Summary
Objective
The p.V433M in cytochrome P450 4F2 (rs2108622, CYP4F2*3) is associated with higher warfarin dose and lower risk of hemorrhage among European Americans. Herein, we evaluate the influence of CYP4F2*3 on warfarin dose, time to target INR and stable dose, proportion of time spent in target range (PTTR), as well as the risk of overanticoagulation and hemorrhage among European and African Americans.
Design
CYP4F2*3 was genotyped in 1238 patients initiated on warfarin in a prospective inception cohort. Multivariable linear regression was used to assess warfarin dose and PTTR; proportional hazards analysis was performed to evaluate time to target INR and stable dose, overanticoagulation, and hemorrhage.
Setting
Two outpatient anticoagulation clinics
Participants
1238 anticoagulated patients
Outcomes
Warfarin dose (mg/day), time to target INR and stable dose, PTTR, over anticoagulation (INR>4), and major hemorrhage.
Results
Minor allele frequency for the CYP4F2*3 variant was 30.3% among European Americans and 8.4% among African Americans. CYP4F2*3 was associated with higher dose among European Americans but not African Americans. Compared to CYP4F2*1/*1, *1/*3 was associated with a statistically nonsignificant increase in dose (4.5%, p=0.22) and *3/*3 was associated with a statistically significant increase in dose (13.2% , p=0.02). CYP4F2 genotype did not influence time to target INR, time to stable dose, or PTTR in either race group. CYP4F2*3/*3 was associated with a 31% lower risk of over anticoagulation (p=0.06). Incidence of hemorrhage was lower among participants with CYP4F2 *3/*3 compared to *1/*3 or *1/*1 (IRR=0.45; 95% CI: 0.14 – 1.11; p=0.09). After controlling for covariates, the CYP4F2 *3/*3 was associated with a 52% lower risk of hemorrhage although this was not statistically significant (p=0.24).
Conclusion
Possession of CYP4F2*3 variant influences warfarin dose among European Americans but not African Americans. The CYP4F2-dose, CYP4F2-overanticoagulation, and CYP4F2-hemorrhage association follows a recessive pattern with possession of CYP4F2*3/*3 genotype likely demonstrating a protective effect. These findings need further confirmation.
Keywords: CYP4F2, race, warfarin dose, PTTR, over anticoagulation, hemorrhage
Introduction
The management of warfarin therapy is complicated by a wide variation in dose and response across patients. Investigations in diverse racial populations have identified the contribution of clinical factors such as age, body mass index (BMI), vitamin K intake, comorbidities, and concurrent medications to variability in warfarin dosing.1, 2 Evaluation of genetic underpinnings have revealed the importance of single nucleotide polymorphism (SNPs) within the cytochrome P450 2C9 (CYP2C9) and vitamin K oxidoreductase complex subunit 1 (VKORC1) genes in influencing warfarin dose. These genes explain significantly more variability in warfarin dose requirements among European Americans than in African Americans.3-12
Investigators have also identified the influence of CYP4F2 on warfarin dose.13 CYP4F2 is a ω-hydroxylase responsible for the hydroxylation of leukotriene B4, and is expressed in the liver, kidneys, lungs, and white blood cells.14 It is also a vitamin K1 mono-oxidase (VK1), catalyzing the hydroxylation of VK1.15 Patients possessing p.V433M (rs2108622; CYP4F2*3) have a reduced capacity to metabolize VK1 in the liver, and the corresponding increase in VK1 levels necessitates a higher warfarin dose to elicit the therapeutic anticoagulant response.15 The CYP4F2*3 variant is associated with higher warfarin dose requirements in European Americans,13, 15-22 Asians 23-26 and Hispanics 27 but not among Brazilians 18 or African Americans8, 27. Among European Americans, studies support the association of CYP4F2*3 with dose variability, attainment of therapeutic INR, and attainment of stable dose.13, 15-22, 28, 29 Recently, CYP4F2 has been associated with a lower hemorrhage risk,30 and increased risk of stent thrombosis31 among patients of European descent.
