Abstract
The Pex1/Pex6 complex is a member of the AAA family of ATPases that is involved in peroxisome biogenesis. Recently, cryo-electron microscopy structures have been determined of the Pex1/Pex6 complex in different nucleotide states. This Structural Snapshot describes the structural features of the complex, their implications for its function, as well as questions that still await answers.
Keywords: Pex1/Pex6 complex, AAA ATPases, peroxisome biogenesis, cryo-electron microscopy, single-particle analysis
Members of the AAA family of ATPases function in a wide variety of cellular processes, including membrane fusion, DNA translocation, protein degradation, and cargo transport along microtubules [1]. In all cases the energy of ATP hydrolysis is used to exert force on a macromolecule. Many AAA ATPases form hexamers that move polypeptide chains. They contain either one or two ATPase domains, arranged in a single or double ring (type I and II, respectively). We best understand single-ring AAA ATPases. For example, in the case of ClpX, up to five of the six subunits bind ATP; hydrolysis starts randomly at one of these subunits and proceeds rapidly through neighboring subunits, thereby moving a polypeptide chain through the central pore into the associated proteolytic chamber of ClpP [2]. A different mechanism has been established for another single-ring ATPase, the F1 ATPase [3] (the F1 ATPase belongs to the “additional strand catalytic E” (ASCE) superfamily of P-loop NTPases, which also includes the AAA ATPases). In this case, the three hydrolysis-competent subunits in the hexamer hydrolyze ATP in a strictly sequential and continuous manner around the ring. This is linked to the rotation of a polypeptide spindle inside the central pore.
While we are beginning to understand single-ring ATPase mechanisms, much less is known about double-ring ATPases. This class of ATPases has important roles in biology. For example, the double-ring ATPase N-ethylmaleimide (NEM)-sensitive fusion protein (NSF) disassembles soluble NSF attachment protein (SNAP) receptor (SNARE) complexes after membrane fusion [4], p97/VCP (Cdc48 in yeast) removes polypeptides from protein complexes or membranes [5], and Hsp104 untangles protein aggregates [6]. It is unclear why these ATPases have a second ring, particularly because some members of the double-ring ATPase family perform functions similar to their single-ring counterparts (e.g., ClpAP versus ClpXP) [7]. Possibly, one of the rings does not actively move polypeptides, but rather stabilizes the hexameric arrangement of the subunits. Consistent with this idea, one of the rings generally hydrolyzes ATP faster than the other. For example, the N-terminal ATPase domains of NSF hydrolyze ATP much more rapidly than the C-terminal subunits of the other ring [8]. The reverse is true for p97 [9, 10]. Whether there is communication between the two rings, and how exactly substrate is moved remains uncertain. Progress on understanding double-ring ATPases has been hampered by a lack of structural information. Crystallography has proven difficult because of conformational heterogeneity and flexibility of some domains. These problems can be circumvented with electron microscopy (EM). Indeed, recent developments in cryo-EM technology have resulted in much better image quality, allowing for better classification of particles in different conformations and for the determination of density maps at near-atomic resolution [11].
Pex1 and Pex6 each contain two ATPase domains (D1 and D2) and are therefore members of the double-ring ATPase family [12, 13]. They form a complex that is involved in the biogenesis of peroxisomes, organelles that house diverse metabolic functions in different organisms, including peroxide detoxification, β-oxidation of fatty acids, and the synthesis of various lipids. Mutations in human Pex1 and Pex6 cause more than 80% of peroxisome biogenesis disorder cases, including Zellweger syndrome [14–16]. Affected children die before puberty as a consequence of neurodevelopmental defects and multiple organ failure [17].
