Abstract
Autism Spectrum Disorders (ASDs) present unique challenges in the fields of genetics and neurobiology because of the clinical and molecular heterogeneity underlying these disorders. Genetic mutations found in ASD patients provide opportunities to dissect the molecular and circuit mechanisms underlying autistic behaviors using animal models. Ongoing studies of genetically modified models have offered critical insight into possible common mechanisms arising from different mutations, but links between molecular abnormalities and behavioral phenotypes remain elusive. The challenges encountered in modeling autism in mice demand a new analytic paradigm that integrates behavioral analysis with circuit-level analysis in genetically modified models with strong construct validity.
Keywords: autism mouse models, Mecp2, Fmr1, Ube3a, Pten, Shank3
Autism Spectrum Disorders (ASDs) are a group of conditions primarily characterized by impairments in social communication and engagement in restricted, repetitive behaviors (American Psychiatric Association, 2013). Common comorbidities include intellectual disability, epilepsy, anxiety, sleep disturbances, abnormal sensory processing, motor impairments, and gastrointestinal complaints (Argyropoulos et al., 2013). ASDs are heterogeneous in nature, as patients display a wide range of symptom severity and prognosis (Howlin et al., 2004; Lord et al., 2000), which is mirrored by hundreds of identified causal or potentially causal genetic variants (Persico and Napolioni, 2013; Willsey and State, 2015). Unfortunately, most genetic mutations are rare or private (i.e. observed only in a single family). Both the phenotypic and genetic heterogeneity present significant obstacles to understanding the disorders and attempts to associate phenotypic severity with genetic differences have had mixed results (Chang et al., 2015; Chaste et al., 2014). While genetics undoubtedly play a substantial role in ASD pathophysiology, the inexplicable phenotypic heterogeneity and incomplete concordance rates between monozygotic twins (Hallmayer et al., 2011), suggest that non-genetic factors may also contribute to the etiology.
A recent survey indicates that 1 in 68 children in the United States are diagnosed with ASDs, a drastic increase from previous estimates over the last few decades (Centers for Disease Control and Prevention, 2014). While there is considerable debate regarding the degree to which the increase in prevalence can be explained by broadened diagnostic criteria (King and Bearman 2009), increased awareness (Liu et al., 2010), or changes in environmental factors (Nevison, 2014), there is nevertheless an ever-increasing urgency to determine the underlying pathophysiology and develop safe, cost-effective interventions for ASDs to improve patient outcomes. Regardless of the source of the rising prevalence, it is an issue of great public health concern, as the lifetime cost of ASD-related care ranges from approximately $1.4 million to $2.2 million per individual (Buescher et al., 2014).
Studies in human clinical populations have been and continue to be critical for understanding the genetic and non-genetic contributions to ASDs (Willsey and State, 2015). However, animal models are needed determine the mechanisms leading to abnormal functioning. Although human brain imaging techniques have identified regions and circuits involved in the disorders (e.g. Karten and Hirsch, 2014), animal models provide opportunities for direct manipulation of these brain regions and circuits to test their precise functions. In current clinical practice, ASDs are defined by behavioral symptoms that are uniquely human and there is no singular neuropathological hallmark identified so far that is pathognomonic, so it is challenging to determine the validity of an animal model of autism. Nevertheless, recent successes in identifying genes implicated in ASDs have paved the way to explore the neurobiology underlying the disorders using animal models.
1. Genetic mutations implicated in ASDs
Substantial progress has been made to understand the genetic causes of ASDs. Genes implicated in syndromic forms of autism were first identified in the 1990s. Subsequent genomic copy number variant (CNV) analysis in the 2000s generated a list of rare but highly penetrant CNVs associated with ASDs. Pathogenic CNVs are estimated to account for ~10% of non-syndromic ASDs (Devlin and Scherer, 2012). Most recently, whole exome and whole genome sequencing techniques have been utilized to identify rare de novo and inherited sequence variants in hundreds of genes from ASD subjects. Despite the inability to establish a causal role for the majority of these sequence variants, a subset of new genes have emerged as strongly causal because de novo loss-of-function and likely-gene-disrupting mutations are found in multiple affected patients and are absent in a large number of controls (Table 1). In other cases, mutations that likely disrupt protein function are found in genes that are implicated in other neuropsychiatric disorders. Functional annotation of these genes immediately suggests the following molecular features: 1) neuronal ion channels and receptors; 2) synapse-related cytoskeleton and scaffolding proteins; 3) epigenetic and transcriptional regulators; 4) post-translational protein modifiers and regulators. An important question is whether or not the mutations in these genes share a common molecular and/or circuit-level mechanism underlying the pathophysiology of ASDs. Modeling these mutations in animal models is essential to address this question.
Table 1.
List of genes with strong evidence for syndromic and non-syndromic ASDs
| Syndromic ASDs | Non-Syndromic ASDs | ||
|---|---|---|---|
| Gene Name | Locus | Gene Name | Locus |
| ADNP | 20q13.13 | ASH1L | 1q22 |
| ADSL | 22q13 | ASXL3 | 18q11 |
| AHI1 | 6q23.3 | CACNA1H | 16p13.3 |
| ALDH5A1 | 6p22 | CACNA2D3 | 3p21.1 |
| ANKRD11 | 16q24.3 | CHD2 | 15q26 |
| ARID1B | 6q25.1 | CHD8 | 14q11.2 |
| ARX | Xp22. | CNTN4 | 3p26 |
| ASXL3 | 18q11 | CNTNAP2 | 7q35 |
| CACNA1C | 12p13.3 | CUL3 | 2q36.2 |
| CDKL5 | Xp22 | DEAF1 | 11p15.5 |
| CHD2 | 15q26 | DSCAM | 21q22.2 |
| CHD7 | 8q12.2 | DYRK1A | 21q22.13 |
| CNTNAP2 | 7q35-q36 | GABRB3 | 15q12 |
| DHCR7 | 11q13 | GRIN2B | 12p12 |
| DYRK1A | 21q22.13 | GRIP1 | 12q14.3 |
| EHMT1 | 9q34.3 | KATNAL2 | 18q21.1 |
| FMR1 | Xq27.3 | KDM5B | 1q32.1 |
| HDAC4 | 2q37.3 | KMT2A | 11q23 |
| KMT2A | 11q23 | KMT2C | 7q36.1 |
| MECP2 | Xq28 | MED13L | 12q24.21 |
| NIPBL | 5p13.2 | MET | 7q31 |
| PTEN | 10q23.3 | MSNP1AS | 5p14.1 |
| RAI1 | 17p11.2 | MYT1L | 2p25.3 |
| SCN1A | 2q24.3 | NRXN1 | 2p16.3 |
| SYNGAP1 | 6p21.3 | POGZ | 1q21.3 |
| TSC1 | 9q34 | PTCHD1 | Xp22.11 |
| TSC2 | 16p13.3 | RELN | 7q22 |
| UBE3A | 15q11.2 | SCN2A | 2q23 |
| VPS13B | 8q22.2 | SETD5 | 3p25.3 |
| SHANK2 | 11q13.3 | ||
| SHANK3 | 22q13.3 | ||
| SUV420H1 | 11q13.2 | ||
| SYNGAP1 | 6p21.3 | ||
| TBR1 | 2q24 | ||
2. What constitutes a valid animal model for ASDs?
Animal models of psychiatric disorders have classically been evaluated on three criteria, which were first applied to mouse models of depression: construct, face, and predictive validity (Willner, 1984). Ever since these criteria were articulated, they have been interpreted in a variety of ways (Belzung and Lemoine, 2011), making it worth elaborating on their precise meanings and their relationships to animal models of ASDs.
Construct validity, for our purposes, refers to the rationale behind the creation of the model and its ability to recapitulate the etiology of the disorder. For instance, a model of ASD with high construct validity mimics a genetic mutation that has been observed in affected human individuals, or at least has the same molecular consequences; this can perhaps more accurately be described as pathogenic validity (Belzung and Lemoine, 2011). The determination of construct validity is difficult for genes that have a complex structure at transcriptional level.
Face validity refers to the model’s resemblance to the clinical features of the disorder, both in terms of behavioral symptoms and physiological biomarkers; this definition of face validity can be separated into ethological validity and biomarker validity (Belzung and Lemoine, 2011). Currently, ASD is diagnosed based solely on behavioral assessments, limiting discussion of face validity to the behavioral realm.
Finally, predictive validity refers to the model’s ability to determine the effectiveness that interventions will have on a clinical population. A model of ASD with high predictive validity will respond (e.g. with a reduction in repetitive behaviors) to pharmaceuticals in similar manner to the patients they are intended to model. This is particularly challenging in autism models because there are no established treatment paradigms for the core symptoms in human autism clinics currently. Conceptually, interventions are not necessarily limited to drugs, but it is more difficult to assess the translational value of non-pharmacological therapies, such as environmental enrichment. Predictive validity can also refer to the penetrance of specific phenotypes, given a specific genetic mutation or other trigger (Belzung and Lemoine, 2011), but these data are seldom reported.
3. Overview of Monogenic Mouse Models of ASDs
By far the most common animals used to model ASDs are mice because of the well-established techniques to manipulate their genome and study brain function at several levels of analysis. As mammals, mice are genetically and biologically similar to humans, but their rapid reproduction and accelerated development allow for the testing of large numbers of animals at a relatively low cost. Various assays have been developed to test mice for behaviors that resemble the core features of autism (Silverman et al., 2010b), allowing researchers to determine the face validity of their models, but the translatability of these assays to humans is unknown.
Much of what we know about the underlying mechanism of ASDs comes from detailed analysis of mouse models of syndromic autism, for which a genetic cause is clearly defined. Unfortunately, while patients with these disorders frequently meet the diagnostic criteria for ASDs, autism is not always present and there are a variety of other symptoms related to abnormal brain function, making the results of these studies not necessarily generalizable to all ASDs. Nevertheless, careful review of common findings in these models contributes to a basic understanding of the pathophysiology contributing to autistic behaviors. The behavioral phenotypes in some of these ASD models have recently been reviewed in a separate paper (Bey and Jiang, 2014), so this review will focus on biochemical, cellular, and synaptic findings from select mouse models of ASDs induced by a mutation in a single gene. It should be noted that there are many models that we excluded, including inbred strains, chromosomal CNVs, and environmentally induced models, due to limited space and the fact that the biological mechanisms in these models are currently less understood. The models we selected to review are grouped by the reported function of the disrupted protein and the findings are summarized in Table 2.
Table 2.
Summary of Cellular, Molecular, and Electrophysiological Findings From Monogenic Mouse Models of ASDs
| Gene | Brain Regions, Cell Types, and Developmental Periods Implicated Through Conditional Mutants |
Molecular Changes | Cellular and Spine Morphology |
Electrophysiology |
|---|---|---|---|---|
| Mecp2 | Excitatory forebrain neurons (CamKII-Cre93) GABAergic neurons (Viaat-Cre) Fully developed mice (CAGCre/ Esr1) |
↓BDNF, ↓RNA splicing |
Mecp2tm1.1Bird CTX: ↓spine density, ↓dendritic complexity Mecp2tm1.1JaeCTX: ↓spine density Mecp2tm1.1Jae CA1: ↓cell size Mecp2tm1.1Jae p7 CA1: ↓spine density Mecp2tm1.1Jae SCX: ↑spine stability (p25–p26) Mecp2tm1.1Bird PNC: ↓glutamatergic synapses |
Mecp2tm1.1Bird CA1: ↓LTP, ↓NMDA LTD Mecp2tm1.1Bird PNC: ↓mEPSC freq, ↓eEPSC amp Mecp2tm1.1Jae CA1: ↓LTP, ↓NMDA LTD Mecp2tm1.1Jae SCX: ↓mEPSC amp Mecp2tm1Hzo CA1: ↓LTP, ↓NMDA LTD Mecp2tm1Hzo SCX: ↓LTP Mecp2tm1Hzo PMC: ↓LTP |
| Fmr1 | None reported (no behavioral phenotype in Nse-Cre mediated deletion) | ↑mGluR5 signaling, ↑mRNA translation, ↓NLGN1, ↑Shank1 |
SCX: ↑spine density, ↓spine stability (p10–p12), ↓mature spines CA1: ↓mature spines |
SCX: ↓LTP CA1: ↓LTP, ↑mGluR LTD p14 CA1: ↓AMPA/NMDA, ↑NMDA LTP PC: ↑mGluR LTD |
| Tsc1/Tsc2 | Purkinje cells (L7-Cre and Pcp2-Cre) Excitatory forebrain neurons (CaMKIIα-Cre) Serotonergic neurons (Slc6a4-Cre) |
↑mTORC1 signaling, ↑activated Rheb |
Tsc1/Tsc2 PNC KD: ↓spine density, ↑spine size, ↑soma size Tsc1fl/fl + viral Cre CA1: ↑soma size Tsc2+/− CTX: ↑spine density, ↑spine stability |
Tsc1 OT KD: ↑mEPSC amp, ↑AMPA/NMDA Tsc1fl/fl + viral Cre CA1: ↓mGluR LTD, ↓mEPSC freq, ↑AMPA, ↑NMDA Tsc1fl/fl + viral Cre PNC GABAergic: ↓eIPSC amp, ↓mIPSC amp Tsc2+/− CA1: ↓mGluR LTD |
| Pten | Cortical and hippocampal neurons (Nse-Cre) Cerebellar granule cells and DG hippocampal neurons (Gfap-Cre) |
↑Akt signaling, ↑mTORC1 signaling, ↓mGluR5, ↑FMRP, ↑p-FMRP |
DG: ↑cell size, ↑axonal processes, ↑ectopic projections, ↑presynaptic vesicles, ↑spine density, ↑spine size, ↑mature spines CTX: ↑cell size, ↑presynaptic vesicles, ↑PSD size CGC: ↑cell size, ↑presynaptic vesicles, ↑spine density Ptenm3m4: ↑cell size |
Pten+/−CA1: ↓LTP, ↓NMDA LTD Ptenfl/fl + CamKIIα-Cre CA1: ↓LTP, ↓NMDA LTD Ptenfl/fl + GFAP-Cre CA1: ↓NMDA LTD Ptenfl/fl + Nse-Cre DG: ↓mGluR LTD |
| Ube3a | Embryonic/early postnatal development (CAG-Cre/Esr1) |
m−/p+: ↑Arc, ↓Mecp2 function, ↓BDNF signaling 2xTg: ↓Arc |
m−/p+ CA1: ↓spine density m−/p+ CTX: ↓apical dendrite length m−/p+ PVC: ↓spine density |
m−/p+ CA1: ↓LTP, ↑mGluR LTD, ↓AMPA/NMDA m−/p+ PVC: ↓LTP, ↓NMDA LTD m−/p+ DMS: ↓mEPSC amp, ↓mEPSC freq 2xTg BC: ↓eEPSC amp, ↓sEPSC amp, ↓sEPSC freq, ↓sIPSC amp, ↓mEPSC amp, ↓mEPSC freq |
| Shank1 | None reported | HS: ↓Homer, ↓SAPAP | CA1: ↓spine density, ↓spine length, ↓PSD thickness | CA1:↓excitatory synaptic strength, ↓mEPSC freq |
| Shank2 | None reported |
Δe7 HS: ↑NMDA Δe7 SS: ↑AMPA, ↑Shank3 Δe6–7 WB: ↑NMDA, ↓NMDA signaling |
Δe7 CA1: ↓spine density |
Δe7 CA1: ↓excitatory synaptic strength, ↓mEPSC frequency, ↓AMPA/NMDA, ↑LTP Δe6–7 CA1: ↑AMPA/NMDA, ↓LTP, ↓NMDA LTD |
| Shank3 | None reported |
Δe4–9J HPSD: ↓Homer1b/c, ↓GluA1, ↓GluN2A, ↓SAPAP1 Δe4–9P SS: ↓Homer1b/c, ↓PSD-95, ↓GluA2, ↓GluA3 Δe11 SS: ↑Shank2, ↑GluN2B Δe13–16 SPSD: ↓SAPAP3, ↓Homer, ↓PSD-93, ↓GluA2, ↓GluN2A, ↓GluN2B Δe21 HS: ↑mGluR5 Δe21 PFC: ↓Rac1/PAK signaling, ↑active cofilin, ↓F-actin |
Δe13–16 DLS: ↓spine density, ↑dendritic complexity, ↓PSD thickness Δe4–9J 4 week CA1:↓spine density, ↑spine length Δe4–9J 10 week CA1: ↓spine length |
Δe9 CA1: ↑mIPSC freq Δe9 PFC: ↓mIPSC freq Δe4–9J CA1: ↓LTP Δe4–9P CA1: ↓LTP Δe4–9P DLS: ↑AMPA/NMDA Δe4–9B CA1: ↓LTP, ↓excitatory synaptic strength, ↓mEPSC amp, ↑mEPSC freq Δe13–16 DLS: ↓excitatory synaptic strength, ↓mEPSC freq, ↓mEPSC amp Δe21 CA1: ↓LTP, ↓excitatory synaptic strength, ↑AMPA/NMDA, ↓mEPSC freq Δe21 PFC: ↑AMPA/NMDA 21insG CA1: : ↓LTD, ↓excitatory synaptic strength, ↑AMPA/NMDA, ↓mEPSC freq |
| Nrxn family | Dominant negative effect in forebrain excitatory neurons of fully developed mice (CaMKIIα-tTa) | ΔNrxn2α HL: ↓Munc18-1 | ΔCntnap4 GABAergic synapses: ↓PSD length, ↑cleft width |
ΔNrxn1α CA1: ↑mEPSC freq, ↓excitatory synaptic strength DN-Nrxn1β SCX: ↓mEPSC freq, ↓mIPSC freq ΔCntnap4 NAc: ↑DA release ΔCntnap4 SCX: ↓sIPSC freq, ↓sIPSC amp, ↓sIPSC kinetics |
| Nlgn family | D1-MSNs (D1-Cre) of the NAc (localized injection of AAV-Cre) |
ΔNlgn2 WB: ↓CSP, ↓Liprin, ↓Munc-18, ↓Nrxnα, ↓Nrxnβ, ↑Nlgn3, ↑Synapsin1a ΔNlgn2 CA1: ↓VGAT R451C Nlgn3 WB: ↑VGAT, ↑Gephyrin R451C Nlgn3 HL: ↓PSD95, ↓SAP102, ↓NR2A, ↓NR2B |
sparse Nlgn2 KD CTX: ↓synaptic density R451C Nlgn3 KI CA1: ↑dendritic complexity, ↓synaptic terminal size, ↓synaptic vesicles, ↓spine size R451C Nlgn3 KI AFC: ↑spine turnover |
ΔNlgn1 CA1: ↓LTP, ↓AMPA EPSCs, ↓NMDA EPSCs, ↑AMPA/NMDA ΔNlgn1 AMG TI: ↓LTP ΔNlgn4: ↓excitatory network excitability, ↓inhibitory network excitability, ↓E/I R451C Nlgn3 KI SCX: ↑mIPSC freq, ↑eIPSC amp, R451C Nlgn3 KI BC: ↓eIPSC amp R451C Nlgn3 KI CA3: ↑GDPs freq, ↑mIPSC freq R451C Nlgn3 KI CA1: ↑LTP, ↓AMPA/NMDA R451C Nlgn3 KI CCK: ↑eIPSC amp, ↓tonic ECB ΔNlgn3 CCK: ↑eIPSC amp, ↓tonic ECB ΔNlgn3 D1-MSNs NAc: ↓mIPSC freq, ↑E/I |
Legend:
↓, decreased;
↑, increased;
Δ, deletion;
Δ4–9B, Shank3 model created by Bozdagi et al., 2010;
Δ4–9J, Shank3 model created by Wang et al., 2011;
Δ4–9P, Shank3 model created by Jaramillo et al., 2015;
AAV, adeno-associated virus
AFC, anterior frontal cortex;
AMG TI, thalamic inputs to the amygdala;
amp, amplitude;
AMPA, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid;
AMPA/NMDA, ratio of AMPA-induced to NMDA-induced current
BC, barrel cortex;
BDNF, brain-derived neurotrophic factor;
CA1, region of the hippocampus;
CA3, region of the hippocampus;
CCK, cholecystokinin basket cells
CGC, cerebellar granule cells;
CTX, cortex;
DA, dopamine;
DG, dentage gyrus;
DLS, dorsolateral striatum;
DMS, dorsomedial striatum;
ECB, endocannabinoid
eEPSC, evoked excitatory postsynaptic current;
eIPSC, evoked inhibitory postsynaptic current;
E/I, excitation-inhibition ratio;
GDPs, giant depolarizing potentials
freq, frequency;
HL, hippocampal lysate;
HPSD, hippocampal postsynaptic density fraction;
HS, hippocampal synaptosomal fraction;
KD, knockdown;
KI, knock-in;
- NMDA LTD, NMDA-dependent LTD
- mGluR5 LTD, mGluR5-dependent LTD
LTP, long-term potentiation;
mEPSC, miniature excitatory postsynaptic current;
mGluR5, metabotropic glutamate receptor;
mIPSC, miniature inhibitory postsynaptic current;
NAc, nucleus accumbens;
NMDA, N-methyl-D-asparate;
OT, organotypic;
p7, postnatal day 7;
p14, postnatal day 14;
PC, Purkinje cells;
PMC, primary motor cortex;
PNC, primary neuron culture;
PVC, primary visual cortex;
sIPSC, spontaneous inhibitory postsynaptic current
SCX, somatosensory cortex;
SPSD, striatal postsynaptic density fraction;
SS, striatal synaptosomal fraction;
WB, whole brain;
3.1 Epigenetic and Transcriptional Regulator: Mecp2 (Rett syndrome)
A number of genes classified as transcriptional or epigenetic regulators have been implicated in ASDs (Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium, 2015). These genetic findings support a molecular mechanism involving transcriptional regulation underlying the pathogenesis of ASDs. One of the best characterized genes in this category is MECP2, a gene encoding a methylated DNA-binding protein (Amir et al., 1999; reviewed in Lyst and Bird, 2015). Although the protein was characterized as a transcriptional repressor when it was first identified (Nan et al., 1997), data from more recent studies indicate that MeCP2 acts as a global transcriptional regulator involved in both the suppression and activation of targeted genes (Chahrour et al., 2008; Ben-Shachar et al., 2009), as well as a regulator of RNA splicing (Young et al., 2005; Maunakea et al. 2013).
