencherif SA, Rivera-Feliciano J, Mooney DJ. Nat Mater. 2010;9:518–526. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang XF, Ha T. Science. 2013;340:991–994. doi: 10.1126/science.1231041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cocco S, Monasson R, Marko JF. Proc Natl Acad Sci U S A. 2001;98:8608–8613. doi: 10.1073/pnas.151257598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatch K, Danilowicz C, Coljee V, Prentiss M. Phys Rev E. 2008:78. doi: 10.1103/PhysRevE.78.011920. [DOI] [PubMed] [Google Scholar]
- 13.Rowley JA, Madlambayan G, Mooney DJ. Biomaterials. 1999;20:45–53. doi: 10.1016/s0142-9612(98)00107-0. [DOI] [PubMed] [Google Scholar]
- 14.Gallant ND, Michael KE, Garcia AJ. Mol Biol Cell. 2005;16:4329–4340. doi: 10.1091/mbc.E05-02-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chowdhury F, Li IT, Leslie BJ, Doganay S, Singh R, Wang X, Seong J, Lee SH, Park S, Wang N, Ha T. Integr Biol. 2015;7:1265–1271. doi: 10.1039/c5ib00080g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coultas L, Chawengsaksophak K, Rossant J. Nature. 2005;438:937–945. doi: 10.1038/nature04479. [DOI] [PubMed] [Google Scholar]
- 17.Yang F, Cho SW, Son SM, Bogatyrev SR, Singh D, Green JJ, Mei Y, Park S, Bhang SH, Kim BS, Langer R, Anderson DG. Proc Natl Acad Sci U S A. 2010;107:3317–3322. doi: 10.1073/pnas.0905432106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.