Abstract
This study was aimed to characterize the distribution of colorectal cancer risk using family history of cancers by data mining. Family histories for 10,066 colorectal cancer cases recruited to population cancer registries of the Colon Cancer Family Registry were analyzed using a data mining framework. A novel index was developed to quantify familial cancer aggregation. Artificial neural network was used to identify distinct categories of familial risk. Standardized incidence ratios (SIRs) and corresponding 95% confidence intervals (CIs) of colorectal cancer were calculated for each category. We identified five major, and sixty-six minor categories of familial risk for developing colorectal cancer. The distribution the major risk categories were: (i) 7% of families (SIR=7.11; 95%CI=6.65–7.59) had a strong family history of colorectal cancer; (ii) 13% of families (SIR=2.94; 95%CI=2.78–3.10) had a moderate family history of colorectal cancer; (iii) 11% of families (SIR=1.23; 95%CI=1.12–1.36) had a strong family history of breast cancer and weak family history of colorectal cancer; (iv) 9% of families (SIR=1.06; 95% CI=0.96–1.18) had a strong family history of prostate cancer and a weak family history of colorectal cancer; and (v) 60% of families (SIR=0.61; 95%CI=0.57–0.65) had weak family history of all cancers. There is a wide variation of colorectal cancer risk that can be categorized by family history of cancer, with a strong gradient of colorectal cancer risk between the highest and lowest risk categories. The risk of colorectal cancer for people with the highest risk category of family history (7% of the population) was 12-times that for people in the lowest risk category (60%) of the population. Data mining was proven an effective approach for gaining insight into the underlying cancer aggregation patterns and for categorizing familial risk of colorectal cancer.
Keywords: Data mining, colorectal cancer, familial risk, familial aggregation
INTRODUCTION
The risk of colorectal cancer is strongly associated with family history of colorectal cancer. First-degree relatives of colorectal cancer cases have, on average, an approximate two-fold risk of the disease compared with those without a family history [1, 2]. Knowledge of this risk factor, has led to the development of guidelines for colorectal cancer screening based on family history and research to identify the genetic factors that contribute to this familial risk. Hereditary cancer syndromes, such as Lynch syndrome and familial adenomatous polyposis, for which underlying genetic mutations have been identified, are major achievements, yet account for the minority of this familial aggregation [3]. A more comprehensive assessment of how familial aggregation of cancer affects colorectal cancer risk, and the distribution of this risk, would provide the basis for formulating new genetic and genomic research, and development of improved clinical management of colorectal cancer families, including refining targeted genetic and cancer screening recommendations. The distribution of familial risk was first analyzed by Fain and Goldgar using a non-parametric test applying to breast cancer family history data [4].
Most studies investigating associations between family history and risk of colorectal cancer have limited their analysis of family history to first-degree relatives only [5–10] with only a few that included up to third-degree relatives [1, 11]. Most have not considered family history of cancers other than colorectal and have used simple counts of cancers without considering the size of the family or degree of relationship. A limitation of this approach is that the underlying association between the constellation of affected relatives and the patterns of aggregation of cancer in relatives is not taken into account. A more comprehensive approach is to address the question:
“What is a person’s risk of colorectal cancer if l of this person’s x first-degree relatives had cancer a, b, c …, and m of this person’s y second-degree relatives had cancer a, b, c, …, and n of this persons’s z third-degree relatives had cancer a, b, c, …?”
To the best of our knowledge, no study has reported the distribution of risk of colorectal cancer, taking into consideration both the underlying association between the patterns of affected relatives (constellations) and the spectrum of co-aggregating cancers (aggregations). We have attempted to fill this gap by conducting a novel data mining approach to identify categories of familial risk by clustering constellations and aggregations simultaneously.
MATERIALS AND METHODS
Study Sample
We studied population-based colorectal cancer families recruited to the Colon Cancer Family Registry [12]. Detail description of recruitment can be found at http://coloncfr.org/. For this analysis, we studied the families that were recruited through recently diagnosed invasive colorectal cancer cases (population-based probands) from state or regional population cancer registries in the USA (Washington, California, Arizona, Minnesota, Colorado, New Hampshire, North Carolina, and Hawaii), Australia (Victoria) and Canada (Ontario) between 1997 and 2007. Population-based probands were recruited regardless of having a family history of cancer. Written informed consent was obtained from all study participants, and the study protocol was approved by the institutional research ethics review board at each study center.
Data Collection
Information on demographics, personal characteristics, personal and family history of cancer, cancer-screening history, and history of polyps, polypectomy, and other surgeries was obtained by questionnaires from all probands and participating relatives. Cancer histories were also cross checked across multiple relatives from within the same family, not just the probands. Participants were followed up approximately every five years after baseline to update this information. The present study was based on all available baseline and follow-up data. Reported cancer diagnoses and age at diagnosis were confirmed, where possible, using pathology reports, medical records, cancer registry reports, and death certificates. Blood samples and permission to access tumor tissue were requested from all participants.