More than 10 distinct lines of Mecp2 mutant mice have been produced. Mice with the deletion of exon 3 (Mecp2tm1.1Jae; Chen et al., 2001), deletion of exons 3 and 4 (Mecp2tm1.1Bird; Guy et al., 2001), and a 308× point mutation (Mecp2tm1Hzo; Shahbazian et al., 2002) are among the best characterized. The complete loss of both MeCP2 protein isoforms were revealed in hemizygous males (Mecp2−/y) of Mecp2tm1.1Jae and Mecp2tm1.1Bird mutants, whereas the 308× mutation in Mecp2tm1Hzo mice introduces a premature stop codon that leads to truncation of the MeCP2 protein (Shahbazian et al., 2002). It should be noted that female heterozygous mice (Mecp2−/+) are the model with best construct validity, as Rett syndrome primarily affects females and is lethal in males in most cases. However, most studies use hemizygous male mice because they develop more severe phenotypes. An important question that remains is why humans are more sensitive than rodents to MeCP2 mutations.
Cellular and molecular abnormalities have been identified in Mecp2 mutant mice that likely contribute to the ASD-like phenotypes. Both Mecp2tm1.1Bird and Mecp2tm1.1Jae mice have cortical neurons with decreased spine density (Belichenko et al., 2009), and Mecp2tm1.1Bird mice have decreased dendritic complexity (Fukuda et al., 2005). Similar results have been reported in Mecp2tm1.1Jae hippocampus (Smrt et al., 2007), but this varies across development: spine density is decreased at postnatal day seven but returns to wild type levels by postnatal day 15 (Chapleau et al., 2012). Moreover, adult Mecp2tm1Hzo mice have normal spine density and dendritic complexity in both cortex and hippocampus (Moretti et al., 2006). More recently, it was demonstrated that neurons in somatosensory cortex of Mecp2tm1.1Jae mice are more stable than controls at postnatal day 25–26 (P25–P26), as assessed by in vivo two-photon imaging (Landi et al., 2011).
The electrophysiological consequences of MeCP2 dysfunction have also been examined. Reduced long-term potentiation (LTP) and long-term depression (LTD) have been reported in hippocampal CA1 synapses in both Mecp2tm1.1Bird and Mecp2tm1.1Jae mice (Asaka et al., 2006), as well as Mecp2tm1Hzo mice (Moretti et al., 2006). Recordings from sensory and motor cortex from Mecp2tm1Hzo mice also revealed reduced LTP (Moretti et al., 2006). Whole-cell patch-clamp recordings from pyramidal neurons in brain slices of somatosensory cortex from Mecp2tm1.1Jae mice revealed reduced spontaneous activity due to a significant reduction in the amplitude of miniature excitatory postsynaptic currents (mEPSCs) and an insignificant increase in the amplitude of miniature inhibitory postsynaptic currents (mIPSCs; Dani et al., 2005). Similarly decreased frequency of mEPSCs and amplitude of evoked EPSCs have also been observed in cultured neurons from Mecp2tm1.1Bird mice and are associated with decreased numbers of glutamatergic synapses on individual neurons (Chao et al., 2007).
How the deficiency of MeCP2 results in abnormal functioning of neuronal synapses as well as ASD-like phenotypes is still not fully understood. Hundreds of genes are dysregulated in both directions in brains of Mecp2 mutant mice. These genes are promising candidates to dissect the molecular mechanism underlying the deficiency of Mecp2 in neurons or synapses. One such candidate is BDNF, a neurotrophic factor that is down-regulated in MeCP2-deficient neurons. Not only do decreases in BDNF expression correlate with symptom severity (Chang et al., 2006), but treatments that increase BDNF also improve symptoms (Ogier et al., 2007; Kline et al., 2010). Additionally, a non-pharmacological intervention – environmental enrichment, which improves synaptic and behavioral phenotypes in Mecp2 mutant mice – is associated with increased expression of BDNF (Lonetti et al., 2010).
A number of research groups have manipulated the expression of MeCP2 in mouse brains temporally and spatially using various Cre lines to determine potential brain regions, cell types, and developmental time periods involved in Rett syndrome pathogenesis. Conditional knockout (KO) of Mecp2 using either the brain-specific Nestin-Cre line in which the recombinase is activated during embryonic development or the forebrain-specific CamK-Cre93 line in which the recombinase is activated postnatally recapitulate the phenotypes observed in the conventional Mecp2 mutant mice (Chen et al., 2001). A subsequent study found that Mecp2 deficiency in excitatory neurons of the forebrain using CamKII-Cre93 mice also produce ASD-like features (Gemelli et al., 2006). However, other studies provide evidence that loss of MeCP2 specifically in GABAergic neurons (Viaat-Cre) is sufficient (Chao et al., 2010). Thus, loss of MeCP2 in a subset of either excitatory or inhibitory neurons can lead to ASD-like features. It remains unclear whether the deficiency of MeCP2 in these excitatory or inhibitory neurons disrupts the same circuits. Using mice engineered with a stop codon surrounded by loxP sites crossed with a ubiquitously-expressed inducible Cre line (CAG-Cre/Esr1), researchers have provided evidence that Rett syndrome phenotypes can be induced in fully developed mice (McGraw et al., 2011) and rescued through gene reactivation (Guy et al., 2007; Robinson et al., 2012; Lang et al., 2014), suggesting that ASDs may arise from persistent abnormal neural functioning, rather than disrupted development. However, this hypothesis needs to be tested in other models of ASD, as the phenotypes reported in these studies are not necessarily generalizable.
3.2 Post-Transcriptional Protein Modifiers or Regulators: Fmr1, Tsc1/2, Ube3a, and Pten
3.2.1 Fmr1 (Fragile X syndrome)
Fragile X syndrome (FXS), primarily affecting males, also has phenotypic overlap with ASDs and is caused by extended CGG trinucleotide repeats in the 5′ untranslated region of FMR1 (Verkerk et al., 1991; Pieretti et al., 1991). Such mutations lead to hypermethylation of the FMR1 promoter, which transcriptionally silences the fragile X mental retardation protein (FMRP), an RNA binding protein involved in suppressing activity-dependent translation of synaptic proteins, many of which have been independently implicated in ASDs (Darnell et al., 2011). Under normal conditions, FMRP is dephosphorylated after activation of metabotropic glutamate receptors (mGluRs), leading to de-repression of local translation (reviewed in Bassell and Warren, 2008). Extended CGG repeats in Fmr1 in mice do not recapitulate the hypermethylation and transcriptional silencing of FMRP that is characteristic of human fragile X (Brouwer et al., 2007). However, researchers have mimicked the molecular consequences of the human mutation by deleting exon 5 (The Dutch-Belgian Fragile X Consortium, 1994) or exon 1 (Mientjes et al., 2006) of Fmr1. Almost all data related to fragile X syndrome mouse models in the literature have been generated from the exon 5 deletion mutant mice, but whereas both models have complete loss of FMRP expression, only the model produced by deleting exon 1 has no detectable Fmr1 mRNA and is conducive to conditional knockout studies.
Many studies have reported differences in dendritic spines in Fmr1 knockouts. The first study reported that the cortical neurons have increased numbers of spines in adult mice, most of which morphologically resemble immature spines (Comery et al., 1997). Since then, there have been conflicting reports regarding spine density in various brain regions and periods of development, but the immature morphology of spines is a relatively consistent finding (reviewed in He and Portera-Cailliau, 2013). Using in vivo two-photon imaging, researchers recently have found increased turnover in dendritic spines from Fmr1 knockout mice in layer 2/3 neurons in the barrel cortex at P10–P12 (Cruz-Martin et al., 2010) and in layer 5 neurons in primary somatosensory cortex at P20–P30 (Pan et al., 2010). Together, these findings suggest that FMRP is required for the development of mature, stable dendritic spines.
In addition to dendritic spine morphology and stability, synaptic plasticity has been well characterized in FXS mouse models (reviewed in Chung et al., 2012; Sidorov et al., 2013). Initial studies reported no change in LTP (Godfraind et al., 1996; Paradee et al., 1999), but a significant enhancement in mGluR-dependent LTD in the hippocampus (Huber et al., 2002). More recently, impaired LTP was reported in hippocampal CA1 synapses using threshold-level stimulation parameters, which are thought to more closely mimic in vivo conditions (Lauterborn et al., 2007; Lee et al., 2011). Furthermore, a transient decrease (at postnatal day 14) in the AMPA/NMDA receptor ratio and an increase in NMDA-dependent LTP in hippocampal neurons has been observed (Pilpel et al., 2009). Reduced LTP has also been reported in somatosensory cortical synapses (Li et al., 2002), and increased mGluR-dependent LTD has been reported in cerebellar Purkinje cells (Koekkoek et al., 2005). Therefore, like spine morphology, electrophysiological consequences of Fmr1 knockout vary across brain regions and development, but increased mGluR-dependent LTD seems to be a relatively consistent finding.
The enhanced mGluR-mediated LTD observed in Fmr1 knockouts (Huber et al., 2002) paved the way for the mGluR theory for FXS pathogenesis, which posits that FXS symptoms are caused by mGluR5 signaling leading to deregulated mRNA translation in the absence of FMRP (Bear et al., 2004). This theory has been supported by the effectiveness of genetic depletion of mGluR5 (Dölen et al., 2007) and mGluR5 negative allosteric modulators (e.g. CTEP; Michalon et al., 2012) in alleviating behavioral and synaptic abnormalities in Fmr1 knockout mice. While neither of these studies included analysis of repetitive behaviors or social interactions, negative allosteric modulation of mGluR5 decreased repetitive behaviors and increased sociability of a different mouse model of ASD: the BTBR inbred strain (Silverman et al., 2012). However, the mGluR5 signaling pathway is not the only molecular cascade that results in FMRP de-repression and subsequent dysregulated translation. For instance, the BDNF/TrkB signaling pathway converges with the mGluR5 pathway to initiate translation (reviewed in Castrén and Castrén, 2014; see Figure 1). It remains to be determined whether dysregulation of the BDNF/TrkB signaling pathway, the mGluR5 pathway, or another pathway entirely contributes to the ASD-like behaviors in FXS mouse models. There is some evidence which suggests that social deficits in mice lacking FMRP are due to decreased neuroligin1 expression and can be rescued by overexpressing neuroligin1 (Dahlhaus and El-Husseini, 2010). Additionally, mice lacking FMRP exhibit elevated postsynaptic Shank1 levels, along with a number of other synaptic proteins (Schütt et al., 2009), but more work needs to be done to confirm these results and determine whether or how these changes may lead to changes in behavior.
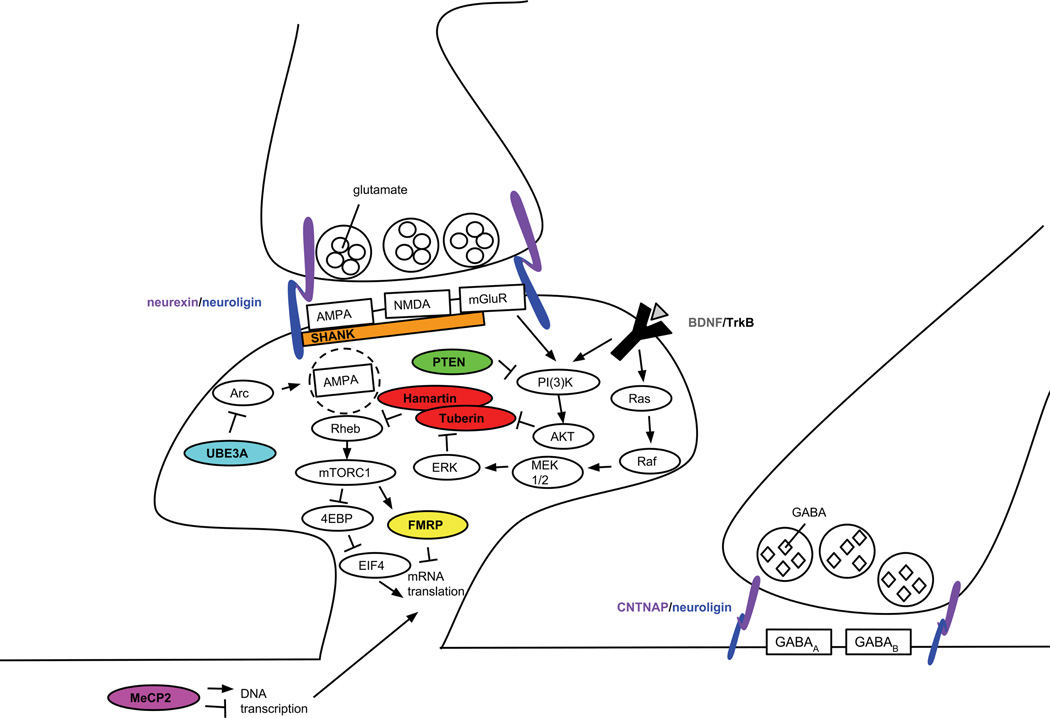
Figure. Monogenic mouse models of ASDs have disruptions in overlapping molecular pathways.
The epigenetic and transcriptional regulator MeCP2 controls the expression of hundreds of different proteins, including BDNF. When BDNF binds to TrkB, its receptor, the resulting signaling pathways converge with pathways known to influence local protein synthesis. The RNA-binding protein FMRP is directly involved in suppressing the translation of mRNA, but other proteins implicated in ASDs, including hamartin and tuberin (the proteins encoded by TSC1 and TSC2), as well as PTEN are upstream signaling molecules that converge on this pathway. The synaptic organizing proteins from the Shank and neurexin/neuroligin families influence these pathways indirectly by affecting the localization and function of glutamate receptors. Similarly, the ubiquitin protein ligase Ube3a normally suppresses the internalization of AMPA receptors, thereby affecting neuronal signaling and plasticity.
Compared to other models, little work has been published utilizing Fmr1 conditional knockout mice to determine the cell types and brain regions underlying the ASD-like phenotypes involved in FMRP dysfunction. One group reported that deletion of Fmr1 in a subset of mature neurons in cortex and hippocampus (Nse-Cre) has no effect on ASD-like behaviors (Amiri et al., 2014), whereas previous work demonstrated that deletion of another ASD susceptibility gene (Pten; see section 3.2.3) using the same Cre line produces ASD-like phenotypes (Kwon et al., 2006a). This finding is somewhat surprising, given that PTEN expression is typically suppressed in FXS, indicating some mechanistic overlap between FXS and PTEN-related ASD (Hagerman et al., 2010). There is some evidence to suggest that, similar to Mecp2 mutant mice, Fmr1 knockout mice have deficits in GABAergic signaling (GABAB reviewed in Braat and Kooy, 2014; GABAA reviewed in Braat and Kooy, 2015), but so far this has not been directly linked to behavior. Clearly, more work needs to be done to determine cell types and brain regions involved in the ASD-like features observed in FXS mouse models.