Two models have been put forward for the function of the Pex1/Pex6 complex. In the first model, the Pex1/Pex6 complex is involved in the import of soluble proteins from the cytosol into peroxisomes. These import substrates are recognized by the import receptor Pex5 and delivered to the peroxisomal membrane [12, 13]. Then, Pex5 is ubiquitinated and returned to the cytosol by the Pex1/Pex6 complex. In this model, the Pex1/Pex6 complex functions analogously to p97/Cdc48 in endoplasmic reticulum (ER)-associated protein degradation (ERAD) [18], in which the ATPase extracts poly-ubiquitinated proteins from the ER membrane [19, 20]. In the alternative model, the Pex1/Pex6 complex is involved in the fusion of peroxisomal precursor vesicles [21] and thus perhaps functions analogously to NSF.
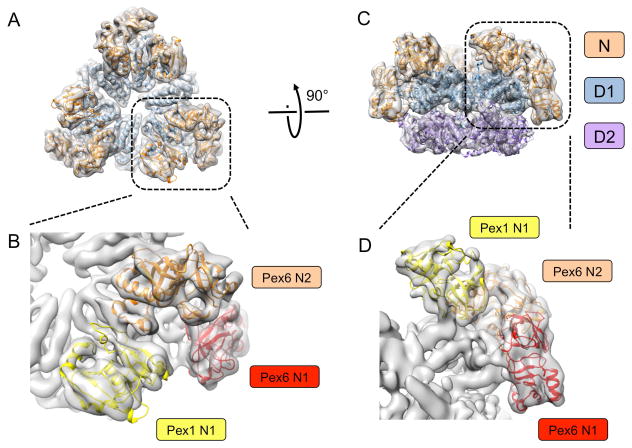
We have recently determined structures of the S. cerevisiae Pex1/Pex6 complex in different nucleotide states by cryo-EM single-particle analysis [22]. Although the overall resolution of the structures is between 6.2 Å and 8.8 Å, molecular models could be generated using novel computational methods that combine the placement of structurally homologous domains into density maps with energy minimization and refinement protocols. The structures show that Pex1 and Pex6 alternate in the hexameric double ring, generating three-fold symmetry (Fig. 1A). As expected, the D1 and D2 rings are stacked on top of each other (Fig. 1C). The N-terminal regions of Pex1 and Pex6 each contain two domains, N1 and N2, that have the same fold as the single N domain in p97 and NSF. The N1 domain of Pex1 is flexibly attached, and therefore not visible in the cryo-EM structures. The other three N domains are packed against the D1 ring (Fig. 1).
Fig. 1. Cryo-EM structure of the Pex1/Pex6 complex.
(A) Models of Pex1 and Pex6 were fitted into the density map (grey envelope) determined in the presence of ATPγS without symmetry imposed [22]. Shown is a top view with the D1 domains colored in blue and the N domains in brown. (B) Magnified view of the N domains in (A). The N domains are distinguished by different colors. (C) As in (A), but side view and including the D2 ring (purple). (D) Magnified view of the N domains in (C), colored as in (B).
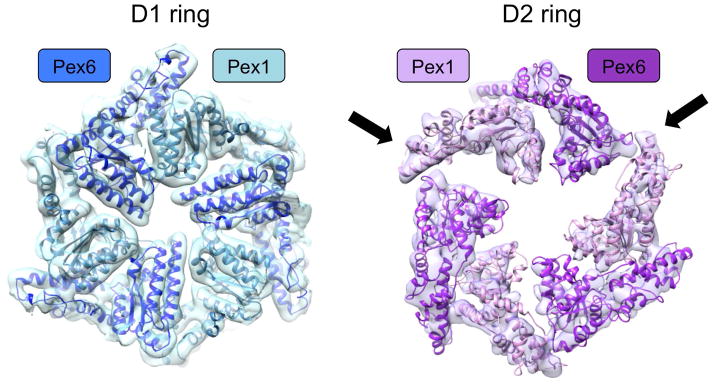
Structures determined in ATPγS or ADP show that the D1 ring has near-perfect six-fold symmetry (Fig. 2). Although not visible in the maps, the nucleotides are likely bound between the subunits, as is the case in other AAA ATPases. Given that the D1 ATPase domains of Pex1 and Pex6 lack typical sequence features required for ATP hydrolysis [22], it is likely that ATP can be bound, but not hydrolyzed, at all subunit interfaces. ATP binding to the D1 ring domains may be required for the stabilization of the hexameric complex, as it is significantly less stable in the presence of ADP and does not assemble in the absence of nucleotide [22, 23].