Mismatch repair (MMR) gene mutation testing
Testing for germline mutations in MLH1, MSH2, MSH6 and PMS2 was performed for all population-based probands who had a colorectal cancer displaying MMR deficiency as evidenced by either tumor microsatellite instability and/or lack of MMR protein expression by immunohistochemistry. Details of germline testing methods have been described elsewhere [13]. A pathogenic mutation was defined as a variant that was predicted to result in a stop codon, a frameshift mutation, a large duplication or deletion, or a missense mutation in the coding region or splice site previously reported within the scientific literature and databases to be pathogenic (InSiGHT database; http://insight-group.org/variants/classifications/). The relatives of probands with a pathogenic MMR germline mutation, who provided a blood sample, underwent testing for the specific mutation identified in the proband.
Definitions
Self-organizing map [14] is a method for projecting high-dimensional data onto low-dimensional output display. A node corresponds to a group of families with similar familial aggregation vectors and is determined using a self-organizing map. K-means [15] is a technique for partitioning data into optimal groups. A cluster is a grouping of nodes on the self-organizing map detected by k-means. A codebook vector refers to the weight vector of each node for the self-organizing map. A prototype vector is used to describe the mean weight vector for a cluster computed by k-means.
Statistical Analysis
Data mining framework
To analyze family history, we used data mining [16] to reveal patterns of familial aggregation that can be used to estimate the colorectal cancer risk of an individual based on their family history of cancers. Every family was represented by a vector modeling family history of cancer. These vectors became the inputs to the clustering algorithms, including the self-organizing map [14] and k-means [15], to identify nodes and clusters. Finally, the risk of colorectal cancer was estimated for each detected nodes and cluster (see Supplementary Figure 1).
(1) Familial aggregation
Family history of each cancer type was transformed into a familial aggregation index that encapsulated the aggregation in each family with the key property that family history of cancer for a person is stronger the larger numbers of relatives with cancer and the closer the genetic relationship they have to these affected relatives. To this end, the vector of familial aggregation of all cancers for each family based on the familial relationship to the proband, was calculated as follows:
where F_Aggci is a familial aggregation index defined as:
with
#FDR, #SDR, #TDR as the numbers of first-, second- and third-degree relatives the proband has, and
#FDRci, #SDRci, #TDRci as the numbers of first-, second- and third-degree relatives of the proband being affected by cancer ci.
(2) Cluster detection
First, the self-organizing map [14] (a non-parametric clustering algorithm from the artificial neural network [17] discipline) was applied to group families with similar family history as defined by their familial aggregation index into nodes so that neighboring nodes are more similar than distant ones. Second, k-means [15] was used to identify global characteristics of familial aggregation by partitioning the nodes into larger clusters so that similarity within cluster and difference between clusters were maximized.
A 6×12 (72 nodes) self-organizing map was used. The number of nodes was initially set to , where n was the number of families, and the vertical and horizontal dimension was defined by the ratio of the two largest eigenvalues of the dataset. The final map size used was chosen by scaling up or down the reference map to achieve maximum map resolution within computational constraints [18].
To find a global perspective for the familial aggregation, families grouped by nodes on the self-organizing map were further clustered using k-means, where k is the number of clusters to be found. K-means is a partitioning clustering algorithm useful for providing groupings with explicit cluster boundaries [15]. All cluster detection tasks were done using the scientific programming package Matlab R2012a [19]. Technical details of the cluster detection step, including (1) clustering families using the self-organizing map, (2) partitioning self-organizing map using k-means, and (3) distance measure for measuring similarity of familial aggregation, are described in the Appendix A, B and C, respectively.
(3) Epidemiologic analysis
To determine the degree of colorectal cancer risk for family members of each node and cluster compared with the general population, we estimated the standardized incidence ratio (SIR) by dividing the numbers of colorectal cancers observed in all family members (excluding the proband) of each node and cluster by the expected numbers [20]. The expected numbers of colorectal cancers for each node and cluster were calculated by multiplying the age-, sex-, and country-specific incidence for the general population by the corresponding observation time (i.e. age) of the family members. Age- and sex-specific cancer incidences for each country in 1988–1992 were obtained from the Cancer Incidence in Five Continents [21]. The corresponding 95% confidence intervals (CIs) were calculated by taking into account familial correlation of cancer risk between relatives using the Jackknife method [22].
To deal with missing age(s) at diagnosis of cancer for affected relatives, we assumed the median age at diagnosis for each cancer in the general population obtained from Surveillance, Epidemiology and End Results Cancer Statistics Review [23]. Unaffected relatives for whom no information on age was available were censored at birth and therefore did not contribute to the analysis. All epidemiologic analysis was done using Stata 11.0 [24].
We conducted a sensitivity analysis to assess the role of an inherited cancer risk syndrome on familial clustering and SIR estimation by excluding the 201 families known to be carrying a DNA mismatch repair gene mutation (Lynch syndrome families). We conducted a sensitivity analysis to assess the role of family history of third-degree relatives by restricting analysis to only first- and second-degree relatives.