3.2.2 Tsc1/Tsc2 (Tuberous sclerosis complex)
Tuberous sclerosis complex (TSC) is a disorder characterized by the formation of benign tumors, intellectual disability, epilepsy, infantile spasm, and ASD (Smalley et al., 1992). Heterozygous mutations in either TSC1 or TSC2 genes that encode the proteins hamartin and tuberin, respectively, are sufficient to cause TSC (van Slegtenhorst et al., 1997; European Chromosome 16 Tuberous Sclerosis Consortium, 1993). Hamartin and tuberin form a heterodimer that negatively regulates the mTORC1 (mammalian target of rapamycin complex 1) signaling cascade by inactivating the small G protein Rheb (Tee et al., 2003). The mTORC1 signaling cascade has been implicated in cell growth and proliferation (Fingar et al., 2002) and in synaptic plasticity (Hoeffer and Klann, 2010).
Homozygous deletion of Tsc1 exons 6–8 (Kobayashi et al., 2001), Tsc1 exons 17–18 (Kwiatkowski et al., 2002), Tsc1 exons 5–7 (Wilson et al., 2005), Tsc2 exon 2 (Onda et al., 1999), or Tsc2 exons 2–5 (Kobayashi et al., 1999) in mice all result in embryonic lethality. However, heterozygous mice (Tsc1+/− or Tsc2+/−) from these constructs recapitulate many of the phenotypes of TSC observed in humans, including ASD-like behaviors (reviewed in Bey and Jiang, 2014).
Depletion of Tsc1 or Tsc2 in cultured post-mitotic hippocampal neurons results in increased soma size, decreased dendritic spine density, and increased spine size (Tavazoie et al., 2005). However, loss of Tsc1 in hippocampal neurons in vivo via viral delivery of Cre recombinase, soma size is increased but there is no significant change in spine density (Bateup et al., 2011). On the other hand, Tsc2+/− mice have normal spine density in cortical projection neurons at P19–P20, but increased spine density at P29–P30, suggesting a synaptic pruning defect (Tang et al., 2014). Thus, whereas the effects of Tsc1/2 loss on spine density vary developmentally and across brain regions, soma size is consistently increased.
Decreased expression of Tsc1 or Tsc2 has drastic impacts on synaptic function as well, but the findings vary between in vivo and in vitro experiments, suggesting non-cell-autonomous and network effects. Organotypically cultured hippocampal neurons lacking Tsc1 have higher amplitudes in mEPSCs and increased AMPA/NMDA current ratios (Tavazoie et al., 2005). However, loss of Tsc1 in hippocampal CA1 neurons in vivo impairs mGluR-dependent LTD, decreases the inter-event-interval (a measure of frequency) in mEPSCs without changing mEPSC amplitude, and increases evoked AMPA and NMDA currents proportionally (Bateup et al., 2011). Mice haploinsufficient for Tsc2 also have impaired mGluR-dependent LTD (Auerbach et al., 2011). Additionally, knockout of Tsc1 in primary hippocampal cultures has no effect on mEPSC amplitude in glutamatergic neurons, but whole cell voltage clamp analysis of GABAergic neurons shows a decrease in evoked IPSC (eIPSC) and mIPSC amplitudes (Weston et al., 2014).
At the cellular level, loss-of-function mutations in Tsc1 and Tsc2 lead to hyperactivation of mTORC1, which is one of the major mechanisms for TSC pathogenesis (reviewed in Crino, 2013). This mechanism overlaps with other known genetic causes of ASDs. For example, long-term potentiation initiated by BDNF is dependent on mTOR signaling (Tang et al., 2002). Additionally, as the mTOR pathway is involved in protein translation, it is hyperactivated in FXS (Sharma et al., 2010). However, Tsc2+/− mice show reduced protein synthesis, whereas Fmr1−/y mice have increased protein synthesis (Auerbach et al., 2011). One possible explanation for the different effects on protein synthesis is that Rheb has functions other than to negatively regulate mTORC1 activity, which is evident by the fact that inhibition of Rheb, but not inhibition of mTORC1, rescues the aberrant spine morphogenesis observed in Tsc2+/− neurons (Yasuda et al., 2014). More work needs to be done to determine whether ASD-like phenotypes in TSC mouse models are the result from disrupted mTORC1-dependent or mTORC1-independent signaling pathways.
Various conditional knockouts of Tsc1 or Tsc2 have been used to determine cell types and brain regions involved in the different components of TSC. Mice with Tsc1 deficiency in cerebellar Purkinje cells by the expression of L7-Cre in either Tsc1flox/+ or Tsc1flox/flox display ASD-like behaviors (Tsai et al., 2012). Moreover, conditional knockout of Tsc1 in Purkinje cells leads to decreased cell numbers, increased soma size, increased spine density, and lower spontaneous spiking rates in these cells (Tsai et al., 2012). Another group crossed Pcp2-Cre mice with Tsc2flox/− mice, resulting in homozygous deletion of Tsc2 in Purkinje cells and heterozygous deletion of Tsc2 in all other cells, and report exacerbated ASD-like phenotypes (Reith et al., 2013). One may argue that the latter study doesn’t make use of the high construct validity provided by the haploinsufficiency of the TSC mouse model. However, loss of heterozygosity (LOH) occurs in a cell-specific manner in TSC (Crino, 2013) and it is possible that LOH in Purkinje cells accounts for some of the ASD symptoms in human TSC patients, but more work needs to be done to confirm this hypothesis. More recently, a group knocked out Tsc1 in all excitatory forebrain neurons (CaMKIIα-Cre) and selectively in serotonergic neurons (Slc6a4-Cre) and each was sufficient to produce ASD-like behaviors (McMahon et al., 2014).
3.2.3 Pten (PTEN hamartoma tumor syndromes and non-syndromic ASDs)
Mutations in phosphatase and tensin homolog (PTEN) were first identified in a number of patients with different conditions (reviewed in Eng, 2003), together called PTEN hamartoma tumor syndromes (PHTS). Although relatively rare, PTEN mutations are consistently identified in non-syndromic ASDs comorbid with significant macrocephaly (Butler et al., 2005; Buxbaum et al., 2007), a feature observed in approximately 20% of all ASD cases (reviewed in Fombonne et al., 1999; Courchesne et al., 2003; Courschesne et al., 2011).The Pten protein normally functions as a protein and lipid phosphatase that negatively regulates the Akt signaling pathway, particularly by dephosphorylating phosphatidylinositol (3, 4, 5)-triphosphate (PIP3; Maehama and Dixon, 1998). Tuberin, the protein product of Tsc2, is one target of the Akt signaling pathway (Potter et al., 2002), which is inhibited following phosphorylation by Akt (Inoki et al., 2002). Thus, the PI3K/Akt signaling pathway increases mTORC1 activity and Pten, like the Tsc1 and Tsc2 products, normally inhibits mTORC1 activity.
The first Pten knockout mouse models showed that homozygous deletion of exons 4 and 5 (Di Cristofano et al., 1998) or exons 3, 4, and 5 (Suzuki et al., 1998) results in embryonic lethality, whereas heterozygous deletions result in widespread tumorigenesis, resembling PHTS. More recently, conditional Pten knockouts have been developed that more closely resemble PTEN-related ASDs. Although these models do not have good construct validity because humans with PTEN-related ASDs have germline missense mutations in PTEN, the various models provide insight into different brain regions and cell types involved in ASDs. Mice with Pten deleted selectively in a subset of mature neurons in cortex and hippocampus (Nse-Cre) show deficits in social interactions (Kwon et al., 2006a) and increased repetitive behaviors (Napoli et al., 2012). Similar behaviors have been observed in mice with Pten deleted in granule cells of the cerebellum and neurons in the dentate gyrus of the hippocampus (Gfap-Cre; Lugo et al., 2014). Another mouse model of PTEN-related ASD (Ptenm3m4) was created by knocking in missense mutations which decrease the amount of nuclear Pten (Tilot et al., 2014). Interestingly, only male mice with the missense mutations display increased social behavior, as opposed to the decreased social behavior observed in other mouse models of ASDs (Tilot et al., 2014). While these particular missense mutations have not yet been reported in humans, mutations that similarly affect the subcellular localization of PTEN have been observed in PTHS patients.
Detailed studies of the PTEN-related ASD mouse models have revealed several cellular structural abnormalities. Generally, neuronal cells lacking Pten have increased size, more abundant and ectopic axonal projections, increased numbers of presynaptic vesicles, increased numbers of dendritic spines, and larger postsynaptic densities (Kwon et al., 2006a; Fraser et al., 2008; Luikart et al., 2011a; Williams et al., 2015). However, one study suggests that overall numbers of dendritic spines are unchanged in neurons lacking Pten, but the spines are larger and there are a greater proportion of spines with mature morphology (Haws et al., 2014). The Ptenm3m4 mice, which have decreased nuclear Pten but normal levels of cytoplasmic Pten, have neurons with increased soma size but normal dendritic thickness (Tilot et al., 2014), indicating the importance of Pten in determining cellular morphology locally.
Not only do neurons lacking Pten have drastic morphological aberrations, but they also have significant changes in firing properties and synaptic plasticity. Pten+/− mice have decreased LTP and completely abolished NMDA-dependent LTD in hippocampal CA1 synapses (Wang et al., 2006). Moreover, Pten conditional knockouts (GFAP-Cre) also have decreased LTP in CA1 synapses (Fraser et al., 2008), whereas dentate granule cell synapses have impaired mGluR-dependent LTD (Takeuchi et al., 2013). Importantly, postnatal deletion of Pten with CamKIIα-Cre reproduced the deficits in LTP and LTD at CA1 synapses but had no effect on neuronal or dendritic morphology, thus indicating these phenotypes are not necessarily associated (Sperow et al., 2012).
Pten is involved in the suppressing the Akt signaling pathway upstream of mTOR activity, which normally leads to dephosphorylation of FMRP and subsequent protein translation. However, there is some evidence that suggests Pten might be acting differently. In particular, deletion of Pten leads to decreased expression of mGluR5, increased expression of FMRP, and increased phosphorylation of FMRP (Lugo et al., 2014). These findings seem to oppose, rather than overlap with, findings from Fmr1 knockout mice. More work needs to be done to determine the molecular consequences of Pten loss-of-function and how this contributes to ASD-like phenotypes.
3.2.4 Ube3a (Angelman syndrome and non-syndromic ASDs)
Patients diagnosed with Angelman syndrome (AS) frequently meet the diagnostic criteria for ASD (Peters et al., 2004). Angelman syndrome is caused by mutations in or lack of expression of the maternal copy of UBE3A (Kishino et al., 1997; Matsuura et al., 1997), the paternal copy of which is normally silenced in neurons (Albrecht et al., 1997; Rougeulle et al., 1997; Yamasaki et al., 2003). Maternally derived duplications and triplications of a genomic region which contains UBE3A have also been identified in patients with non-syndromic ASDs (Cook et al., 1997; Glessner et al., 2009; Hogart et al. 2010). The protein encoded by UBE3A is the ubiquitin-protein ligase (UBE3A), also known as E6AP ubiquitin-protein ligase (E6AP; Huibregtse et al., 1993). The ubiquitin-proteasome degradation system has been implicated in synapse function and plasticity (reviewed in Mabb and Ehlers, 2010).
Mouse models of AS that recapitulate the major features of the disorder, including some ASD-like phenotypes, have been created and studied extensively. The first Ube3a mutant mice were created by making a targeted deletion of coding exon 2 (Jiang et al., 1998) and another model was created by disrupting last two coding exons 15–16 (Miura et al., 2002), but only the model with exon 2 deletion has been extensively studied subsequently. The brains of the maternally deficient (Ube3am−/p+) mice have significantly low or no detectable Ube3a protein, conferring high construct validity. Another group created mice that express either one extra (1xTg) or two extra copies (2xTg) of Ube3a, with reasonable construct validity for patients with maternally derived duplications and triplications in the genomic region encompassing UBE3A (Smith et al., 2011).
Maternal deficiency of Ube3a affects dendritic and spine morphology. Specifically, Ube3am−/p+ mice have decreased spine density in hippocampal CA1 and cortical layer 3–5 pyramidal neurons (Dindot et al., 2008). In contrast, Ube3am−/p+ mice raised in darkness have similar spine densities in visual cortex, suggesting a deficit specifically in experience-dependent remodeling of synapses (Yashiro et al., 2009). Knockdown of Ube3a isoforms via in utero electroporation of shRNA revealed that Ube3a is also required for apical, but not basal, dendrite outgrowth (Miao et al., 2013). On the other hand, increased Ube3a gene dosage does not affect glutamatergic synapse number in layer 2/3 barrel cortex, as assessed by electron microscopy, immunofluorescence, and golgi staining (Smith et al., 2011).
Perhaps the best studied aspect of AS and other Ube3a mouse models are the electrophysiological properties of affected neurons. Reduced LTP has been observed in hippocampal CA synapses (Jiang et al., 1998; Weeber et al., 2003) as well as in visual cortex (Yashiro et al., 2009; Sato and Stryker, 2010) of AS model mice. Like dendritic spine density, LTP in visual cortex is not significantly different between wild-type and Ube3am−/p+ mice that were raised in darkness (Yashiro et al., 2009). In addition, reduced NMDA-dependent LTD has been reported in the visual cortex of Ube3am−/p+ mice raised in normal conditions (Yashiro et al., 2009), whereas enhanced mGluR-dependent LTD has been reported in hippocampal CA1 synapses (Pignatelli et al., 2014). A decreased AMPA/NMDA current ratio has also been reported in Ube3am−/p+ CA1 hippocampal neurons (Greer et al., 2010), but this ratio is unchanged in Ube3a2xTg barrel cortex (Smith et al., 2011). Whole-cell patch clamp recording revealed reduced frequency and amplitude of mEPSCs in the dorsomedial striatum of Ube3am−/p+ mice, but not in the dorsolateral striatum (Hayrapetyan et al., 2013). Layer 2/3 pyramidal neurons in Ube3a2xTg barrel cortex display reduced eEPSC amplitude, reduced sEPSC amplitude and frequency, reduced sIPSC amplitude, and reduced mEPSC amplitude and frequency, along with decreased release probability and synaptic glutamate concentration (Smith et al., 2011). In general, the mechanisms underlying the different impairments in synapses of different brain regions remains poorly understood.
Currently, no conditional knockouts of Ube3a have been reported, so there is limited knowledge about what cell types and brain regions are involved in the behavioral abnormalities in Ube3am−/p+ mice. However, using mice engineered with a stop codon surrounded by loxP sites that prevents maternal Ube3a expression crossed with the commonly used ubiquitously-expressed inducible Cre line (CAG-Cre/Esr1), it was recently demonstrated that Ube3a reinstatement during embryonic or early postnatal development can prevent the expression of some ASD-like phenotypes, but reinstatement in mice later in development is not effective (Silva-Santos et al., 2015). Interestingly, these results are in contrast with those reported in Rett syndrome mice (see section 3.1). This could be due to the different behaviors reported in these studies, or it could indicate that some ASD susceptibility genes have tightly-regulated functions during development whereas others are required during the entire lifespan. Again, these types of studies need to be investigated using other models of ASDs before making any conclusions about the developmental time course of ASD pathogenesis.
Numerous targets of Ube3a-dependent ubiquitination have been identified (Kumar et al., 1999; Mani et al., 2006; Mulherkar et al., 2009; Louria-Hayon et al., 2009; Zaaroor-Regev et al., 2010) in attempts to dissect the molecular mechanisms underlying Angelman syndrome. One target is activity-regulated cytoskeleton protein (Arc), which promotes the internalization of AMPA receptors, and is interesting considering the functional defect of AMPA receptor mediated synaptic transmission (Greer et al., 2010). More recent evidence suggests that Ube3a regulates Arc expression through estradiol-induced transcription rather than through ubiquitination (Kühnle et al., 2013). Recent findings also indicate that in Drosophila Ube3a acts as a cofactor for some MeCP2 functions (Kim et al., 2013) and Ube3am−/p+ mice demonstrate deficits in BDNF signaling (Cao et al., 2013). Additionally, Ube3am−/p+ mice have presynaptic vesicle cycling defects specifically in inhibitory interneurons (Wallace et al., 2012). These observations clearly demand further investigation on some very basic questions. For instance, is the dosage of Ube3a in excitatory or inhibitory neurons responsible for the profound neurobehavioral impairment? Moreover, is the dosage of Ube3a primarily affecting pre- or postsynaptic sites?
3.3 Synaptic Organizing and Scaffolding: Shanks, Neurexins/Neuroligins
3.3.1 Shanks (Phelan-McDermid syndrome and non-syndromic ASDs)
Mutations in the SHANK/ProSAP family genes (SHANK1–3), particularly SHANK2 and SHANK3, have been identified recently as pathogenic for non-syndromic ASDs (Durand et al., 2007; Berkel et al., 2010; Sato et al., 2012). In addition, loss of one copy of SHANK3 in Phelan-McDermid syndrome is thought to contribute to the neurobehavioral features of the disorder, including ASD (Bonaglia et al., 2001; Wilson et al., 2003). The Shank proteins function as scaffolds that organize postsynaptic densities (PSDs) in glutamatergic synapses and link receptors to cytoskeletal signaling molecules (Sheng and Kim 2000). Shank proteins have also been implicated in spinogenesis and synapse development. In particular, transfection of Shank3 is sufficient to induce dendritic spine formation in otherwise aspiny neurons (Roussignol et al., 2005). Another study provides evidence that Shank2 and Shank3 are involved in spine formation, whereas Shank1 is involved in synapse maturation and stability (Grabrucker et al., 2011).
Mouse models for each of the three Shank proteins have been created to better understand the role of these proteins in vivo and their contributions to ASDs. The first model was created by deleting exons 14–15 of Shank1 (Hung et al., 2008) where homozygous deletion (Shank1−/−) results in some ASD-like features (Silverman et al., 2011; Wöhr et al., 2011; Sungur et al, 2014). Two different lines of Shank2 mutant mice have been reported, which involved deletion of exon 7 (Schmeisser et al., 2012) or exons 6–7 (Won et al., 2012) of the Shank2a isoform, corresponding to exon 17 and exons 16–17, respectively, of full length Shank2 (Jiang and Ehlers, 2013). Both lines are assumed to have disrupted function of all Shank2 isoforms and display stereotypy, diminished social interactions, and impaired vocalizations (Schmeisser et al., 2012; Won et al., 2012; Ey et al., 2013).