Fig. 2. Symmetry of the D1 and D2 rings.
The D1 ring is symmetric, while the D2 ring shows pairs of Pex1/Pex6 ATPase domains with varying spacing. The arrows indicate large gaps adjacent to one of the pairs.
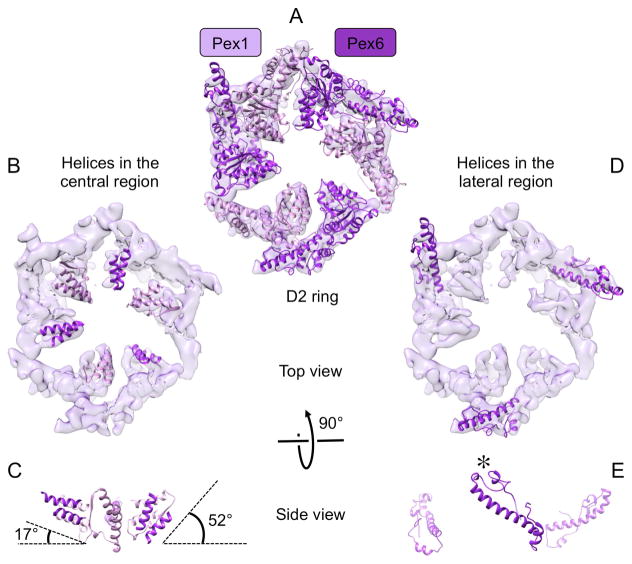
The D2 ring is clearly asymmetric, with the domains of Pex1 and Pex6 being arranged in three pairs (Fig. 2 and 3A). The distance between the pairs in the D2 ring varies, such that there is one narrow and two wide interfaces (Fig. 2). The large gaps are caused by one pair of Pex1/Pex6 D2 domains tilting downwards, away from the D1 ring. While the central parts of these domains move downwards (Fig. 3B,C), the lateral regions move upwards and contact the D1 domain (Fig. 3D,E). Such inter-ring connections are not seen anywhere else around the rings. To our knowledge, this is the first example of structural communication between the rings of a type II AAA ATPase.
Fig. 3. Structural elements of the D2 ring.
(A) Top view of the D2 ring with homology models fitted into the density map. (B) Top view of the D2 ring with secondary structure elements shown in the central region. (C) A side view shows that the two helices of the Pex1/Pex6 pair in the front tilt down by ~35°. For clarity, the helix pair of the Pex6 subunit in the back is not shown. The angles were measured between one of the helices of the Pex6 subunit and the plane of the D2 ring. (D) Top view of the D2 ring showing secondary structure elements in the lateral region of the D2 ring. For clarity, only the structural elements of the Pex6 subunits are shown. (E) The side view shows that the long helix of the Pex6 subunit in the front tilts upwards, contacting the D1 ring at the position indicated by an asterisk.
Although the resolution of the structures is too low to see nucleotides, the interfaces with wide spacing likely cannot bind nucleotides. Thus, only four out of six potential binding sites may be occupied at any given moment. In contrast to the D1 domains, the D2 domains of both Pex1 and Pex6 contain all the typical sequence elements required for ATP hydrolysis; their mutation leads to the total loss of ATPase activity. Interestingly, the D2 domains of Pex1 and Pex6 are not equivalent: Pex1 subunits cannot hydrolyze ATP when Pex6 subunits are in the ATP-bound state, whereas the reverse is not true. Mutations at interfaces within the Pex1/Pex6 pairs abolish all ATPase activity, whereas mutations at the other interface have only a moderate effect [22, 24, 25].