RESULTS
Of the 10,407 families identified from population-based resources of the Colon Cancer Family Registry, 341 families (3%) were excluded because of having less than four family members. The remaining 10,066 families that contained 181,555 individuals (90,188 women) were included. On average, each family had 7 first-degree relatives, 9 second-degree relatives and 5 third-degree relatives (Table 1). We identified sixty-six minor clusters of familial risk for developing colorectal cancer (Figure 1). There were 5 major clusters corresponding to an optimal partition capturing the most representative family history patterns associated with various degrees of risk for developing colorectal cancer (Supplementary Figure 2). Demographic characteristics of each cluster are summarized in Table 2.
Table 1.
Baseline characteristics of population-based families from the Colon Cancer Family Registry
| Characteristics | Number |
|---|---|
| No. of families | 10,066 |
| No. of persons | 181,555 |
| Canada | 60,875 (34%) |
| Australia | 34,669 (19%) |
| USA | 86,011 (47%) |
| Male : Female | 1 : 1.01 |
| Age (median) | 57 |
| Cancers | |
| Colorectal cancer | 6,808 |
| Endometrial cancer | 858 |
| Stomach cancer | 1,526 |
| Small bowel cancer | 46 |
| Hepatobiliary tract cancer | 754 |
| Pancreatic cancer | 680 |
| Renal cancer | 478 |
| Ureter cancer | 32 |
| Bladder cancer | 504 |
| Brain cancer | 806 |
| Cervical cancer | 586 |
| Ovarian cancer | 610 |
| Breast cancer | 3,841 |
| Prostate cancer | 2,470 |
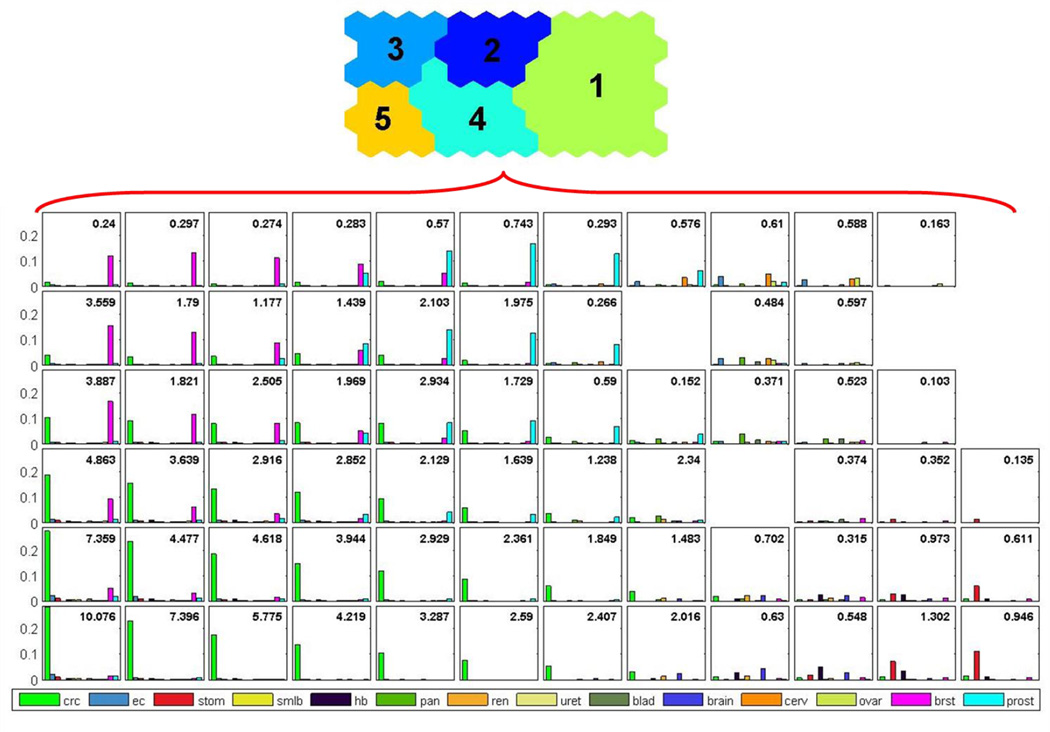
Figure 1.
Familial risks for population-based colorectal cancer families from the Colon Cancer Family Registry. The number in the right top corner represents the standardized incidence ratio of colorectal cancer. Node without a bar chart in the diagram is an empty node with no families mapped to it. crc = colorectal cancer, ec = endometrial cancer, stom = stomach cancer, smlb = small bowel cancer, hb = hepatobiliary cancer, pan = pancreatic cancer, ren = renal cancer, uret = ureteric cancer, blad = urinary bladder cancer, brain = brain cancer, cerv = cervical cancer, ovar = ovarian cancer, brst = breast cancer, prost = prostate cancer
Table 2.