Ten different lines of Shank3 mutant mice have been reported, but due to the transcriptional complexity of Shank3 (Wang et al., 2014), none of the published lines disrupt all isoforms. Still, deletion of exons 4–7 (Peça et al., 2011), exons 4–9 (Bozdagi et al., 2010; Wang et al., 2011; Jaramillo et al., 2015), exon 9 (Lee et al., 2015), exon 11 (Schmeisser et al., 2012), exons 13–16 (Peça et al., 2011), or exon 21 (Kouser et al., 2013; Duffney et al., 2015), and an insertion mutation in exon 21 (Speed et al., 2015) all result in ASD-like behaviors to various degrees (reviewed in Bey and Jiang, 2014). Whereas humans with SHANK3-related ASDs are either haploinsufficient due to a large chromosomal deletion or have point mutations in one copy of their SHANK genes, many of the studies utilizing mouse models only report behavioral abnormalities in homozygous mutants. This, along with the fact that there currently is no model in which all Shank3 isoforms are disrupted, limits the construct validity of these models.
Although there are no published studies that have explored the role of the Shank proteins in different cell types and brain regions using conditional knockouts, there have been considerable efforts to characterize the cellular phenotypes that result from the loss of these proteins. Deletion of Shank1 results in decreased spine density, shorter spines, and thinner PSDs in CA1 pyramidal neurons (Hung et al., 2008). However, both normal spine density (Won et al., 2012) and decreased spine density (Schmeisser et al., 2012) have been reported in hippocampal CA1 neurons from Shank2 mutants. Moreover, PSD length and thickness are not affected in CA1 neurons from either Shank2 line (Won et al., 2012; Schmeisser et al., 2012). Deletion of exons 13–16 of Shank3 led to decreased spine density, increased dendritic complexity, and thinner PSDs in medium spiny neurons (MSNs) of the striatum (Peça et al., 2011). However, deletion of exon 11 (Schmeisser et al., 2012) or exon 21 (Kouser et al, 2013) of Shank3 had no effect on either spine density nor on dendritic complexity in CA1 neurons. This may have been due to the age of the mice tested, as deletion of exons 4–9 of Shank3 had an age-dependent effect on spine density and morphology: CA1 neurons from 4 week old mice had decreased spine density and increased spine length, whereas CA1 neurons from 10 week old mice had normal spine density, but decreased spine length (Wang et al., 2011).
Accumulating evidence in vivo suggests that Shank proteins are required for proper synaptic function as well, but the findings are somewhat inconsistent. Extracellular recordings in the stratum radiatum of Shank1−/− mice revealed a significantly decreased excitatory synaptic strength, as assessed by the input-output relationship of field excitatory postsynaptic potentials, and recordings from CA1 hippocampal neurons in these mice revealed decreased mEPSC frequency (Hung et al, 2008). The two lines of Shank2 mutant mice have distinct electrophysiological phenotypes, despite the two mutations having presumably the same effect on Shank2 protein expression. Deletion of Shank2 exon 7 results in decreased excitatory synaptic strength, decreased mEPSC frequency, a decreased AMPA/NMDA current ratio, and increased LTP in the hippocampus (Schmeisser et al., 2012). However, deletion of Shank2 exons 6–7 results in no significant changes in excitatory synaptic transmission, an increase in the AMPA/NMDA current ratio, and impaired LTP and NMDA-dependent LTD (Won et al., 2012).
The findings from Shank3 mutants are also somewhat inconsistent, but many of the differences can be attributed to different isoforms disrupted due to the transcriptional complexity of the gene. Deletion of exon 9 leads to increased mIPSC frequency in the hippocampus and decreased mIPSC frequency in the prefrontal cortex (Lee et al., 2015). All three models which have exons 4–9 deleted have reduced LTP in the CA1 region of the hippocampus (Bozdagi et al., 2010; Wang et al., 2011; Jaramillo et al., 2015). Whereas the model produced by Bozdagi et al. has reduced excitatory synaptic strength, decreased mEPSC amplitude, increased mEPSC frequency, and a decreased paired-pulse ratio, the other models exhibit none of these phenotypes, but the model produced by Jaramillo et al. did have an increased AMPA/NMDA ratio in the striatum. Deletion of exons 13–16 had similar results to those observed by Wang et al. and Jaramillo et al. in the hippocampus, but resulted in decreased excitatory synaptic strength, reduced mEPSC frequency, and reduced mEPSC amplitude in MSNs of the dorsolateral striatum (Peça et al., 2011). On the other hand, deletion of exon 21 results in decreased LTP, an increased AMPA/NMDA current ratio, decreased mEPSC frequency, and reduced excitatory synaptic strength in the CA1 region of the hippocampus (Kouser et al., 2013) as well as an increased AMPA/NMDA current ratio in layer 5 prefrontal cortex pyramidal neurons (Duffney et al., 2015). The insertion mutation in exon 21 is similar to deletion of exon 21 in terms of hippocampal electrophysiology, but one difference is that LTD, and not LTP, is impaired (Speed et al., 2015).
The mechanism through which deletion of Shank proteins leads to synaptic dysfunction is likely due to impaired interactions between glutamate receptors and postsynaptic density proteins leading to impaired signaling in dendritic spines. For instance, rescue of NMDA receptor hypofunction with an actin stabilizer after knockdown of Shank3 in cultured rat cortical neurons suggests that some of the synaptic defects are due to disrupted cytoskeleton signaling (Duffney et al., 2013). Similarly, treating Shank3 exon 21 deletion mice with an actin stabilizer rescues both behavioral phenotypes and synaptic deficits (Duffney et al., 2015).
Moreover, changes in the densities of receptors and synaptic proteins have been reported in vivo. Mice lacking Shank1 have decreased expression of Homer and GKAP/SAPAP in hippocampal PSD fractions (Hung et al., 2008). Deletion of Shank2 exon 7 results in significantly increased expression of NMDAR subunits in the hippocampus and significantly increased expression of both NMDAR and AMPAR subunits as well as Shank3 in the striatum (Schmeisser et al., 2012). Deletion of Shank2 exons 6–7 also results in increased NMDAR subunit expression, but reduces NMDAR-associated signaling, as assessed by the proportion of phosphorylated CaMKII, ERK1/2, and p38 in whole brain lysates (Won et al., 2012).
Mice with different Shank3 mutations have varying degrees of altered expression of receptors, synaptic proteins, and downstream signaling molecules. Deletion of exon 21 of Shank3 results in increased mGluR5 expression in hippocampal synaptosome and PSD fractions (Kouser et al., 2013) as well as decreased Rac1/PAK signaling, increased activated cofilin, and decreased expression of F-actin in synapses (Duffney et al., 2015). Deletion of exons 4–9 of Shank3 reduces Homer1b/c, GluA1, GluN2A, and SAPAP1 in hippocampal PSD fractions (Wang et al., 2011) as well as reduces Homer1b/c, PSD-95, GluA2, and GluA3 in striatal synaptosomes (Jaramillo et al., 2015). Finally, deletion of exons 13–16 results in reduced expression of SAPAP3, Homer, PSD-93, GluA2, GluN2A, and GluN2B in striatal PSD fractions (Peça et al., 2011) whereas deletion of exon 11 results in increased expression of GluN2B and Shank2 in striatal synaptosomal fractions (Schmeisser et al., 2012). It is currently unclear how these molecular changes lead to synaptic dysfunction and how synaptic dysfunction leads to changes in behavior.
3.3.2 Neurexins/Neuroligins (non-syndromic ASDs)
Mutations in the genes encoding the presynaptic cell-adhesion molecules, neurexins, and their postsynaptic binding partners, neuroligins, have been implicated in non-syndromic ASDs (Gauthier et al., 2011; Camacho-Garcia et al., 2013; Vaags et al., 2012; Steinberg et al., 2012; Jamain et al., 2003; Feng et al., 2006). Additionally, mutations in genes coding for other members of the neurexin superfamily, contactin associated protein-2 (CNTNAP2; Alarcón et al., 2008; Arking et al., 2008) and contactin associated protein-4 (CNTNAP4; Karayannis et al., 2014), have been identified in cases of ASD. Each of the three neurexin genes (NRXN1, NRXN2, NRXN3) contains two promoters, which generate a longer α-NRXN or a shorter β-NRXN, respectively (Tabuchi and Südhof, 2002). Moreover, the neurexins undergo extensive alternative splicing, resulting in thousands of different isoforms (Ullrich et al., 1995; Treutlein et al., 2014). On the other hand, the neuroligin genes (NLGN1, NLGN2, NLGN3, NLGN4X, NLGN4Y) contain one promoter and two alternatively spliced regions, allowing for up to four different isoforms per gene (Ichtchenko et al., 1996). Together, neurexins and neuroligins form trans-synaptic complexes that facilitate synapse formation and maturation, as well as excitatory and inhibitory transmission (reviewed in Bang and Owczarek, 2013).
A multitude of mice with mutations in the neurexins, neuroligins, and related genes have been generated to understand the roles of these cell adhesion molecules in normal development and in ASDs. Mice lacking either Nrxn1α (Etherton et al., 2009; Grayton et al., 2013) or Nrxn2α (Dachtler et al., 2014) show some ASD-like features. Additionally, mice with a subset of forebrain neurons overexpressing of a dominant-negative form of Nrxn1β missing its cytoplasmic tail display ASD-like behaviors (Rabaneda et al., 2014). Mice lacking Cntnap2 (Peñagarikano et al., 2011) or Cntnap4 (Karayannis et al., 2014) also have phenotypes which resemble ASD. Likewise, mice lacking Nlgn1 (Blundell et al., 2010), Nlgn2 (Blundell et al., 2009; Wöhr et al., 2013), Nlgn3 (Radyushkin et al., 2009; Rothwell et al., 2014), or Nlgn4 (Jamain et al., 2008; El-Kordi et al., 2013; Ju et al., 2014) all have ASD-like behaviors to various degrees. Mice with a knock-in mutation of NLGN3 (R451C) found in human populations have also been generated and recapitulate some ASD-like features (Tabuchi et al., 2007; Jaramillo et al., 2014; Rothwell et al., 2014).
In addition to the study that reported ASD-like behaviors in mice overexpressing a dominant-negative form of Nrxn1β in a subset of excitatory forebrain neurons (CaMKIIα-tTa; Rabaneda et al., 2014), another study involved various conditional knockouts of Nlgn3 to determine the cell types and brain regions involved in the behaviors (Rothwell et al., 2014). The authors of the latter study found that depletion of Nlgn3 in the D1-MSNs (D1-Cre) of the nucleus accumbens (localized injection of AAV-Cre) was sufficient to drive repetitive motor routines on the rotarod test, whereas depletion of Nlgn3 in D2-MSNs (A2a-Cre) or in the dorsal striatum (localized injection of AAV-Cre) was not (Rothwell et al., 2014). Moreover, depletion of Nlgn3 in Purkinje cells (P7-Cre) or parvalbumin-positive interneurons (PV-Cre) did not affect motor performance, but increased and decreased activity in the open field, respectively (Rothwell et al., 2014).
Unlike the other mouse models of ASDs, individual neurexin and neuroligin mutants do not have many reported changes in dendritic spine density, which is somewhat surprising, given that neurexins (Graf et al., 2004) and neuroligins (Scheiffele et al., 2000) induce synapse formation in cultured cells in vitro and that knockdown of neuroligins in cultured neurons reduces spine density (Chih et al., 2005). This suggests that there may be functional redundancy in the different proteins that can compensate for one another during development in vivo. However, even mice lacking either all three α-Nrxn proteins (Missler et al., 2003) or Nlgn1, Nlgn2, and Nlgn3 (Varoqueaux et al., 2006) have normal synapse numbers, suggesting that these molecules may not be required for synapse formation at all. A more recent study showed that knockdown of Nlgn1 in vivo reduces synaptic density when neighboring neurons still express Nlgn1 (Kwon et al., 2012). Whereas mice lacking Nlgn2 have normal numbers of both excitatory and inhibitory synapses, they have decreased expression of VGAT, which represents an impairment in recruiting GABAergic synaptic vesicles to presynaptic terminals (Blundell et al., 2009). The Nlgn3R451C mice also have normal numbers of both excitatory and inhibitory synapses, but have increased expression of VGAT (Tabuchi et al., 2007). These mice have increased dendritic complexity in the stratum radiatum, decreased synaptic terminal size, fewer vesicles per terminal, and decreased spine size (Etherton et al., 2011a). Additionally, the Nlgn3R451C mice have increased dendritic spine turnover in pyramidal neurons in the anterior frontal cortex (Isshiki et al., 2014). Moreover, mice lacking Cntnap4 have increased width of synaptic clefts and decreased PSD length in GABAergic synapses (Karayannis et al., 2014).
Although changes in cellular and spine morphology in neurexin and neuroligin mouse models of ASD are somewhat minor, synaptic function is drastically altered. Deletion of Nrxn1α leads to reduced mEPSC frequency and decreased excitatory synaptic strength in the CA1 region of the hippocampus (Etherton et al., 2009). Mice with the dominant-negative form of Nrxn1β have decreased mEPSC frequency and mIPSC frequncy in L5/6 pyramidal neurons from the somatosensory cortex (Rabaneda et al., 2014). Mice lacking Cntnap4 have increased dopamine signaling through a presynaptic mechanism in the nucleus accumbens but fewer, smaller, and slower sIPSCs in PV-positive cells of the somatosensory cortex (Karayannis et al., 2014). Deletion of Nlgn1 results in a decrease in AMPAR EPSCs, NMDA EPSCs, and an increase in the AMPA/NMDA current ratio in the CA1 region of the hippocampus (Budreck et al., 2013) as well as impaired LTP at thalamic inputs to the amygdala (Jung et al., 2010). Impaired LTP has also been observed the hippocampi of Nlgn1 mutants in vitro (Blundell et al., 2010) and in vivo (Jedlicka et al., 2015). Mice lacking Nlgn4 have decreased network excitability in both excitatory and inhibitory circuits in layer 2/3 pyramidal cells, but the excitatory circuits are affected more greatly such that the excitation-inhibition ratio is decreased in the mutants (Delattre et al., 2013).
The electrophysiological consequences of the Nlgn3R451C mutation in particular have been studied extensively. These mice have increased mIPSC frequency and eIPSC amplitude in layer 2/3 somatosensory cortex neurons (Tabuchi et al., 2007). However, they have decreased eIPSC amplitude in spiny neurons that receive input from PV-expressing basket cells in layer IV barrel cortex (Cellot and Cherubini, 2014). The R451C mutation also leads to an increased frequency of giant depolarizing potentials (GDPs) and mIPSCs in the CA3 region of the hippocampus starting during the first two weeks of postnatal life (Pizzarelli and Cherubini, 2013). These mice also exhibit increased excitatory transmission, increased LTP, and a decreased AMPA/NMDA current ratio in the CA1 region of the hippocampus (Etherton et al., 2011a). While most studies failed to find similar deficits in Nlgn3 knockout mice, one study found that both the Nlgn3R451C mice and Nlgn3 knockout mice have increased GABAergic synaptic transmission (eIPSCs) in cholecystokinin (CCK) basket cells and impair tonic endocannabinoid (ECB) signaling (Földy et al., 2013). Mice lacking Nlgn3 also have decreased frequency of mIPSCs in D1-MSNs of the NAc while having normal frequency of mEPSCs, thereby increasing the excitatory/inhibitory current ratio (Rothwell et al., 2014).
Neuroligins interact with multiple postsynaptic proteins, including Shank3 (Meyer et al., 2004). Moreover, it was shown more recently that Shank3 expression mediates transsynaptic changes in synaptic proteins and that this function depends on the formation of neurexin/neuroligin complexes (Arons et al., 2012). Accordingly, neurexin and neuroligin mutants, like Shank mutants, exhibit changes in the expression of synaptic proteins. Specifically, mice lacking Nrxn2α have reduced Munc-18 expression in hippocampal lysates (Dachtler et al., 2014). Mice lacking Nlgn1 have decreased expression of CSP, Liprin, Munc-18, Nxnα, and Nrxnβ, but increased expression of Nlgn3 and Synapsin1a in whole brain lysates (Blundell et al., 2010). As mentioned previously, Nlgn2 have decreased VGAT expression in hippocampus (Blundell et al., 2009), whereas Nlgn3R451C mutants increased VGAT and Gephyrin expression in whole brain lysates (Tabuchi et al., 2007). The Nlgn3R451C mutants also have decreased expression of PSD95, SAP102, NR2A, and NR2B in hippocampal lysates (Etherton et al., 2011a). Analysis of synaptosome and PSD fractions from neurexin and neuroligin mutant mice could be performed to determine if the changes more similar to changes observed in Shank mutants.
4. Concluding Remarks
4.1 Convergent Molecular Pathways and Mechanisms
Although much remains to be investigated in various directions, it is tempting to generalize these findings on a number of levels. Etiologically, rare and private mutations clustered in select molecular classes appear to be a major driver for a genetic basis in a sub-set of ASD patients. At the molecular level, several pathways appear to emerge from analyzing the existing ASD mouse models. These include disruption of overlapping signaling pathways mediated by mGluR5, BDNF, and mTOR (see Figure 1 and Table 2), although the degree of evidence varies depending on the model.
Moreover, the available evidence strongly supports dysfunctional synapses as a component of autism pathophysiology. However, although there are widespread disruptions in synaptic function across the mouse models of ASD, the direction of change and magnitude of effect are inconsistent between different models, as well as between different types of synapses within any given model. Although there are many apparent differences between models, it is difficult to compare results that were reported in different brain regions or at different times during development. For instance, Fmr1 knockout mice have decreased spine stability somatosensory cortex (Cruz-Martin et al., 2010), whereas Mecp2tm1.1Jae mice have increased spine stability in the same region (Landi et al., 2011), but the former study used mice that were postnatal day 10–12, whereas the latter study used mice that were postnatal day 25–26. Therefore, more side-by-side comparisons of different mouse models of ASDs would be valuable. One interesting discovery from genetics studies of ASDs is the frequent mutations in genes encoding epigenetic machinery. However, it is not immediately clear how deficiency of these proteins contributes to autism pathophysiology. As has been demonstrated in Rett syndrome mouse models, the dysfunction of synapses could still be the major mechanism. However, a mechanism that is independent from synaptic dysfunction may also be possible.