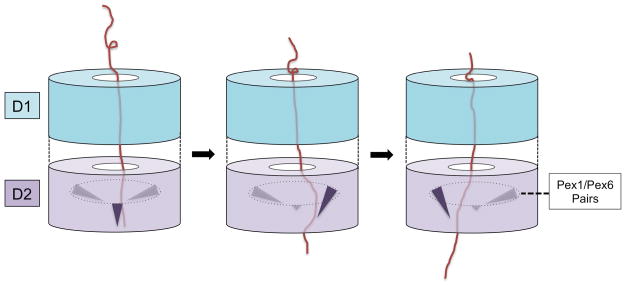
The current data are insufficient to derive a specific molecular model for the function of the Pex1/Pex6 complex. One problem is the identity of the substrate(s). The Pex1/Pex6 complex has been proposed to act solely on ubiquitinated Pex5 [13, 18], but a direct interaction between these proteins has not been demonstrated, and other substrates have not been excluded. Regardless of what the substrate is, the analogy to other AAA ATPases suggests that it is moved into and maybe even through the central pore. Movement of the polypeptide would be caused by ATP hydrolysis-dependent conformational changes in the D2 ring. Our structural data suggest that the tilting of a central segment of a Pex1/Pex6 pair is responsible for polypeptide movement. Indeed, this segment contains a conserved and essential aromatic residue [24] that in other AAA ATPases is required to push polypeptide substrates [16, 26–28]. In one scenario, the three pairs of Pex1/Pex6 domains would undergo the tilting movement in a sequential manner around the D2 ring (Fig. 4). However, given that ATP hydrolysis occurs within Pex1/Pex6 pairs [22], it seems likely that each pair has to separate transiently during the ATP hydrolysis cycle. Other scenarios, in which the D2 subunits fire more randomly, cannot strictly be excluded.
Fig. 4. Model for polypeptide chain movement by Pex1/Pex6.
A polypeptide chain moves through the D1 ring into the D2 ring. Pairs of Pex1/Pex6 ATPase domains in the D2 ring undergo a tilting motion in response to ATP hydrolysis, dragging the polypeptide chain through the central pore. At any given moment, one of the Pex1/Pex6 pair is tilted (shown in black), while the other two pairs are not (shown in grey). The tilting motion occurs in a circular manner around the ring.
The division of labor between the D1 and D2 rings of Pex1 and Pex6 is reminiscent of that in p97/Cdc48, although in this case, the D1 ring is not entirely inactive [9, 10]. Other similarities also support the idea that the Pex1/Pex6 complex functions analogously to p97/Cdc48. When operating in ERAD, this ATPase associates with the cofactor Ufd1/Npl4 to recognize ubiquitinated protein substrates [10]. Interestingly, recognition requires a domain in Ufd1, which has the same fold as the N-terminal domains of p97/Cdc48, Pex1, and Pex6 [29]. It is therefore possible that, together, the N1 and N2 domains of Pex1 and Pex6 would function analogously to the complex of the corresponding folds in p97/Cdc48 and Ufd1/Npl4. However, whether the N1 and N2 domains of Pex1 and/or Pex6 bind ubiquitin remains unclear.
In p97/Cdc48 the N domains are thought to move in an ATP-dependent manner; they would be in an “up” conformation when the D1 domains are in the ATP-bound state, a conformation stabilized by disease mutations [30], and in a “down” conformation in the ADP-bound state [31]. In the Pex1/Pex6 complex, the three visible N domains occupy positions corresponding to both “up” and “down” conformations. These domains are at the same positions in the ATPγS and ADP structures and, because they also make extensive contacts with one another and the D1 domains (Fig. 1), they are unlikely to move during the ATP-hydrolysis cycle. The flexibly attached N1 domains of Pex1 probably need an interaction partner to adopt a defined conformation. Thus, while there are several similarities between the Pex1/Pex6 complex and p97/Cdc48, there are also fundamental differences.