The CYP4F2*3 polymorphism has been incorporated in the warfarin dosing algorithm offered at warfarindosing.org, although it is not yet a part of the guidelines provided by the Clinical Pharmacogenetics Implementation Consortium (CPIC).32 Recent clinical trials such as Clarification of Optimal Anticoagulation through Genetics (COAG) and European Pharmacogenetics of Anticoagulant Therapy (EU-PACT) do not include CYP4F2*3 in their dosing algorithms.33, 34 Although, there is enough evidence of the influence of CYP4F2*3 on warfarin dose, its influence on warfarin response, especially among African Americans has not been extensively evaluated. Herein, we evaluate the influence of CYP4F2 on warfarin dose, time to attain therapeutic INR and stable dose, anticoagulation control (assessed as proportion of time spent in target range; PTTR), over anticoagulation (INR>4), and major hemorrhage among European and African American warfarin users.
Methods
The prospective Warfarin Pharmacogenetics Cohort (WPC) recruited patients ≥20 years of age initiating warfarin therapy and managed at an anticoagulation clinic under the approval of the Institutional Review Board at the University of Alabama at Birmingham and at Emory University. Warfarin therapy requiring a target international normalized ratio (INR) range of 2-3 was initiated in patients with venous thromboembolism, stroke/transient ischemic attacks, atrial fibrillation, myocardial infarction, peripheral arterial disease, and others (i.e., cardiac thrombus). Patients requiring a higher (INR 2.5 to 3.5; e.g. mechanical heart valves) or lower (INR 1.5-2.5; e.g. pulmonary arterial hypertension) intensity of anticoagulation were excluded. The goals of the study were to identify the influence of clinical and genetic factors on warfarin dose, anticoagulation control, and risk of hemorrhage.
Patients were followed monthly from initiation of warfarin therapy for up to 2 years. A detailed history was obtained that including self-reported race, education, income, medical insurance, height and weight, blood urea nitrogen, serum creatinine, hemoglobin and hematocrit, indication for warfarin therapy, co-morbid conditions, medications, smoking status, alcohol use, and vitamin K intake as detailed in previous publications.3, 4, 35, 36 At each clinic visit, the INR, warfarin dose, concurrent medications, level of physical activity, dietary vitamin K intake, alcohol intake, and medication compliance were recorded. Changes in concomitant medications that influence warfarin pharmacodynamics (antiplatelet agents) or pharmacokinetics (including CYP2C9 inhibitors [e.g. amiodarone and statins] and CYP2C9 inducers [e.g. rifampin]) were documented at each visit and verified through medical record review and pharmacy refill records.
In addition to VKORC1 (rs9923231) and CYP2C9 (*2 [rs1799853], *3 [rs1057910]), we assessed CYP4F2*3 (rs2108622), and the African American specific CYP2C9 SNPs (*5 [rs28371686], *6 [rs9332131], *11 [rs28371685]), and the CYP2C SNP (rs12777823).3, 4, 35, 36 The assumption of Hardy Weinberg Equilibrium was met for all SNPs (p>0.20).
Outcome definitions
Warfarin dose (mg/day; log transformed to attain normality of residuals) was defined as the average dose required for maintenance of therapeutic anticoagulation (INR 2-3).
Time to attaining a therapeutic INR was calculated as number of days from warfarin initiation to the first INR of >2. Time required to attain stable warfarin dose was defined based on attainment of three consecutive target INRs in target INR range (2-3) with the INRs assessed at least 2 weeks apart.
Proportion of time spent in target range (PTTR) was calculated for each patient using the Rosendaal linear interpolation method.37 This method assumes that a linear relationship exists between two consecutively measured INR values and allows one to allocate a specific INR value to each day for each patient. Time in target range (TTR) for each patient was assessed by the percentage of interpolated INR values within the target range of 2.0–3.0 after attainment of first INR in target range.
Overanticoagulation was defined as INR>4. Major hemorrhages included serious, life threatening and fatal bleeding episodes as per Schulman et al.38 For all major hemorrhagic complications during the 2-year follow-up, the complication site (e.g. gastrointestinal tract), gravity of the event (e.g. requiring transfusion, surgical intervention, etc.), and laboratory findings (INR, hemoglobin/hematocrit, etc.) at the time of the event were objectively documented. Isolated subtherapeutic or supratherapeutic INRs in the absence of bleeding were not classified as events.