4.2 Missing Links and Challenges
There are some glaring missing links in our understanding of the pathophysiology of ASDs. Specifically, how the mutation of an individual gene, a disrupted molecular pathway, and dysfunctional synapses affect the circuitry and produce ASD-like behavioral manifestations is unclear. Understanding the circuitry underlying autism, both anatomically and functionally, is critical to the development of effective clinical intervention. Several competing hypotheses regarding the circuit-level mechanisms have been proposed in the human literature. One widely tested hypothesis is altered structural and functional brain connectivity (Belmonte et al., 2004; Geschwind and Levitt, 2007; Kana et al., 2014). Structural connectivity is the physical connections between different brain regions, while functional connectivity refers to the integrated relationship between spatially separated brain regions. It is believed that structural connections within the brain give rise to functional network activity as measured by coherence or information flow. Neuroimaging investigations indicate that ASDs are associated with perturbed connectivity at both structural and functional levels (Minshew and Keller, 2010; Vissers et al., 2012); however, the exact nature and pattern of this aberrant neural connectivity remains uncertain due to inconsistent findings from neuroimaging studies in patients (Di Martino et al., 2014; Kana et al., 2014; Uddin et al., 2013). While early studies reported reduced functional connectivity (Just et al., 2004), recent investigations implicate hyper-connectivity in multiple brain regions and across neural circuits (Keown et al., 2013; Supekar et al., 2013). In addition to methodological and conceptual controversy, this uncertainty reflects the substantial molecular heterogeneity of human patients. Notably, these studies were conducted primarily in high-functioning ASD patients for whom the etiologies are mostly unknown. For these reasons, autism animal models offer a unique opportunity to test the functional connectivity hypothesis because of homogenous genetic defects. Using optogenetic tools, Gunaydin et al. have shown that the activity of ventral tegmental area (VTA) to nucleus accumbens (NAc) projections could encode and predict key features of social interaction in wild type mice (Gunaydin et al., 2014). Similarly, Felix-Ortix and Tye revealed the role of projections from the basolateral complex of the amygdala (BLA) to the ventral hippocampus (vHPC) in two different social interaction tests (Felix-Ortiz and Tye, 2014). Few reports have investigated the circuit-level mechanisms underlying autism-like behaviors in genetically modified autism models with strong construct validity. Therefore, the combination of optogenetics and CRISPR/Cas9 genome editing tools in ASD models is expected to produce significant insight into whether there are common circuits disrupted in ASD models with different genetic defects.
4.3 Future Directions
Several critical questions remain poorly understood to understand the pathophysiology of ASDs. The molecular and circuit-level mechanisms underlying autism behaviors are mostly unknown. Moreover, the developmental origin of autistic behaviors has not been delineated. Numerous challenges have been encountered in modeling autism mutations in mice. First, recent genetics studies have identified mutations in a long list of genes implicated in ASDs. However, most of these mutations are rare and private. The full spectrum of clinical features associated with these genetic defects is not known, posing a significant challenge for experimental design and potential to overly generalize findings from individual models. Second, the lack of robust and strong behavioral assays for measuring behaviors that resemble human ASDs remains a significant obstacle to dissect the circuit-level mechanisms. Third, various effects on synapses in different brain regions and different stages of development makes it difficult to establish a link between synaptic dysfunction and behavioral impairments. These challenges thus demand a shift from the current analytic paradigm that combines behavior, synaptic electrophysiology, and biochemical approaches. We propose that an analytic paradigm that integrates the behavior and in vivo physiological recordings in multiple sites is a more productive approach to model autism in mice. Furthermore, manipulation of ASD candidate genes spatially and temporally will further delineate the circuits underlying ASDs. In the future, manipulation of the genomes in other species such as rat or non-human primate models may overcome the many limitations of mouse models. A more complete understanding of the current genetic models of ASDs will allow researchers to study the role of non-genetic factors and their biological underpinnings. Ultimately, knowledge learned from animal models will lead to the development of effective clinical intervention that targets the specific molecular pathways and neural circuits.
Highlights.
Disrupted synaptic function is common across mouse models of ASDs
The synapses affected and the magnitude and direction of effects are inconsistent
Overlapping molecular cascades include mGluR5, BDNF/TrkB, and mTOR signaling.
The developmental origin and circuits underlying the disorders remain largely unknown
Acknowledgments
We would like to thank the Jiang lab members for their contribution to this review and particularly Lara J. Duffney for a critical reading of the manuscript. The research in the Jiang lab is supported by NIH grants 5R01-MH098114-03, 1R21-HD077197-01, and 1R21-MH104316-01, a Duke Institute for Brain Science Incubator award, Autism Speaks, Marcus Foundation, Foundation for Prader-Willi Research, Phelan-McDermid Syndrome Foundation, the Ruth. K. Broad Foundation, and Roche. SWH is supported by a Weatherstone Predoctoral Fellowship from Autism Speaks.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders: DSM-5™. 5th. Arlington, VA: American Psychiatric Publishing, Inc; 2013. [Google Scholar]
- Alarcon M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bomar JM, Sebat J, Wigler M, Martin CL, Ledbetter DH, Nelson SF, Cantor RM, Geschwind DH. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82:150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht U, Sutcliffe JS, Cattanach BM, Beechey CV, Armstrong D, Eichele G, Beaudet AL. Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nat Genet. 1997;17:75–78. doi: 10.1038/ng0997-75. [DOI] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Amiri A, Sanchez-Ortiz E, Cho W, Birnbaum SG, Xu J, McKay RM, Parada LF. Analysis of FMR1 deletion in a subpopulation of post-mitotic neurons in mouse cortex and hippocampus. Autism Res. 2014;7:60–71. doi: 10.1002/aur.1342. [DOI] [PubMed] [Google Scholar]
- Argyropoulos A, Gilby KL, Hill-Yardin EL. Studying autism in rodent models: reconciling endophenotypes with comorbidities. Front Hum Neurosci. 2013;7:417. doi: 10.3389/fnhum.2013.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arking DE, Cutler DJ, Brune CW, Teslovich TM, West K, Ikeda M, Rea A, Guy M, Lin S, Cook EH, Chakravarti A. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet. 2008;82:160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arons MH, Thynne CJ, Grabrucker AM, Li D, Schoen M, Cheyne JE, Boeckers TM, Montgomery JM, Garner CC. Autism-associated mutations in ProSAP2/Shank3 impair synaptic transmission and neurexin-neuroligin-mediated transsynaptic signaling. J Neurosci. 2012;32:14966–14978. doi: 10.1523/JNEUROSCI.2215-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaka Y, Jugloff DG, Zhang L, Eubanks JH, Fitzsimonds RM. Hippocampal synaptic plasticity is impaired in the Mecp2-null mouse model of Rett syndrome. Neurobiol Dis. 2006;21:217–227. doi: 10.1016/j.nbd.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Auerbach BD, Osterweil EK, Bear MF. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature. 2011;480:63–68. doi: 10.1038/nature10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang ML, Owczarek S. A matter of balance: role of neurexin and neuroligin at the synapse. Neurochem Res. 2013;38:1174–1189. doi: 10.1007/s11064-013-1029-9. [DOI] [PubMed] [Google Scholar]
- Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateup HS, Takasaki KT, Saulnier JL, Denefrio CL, Sabatini BL. Loss of Tsc1 in vivo impairs hippocampal mGluR-LTD and increases excitatory synaptic function. J Neurosci. 2011;31:8862–8869. doi: 10.1523/JNEUROSCI.1617-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Belichenko PV, Wright EE, Belichenko NP, Masliah E, Li HH, Mobley WC, Francke U. Widespread changes in dendritic and axonal morphology in Mecp2-mutant mouse models of Rett syndrome: evidence for disruption of neuronal networks. J Comp Neurol. 2009;514:240–258. doi: 10.1002/cne.22009. [DOI] [PubMed] [Google Scholar]
- Belmonte MK, Allen G, Beckel-Mitchener A, Boulanger LM, Carper RA, Webb SJ. Autism and abnormal development of brain connectivity. J Neurosci. 2004;24:9228–9231. doi: 10.1523/JNEUROSCI.3340-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belzung C, Lemoine M. Criteria of validity for animal models of psychiatric disorders: focus on anxiety disorders and depression. Biol Mood Anxiety Disord. 2011;1 doi: 10.1186/2045-5380-1-9. 9-5380-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shachar S, Chahrour M, Thaller C, Shaw CA, Zoghbi HY. Mouse models of MeCP2 disorders share gene expression changes in the cerebellum and hypothalamus. Hum Mol Genet. 2009;18(13):2431–2442. doi: 10.1093/hmg/ddp181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkel S, Marshall CR, Weiss B, Howe J, Roeth R, Moog U, Endris V, Roberts W, Szatmari P, Pinto D, Bonin M, Riess A, Engels H, Sprengel R, Scherer SW, Rappold GA. Mutations in the SHANK2 synaptic scaffolding gene in autism spectrum disorder and mental retardation. Nat Genet. 2010;42:489–491. doi: 10.1038/ng.589. [DOI] [PubMed] [Google Scholar]
- Bey AL, Jiang YH. Overview of mouse models of autism spectrum disorders. Curr Protoc Pharmacol. 2014;66:5.66.1–5.66.26. doi: 10.1002/0471141755.ph0566s66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell J, Blaiss CA, Etherton MR, Espinosa F, Tabuchi K, Walz C, Bolliger MF, Sudhof TC, Powell CM. Neuroligin-1 deletion results in impaired spatial memory and increased repetitive behavior. J Neurosci. 2010;30:2115–2129. doi: 10.1523/JNEUROSCI.4517-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell J, Tabuchi K, Bolliger MF, Blaiss CA, Brose N, Liu X, Sudhof TC, Powell CM. Increased anxiety-like behavior in mice lacking the inhibitory synapse cell adhesion molecule neuroligin 2. Genes Brain Behav. 2009;8:114–126. doi: 10.1111/j.1601-183X.2008.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaglia MC, Giorda R, Borgatti R, Felisari G, Gagliardi C, Selicorni A, Zuffardi O. Disruption of the ProSAP2 gene in a t(12;22)(q24.1;q13.3) is associated with the 22q13.3 deletion syndrome. Am J Hum Genet. 2001;69:261–268. doi: 10.1086/321293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdagi O, Sakurai T, Papapetrou D, Wang X, Dickstein DL, Takahashi N, Kajiwara Y, Yang M, Katz AM, Scattoni ML, Harris MJ, Saxena R, Silverman JL, Crawley JN, Zhou Q, Hof PR, Buxbaum JD. Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Mol Autism. 2010;1 doi: 10.1186/2040-2392-1-15. 15-2392-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braat S, Kooy RF. Insights into GABAAergic system deficits in fragile X syndrome lead to clinical trials. Neuropharmacology. 2015;88:48–54. doi: 10.1016/j.neuropharm.2014.06.028. [DOI] [PubMed] [Google Scholar]
- Braat S, Kooy RF. Fragile X syndrome neurobiology translates into rational therapy. Drug Discov Today. 2014;19:510–519. doi: 10.1016/j.drudis.2014.01.013. [DOI] [PubMed] [Google Scholar]
- Brouwer JR, Mientjes EJ, Bakker CE, Nieuwenhuizen IM, Severijnen LA, Van der Linde HC, Nelson DL, Oostra BA, Willemsen R. Elevated Fmr1 mRNA levels and reduced protein expression in a mouse model with an unmethylated Fragile X full mutation. Exp Cell Res. 2007;313:244–253. doi: 10.1016/j.yexcr.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budreck EC, Kwon OB, Jung JH, Baudouin S, Thommen A, Kim HS, Fukazawa Y, Harada H, Tabuchi K, Shigemoto R, Scheiffele P, Kim JH. Neuroligin-1 controls synaptic abundance of NMDA-type glutamate receptors through extracellular coupling. Proc Natl Acad Sci USA. 2013;110:725–730. doi: 10.1073/pnas.1214718110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buescher AV, Cidav Z, Knapp M, Mandell DS. Costs of autism spectrum disorders in the United Kingdom and the United States. JAMA Pediatr. 2014;168:721–728. doi: 10.1001/jamapediatrics.2014.210. [DOI] [PubMed] [Google Scholar]
- Butler MG, Dasouki MJ, Zhou XP, Talebizadeh Z, Brown M, Takahashi TN, Miles JH, Wang CH, Stratton R, Pilarski R, Eng C. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J Med Genet. 2005;42:318–321. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum JD, Cai G, Chaste P, Nygren G, Goldsmith J, Reichert J, Anckarsater H, Rastam M, Smith CJ, Silverman JM, Hollander E, Leboyer M, Gillberg C, Verloes A, Betancur C. Mutation screening of the PTEN gene in patients with autism spectrum disorders and macrocephaly. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:484–491. doi: 10.1002/ajmg.b.30493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho-Garcia RJ, Hervas A, Toma C, Balmana N, Cormand B, Martinez-Mir A, Scholl FG. Rare variants analysis of neurexin-1beta in autism reveals a novel start codon mutation affecting protein levels at synapses. Psychiatr Genet. 2013;23:262–266. doi: 10.1097/YPG.0000000000000013. [DOI] [PubMed] [Google Scholar]
- Cao C, Rioult-Pedotti MS, Migani P, Yu CJ, Tiwari R, Parang K, Spaller MR, Goebel DJ, Marshall J. Impairment of TrkB-PSD-95 signaling in Angelman syndrome. PLoS Biol. 2013;11:e1001478. doi: 10.1371/journal.pbio.1001478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castren ML, Castren E. BDNF in fragile X syndrome. Neuropharmacology. 2014;76(Pt C):729–736. doi: 10.1016/j.neuropharm.2013.05.018. [DOI] [PubMed] [Google Scholar]
- Cellot G, Cherubini E. Reduced inhibitory gate in the barrel cortex of Neuroligin3R451C knock-in mice, an animal model of autism spectrum disorders. Physiol Rep. 2014;2 doi: 10.14814/phy2.12077. 10.14814/phy2.12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Gilman SR, Chiang AH, Sanders SJ, Vitkup D. Genotype to phenotype relationships in autism spectrum disorders. Nat Neurosci. 2015;18:191–198. doi: 10.1038/nn.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Q, Khare G, Dani V, Nelson S, Jaenisch R. The disease progression of Mecp2 mutant mice is affected by the level of BDNF expression. Neuron. 2006;49:341–348. doi: 10.1016/j.neuron.2005.12.027. [DOI] [PubMed] [Google Scholar]
- Chao HT, Chen H, Samaco RC, Xue M, Chahrour M, Yoo J, Neul JL, Gong S, Lu HC, Heintz N, Ekker M, Rubenstein JL, Noebels JL, Rosenmund C, Zoghbi HY. Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature. 2010;468:263.C–269.C. doi: 10.1038/nature09582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao HT, Zoghbi HY, Rosenmund C. MeCP2 controls excitatory synaptic strength by regulating glutamatergic synapse number. Neuron. 2007;56:58–65. doi: 10.1016/j.neuron.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapleau CA, Boggio EM, Calfa G, Percy AK, Giustetto M, Pozzo-Miller L. Hippocampal CA1 pyramidal neurons of Mecp2 mutant mice show a dendritic spine phenotype only in the presymptomatic stage. Neural Plast. 2012;2012:976164. doi: 10.1155/2012/976164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaste P, Klei L, Sanders SJ, Hus V, Murtha MT, Lowe JK, Willsey AJ, Moreno-De-Luca D, Yu TW, Fombonne E, Geschwind D, Grice DE, Ledbetter DH, Mane SM, Martin DM, Morrow EM, Walsh CA, Sutcliffe JS, Lese Martin C, Beaudet AL, Lord C, State MW, Cook EH, Jr, Devlin B. A Genome-wide Association Study of Autism Using the Simons Simplex Collection: Does Reducing Phenotypic Heterogeneity in Autism Increase Genetic Homogeneity? Biol Psychiatry. 2014 doi: 10.1016/j.biopsych.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- Chih B, Engelman H, Scheiffele P. Control of excitatory and inhibitory synapse formation by neuroligins. Science. 2005;307:1324–1328. doi: 10.1126/science.1107470. [DOI] [PubMed] [Google Scholar]
- Chung L, Bey AL, Jiang YH. Synaptic plasticity in mouse models of autism spectrum disorders. Korean J Physiol Pharmacol. 2012;16:369–378. doi: 10.4196/kjpp.2012.16.6.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comery TA, Harris JB, Willems PJ, Oostra BA, Irwin SA, Weiler IJ, Greenough WT. Abnormal dendritic spines in fragile X knockout mice: maturation and pruning deficits. Proc Natl Acad Sci USA. 1997;94:5401–5404. doi: 10.1073/pnas.94.10.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook EH, Jr, Lindgren V, Leventhal BL, Courchesne R, Lincoln A, Shulman C, Lord C, Courchesne E. Autism or atypical autism in maternally but not paternally derived proximal 15q duplication. Am J Hum Genet. 1997;60:928–934. [PMC free article] [PubMed] [Google Scholar]