Two other Pex1/Pex6 structures have recently been reported, both based on negative-stain EM and therefore of significantly lower resolution (~20Å) [24, 25]. Nevertheless, these studies arrived at the same alternating arrangement of Pex1 and Pex6 around the ring, with the proteins assigned on the basis of fusions with glutathione S-transferase or maltose binding protein. The low resolution did not permit identification of the folds of the individual N domains. Ciniawsky et al. reported a very large opening of the central pore in the D1 ring in the ADP-bound state [24], clearly in contrast to the almost undetectable differences between our cryo-EM structures of the complex in the ATPγS and ADP states [22]. The drastic changes in the D1 ring are similar to those reported for an Hsp104 structure that has since been corrected [32–34], and they are difficult to reconcile with the known mode of interaction of subunits in other AAA ATPases. One possible explanation for the results obtained with negative-stain EM is the instability of the Pex1/Pex6 complex in ADP, which results in considerable structural heterogeneity in our hands. We coped with the heterogeneity by careful particle classification. The best class refined to a resolution at which individual helices could be recognized, while other classes showed features reminiscent of those reported in the negative-stain EM analysis [24], but these did not refine to higher resolution. Thus, although the Pex1/Pex6 complex must undergo ATP hydrolysis-dependent conformational changes, these have yet to be visualized.
Besides our cryo-EM structure of the Pex1/Pex6 complex, there is only one other structure of a double-ring ATPase at a comparable resolution, the recently determined structure of NSF [35]. NSF appears to undergo a rather dramatic change during the transition from the ATP to ADP state. Surprisingly, in both states, the ATP-hydrolyzing D1 ring does not form a closed, planar circle. Rather, it forms a split washer in the ATP state, and an open, flat “Pacman” structure in the ADP state. Several of the N domains seem to be flexible, as they are ill-defined or invisible. How the observed structural differences between these nucleotide states relate to the mechanism of ATP hydrolysis remains to be determined.
Clearly, we are just beginning to understand these complicated machines. Although cryo-EM single-particle analysis is the method of choice to gain structural insight into AAA ATPases, the determination of high-resolution structures in different conformations remains a huge challenge. So far, the focus has been on trapping all subunits in the same nucleotide state. However, this is likely a non-physiological condition, as different subunits in an ATP-hydrolyzing ring are probably in different states at any given moment. It may thus be necessary to determine high-resolution structures while the ATPases undergo active ATP hydrolysis. This will require the collection of a large number of particles and their careful classification to identify distinct conformations. In addition, future work needs to address structural aspects of how cofactors and substrates bind to the ATPases. The next years should bring exciting progress on these intriguing machines.
Acknowledgments
The work in the authors’ laboratories was supported by grants from the National Institutes of Health (GM05586 and GM007753). T.A.R. is a Howard Hughes Medical Institute investigator.