Demographic characteristics of the five clusters
| Cluster | No of families | No of persons | Male : Female | Age (years) median (inter-quartile range) |
|---|---|---|---|---|
| 1 | 5,969 (59%) | 96,326 | 1 : 1.02 | 56 (40–72) |
| 2 | 915 (9%) | 19,544 | 1 : 1.02 | 59 (43–74) |
| 3 | 1,087 (11%) | 19,916 | 1 : 0.95 | 59 (43–73) |
| 4 | 1,353 (13%) | 33,938 | 1 : 1.01 | 57 (40–73) |
| 5 | 742 (7%) | 11,831 | 1 : 0.99 | 58 (42–72) |
Identification of the familial risk
There was a moderate variation in colorectal cancer risk across nodes within each cluster (Figure 1) and a wide variation in colorectal cancer risk across clusters in terms of both the type and strength of aggregation of extracolonic cancers and the strength of aggregation of colorectal cancer (Figure 2).
Figure 2.
Familial risk of colorectal cancer by each cluster. SIR = standardized incidence ratio of colorectal cancer (corresponding 95% confidence interval), ageDX = age at diagnosis of colorectal cancer (years), crc = colorectal cancer, ec = endometrial cancer, stom = stomach cancer, smlb = small bowel cancer, hb = hepatobiliary cancer, pan = pancreatic cancer, ren = renal cancer, uret = ureteric cancer, blad = urinary bladder cancer, brain = brain cancer, cerv = cervical cancer, ovar = ovarian cancer, brst = breast cancer, prost = prostate cancer
Familial risk for each cluster
Members of families with the strongest aggregation of colorectal cancer had the highest risk of the disease (Cluster 5 in Figure 2), with an average seven-fold increased risk of colorectal cancer compared with the general population (SIR = 7.11; 95% CI = 6.65 to 7.59). Within this cluster, the increased risk of colorectal cancer ranged from 1.64 to 4.22 (between the 13 nodes). This cluster comprised 1,353 families, which was 13% of all families, and the median age of disease onset was 69 (inter-quartile range 52–72) years. Members of families with the second highest aggregation of colorectal cancer (Cluster 4 in Figure 2), had an average three-fold increased risk of colorectal cancer (SIR = 2.94; 95% CI = 2.78 to 3.10). Within this cluster, the increased risk of colorectal cancer ranged from 1.64 to 4.22 (between the 13 nodes). This cluster comprised 1,353 families, which was 13% of all families, and the median age of disease onset is 69 (inter-quartile range 52–72) years. In contrast, colorectal cancer risk was the lowest for members of families with weak aggregation of colorectal cancer (Cluster 1 in Figure 2). They had an average risk of colorectal cancer 40% lower than the general population (SIR = 0.61; 95% CI = 0.57 to 0.65). Within this cluster, the increased risk of colorectal cancer ranged from 0.1 to 2.34 (between the 24 nodes). This cluster comprised 5,969 families, which was 60% of all families, and the median age of disease onset was 70 (inter-quartile range 56–72) years.
Members of families with a strong aggregation of breast cancer and weak aggregation of colorectal cancer (Cluster 3 in Figure 2) had a modest increased risk of colorectal cancer (SIR = 1.23; 95% CI = 1.12 to 1.36) i.e., familial aggregation of breast cancer was a risk factor for colorectal cancer. Within this cluster, the increased risk of colorectal cancer ranged from 0.24 to 3.89 (between the 10 nodes). This cluster comprised 1,087 families, which was 11% of all families, and the median age of disease onset was 71 (inter-quartile range 61–72) years. Members of families with a strong aggregation of prostate cancer and weak aggregation of colorectal cancer (Cluster 2 in Figure 2) had no increased risk of colorectal cancer (SIR = 1.06; 95% CI = 0.96 to 1.18) i.e., familial aggregation of prostate cancer was not a risk factor for colorectal cancer. Within this cluster, the increased risk of colorectal cancer ranged from 0.27 to 2.93 (between the 11 nodes). This cluster comprised 915 families, which was 9% of all families, and the median age of disease onset was 71 (inter-quartile range 43–74) years.
Extracolonic cancers co-aggregated with colorectal cancer in all five clusters but the aggregations were all weak and no particular cancer appeared to be a risk factor for colorectal cancer except in Cluster 3. Averaged across all nodes in all clusters weighted by the number of families in each node, the SIR was 1.59 (95% CI = 1.54 to 1.65).