- Courchesne E, Campbell K, Solso S. Brain growth across the life span in autism: age-specific changes in anatomical pathology. Brain Res. 2011;1380:138–145. doi: 10.1016/j.brainres.2010.09.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne E, Carper R, Akshoomoff N. Evidence of brain overgrowth in the first year of life in autism. JAMA. 2003;290:337–344. doi: 10.1001/jama.290.3.337. [DOI] [PubMed] [Google Scholar]
- Crino PB. Evolving neurobiology of tuberous sclerosis complex. Acta Neuropathol. 2013;125:317–332. doi: 10.1007/s00401-013-1085-x. [DOI] [PubMed] [Google Scholar]
- Cruz-Martin A, Crespo M, Portera-Cailliau C. Delayed stabilization of dendritic spines in fragile X mice. J Neurosci. 2010;30:7793–7803. doi: 10.1523/JNEUROSCI.0577-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dachtler J, Glasper J, Cohen RN, Ivorra JL, Swiffen DJ, Jackson AJ, Harte MK, Rodgers RJ, Clapcote SJ. Deletion of alpha-neurexin II results in autism-related behaviors in mice. Transl Psychiatry. 2014;4:e484. doi: 10.1038/tp.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlhaus R, El-Husseini A. Altered neuroligin expression is involved in social deficits in a mouse model of the fragile X syndrome. Behav Brain Res. 2010;208:96–105. doi: 10.1016/j.bbr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- Dani VS, Chang Q, Maffei A, Turrigiano GG, Jaenisch R, Nelson SB. Reduced cortical activity due to a shift in the balance between excitation and inhibition in a mouse model of Rett syndrome. Proc Natl Acad Sci USA. 2005;102:12560–12565. doi: 10.1073/pnas.0506071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, Licatalosi DD, Richter JD, Darnell RB. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delattre V, La Mendola D, Meystre J, Markram H, Markram K. Nlgn4 knockout induces network hypo-excitability in juvenile mouse somatosensory cortex in vitro. Sci Rep. 2013;3:2897. doi: 10.1038/srep02897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Developmental Disabilities Monitoring Network Surveillance Year 2010 Principal Investigators, Centers for Disease Control and Prevention (CDC) Prevalence of autism spectrum disorder among children aged 8 years - autism and developmental disabilities monitoring network, 11 sites, United States, 2010. MMWR Surveill Summ. 2014;63:1–21. [PubMed] [Google Scholar]
- Devlin B, Scherer SW. Genetic architecture in autism spectrum disorder. Curr Opin Genet Dev. 2012;22:229–237. doi: 10.1016/j.gde.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nat Genet. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- Di Martino A, Yan CG, Li Q, Denio E, Castellanos FX, Alaerts K, Anderson JS, Assaf M, Bookheimer SY, Dapretto M, Deen B, Delmonte S, Dinstein I, Ertl-Wagner B, Fair DA, Gallagher L, Kennedy DP, Keown CL, Keysers C, Lainhart JE, Lord C, Luna B, Menon V, Minshew NJ, Monk CS, Mueller S, Müller RA, Nebel MB, Nigg JT, O'Hearn K, Pelphrey KA, Peltier SJ, Rudie JD, Sunaert S, Thioux M, Tyszka JM, Uddin LQ, Verhoeven JS, Wenderoth N, Wiggins JL, Mostofsky SH, Milham MP. The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol Psychiatry. 2014;19:659–667. doi: 10.1038/mp.2013.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dindot SV, Antalffy BA, Bhattacharjee MB, Beaudet AL. The Angelman syndrome ubiquitin ligase localizes to the synapse and nucleus, and maternal deficiency results in abnormal dendritic spine morphology. Hum Mol Genet. 2008;17:111–118. doi: 10.1093/hmg/ddm288. [DOI] [PubMed] [Google Scholar]
- Dolen G, Osterweil E, Rao BS, Smith GB, Auerbach BD, Chattarji S, Bear MF. Correction of fragile X syndrome in mice. Neuron. 2007;56:955–962. doi: 10.1016/j.neuron.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffney LJ, Wei J, Cheng J, Liu W, Smith KR, Kittler JT, Yan Z. Shank3 deficiency induces NMDA receptor hypofunction via an actin-dependent mechanism. J Neurosci. 2013;33:15767–15778. doi: 10.1523/JNEUROSCI.1175-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffney LJ, Zhong P, Wei J, Matas E, Cheng J, Qin L, Ma K, Dietz DM, Kajiwara Y, Buxbaum JD, Yan Z. Autism-like deficits in Shank3-deficient mice are rescued by targeting actin regulators. Cell Rep. 2015;11:1400–1413. doi: 10.1016/j.celrep.2015.04.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, Nygren G, Rastam M, Gillberg IC, Anckarsater H, Sponheim E, Goubran-Botros H, Delorme R, Chabane N, Mouren-Simeoni MC, de Mas P, Bieth E, Roge B, Heron D, Burglen L, Gillberg C, Leboyer M, Bourgeron T. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Dutch-Belgian Fragile X Consortium. Fmr1 knockout mice: a model to study fragile X mental retardation. Cell. 1994;78:23–33. [PubMed] [Google Scholar]
- Edmonson C, Ziats MN, Rennert OM. Altered glial marker expression in autistic post-mortem prefrontal cortex and cerebellum. Mol Autism. 2014;5 doi: 10.1186/2040-2392-5-3. 3-2392-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Kordi A, Winkler D, Hammerschmidt K, Kastner A, Krueger D, Ronnenberg A, Ritter C, Jatho J, Radyushkin K, Bourgeron T, Fischer J, Brose N, Ehrenreich H. Development of an autism severity score for mice using Nlgn4 null mutants as a construct-valid model of heritable monogenic autism. Behav Brain Res. 2013;251:41–49. doi: 10.1016/j.bbr.2012.11.016. [DOI] [PubMed] [Google Scholar]
- Eng C. PTEN: one gene, many syndromes. Hum Mutat. 2003;22:183–198. doi: 10.1002/humu.10257. [DOI] [PubMed] [Google Scholar]
- Etherton M, Foldy C, Sharma M, Tabuchi K, Liu X, Shamloo M, Malenka RC, Sudhof TC. Autism-linked neuroligin-3 R451C mutation differentially alters hippocampal and cortical synaptic function. Proc Natl Acad Sci USA. 2011;108:13764–13769. doi: 10.1073/pnas.1111093108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etherton MR, Blaiss CA, Powell CM, Sudhof TC. Mouse neurexin-1alpha deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proc Natl Acad Sci USA. 2009;106:17998–18003. doi: 10.1073/pnas.0910297106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Chromosome 16 Tuberous Sclerosis Consortium. Identification and characterization of the tuberous sclerosis gene on chromosome 16. Cell. 1993;75:1305–1315. doi: 10.1016/0092-8674(93)90618-z. [DOI] [PubMed] [Google Scholar]
- Ey E, Torquet N, Le Sourd AM, Leblond CS, Boeckers TM, Faure P, Bourgeron T. The Autism ProSAP1/Shank2 mouse model displays quantitative and structural abnormalities in ultrasonic vocalisations. Behav Brain Res. 2013;256:677–689. doi: 10.1016/j.bbr.2013.08.031. [DOI] [PubMed] [Google Scholar]
- Felix-Ortiz AC, Tye KM. Amygdala inputs to the ventral hippocampus bidirectionally modulate social behavior. J Neurosci. 2014;34:586–595. doi: 10.1523/JNEUROSCI.4257-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Schroer R, Yan J, Song W, Yang C, Bockholt A, Cook EH, Jr, Skinner C, Schwartz CE, Sommer SS. High frequency of neurexin 1beta signal peptide structural variants in patients with autism. Neurosci Lett. 2006;409:10–13. doi: 10.1016/j.neulet.2006.08.017. [DOI] [PubMed] [Google Scholar]
- Fingar DC, Salama S, Tsou C, Harlow E, Blenis J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 2002;16:1472–1487. doi: 10.1101/gad.995802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foldy C, Malenka RC, Sudhof TC. Autism-associated neuroligin-3 mutations commonly disrupt tonic endocannabinoid signaling. Neuron. 2013;78:498–509. doi: 10.1016/j.neuron.2013.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fombonne E, Roge B, Claverie J, Courty S, Fremolle J. Microcephaly and macrocephaly in autism. J Autism Dev Disord. 1999;29:113–119. doi: 10.1023/a:1023036509476. [DOI] [PubMed] [Google Scholar]
- Fraser MM, Bayazitov IT, Zakharenko SS, Baker SJ. Phosphatase and tensin homolog, deleted on chromosome 10 deficiency in brain causes defects in synaptic structure, transmission and plasticity, and myelination abnormalities. Neuroscience. 2008;151:476–488. doi: 10.1016/j.neuroscience.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T, Itoh M, Ichikawa T, Washiyama K, Goto Y. Delayed maturation of neuronal architecture and synaptogenesis in cerebral cortex of Mecp2-deficient mice. J Neuropathol Exp Neurol. 2005;64:537–544. doi: 10.1093/jnen/64.6.537. [DOI] [PubMed] [Google Scholar]
- Gauthier J, Siddiqui TJ, Huashan P, Yokomaku D, Hamdan FF, Champagne N, Lapointe M, Spiegelman D, Noreau A, Lafreniere RG, Fathalli F, Joober R, Krebs MO, DeLisi LE, Mottron L, Fombonne E, Michaud JL, Drapeau P, Carbonetto S, Craig AM, Rouleau GA. Truncating mutations in NRXN2 and NRXN1 in autism spectrum disorders and schizophrenia. Hum Genet. 2011;130:563–573. doi: 10.1007/s00439-011-0975-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemelli T, Berton O, Nelson ED, Perrotti LI, Jaenisch R, Monteggia LM. Postnatal loss of methyl-CpG binding protein 2 in the forebrain is sufficient to mediate behavioral aspects of Rett syndrome in mice. Biol Psychiatry. 2006;59:468–476. doi: 10.1016/j.biopsych.2005.07.025. [DOI] [PubMed] [Google Scholar]
- Geschwind DH, Levitt P. Autism spectrum disorders: developmental disconnection syndromes. Curr Opin Neurobiol. 2007;17:103–111. doi: 10.1016/j.conb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, Zhang H, Estes A, Brune CW, Bradfield JP, Imielinski M, Frackelton EC, Reichert J, Crawford EL, Munson J, Sleiman PM, Chiavacci R, Annaiah K, Thomas K, Hou C, Glaberson W, Flory J, Otieno F, Garris M, Soorya L, Klei L, Piven J, Meyer KJ, Anagnostou E, Sakurai T, Game RM, Rudd DS, Zurawiecki D, McDougle CJ, Davis LK, Miller J, Posey DJ, Michaels S, Kolevzon A, Silverman JM, Bernier R, Levy SE, Schultz RT, Dawson G, Owley T, McMahon WM, Wassink TH, Sweeney JA, Nurnberger JI, Coon H, Sutcliffe JS, Minshew NJ, Grant SF, Bucan M, Cook EH, Buxbaum JD, Devlin B, Schellenberg GD, Hakonarson H. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfraind JM, Reyniers E, De Boulle K, D'Hooge R, De Deyn PP, Bakker CE, Oostra BA, Kooy RF, Willems PJ. Long-term potentiation in the hippocampus of fragile X knockout mice. Am J Med Genet. 1996;64:246–251. doi: 10.1002/(SICI)1096-8628(19960809)64:2<246::AID-AJMG2>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Grabrucker AM, Knight MJ, Proepper C, Bockmann J, Joubert M, Rowan M, Nienhaus GU, Garner CC, Bowie JU, Kreutz MR, Gundelfinger ED, Boeckers TM. Concerted action of zinc and ProSAP/Shank in synaptogenesis and synapse maturation. EMBO J. 2011;30:569–581. doi: 10.1038/emboj.2010.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf ER, Zhang X, Jin SX, Linhoff MW, Craig AM. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell. 2004;119:1013–1026. doi: 10.1016/j.cell.2004.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayton HM, Missler M, Collier DA, Fernandes C. Altered social behaviours in neurexin 1alpha knockout mice resemble core symptoms in neurodevelopmental disorders. PLoS One. 2013;8:e67114. doi: 10.1371/journal.pone.0067114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer PL, Hanayama R, Bloodgood BL, Mardinly AR, Lipton DM, Flavell SW, Kim TK, Griffith EC, Waldon Z, Maehr R, Ploegh HL, Chowdhury S, Worley PF, Steen J, Greenberg ME. The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell. 2010;140:704–716. doi: 10.1016/j.cell.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunaydin LA, Grosenick L, Finkelstein JC, Kauvar IV, Fenno LE, Adhikari A, Lammel S, Mirzabekov JJ, Airan RD, Zalocusky KA, Tye KM, Anikeeva P, Malenka RC, Deisseroth K. Natural neural projection dynamics underlying social behavior. Cell. 2014;157:1535–1551. doi: 10.1016/j.cell.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315:1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- Hagerman R, Hoem G, Hagerman P. Fragile X and autism: Intertwined at the molecular level leading to targeted treatments. Mol Autism. 2010;1 doi: 10.1186/2040-2392-1-12. 12-2392-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallmayer J, Cleveland S, Torres A, Phillips J, Cohen B, Torigoe T, Miller J, Fedele A, Collins J, Smith K, Lotspeich L, Croen LA, Ozonoff S, Lajonchere C, Grether JK, Risch N. Genetic heritability and shared environmental factors among twin pairs with autism. Arch Gen Psychiatry. 2011;68:1095–1102. doi: 10.1001/archgenpsychiatry.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haws ME, Jaramillo TC, Espinosa F, Widman AJ, Stuber GD, Sparta DR, Tye KM, Russo SJ, Parada LF, Stavarache M, Kaplitt M, Bonci A, Powell CM. PTEN knockdown alters dendritic spine/protrusion morphology, not density. J Comp Neurol. 2014;522:1171–1190. doi: 10.1002/cne.23488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayrapetyan V, Castro S, Sukharnikova T, Yu C, Cao X, Jiang YH, Yin HH. Region-specific impairments in striatal synaptic transmission and impaired instrumental learning in a mouse model of Angelman syndrome. Eur J Neurosci. 2014;39:1018–1025. doi: 10.1111/ejn.12442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He CX, Portera-Cailliau C. The trouble with spines in fragile X syndrome: density, maturity and plasticity. Neuroscience. 2013;251:120–128. doi: 10.1016/j.neuroscience.2012.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffer CA, Klann E. mTOR signaling: at the crossroads of plasticity, memory and disease. Trends Neurosci. 2010;33:67–75. doi: 10.1016/j.tins.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogart A, Wu D, LaSalle JM, Schanen NC. The comorbidity of autism with the genomic disorders of chromosome 15q11.2-q13. Neurobiol Dis. 2010;38:181–191. doi: 10.1016/j.nbd.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlin P, Goode S, Hutton J, Rutter M. Adult outcome for children with autism. J Child Psychol Psychiatry. 2004;45:212–229. doi: 10.1111/j.1469-7610.2004.00215.x. [DOI] [PubMed] [Google Scholar]
- Huber KM, Gallagher SM, Warren ST, Bear MF. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc Natl Acad Sci USA. 2002;99:7746–7750. doi: 10.1073/pnas.122205699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Howley PM. Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol Cell Biol. 1993;13:775–784. doi: 10.1128/mcb.13.2.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung AY, Futai K, Sala C, Valtschanoff JG, Ryu J, Woodworth MA, Kidd FL, Sung CC, Miyakawa T, Bear MF, Weinberg RJ, Sheng M. Smaller dendritic spines, weaker synaptic transmission, but enhanced spatial learning in mice lacking Shank1. J Neurosci. 2008;28:1697–1708. doi: 10.1523/JNEUROSCI.3032-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichtchenko K, Nguyen T, Sudhof TC. Structures, alternative splicing, and neurexin binding of multiple neuroligins. J Biol Chem. 1996;271:2676–2682. doi: 10.1074/jbc.271.5.2676. [DOI] [PubMed] [Google Scholar]
- Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- Isshiki M, Tanaka S, Kuriu T, Tabuchi K, Takumi T, Okabe S. Enhanced synapse remodelling as a common phenotype in mouse models of autism. Nat Commun. 2014;5:4742. doi: 10.1038/ncomms5742. [DOI] [PubMed] [Google Scholar]
- Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, Gillberg IC, Soderstrom H, Giros B, Leboyer M, Gillberg C, Bourgeron T Paris Autism Research International Sibpair Study. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamain S, Radyushkin K, Hammerschmidt K, Granon S, Boretius S, Varoqueaux F, Ramanantsoa N, Gallego J, Ronnenberg A, Winter D, Frahm J, Fischer J, Bourgeron T, Ehrenreich H, Brose N. Reduced social interaction and ultrasonic communication in a mouse model of monogenic heritable autism. Proc Natl Acad Sci USA. 2008;105:1710–1715. doi: 10.1073/pnas.0711555105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo TC, Liu S, Pettersen A, Birnbaum SG, Powell CM. Autism-related neuroligin-3 mutation alters social behavior and spatial learning. Autism Res. 2014;7:264–272. doi: 10.1002/aur.1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo TC, Speed HE, Xuan Z, Reimers JM, Liu S, Powell CM. Altered Striatal Synaptic Function and Abnormal Behaviour in Shank3 Exon4–9 Deletion Mouse Model of Autism. Autism Res. 2015 doi: 10.1002/aur.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedlicka P, Vnencak M, Krueger DD, Jungenitz T, Brose N, Schwarzacher SW. Neuroligin-1 regulates excitatory synaptic transmission, LTP and EPSP-spike coupling in the dentate gyrus in vivo. Brain Struct Funct. 2015;220:47–58. doi: 10.1007/s00429-013-0636-1. [DOI] [PubMed] [Google Scholar]
- Jiang YH, Armstrong D, Albrecht U, Atkins CM, Noebels JL, Eichele G, Sweatt JD, Beaudet AL. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21:799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- Jiang YH, Ehlers MD. Modeling autism by SHANK gene mutations in mice. Neuron. 2013;78:8–27. doi: 10.1016/j.neuron.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju A, Hammerschmidt K, Tantra M, Krueger D, Brose N, Ehrenreich H. Juvenile manifestation of ultrasound communication deficits in the neuroligin-4 null mutant mouse model of autism. Behav Brain Res. 2014;270:159–164. doi: 10.1016/j.bbr.2014.05.019. [DOI] [PubMed] [Google Scholar]