References
- 1.Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 2.Baker TA, Sauer RT. ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim Biophys Acta. 2012;1823:15–28. doi: 10.1016/j.bbamcr.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leslie AG, Walker JE. Structural model of F1-ATPase and the implications for rotary catalysis. Philos Trans R Soc Lond B Biol Sci. 2000;355:465–471. doi: 10.1098/rstb.2000.0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao C, Smith EC, Whiteheart SW. Requirements for the catalytic cycle of the N-ethylmaleimide-Sensitive Factor (NSF) Biochim Biophys Acta. 2012;1823:159–171. doi: 10.1016/j.bbamcr.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer H, Bug M, Bremer S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol. 2012;14:117–123. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- 6.Bosl B, Grimminger V, Walter S. The molecular chaperone Hsp104–a molecular machine for protein disaggregation. J Struct Biol. 2006;156:139–148. doi: 10.1016/j.jsb.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Olivares AO, Nager AR, Iosefson O, Sauer RT, Baker TA. Mechanochemical basis of protein degradation by a double-ring AAA+ machine. Nat Struct Mol Biol. 2014;21:871–875. doi: 10.1038/nsmb.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whiteheart SW, Rossnagel K, Buhrow SA, Brunner M, Jaenicke R, Rothman JE. N-ethylmaleimide-sensitive fusion protein: a trimeric ATPase whose hydrolysis of ATP is required for membrane fusion. J Cell Biol. 1994;126:945–954. doi: 10.1083/jcb.126.4.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song C, Wang Q, Li CC. ATPase activity of p97-valosin-containing protein (VCP). D2 mediates the major enzyme activity, and D1 contributes to the heat-induced activity. J Biol Chem. 2003;278:3648–3655. doi: 10.1074/jbc.M208422200. [DOI] [PubMed] [Google Scholar]
- 10.Ye Y, Meyer HH, Rapoport TA. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J Cell Biol. 2003;162:71–84. doi: 10.1083/jcb.200302169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng Y. Single-particle cryo-EM at crystallographic resolution. Cell. 2015;161:450–457. doi: 10.1016/j.cell.2015.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grimm I, Saffian D, Platta HW, Erdmann R. The AAA-type ATPases Pex1p and Pex6p and their role in peroxisomal matrix protein import in Saccharomyces cerevisiae. Biochim Biophys Acta. 2012;1823:150–158. doi: 10.1016/j.bbamcr.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Liu X, Ma C, Subramani S. Recent advances in peroxisomal matrix protein import. Curr Opin Cell Biol. 2012;24:484–489. doi: 10.1016/j.ceb.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geisbrecht BV, Collins CS, Reuber BE, Gould SJ. Disruption of a PEX1-PEX6 interaction is the most common cause of the neurologic disorders Zellweger syndrome, neonatal adrenoleukodystrophy, and infantile Refsum disease. Proc Natl Acad Sci U S A. 1998;95:8630–8635. doi: 10.1073/pnas.95.15.8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reuber BE, Germain-Lee E, Collins CS, Morrell JC, Ameritunga R, Moser HW, Valle D, Gould SJ. Mutations in PEX1 are the most common cause of peroxisome biogenesis disorders. Nat Genet. 1997;17:445–448. doi: 10.1038/ng1297-445. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z, Suzuki Y, Shimozawa N, Fukuda S, Imamura A, Tsukamoto T, Osumi T, Fujiki Y, Orii T, Wanders RJ, Barth PG, Moser HW, Paton BC, Besley GT, Kondo N. Genomic structure and identification of 11 novel mutations of the PEX6 (peroxisome assembly factor-2) gene in patients with peroxisome biogenesis disorders. Hum Mutat. 1999;13:487–496. doi: 10.1002/(SICI)1098-1004(1999)13:6<487::AID-HUMU9>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 17.Braverman NE, D’Agostino MD, Maclean GE. Peroxisome biogenesis disorders: biological, clinical and pathophysiological perspectives. Dev Disabil Res Rev. 2013;17:187–196. doi: 10.1002/ddrr.1113. [DOI] [PubMed] [Google Scholar]
- 18.Schliebs W, Girzalsky W, Erdmann R. Peroxisomal protein import and ERAD: variations on a common theme. Nat Rev Mol Cell Biol. 2010;11:885–890. doi: 10.1038/nrm3008. [DOI] [PubMed] [Google Scholar]