Distribution of familial risks by clusters
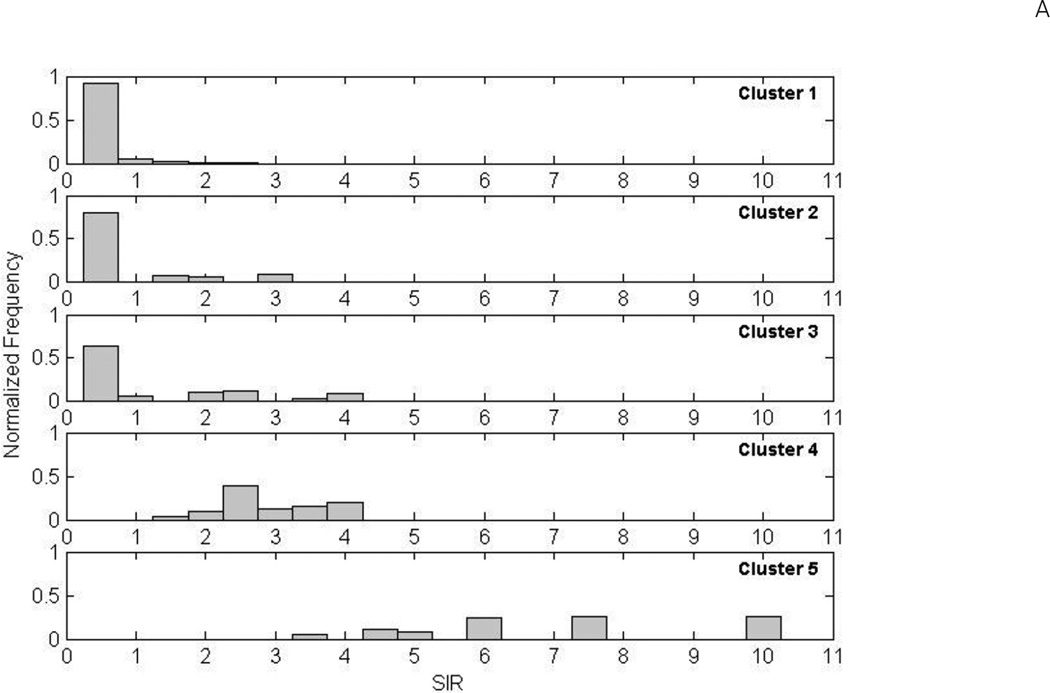
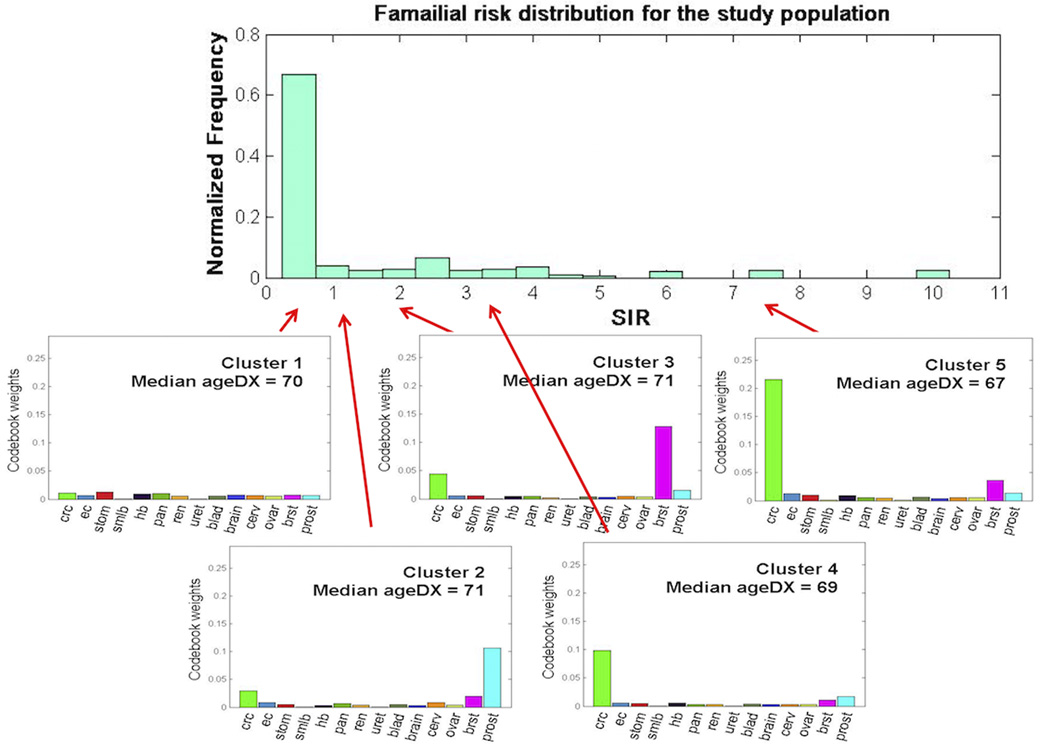
Figure 3A shows the distribution of SIRs of colorectal cancer for all family members within each node for each of the five clusters. Familial risk distributions were similarly right skewed for Clusters 1, 2 and 3): about 90% of families in Cluster 1 had a lower risk of colorectal cancer than the general population; about 80% in Cluster 2; and about 60% in Cluster 3. When we compared the familial risks (expressed as SIRs) between different clusters, there was a strong gradient between them (Figure 3B). The ratio of the SIRs between the highest risk category of family history (Cluster 5) and the lowest risk category (Cluster 1), was 11.67. When combined across all nodes in all clusters, i.e. all families, the overall distribution of familial risks in the whole study sample was right skewed with a long tail (Figure 4).