- Jung SY, Kim J, Kwon OB, Jung JH, An K, Jeong AY, Lee CJ, Choi YB, Bailey CH, Kandel ER, Kim JH. Input-specific synaptic plasticity in the amygdala is regulated by neuroligin-1 via postsynaptic NMDA receptors. Proc Natl Acad Sci USA. 2010;107:4710–4715. doi: 10.1073/pnas.1001084107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Just MA, Cherkassky VL, Keller TA, Minshew NJ. Cortical activation and synchronization during sentence comprehension in high-functioning autism: evidence of underconnectivity. Brain. 2004;127:1811–1821. doi: 10.1093/brain/awh199. [DOI] [PubMed] [Google Scholar]
- Kana RK, Uddin LQ, Kenet T, Chugani D, Muller RA. Brain connectivity in autism. Front Hum Neurosci. 2014;8:349. doi: 10.3389/fnhum.2014.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karayannis T, Au E, Patel JC, Kruglikov I, Markx S, Delorme R, Heron D, Salomon D, Glessner J, Restituito S, Gordon A, Rodriguez-Murillo L, Roy NC, Gogos JA, Rudy B, Rice ME, Karayiorgou M, Hakonarson H, Keren B, Huguet G, Bourgeron T, Hoeffer C, Tsien RW, Peles E, Fishell G. Cntnap4 differentially contributes to GABAergic and dopaminergic synaptic transmission. Nature. 2014;511:236–240. doi: 10.1038/nature13248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karten A, Hirsch J. Brief Report: Anomalous Neural Deactivations and Functional Connectivity During Receptive Language in Autism Spectrum Disorder: A Functional MRI Study. J Autism Dev Disord. 2014;45:1905–1914. doi: 10.1007/s10803-014-2344-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keown CL, Shih P, Nair A, Peterson N, Mulvey ME, Muller RA. Local functional overconnectivity in posterior brain regions is associated with symptom severity in autism spectrum disorders. Cell reports. 2013;5:567–572. doi: 10.1016/j.celrep.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Chahrour M, Ben-Shachar S, Lim J. Ube3a/E6AP is involved in a subset of MeCP2 functions. Biochem Biophys Res Commun. 2013;437:67–73. doi: 10.1016/j.bbrc.2013.06.036. [DOI] [PubMed] [Google Scholar]
- King M, Bearman P. Diagnostic change and the increased prevalence of autism. Int J Epidemiol. 2009;38:1224–1234. doi: 10.1093/ije/dyp261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- Kline DD, Ogier M, Kunze DL, Katz DM. Exogenous brain-derived neurotrophic factor rescues synaptic dysfunction in Mecp2-null mice. J Neurosci. 2010;30:5303–5310. doi: 10.1523/JNEUROSCI.5503-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Minowa O, Kuno J, Mitani H, Hino O, Noda T. Renal carcinogenesis, hepatic hemangiomatosis, and embryonic lethality caused by a germ-line Tsc2 mutation in mice. Cancer Res. 1999;59:1206–1211. [PubMed] [Google Scholar]
- Kobayashi T, Minowa O, Sugitani Y, Takai S, Mitani H, Kobayashi E, Noda T, Hino O. A germ-line Tsc1 mutation causes tumor development and embryonic lethality that are similar, but not identical to, those caused by Tsc2 mutation in mice. Proc Natl Acad Sci USA. 2001;98:8762–8767. doi: 10.1073/pnas.151033798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koekkoek SK, Yamaguchi K, Milojkovic BA, Dortland BR, Ruigrok TJ, Maex R, De Graaf W, Smit AE, VanderWerf F, Bakker CE, Willemsen R, Ikeda T, Kakizawa S, Onodera K, Nelson DL, Mientjes E, Joosten M, De Schutter E, Oostra BA, Ito M, De Zeeuw CI. Deletion of FMR1 in Purkinje cells enhances parallel fiber LTD, enlarges spines, and attenuates cerebellar eyelid conditioning in Fragile X syndrome. Neuron. 2005;47:339–352. doi: 10.1016/j.neuron.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Kouser M, Speed HE, Dewey CM, Reimers JM, Widman AJ, Gupta N, Liu S, Jaramillo TC, Bangash M, Xiao B, Worley PF, Powell CM. Loss of predominant Shank3 isoforms results in hippocampus-dependent impairments in behavior and synaptic transmission. J Neurosci. 2013;33:18448–18468. doi: 10.1523/JNEUROSCI.3017-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhnle S, Mothes B, Matentzoglu K, Scheffner M. Role of the ubiquitin ligase E6AP/UBE3A in controlling levels of the synaptic protein Arc. Proc Natl Acad Sci USA. 2013;110:8888–8893. doi: 10.1073/pnas.1302792110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Talis AL, Howley PM. Identification of HHR23A as a substrate for E6-associated protein-mediated ubiquitination. J Biol Chem. 1999;274:18785–18792. doi: 10.1074/jbc.274.26.18785. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski DJ, Zhang H, Bandura JL, Heiberger KM, Glogauer M, el-Hashemite N, Onda H. A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum Mol Genet. 2002;11:525–534. doi: 10.1093/hmg/11.5.525. [DOI] [PubMed] [Google Scholar]
- Kwon CH, Luikart BW, Powell CM, Zhou J, Matheny SA, Zhang W, Li Y, Baker SJ, Parada LF. Pten regulates neuronal arborization and social interaction in mice. Neuron. 2006;50:377– 388. doi: 10.1016/j.neuron.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HB, Kozorovitskiy Y, Oh WJ, Peixoto RT, Akhtar N, Saulnier JL, Gu C, Sabatini BL. Neuroligin-1-dependent competition regulates cortical synaptogenesis and synapse number. Nat Neurosci. 2012;15:1667–1674. doi: 10.1038/nn.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landi S, Putignano E, Boggio EM, Giustetto M, Pizzorusso T, Ratto GM. The short-time structural plasticity of dendritic spines is altered in a model of Rett syndrome. Sci Rep. 2011;1:45. doi: 10.1038/srep00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang M, Wither RG, Colic S, Wu C, Monnier PP, Bardakjian BL, Zhang L, Eubanks JH. Rescue of behavioral and EEG deficits in male and female Mecp2-deficient mice by delayed Mecp2 gene reactivation. Hum Mol Genet. 2014;23:303–318. doi: 10.1093/hmg/ddt421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauterborn JC, Rex CS, Kramar E, Chen LY, Pandyarajan V, Lynch G, Gall CM. Brain-derived neurotrophic factor rescues synaptic plasticity in a mouse model of fragile X syndrome. J Neurosci. 2007;27:10685–10694. doi: 10.1523/JNEUROSCI.2624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Chung C, Ha S, Lee D, Kim DY, Kim H, Kim E. Shank3-mutant mice lacking exon 9 show altered excitation/inhibition balance, enhanced rearing, and spatial memory deficit. Front Cell Neurosci. 2015;9:94. doi: 10.3389/fncel.2015.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Ge WP, Huang W, He Y, Wang GX, Rowson-Baldwin A, Smith SJ, Jan YN, Jan LY. Bidirectional regulation of dendritic voltage-gated potassium channels by the fragile X mental retardation protein. Neuron. 2011;72:630–642. doi: 10.1016/j.neuron.2011.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Pelletier MR, Perez Velazquez JL, Carlen PL. Reduced cortical synaptic plasticity and GluR1 expression associated with fragile X mental retardation protein deficiency. Mol Cell Neurosci. 2002;19:138–151. doi: 10.1006/mcne.2001.1085. [DOI] [PubMed] [Google Scholar]
- Liu KY, King M, Bearman PS. Social influence and the autism epidemic. AJS. 2010;115:1387–1434. doi: 10.1086/651448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonetti G, Angelucci A, Morando L, Boggio EM, Giustetto M, Pizzorusso T. Early environmental enrichment moderates the behavioral and synaptic phenotype of MeCP2 null mice. Biol Psychiatry. 2010;67:657–665. doi: 10.1016/j.biopsych.2009.12.022. [DOI] [PubMed] [Google Scholar]
- Lord C, Cook EH, Leventhal BL, Amaral DG. Autism spectrum disorders. Neuron. 2000;28:355–363. doi: 10.1016/s0896-6273(00)00115-x. [DOI] [PubMed] [Google Scholar]
- Louria-Hayon I, Alsheich-Bartok O, Levav-Cohen Y, Silberman I, Berger M, Grossman T, Matentzoglu K, Jiang YH, Muller S, Scheffner M, Haupt S, Haupt Y. E6AP promotes the degradation of the PML tumor suppressor. Cell Death Differ. 2009;16:1156–1166. doi: 10.1038/cdd.2009.31. [DOI] [PubMed] [Google Scholar]
- Lugo JN, Smith GD, Arbuckle EP, White J, Holley AJ, Floruta CM, Ahmed N, Gomez MC, Okonkwo O. Deletion of PTEN produces autism-like behavioral deficits and alterations in synaptic proteins. Front Mol Neurosci. 2014;7:27. doi: 10.3389/fnmol.2014.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikart BW, Schnell E, Washburn EK, Bensen AL, Tovar KR, Westbrook GL. Pten knockdown in vivo increases excitatory drive onto dentate granule cells. J Neurosci. 2011;31:4345–4354. doi: 10.1523/JNEUROSCI.0061-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyst MJ, Bird A. Rett syndrome: a complex disorder with simple roots. Nat Rev Genet. 2015;16:261–275. doi: 10.1038/nrg3897. [DOI] [PubMed] [Google Scholar]
- Mabb AM, Ehlers MD. Ubiquitination in postsynaptic function and plasticity. Annu Rev Cell Dev Biol. 2010;26:179–210. doi: 10.1146/annurev-cellbio-100109-104129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- Mani A, Oh AS, Bowden ET, Lahusen T, Lorick KL, Weissman AM, Schlegel R, Wellstein A, Riegel AT. E6AP mediates regulated proteasomal degradation of the nuclear receptor coactivator amplified in breast cancer 1 in immortalized cells. Cancer Res. 2006;66:8680–8686. doi: 10.1158/0008-5472.CAN-06-0557. [DOI] [PubMed] [Google Scholar]
- Matsuura T, Sutcliffe JS, Fang P, Galjaard RJ, Jiang YH, Benton CS, Rommens JM, Beaudet AL. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet. 1997;15:74–77. doi: 10.1038/ng0197-74. [DOI] [PubMed] [Google Scholar]
- Maunakea AK, Chepelev I, Cui K, Zhao K. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 2013;23:1256–1269. doi: 10.1038/cr.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw CM, Samaco RC, Zoghbi HY. Adult neural function requires MeCP2. Science. 2011 Jul 8;333(6039):186. doi: 10.1126/science.1206593. Epub 2011 Jun 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon JJ, Yu W, Yang J, Feng H, Helm M, McMahon E, Zhu X, Shin D, Huang Y. Seizure-dependent mTOR activation in 5-HT neurons promotes autism-like behaviors in mice. Neurobiol Dis. 2014;73C:296–306. doi: 10.1016/j.nbd.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer G, Varoqueaux F, Neeb A, Oschlies M, Brose N. The complexity of PDZ domain-mediated interactions at glutamatergic synapses: a case study on neuroligin. Neuropharmacology. 2004;47:724–733. doi: 10.1016/j.neuropharm.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Miao S, Chen R, Ye J, Tan GH, Li S, Zhang J, Jiang YH, Xiong ZQ. The Angelman syndrome protein Ube3a is required for polarized dendrite morphogenesis in pyramidal neurons. J Neurosci. 2013;33:327–333. doi: 10.1523/JNEUROSCI.2509-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalon A, Sidorov M, Ballard TM, Ozmen L, Spooren W, Wettstein JG, Jaeschke G, Bear MF, Lindemann L. Chronic pharmacological mGlu5 inhibition corrects fragile X in adult mice. Neuron. 2012;74:49–56. doi: 10.1016/j.neuron.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mientjes EJ, Nieuwenhuizen I, Kirkpatrick L, Zu T, Hoogeveen-Westerveld M, Severijnen L, Rife M, Willemsen R, Nelson DL, Oostra BA. The generation of a conditional Fmr1 knock out mouse model to study Fmrp function in vivo. Neurobiol Dis. 2006;21:549–555. doi: 10.1016/j.nbd.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Minshew NJ, Keller TA. The nature of brain dysfunction in autism: functional brain imaging studies. Curr Opin Neurol. 2010;23:124–130. doi: 10.1097/WCO.0b013e32833782d4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missler M, Zhang W, Rohlmann A, Kattenstroth G, Hammer RE, Gottmann K, Sudhof TC. Alpha-neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature. 2003;423:939–948. doi: 10.1038/nature01755. [DOI] [PubMed] [Google Scholar]
- Miura K, Kishino T, Li E, Webber H, Dikkes P, Holmes GL, Wagstaff J. Neurobehavioral and electroencephalographic abnormalities in Ube3a maternal-deficient mice. Neurobiol Dis. 2002;9:149–159. doi: 10.1006/nbdi.2001.0463. [DOI] [PubMed] [Google Scholar]
- Moretti P, Levenson JM, Battaglia F, Atkinson R, Teague R, Antalffy B, Armstrong D, Arancio O, Sweatt JD, Zoghbi HY. Learning and memory and synaptic plasticity are impaired in a mouse model of Rett syndrome. J Neurosci. 2006;26:319–327. doi: 10.1523/JNEUROSCI.2623-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulherkar SA, Sharma J, Jana NR. The ubiquitin ligase E6-AP promotes degradation of alpha-synuclein. J Neurochem. 2009;110:1955–1964. doi: 10.1111/j.1471-4159.2009.06293.x. [DOI] [PubMed] [Google Scholar]
- Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88(4):471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- Napoli E, Ross-Inta C, Wong S, Hung C, Fujisawa Y, Sakaguchi D, Angelastro J, Omanska-Klusek A, Schoenfeld R, Giulivi C. Mitochondrial dysfunction in Pten haplo-insufficient mice with social deficits and repetitive behavior: interplay between Pten and p53. PLoS One. 2012;7:e42504. doi: 10.1371/journal.pone.0042504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevison CD. A comparison of temporal trends in United States autism prevalence to trends in suspected environmental factors. Environ Health. 2014;13 doi: 10.1186/1476-069X-13-73. 73-069X-13-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci. 2015;18(2):199–209. doi: 10.1038/nn.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogier M, Wang H, Hong E, Wang Q, Greenberg ME, Katz DM. Brain-derived neurotrophic factor expression and respiratory function improve after ampakine treatment in a mouse model of Rett syndrome. J Neurosci. 2007;27:10912–10917. doi: 10.1523/JNEUROSCI.1869-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onda H, Lueck A, Marks PW, Warren HB, Kwiatkowski DJ. Tsc2(+/−) mice develop tumors in multiple sites that express gelsolin and are influenced by genetic background. J Clin Invest. 1999;104:687–695. doi: 10.1172/JCI7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F, Aldridge GM, Greenough WT, Gan WB. Dendritic spine instability and insensitivity to modulation by sensory experience in a mouse model of fragile X syndrome. Proc Natl Acad Sci USA. 2010;107:17768–17773. doi: 10.1073/pnas.1012496107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradee W, Melikian HE, Rasmussen DL, Kenneson A, Conn PJ, Warren ST. Fragile X mouse: strain effects of knockout phenotype and evidence suggesting deficient amygdala function. Neuroscience. 1999;94:185–192. doi: 10.1016/s0306-4522(99)00285-7. [DOI] [PubMed] [Google Scholar]
- Peca J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, Lascola CD, Fu Z, Feng G. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penagarikano O, Abrahams BS, Herman EI, Winden KD, Gdalyahu A, Dong H, Sonnenblick LI, Gruver R, Almajano J, Bragin A, Golshani P, Trachtenberg JT, Peles E, Geschwind DH. Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell. 2011;147:235–246. doi: 10.1016/j.cell.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persico AM, Napolioni V. Autism genetics. Behav Brain Res. 2013;251:95–112. doi: 10.1016/j.bbr.2013.06.012. [DOI] [PubMed] [Google Scholar]
- Peters SU, Beaudet AL, Madduri N, Bacino CA. Autism in Angelman syndrome: implications for autism research. Clin Genet. 2004;66:530–536. doi: 10.1111/j.1399-0004.2004.00362.x. [DOI] [PubMed] [Google Scholar]
- Pieretti M, Zhang FP, Fu YH, Warren ST, Oostra BA, Caskey CT, Nelson DL. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991;66:817–822. doi: 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- Pignatelli M, Piccinin S, Molinaro G, Di Menna L, Riozzi B, Cannella M, Motolese M, Vetere G, Catania MV, Battaglia G, Nicoletti F, Nistico R, Bruno V. Changes in mGlu5 receptor-dependent synaptic plasticity and coupling to homer proteins in the hippocampus of Ube3A hemizygous mice modeling angelman syndrome. J Neurosci. 2014;34:4558–4566. doi: 10.1523/JNEUROSCI.1846-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilpel Y, Kolleker A, Berberich S, Ginger M, Frick A, Mientjes E, Oostra BA, Seeburg PH. Synaptic ionotropic glutamate receptors and plasticity are developmentally altered in the CA1 field of Fmr1 knockout mice. J Physiol. 2009;587:787–804. doi: 10.1113/jphysiol.2008.160929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzarelli R, Cherubini E. Developmental regulation of GABAergic signalling in the hippocampus of neuroligin 3 R451C knock-in mice: an animal model of Autism. Front Cell Neurosci. 2013;7:85. doi: 10.3389/fncel.2013.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CJ, Pedraza LG, Xu T. Akt regulates growth by directly phosphorylating Tsc2. Nat Cell Biol. 2002;4:658–665. doi: 10.1038/ncb840. [DOI] [PubMed] [Google Scholar]
- Rabaneda LG, Robles-Lanuza E, Nieto-Gonzalez JL, Scholl FG. Neurexin dysfunction in adult neurons results in autistic-like behavior in mice. Cell Rep. 2014;8:338–346. doi: 10.1016/j.celrep.2014.06.022. [DOI] [PubMed] [Google Scholar]
- Radyushkin K, Hammerschmidt K, Boretius S, Varoqueaux F, El-Kordi A, Ronnenberg A, Winter D, Frahm J, Fischer J, Brose N, Ehrenreich H. Neuroligin-3-deficient mice: model of a monogenic heritable form of autism with an olfactory deficit. Genes Brain Behav. 2009;8:416–425. doi: 10.1111/j.1601-183X.2009.00487.x. [DOI] [PubMed] [Google Scholar]
- Reith RM, McKenna J, Wu H, Hashmi SS, Cho SH, Dash PK, Gambello MJ. Loss of Tsc2 in Purkinje cells is associated with autistic-like behavior in a mouse model of tuberous sclerosis complex. Neurobiol Dis. 2013;51:93–103. doi: 10.1016/j.nbd.2012.10.014. [DOI] [PubMed] [Google Scholar]