- 19.Bagola K, Mehnert M, Jarosch E, Sommer T. Protein dislocation from the ER. Biochim Biophys Acta. 2011;1808:925–936. doi: 10.1016/j.bbamem.2010.06.025. [DOI] [PubMed] [Google Scholar]
- 20.Christianson JC, Ye Y. Cleaning up in the endoplasmic reticulum: ubiquitin in charge. Nat Struct Mol Biol. 2014;21:325–335. doi: 10.1038/nsmb.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Zand A, Gent J, Braakman I, Tabak HF. Biochemically distinct vesicles from the endoplasmic reticulum fuse to form peroxisomes. Cell. 2012;149:397–409. doi: 10.1016/j.cell.2012.01.054. [DOI] [PubMed] [Google Scholar]
- 22.Blok NB, Tan D, Wang RY, Penczek PA, Baker D, DiMaio F, Rapoport TA, Walz T. Unique double-ring structure of the peroxisomal Pex1/Pex6 ATPase complex revealed by cryo-electron microscopy. Proc Natl Acad Sci U S A. 2015;112:E4017–4025. doi: 10.1073/pnas.1500257112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saffian D, Grimm I, Girzalsky W, Erdmann R. ATP-dependent assembly of the heteromeric Pex1p-Pex6p-complex of the peroxisomal matrix protein import machinery. J Struct Biol. 2012;179:126–132. doi: 10.1016/j.jsb.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 24.Ciniawsky S, Grimm I, Saffian D, Girzalsky W, Erdmann R, Wendler P. Molecular snapshots of the Pex1/6 AAA+ complex in action. Nat Commun. 2015;6:7331. doi: 10.1038/ncomms8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gardner BM, Chowdhury S, Lander GC, Martin A. The Pex1/Pex6 complex is a heterohexameric AAA+ motor with alternating and highly coordinated subunits. J Mol Biol. 2015;427:1375–1388. doi: 10.1016/j.jmb.2015.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siddiqui SM, Sauer RT, Baker TA. Role of the processing pore of the ClpX AAA+ ATPase in the recognition and engagement of specific protein substrates. Genes Dev. 2004;18:369–374. doi: 10.1101/gad.1170304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinnerwisch J, Fenton WA, Furtak KJ, Farr GW, Horwich AL. Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell. 2005;121:1029–1041. doi: 10.1016/j.cell.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 28.Martin A, Baker TA, Sauer RT. Diverse pore loops of the AAA+ ClpX machine mediate unassisted and adaptor-dependent recognition of ssrA-tagged substrates. Mol Cell. 2008;29:441–450. doi: 10.1016/j.molcel.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park S, Isaacson R, Kim HT, Silver PA, Wagner G. Ufd1 exhibits the AAA-ATPase fold with two distinct ubiquitin interaction sites. Structure. 2005;13:995–1005. doi: 10.1016/j.str.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 30.Tang WK, Li D, Li CC, Esser L, Dai R, Guo L, Xia D. A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants. EMBO J. 2010;29:2217–2229. doi: 10.1038/emboj.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies JM, Brunger AT, Weis WI. Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change. Structure. 2008;16:715–726. doi: 10.1016/j.str.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 32.Wendler P, Shorter J, Plisson C, Cashikar AG, Lindquist S, Saibil HR. Atypical AAA+ subunit packing creates an expanded cavity for disaggregation by the protein-remodeling factor Hsp104. Cell. 2007;131:1366–1377. doi: 10.1016/j.cell.2007.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee S, Sielaff B, Lee J, Tsai FT. CryoEM structure of Hsp104 and its mechanistic implication for protein disaggregation. Proc Natl Acad Sci U S A. 2010;107:8135–8140. doi: 10.1073/pnas.1003572107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carroni M, Kummer E, Oguchi Y, Wendler P, Clare DK, Sinning I, Kopp J, Mogk A, Bukau B, Saibil HR. Head-to-tail interactions of the coiled-coil domains regulate ClpB activity and cooperation with Hsp70 in protein disaggregation. Elife. 2014;3:e02481. doi: 10.7554/eLife.02481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao M, Wu S, Zhou Q, Vivona S, Cipriano DJ, Cheng Y, Brunger AT. Mechanistic insights into the recycling machine of the SNARE complex. Nature. 2015;518:61–67. doi: 10.1038/nature14148. [DOI] [PMC free article] [PubMed] [Google Scholar]