Figure 3.
(A) Distribution of familial risk of colorectal cancer by family clusters (expressed as the standardized incidence ratios (SIRs)). The normalized frequency, corresponding to the height of each bar, indicates the percentage of families with the same colorectal cancer risk. (B) SIRs and their 95% confidence intervals (CIs) for each family cluster. The dot points represent the estimates of SIR and the vertical lines represent 95% CIs.
Figure 4.
Mapping between familial risk distribution and cancer aggregation patterns. SIR = standardized incidence ratio of colorectal cancer, ageDX = age at diagnosis of colorectal cancer (years), crc = colorectal cancer, ec = endometrial cancer, stom = stomach cancer, smlb = small bowel cancer, hb = hepatobiliary cancer, pan = pancreatic cancer, ren = renal cancer, uret = ureteric cancer, blad = urinary bladder cancer, brain = brain cancer, cerv = cervical cancer, ovar = ovarian cancer, brst = breast cancer, prost = prostate cancer
Sensitivity analyses
There were 201 (2%) families in which at least one family member was identified as having a MMR gene mutation (Lynch syndrome). Excluding the Lynch syndrome families made no substantial difference to the findings in terms of self-organizing map patterns, distributions of aggregating cancers in families, age at diagnosis, or SIR of colorectal cancer in any cluster (Supplementary Figure 3). The distribution of familial clusters for Lynch syndrome families, median age at diagnosis and SIR of colorectal cancer for each cluster are shown in Supplementary Table 1. Similarly, excluding all third-degree relatives made no substantial difference (detail data not shown).
DISCUSSION
In this study, we identified patterns of familial aggregation and simultaneously assessed risk of colorectal cancer for these familial aggregation patterns, instead of estimating risk only based on numbers of affected relatives and degree of relatedness as in previous studies [25–28]. We used a novel data mining approach, which facilitated the clustering of both constellation patterns and familial cancer aggregation. The multivariable nature of this approach is particularly relevant for examining risk where cancers at multiple organ sites can co-aggregate within the family.
We found that the overall distribution of the familial risks of colorectal cancer across the whole study sample (all of whom had at least one relative with colorectal cancer, i.e. the proband) to be right skewed (Figure 4). This is consistent with the description of familial risks of people in the general population by Hopper [29] (see Figure 2 in [29]): under the assumption of a multiplicative polygenic risk model, the distribution of familial risks for colorectal cancer for the general population (and in our case, families with at least one colorectal cancer) is skewed with a long tail, and the mode of the familial risk is substantially below the population average of 5% risk. Moreover, a trend parallel to Hopper’s description was clearly observed in our cluster familial risk distributions. The average familial risk distributions spread further to the right as more close relatives are affected [29, 30]. Starting from the lowest risk category (Cluster 1), as the average familial risk increases (Cluster 2 and 3), the distribution of the familial risks moves to the right (Cluster 4 and 5). We, therefore, provide empirical evidence for the existence of a wide variation of familial risks (even within a set of families ascertained because of a colorectal cancer case), and a strong gradient of colorectal cancer risk between the highest and lowest risk groups for colorectal cancer. We also found that the types and aggregation of extracolonic cancers and strength of the aggregation of colorectal cancer in a family might explain this wide variation of familial risks of colorectal cancer.
This study has some limitations. The clustering results of the self-organizing map might not be identical every time it is run though they were very similar consistently. This is a common issue for neural network models and optimization problems in general. However, it is not a major concern here because the focus is in identifying a small finite set of representative prototypes (i.e. the familial risk categories) by minimizing the quantization error of the neural network model. These prototypes are formed as averages of the data within a cluster. Variations of these averages are insignificant when the number of training data is sufficiently large, as in our case.
The contribution this study could make to the field is two-fold. First, the findings about the familial risk distribution will serve as empirical evidence supporting existing familial risk models confirming that, for colorectal cancer, there exist a wide variation of familial risks, and a very strong risk gradient between the highest and lowest risk groups [29, 31]. Second, the data mining approach will advance current familial studies which are limited to univariable analysis to addressing the multivariable question, relevant to the studies of other familial cancer syndromes.
In conclusion, our data mining approach showed a wide variation of familial colorectal cancer risk and a strong risk gradient between the highest risk and lowest risk families. The types and aggregation of extracolonic cancers and strength of the aggregation of colorectal cancer in a family might explain this wide variation of familial risks. A family history of colorectal cancer with a broad spectrum of co-aggregating cancers could substantially elevate the risk of colorectal cancer; but co-aggregating extracolonic cancers may not influence the risk of colorectal cancer when the familial aggregation of colorectal cancer is not strong. Data mining was proven an effective approach for gaining insight to the underlying cancer aggregation patterns and for categorizing familial risk of colorectal cancer.