- Robinson L, Guy J, McKay L, Brockett E, Spike RC, Selfridge J, De Sousa D, Merusi C, Riedel G, Bird A, Cobb SR. Morphological and functional reversal of phenotypes in a mouse model of Rett syndrome. Brain. 2012;135:2699–2710. doi: 10.1093/brain/aws096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell PE, Fuccillo MV, Maxeiner S, Hayton SJ, Gokce O, Lim BK, Fowler SC, Malenka RC, Sudhof TC. Autism-associated neuroligin-3 mutations commonly impair striatal circuits to boost repetitive behaviors. Cell. 2014;158:198–212. doi: 10.1016/j.cell.2014.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougeulle C, Glatt H, Lalande M. The Angelman syndrome candidate gene, UBE3A/E6-AP, is imprinted in brain. Nat Genet. 1997;17:14–15. doi: 10.1038/ng0997-14. [DOI] [PubMed] [Google Scholar]
- Roussignol G, Ango F, Romorini S, Tu JC, Sala C, Worley PF, Bockaert J, Fagni L. Shank expression is sufficient to induce functional dendritic spine synapses in aspiny neurons. J Neurosci. 2005;25:3560–3570. doi: 10.1523/JNEUROSCI.4354-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato D, Lionel AC, Leblond CS, Prasad A, Pinto D, Walker S, O'Connor I, Russell C, Drmic IE, Hamdan FF, Michaud JL, Endris V, Roeth R, Delorme R, Huguet G, Leboyer M, Rastam M, Gillberg C, Lathrop M, Stavropoulos DJ, Anagnostou E, Weksberg R, Fombonne E, Zwaigenbaum L, Fernandez BA, Roberts W, Rappold GA, Marshall CR, Bourgeron T, Szatmari P, Scherer SW. SHANK1 Deletions in Males with Autism Spectrum Disorder. Am J Hum Genet. 2012;90:879–887. doi: 10.1016/j.ajhg.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Stryker MP. Genomic imprinting of experience-dependent cortical plasticity by the ubiquitin ligase gene Ube3a. Proc Natl Acad Sci USA. 2010;107:5611–5616. doi: 10.1073/pnas.1001281107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiffele P, Fan J, Choih J, Fetter R, Serafini T. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell. 2000;101:657–669. doi: 10.1016/s0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- Schmeisser MJ, Ey E, Wegener S, Bockmann J, Stempel AV, Kuebler A, Janssen AL, Udvardi PT, Shiban E, Spilker C, Balschun D, Skryabin BV, Dieck S, Smalla KH, Montag D, Leblond CS, Faure P, Torquet N, Le Sourd AM, Toro R, Grabrucker AM, Shoichet SA, Schmitz D, Kreutz MR, Bourgeron T, Gundelfinger ED, Boeckers TM. Autistic-like behaviours and hyperactivity in mice lacking ProSAP1/Shank2. Nature. 2012;486:256–260. doi: 10.1038/nature11015. [DOI] [PubMed] [Google Scholar]
- Schutt J, Falley K, Richter D, Kreienkamp HJ, Kindler S. Fragile X mental retardation protein regulates the levels of scaffold proteins and glutamate receptors in postsynaptic densities. J Biol Chem. 2009;284:25479–25487. doi: 10.1074/jbc.M109.042663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian M, Young J, Yuva-Paylor L, Spencer C, Antalffy B, Noebels J, Armstrong D, Paylor R, Zoghbi H. Mice with truncated MeCP2 recapitulate many Rett syndrome features and display hyperacetylation of histone H3. Neuron. 2002;35:243–254. doi: 10.1016/s0896-6273(02)00768-7. [DOI] [PubMed] [Google Scholar]
- Sharma A, Hoeffer CA, Takayasu Y, Miyawaki T, McBride SM, Klann E, Zukin RS. Dysregulation of mTOR signaling in fragile X syndrome. J Neurosci. 2010;30:694–702. doi: 10.1523/JNEUROSCI.3696-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, Kim E. The Shank family of scaffold proteins. J Cell Sci. 2000;113(Pt 11):1851–1856. doi: 10.1242/jcs.113.11.1851. [DOI] [PubMed] [Google Scholar]
- Sidorov MS, Auerbach BD, Bear MF. Fragile X mental retardation protein and synaptic plasticity. Mol Brain. 2013;8;6:15. doi: 10.1186/1756-6606-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Santos S, van Woerden GM, Bruinsma CF, Mientjes E, Jolfaei MA, Distel B, Kushner SA, Elgersma Y. Ube3a reinstatement identifies distinct developmental windows in a murine Angelman syndrome model. J Clin Invest. 2015;125:2069–2076. doi: 10.1172/JCI80554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Smith DG, Rizzo SJ, Karras MN, Turner SM, Tolu SS, Bryce DK, Smith DL, Fonseca K, Ring RH, Crawley JN. Negative allosteric modulation of the mGluR5 receptor reduces repetitive behaviors and rescues social deficits in mouse models of autism. Sci Transl Med. 2012;4:131ra51. doi: 10.1126/scitranslmed.3003501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Turner SM, Barkan CL, Tolu SS, Saxena R, Hung AY, Sheng M, Crawley JN. Sociability and motor functions in Shank1 mutant mice. Brain Res. 2011;1380:120–137. doi: 10.1016/j.brainres.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Yang M, Lord C, Crawley JN. Behavioural phenotyping assays for mouse models of autism. Nat Rev Neurosci. 2010;11:490–502. doi: 10.1038/nrn2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley SL, Tanguay PE, Smith M, Gutierrez G. Autism and tuberous sclerosis. J Autism Dev Disord. 1992;22:339–355. doi: 10.1007/BF01048239. [DOI] [PubMed] [Google Scholar]
- Smith SE, Zhou YD, Zhang G, Jin Z, Stoppel DC, Anderson MP. Increased gene dosage of Ube3a results in autism traits and decreased glutamate synaptic transmission in mice. Sci Transl Med. 2011;3:103ra97. doi: 10.1126/scitranslmed.3002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrt RD, Eaves-Egenes J, Barkho BZ, Santistevan NJ, Zhao C, Aimone JB, Gage FH, Zhao X. Mecp2 deficiency leads to delayed maturation and altered gene expression in hippocampal neurons. Neurobiol Dis. 2007;27:77–89. doi: 10.1016/j.nbd.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speed HE, Kouser M, Xuan Z, Reimers JM, Ochoa CF, Gupta N, Liu S, Powell CM. Autism-Associated Insertion Mutation (InsG) of Shank3 Exon 21 Causes Impaired Synaptic Transmission and Behavioral Deficits. J Neurosci. 2015;35:9648–9665. doi: 10.1523/JNEUROSCI.3125-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperow M, Berry RB, Bayazitov IT, Zhu G, Baker SJ, Zakharenko SS. Phosphatase and tensin homologue (PTEN) regulates synaptic plasticity independently of its effect on neuronal morphology and migration. J Physiol. 2012;590:777–792. doi: 10.1113/jphysiol.2011.220236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg KM, Ramachandran D, Patel VC, Shetty AC, Cutler DJ, Zwick ME. Identification of rare X-linked neuroligin variants by massively parallel sequencing in males with autism spectrum disorder. Mol Autism. 2012;3 doi: 10.1186/2040-2392-3-8. 8-2392-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungur AO, Vorckel KJ, Schwarting RK, Wohr M. Repetitive behaviors in the Shank1 knockout mouse model for autism spectrum disorder: developmental aspects and effects of social context. J Neurosci Methods. 2014;234:92–100. doi: 10.1016/j.jneumeth.2014.05.003. [DOI] [PubMed] [Google Scholar]
- Supekar K, Uddin LQ, Khouzam A, Phillips J, Gaillard WD, Kenworthy LE, Yerys BE, Vaidya CJ, Menon V. Brain hyperconnectivity in children with autism and its links to social deficits. Cell Rep. 2013;5:738–747. doi: 10.1016/j.celrep.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, de la Pompa JL, Stambolic V, Elia AJ, Sasaki T, del Barco Barrantes I, Ho A, Wakeham A, Itie A, Khoo W, Fukumoto M, Mak TW. High cancer susceptibility and embryonic lethality associated with mutation of the PTEN tumor suppressor gene in mice. Curr Biol. 1998;8:1169–1178. doi: 10.1016/s0960-9822(07)00488-5. [DOI] [PubMed] [Google Scholar]
- Tabuchi K, Blundell J, Etherton MR, Hammer RE, Liu X, Powell CM, Sudhof TC. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science. 2007;318:71–76. doi: 10.1126/science.1146221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi K, Sudhof TC. Structure and evolution of neurexin genes: insight into the mechanism of alternative splicing. Genomics. 2002;79:849–859. doi: 10.1006/geno.2002.6780. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Gertner MJ, Zhou J, Parada LF, Bennett MV, Zukin RS. Dysregulation of synaptic plasticity precedes appearance of morphological defects in a Pten conditional knockout mouse model of autism. Proc Natl Acad Sci USA. 2013;110:4738–4743. doi: 10.1073/pnas.1222803110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Gudsnuk K, Kuo SH, Cotrina ML, Rosoklija G, Sosunov A, Sonders MS, Kanter E, Castagna C, Yamamoto A, Yue Z, Arancio O, Peterson BS, Champagne F, Dwork AJ, Goldman J, Sulzer D. Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron. 2014;83:1131–1143. doi: 10.1016/j.neuron.2014.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang SJ, Reis G, Kang H, Gingras AC, Sonenberg N, Schuman EM. A rapamycin-sensitive signaling pathway contributes to long-term synaptic plasticity in the hippocampus. Proc Natl Acad Sci USA. 2002;99:467–472. doi: 10.1073/pnas.012605299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie SF, Alvarez VA, Ridenour DA, Kwiatkowski DJ, Sabatini BL. Regulation of neuronal morphology and function by the tumor suppressors Tsc1 and Tsc2. Nat Neurosci. 2005;8:1727–1734. doi: 10.1038/nn1566. [DOI] [PubMed] [Google Scholar]
- Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- Tilot AK, Gaugler MK, Yu Q, Romigh T, Yu W, Miller RH, Frazier TW, 2nd, Eng C. Germline disruption of Pten localization causes enhanced sex-dependent social motivation and increased glial production. Hum Mol Genet. 2014;23:3212–3227. doi: 10.1093/hmg/ddu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein B, Gokce O, Quake SR, Sudhof TC. Cartography of neurexin alternative splicing mapped by single-molecule long-read mRNA sequencing. Proc Natl Acad Sci USA. 2014;111:E1291–E1299. doi: 10.1073/pnas.1403244111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai PT, Hull C, Chu Y, Greene-Colozzi E, Sadowski AR, Leech JM, Steinberg J, Crawley JN, Regehr WG, Sahin M. Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature. 2012;488:647–651. doi: 10.1038/nature11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin LQ, Supekar K, Menon V. Reconceptualizing functional brain connectivity in autism from a developmental perspective. Front Hum Neurosci. 2013;7:458. doi: 10.3389/fnhum.2013.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich B, Ushkaryov YA, Sudhof TC. Cartography of neurexins: more than 1000 isoforms generated by alternative splicing and expressed in distinct subsets of neurons. Neuron. 1995;14:497–507. doi: 10.1016/0896-6273(95)90306-2. [DOI] [PubMed] [Google Scholar]
- Vaags AK, Lionel AC, Sato D, Goodenberger M, Stein QP, Curran S, Ogilvie C, Ahn JW, Drmic I, Senman L, Chrysler C, Thompson A, Russell C, Prasad A, Walker S, Pinto D, Marshall CR, Stavropoulos DJ, Zwaigenbaum L, Fernandez BA, Fombonne E, Bolton PF, Collier DA, Hodge JC, Roberts W, Szatmari P, Scherer SW. Rare deletions at the neurexin 3 locus in autism spectrum disorder. Am J Hum Genet. 2012;90:133–141. doi: 10.1016/j.ajhg.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Slegtenhorst M, de Hoogt R, Hermans C, Nellist M, Janssen B, Verhoef S, Lindhout D, van den Ouweland A, Halley D, Young J, Burley M, Jeremiah S, Woodward K, Nahmias J, Fox M, Ekong R, Osborne J, Wolfe J, Povey S, Snell RG, Cheadle JP, Jones AC, Tachataki M, Ravine D, Sampson JR, Reeve MP, Richardson P, Wilmer F, Munro C, Hawkins TL, Sepp T, Ali JB, Ward S, Green AJ, Yates JR, Kwiatkowska J, Henske EP, Short MP, Haines JH, Jozwiak S, Kwiatkowski DJ. Identification of the tuberous sclerosis gene TSC1 on chromosome 9q34. Science. 1997;277:805–808. doi: 10.1126/science.277.5327.805. [DOI] [PubMed] [Google Scholar]
- Varoqueaux F, Aramuni G, Rawson RL, Mohrmann R, Missler M, Gottmann K, Zhang W, Sudhof TC, Brose N. Neuroligins determine synapse maturation and function. Neuron. 2006;51:741–754. doi: 10.1016/j.neuron.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Vissers ME, Cohen MX, Geurts HM. Brain connectivity and high functioning autism: a promising path of research that needs refined models, methodological convergence, and stronger behavioral links. Neurosci Biobehav Rev. 2012;36:604–625. doi: 10.1016/j.neubiorev.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Wallace ML, Burette AC, Weinberg RJ, Philpot BD. Maternal loss of Ube3a produces an excitatory/inhibitory imbalance through neuron type-specific synaptic defects. Neuron. 2012;74:793–800. doi: 10.1016/j.neuron.2012.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, McCoy PA, Rodriguiz RM, Pan Y, Je HS, Roberts AC, Kim CJ, Berrios J, Colvin JS, Bousquet-Moore D, Lorenzo I, Wu G, Weinberg RJ, Ehlers MD, Philpot BD, Beaudet AL, Wetsel WC, Jiang YH. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum Mol Genet. 2011;20:3093–3108. doi: 10.1093/hmg/ddr212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Xu Q, Bey AL, Lee Y, Jiang YH. Transcriptional and functional complexity of Shank3 provides a molecular framework to understand the phenotypic heterogeneity of SHANK3 causing autism and Shank3 mutant mice. Mol Autism. 2014;5 doi: 10.1186/2040-2392-5-30. 30-2392-5-30. eCollection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cheng A, Mattson MP. The PTEN phosphatase is essential for long-term depression of hippocampal synapses. Neuromolecular Med. 2006;8:329–336. doi: 10.1385/NMM:8:3:329. [DOI] [PubMed] [Google Scholar]
- Weeber EJ, Jiang YH, Elgersma Y, Varga AW, Carrasquillo Y, Brown SE, Christian JM, Mirnikjoo B, Silva A, Beaudet AL, Sweatt JD. Derangements of hippocampal calcium/calmodulin-dependent protein kinase II in a mouse model for Angelman mental retardation syndrome. J Neurosci. 2003;23:2634–2644. doi: 10.1523/JNEUROSCI.23-07-02634.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston MC, Chen H, Swann JW. Loss of mTOR repressors Tsc1 or Pten has divergent effects on excitatory and inhibitory synaptic transmission in single hippocampal neuron cultures. Front Mol Neurosci. 2014;7:1. doi: 10.3389/fnmol.2014.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams MR, DeSpenza T, Jr, Li M, Gulledge AT, Luikart BW. Hyperactivity of Newborn Pten Knock-out Neurons Results from Increased Excitatory Synaptic Drive. J Neurosci. 2015;35:943–959. doi: 10.1523/JNEUROSCI.3144-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willner P. The validity of animal models of depression. Psychopharmacology (Berl) 1984;83:1–16. doi: 10.1007/BF00427414. [DOI] [PubMed] [Google Scholar]
- Willsey AJ, State MW. Autism spectrum disorders: from genes to neurobiology. Curr Opin Neurobiol. 2015;30:92–99. doi: 10.1016/j.conb.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C, Idziaszczyk S, Parry L, Guy C, Griffiths DF, Lazda E, Bayne RA, Smith AJ, Sampson JR, Cheadle JP. A mouse model of tuberous sclerosis 1 showing background specific early post-natal mortality and metastatic renal cell carcinoma. Hum Mol Genet. 2005;14:1839–1850. doi: 10.1093/hmg/ddi190. [DOI] [PubMed] [Google Scholar]
- Wilson HL, Wong AC, Shaw SR, Tse WY, Stapleton GA, Phelan MC, Hu S, Marshall J, McDermid HE. Molecular characterisation of the 22q13 deletion syndrome supports the role of haploinsufficiency of SHANK3/PROSAP2 in the major neurological symptoms. J Med Genet. 2003;40:575–584. doi: 10.1136/jmg.40.8.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohr M, Roullet FI, Hung AY, Sheng M, Crawley JN. Communication impairments in mice lacking Shank1: reduced levels of ultrasonic vocalizations and scent marking behavior. PLoS One. 2011;6:e20631. doi: 10.1371/journal.pone.0020631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohr M, Silverman JL, Scattoni ML, Turner SM, Harris MJ, Saxena R, Crawley JN. Developmental delays and reduced pup ultrasonic vocalizations but normal sociability in mice lacking the postsynaptic cell adhesion protein neuroligin2. Behav Brain Res. 2013;251:50–64. doi: 10.1016/j.bbr.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won H, Lee HR, Gee HY, Mah W, Kim JI, Lee J, Ha S, Chung C, Jung ES, Cho YS, Park SG, Lee JS, Lee K, Kim D, Bae YC, Kaang BK, Lee MG, Kim E. Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature. 2012;486:261–265. doi: 10.1038/nature11208. [DOI] [PubMed] [Google Scholar]
- Yamasaki K, Joh K, Ohta T, Masuzaki H, Ishimaru T, Mukai T, Niikawa N, Ogawa M, Wagstaff J, Kishino T. Neurons but not glial cells show reciprocal imprinting of sense and antisense transcripts of Ube3a. Hum Mol Genet. 2003;12:837–847. doi: 10.1093/hmg/ddg106. [DOI] [PubMed] [Google Scholar]
- Yashiro K, Riday TT, Condon KH, Roberts AC, Bernardo DR, Prakash R, Weinberg RJ, Ehlers MD, Philpot BD. Ube3a is required for experience-dependent maturation of the neocortex. Nat Neurosci. 2009;12:777–783. doi: 10.1038/nn.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda S, Sugiura H, Katsurabayashi S, Shimada T, Tanaka H, Takasaki K, Iwasaki K, Kobayashi T, Hino O, Yamagata K. Activation of Rheb, but not of mTORC1, impairs spine synapse morphogenesis in tuberous sclerosis complex. Sci Rep. 2014;4:5155. doi: 10.1038/srep05155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JI, Hong EP, Castle JC, Crespo-Barreto J, Bowman AB, Rose MF, Kang D, Richman R, Johnson JM, Berget S, Zoghbi HY. Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proc Natl Acad Sci USA. 2005;102:17551–17558. doi: 10.1073/pnas.0507856102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaaroor-Regev D, de Bie P, Scheffner M, Noy T, Shemer R, Heled M, Stein I, Pikarsky E, Ciechanover A. Regulation of the polycomb protein Ring1B by self-ubiquitination or by E6-AP may have implications to the pathogenesis of Angelman syndrome. Proc Natl Acad Sci USA. 2010;107:6788–6793. doi: 10.1073/pnas.1003108107. [DOI] [PMC free article] [PubMed] [Google Scholar]