Supplementary Material
Acknowledgments
FUNDING
This work was supported by grant UM1 CA167551 from the National Cancer Institute, National Institutes of Health (NIH) and through cooperative agreements with the following Colon Cancer Family Registry (CCFR) centers: Australasian Colorectal Cancer Family Registry (U01/U24 CA097735), Mayo Clinic Cooperative Family Registry for Colon Cancer Studies (U01/U24 CA074800), Ontario Familial Colorectal Cancer Registry (U01/U24 CA074783), Seattle Colorectal Cancer Family Registry (U01/U24 CA074794), and USC Consortium Colorectal Cancer Family Registry (U01/U24 CA074799).
Seattle CCFR research was also supported by the Cancer Surveillance System of the Fred Hutchinson Cancer Research Center, which was funded by Control Nos. N01-CN-67009 (1996–2003) and N01-PC-35142 (2003–2010) and Contract No. HHSN2612013000121 (2010–2017) from the Surveillance, Epidemiology and End Results (SEER) Program of the National Cancer Institute with additional support from the Fred Hutchinson Cancer Research Center.
The collection of cancer incidence data used in this study was supported by the California Department of Public Health as part of the statewide cancer reporting program mandated by California Health and Safety Code Section 103885; the National Cancer Institute’s Surveillance, Epidemiology and End Results Program under contract HHSN261201000035C awarded to the University of Southern California, and contract HHSN261201000034C awarded to the Public Health Institute; and the Centers for Disease Control and Prevention’s National Program of Cancer Registries, under agreement U58DP003862-01 awarded to the California Department of Public Health. The ideas and opinions expressed herein are those of the author(s) and endorsement by the State of California, Department of Public Health the National Cancer Institute, and the Centers for Disease Control and Prevention or their Contractors and Subcontractors is not intended nor should be inferred.
This work is also supported by Centre for Research Excellence grant APP1042021 and Program grant APP1074383 from National Health and Medical Research Council (NHMRC), Australia. AKW is a NHMRC Early Career Fellow. MAJ is an NHMRC Senior Research Fellow. JLH is a NHMRC Senior Principal Research Fellow. DDB is a University of Melbourne Research at Melbourne Accelerator Program (R@MAP) Senior Research Fellow.
The authors thank all study participants of the Colon Cancer Family Registry and staff for their contributions to this project.
Footnotes
DISCLAIMER
The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the CFRs, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government or the CFR. Authors had full responsibility for the design of the study, the collection of the data, the analysis and interpretation of the data, the decision to submit the manuscript for publication, and the writing of the manuscript.
DISCLOSURE
The authors have no conflict of interest to declare with respect to this manuscript.
REFERENCES
- 1.Taylor DP, Burt RW, Williams MS, Haug PJ, Cannon-Albright LA. Population-Based Family History-Specific Risks for Colorectal Cancer: A Constellation Approach. Gastroenterology. 2010;138(3):877–885. doi: 10.1053/j.gastro.2009.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baglietto L, Jenkins MA, Severi G, et al. Measures of familial aggregation depend on definition of family history: meta-analysis for colorectal cancer. J Clin Epidemiol. 2006;59(2):114–124. doi: 10.1016/j.jclinepi.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 3.Al-Sukhni W, Aronson M, Gallinger S. Hereditary colorectal cancer syndromes: familial adenomatous polyposis and lynch syndrome. Surg Clin North Am. 2008;88(4):819–844. vii. doi: 10.1016/j.suc.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 4.Fain PR, Goldgar DE. A nonparametric test of heterogeneity of family risk. Genetic epidemiology. 1986;(Supplement 1):61–66. doi: 10.1002/gepi.1370030710. [DOI] [PubMed] [Google Scholar]
- 5.Negri E, Braga C, La Vecchia C, et al. Family history of cancer and risk of colorectal cancer in Italy. Br J Cancer. 1998;77(1):174–179. doi: 10.1038/bjc.1998.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johns LE, Houlston RS. A systematic review and meta-analysis of familial colorectal cancer risk. Am J Gastroenterol. 2001;96(10):2992–3003. doi: 10.1111/j.1572-0241.2001.04677.x. [DOI] [PubMed] [Google Scholar]
- 7.Fuchs CS, Giovannucci EL, Colditz GA, Hunter DJ, Speizer FE, Willett WC. A prospective study of family history and the risk of colorectal cancer. N Engl J Med. 1994;331(25):1669–1674. doi: 10.1056/NEJM199412223312501. [DOI] [PubMed] [Google Scholar]
- 8.Goldgar DE, Easton DF, Cannon-Albright LA, Skolnick MH. Systematic population-based assessment of cancer risk in first-degree relatives of cancer probands. Journal of the National Cancer Institute. 1994;86(21):1600–1608. doi: 10.1093/jnci/86.21.1600. [DOI] [PubMed] [Google Scholar]
- 9.Ahsan H, Neugut AI, Garbowski GC, et al. Family history of colorectal adenomatous polyps and increased risk for colorectal cancer. Ann Intern Med. 1998;128(11):900–905. doi: 10.7326/0003-4819-128-11-199806010-00006. [DOI] [PubMed] [Google Scholar]
- 10.Winawer SJ, Zauber AG, Gerdes H, et al. Risk of colorectal cancer in the families of patients with adenomatous polyps. National Polyp Study Workgroup. N Engl J Med. 1996;334(2):82–87. doi: 10.1056/NEJM199601113340204. [DOI] [PubMed] [Google Scholar]
- 11.Slattery ML, Kerber RA. Family history of cancer and colon cancer risk: the Utah Population Database. J Natl Cancer Inst. 1994;86(21):1618–1626. doi: 10.1093/jnci/86.21.1618. [DOI] [PubMed] [Google Scholar]
- 12.Newcomb PA, Baron J, Cotterchio M, et al. Colon Cancer Family Registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol Biomarkers Prev. 2007;16(11):2331–2343. doi: 10.1158/1055-9965.EPI-07-0648. [DOI] [PubMed] [Google Scholar]
- 13.Win AK, Lindor NM, Young JP, et al. Risks of primary extracolonic cancers following colorectal cancer in Lynch syndrome. J Natl Cancer Inst. 2012;104(18):1363–1372. doi: 10.1093/jnci/djs351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kohonen T. Self-organizing maps. 3rd. Berlin ; New York: Springer; 2001. [Google Scholar]
- 15.Jain AK, Murty MN, Flynn PJ. Data clustering: a review. ACM Comput Surv. 1999;31(3):264–323. [Google Scholar]
- 16.Hastie T, Tibshirani R, Friedman JH. The elements of statistical learning : data mining, inference, and prediction. 2nd. New York: Springer; 2009. [Google Scholar]
- 17.Haykin SS. Neural networks and learning machines. 3rd. New York: Prentice Hall; 2009. [Google Scholar]
- 18.Vesanto J, Himberg J, Alhoniemi E, Parhankangas J. SOM toolbox for Matlab. Tech Rep Laboratory of Computer and Information Science, Helsinki University of Technology; 2000. [Google Scholar]
- 19.The MathWorks I MATLAB version 7.10.0. Natick, Massachusetts: 2010. [Google Scholar]
- 20.Breslow NE, Day NE. Statistical methods in cancer research. Volume II--The design and analysis of cohort studies. IARC scientific publications. 1987;(82):1–406. [PubMed] [Google Scholar]
- 21.Parkin DMWS, Ferlay J, Storm H. Cancer incidence in five continents. Volume VII. IARC scientific publications. 1997;(143):i–xxxiv. 1–1240. [PubMed] [Google Scholar]
- 22.Gould W. Jackknife estimation. Stata Technical Bulletin. 1995;(4):25–29. [Google Scholar]
- 23.Ries L, Eisner M, Kosary C, et al. SEER Cancer Statistics Review, 1975–2000. Bethesda, MD: National Cancer Institute; 2003. [Google Scholar]
- 24.StataCorp. Stata Statistical Software: Release 11. College Station, TX: StataCorp LP; 2009. [Google Scholar]
- 25.Taylor DP, Burt RW, Williams MS, Haug PJ, Cannon-Albright LA. Population-based family history-specific risks for colorectal cancer: a constellation approach. Gastroenterology. 2010;138(3):877–885. doi: 10.1053/j.gastro.2009.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerber RA, O'Brien E. A cohort study of cancer risk in relation to family histories of cancer in the Utah population database. Cancer. 2005;103(9):1906–1915. doi: 10.1002/cncr.20989. [DOI] [PubMed] [Google Scholar]
- 27.Teerlink CC, Albright FS, Lins L, Cannon-Albright LA. A comprehensive survey of cancer risks in extended families. Genet Med. 2012;14(1):107–114. doi: 10.1038/gim.2011.2. [DOI] [PubMed] [Google Scholar]
- 28.Andrieu N, Launoy G, Guillois R, Ory-Paoletti C, Gignoux M. Estimation of the familial relative risk of cancer by site from a French population based family study on colorectal cancer (CCREF study) Gut. 2004;53(9):1322–1328. doi: 10.1136/gut.2003.036376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hopper JL. Disease-specific prospective family study cohorts enriched for familial risk. Epidemiol Perspect Innov. 2011;8(1):2. doi: 10.1186/1742-5573-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Win AK, Ait Ouakrim D, Jenkins MA. Risk profiling: familial colorectal cancer. Cancer Forum. 2014;38(1):15–25. [Google Scholar]
- 31.Hopper JL, Carlin JB. Familial Aggregation of a Disease Consequent upon Correlation between Relatives in a Risk Factor Measured on a Continuous Scale. Am J Epidemiol. 1992;136(9):1138–1147. doi: 10.1093/oxfordjournals.aje.a116580. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.