During follow-up, all major hemorrhagic complications were captured and verified through review of admissions and emergency department visits. Only medically documented events were included in the analyses. The Alabama Center for Health Statistics was queried to verify cause of death for all deceased to ensure inclusion of deaths due to hemorrhagic complications. All complications were documented by the study nurse, confirmed by the principal investigator, and finally reviewed independently by the Medical Director (TMB) of the Anticoagulation Clinic.
Statistical methods
Analysis of variance was used to assess group differences for continuous variables and chi-square (χ2) test of independence for categorical variables. The Hardy-Weinberg equilibrium assumption was tested using the χ2 test. Multivariable linear regression analysis was performed to evaluate the association between CYP4F2*3 on warfarin dose and PTTR after accounting for sociodemographic factors (age, race, gender, body surface area [BSA]), clinical factors (e.g. kidney impairment, based on estimated glomerular filtration rate eGFR >60, 30-59, <30ml/min/1.73 m2 calculated using the Modification of Diet in Renal Disease Study equation), 39 concurrent medications (e.g. amiodarone, statins), and variants in CYP2C9, VKORC1 and rs12777823.
The influence of CYP4F2*3 on time to target INR (2-3), time to stable dose, the risk of over anticoagulation (INR>4) and major hemorrhage was assessed using the counting process format in the proportional hazard (PH) model. Robust variance estimation was used to correct for the dependence among multiple events per individual and provide 95% confidence intervals (CIs) for the hazard ratios (HRs) of interest. Departures from the PH assumption were assessed by evaluating interactions of the predictors and a function of survival time. All analyses were performed using SAS version 9.3 at a non-directional alpha level of 0.05 accounting for demographic, clinical, and genetic factors. As the current evidence suggests that the influence of CYP4F2*3 on warfarin response may be dependent on race, we evaluated the CYP4F2-dose, anticoagulation control, and hemorrhage risk after stratifying by race. We evaluated the influence of CYP4F2*3 using additive (*1/*1 versus *1/*3 versus *3/*3), dominant (*1/*1 versus *1/*3 or *3/*3), and recessive (*1/*1 or *1/*3 versus *3/*3) models.
Role of the funding source
The study was funded by the NIH and the funders did not have any role in the study design, data collection, data analysis, data interpretation, or writing of the report.
Results
A total of 1238 patients were genotyped for the CYP4F2*3 variant (Table 1). The minor allele frequency (MAF) for the CYP4F2*3 variant was 30.3% among European Americans and 8.4% among African Americans. The prevalence of CYP2C9*2 and VKORC1 was higher among patients with the CYP4F2*3 variant compared to the wild-type allele. However, the prevalence of CYP2C9*3, *5, *6, and *11, and rs12777823 did not differ by CYP4F2 genotype. Patients with the CYP4F2*3 variant were older and more likely to be on warfarin for atrial fibrillation, whereas patients with CYP4F2 wild-type genotype were more likely to be on warfarin for venous thromboembolism and to have chronic kidney disease. The prevalence of comorbid conditions such as hypertension, hyperlipidemia, and diabetes mellitus did not differ by CYP4F2 genotype nor did the use of statins, amiodarone or antiplatelet therapy.
Table 1. Clinical, genetic and socio-demographic characteristics of patients by CYP4F2 genotypes.
| Characteristics |
CYP4F2 *1/*1 n=784 |
CYP4F2 *1/*3 n=390 |
CYP4F2 *3/*3 n=64 |
P Value |
|---|---|---|---|---|
|
| ||||
| Mean ± SD | Mean ± SD | Mean ± SD | ||
| Follow-up, years | 1.5 ± 0.9 | 1.3 ± 0.9 | 1.3 ± 0.9 | 0.06 |
| Age, years | 60.1 ± 15.6 | 63.0 ± 15.7 | 63.5 ± 15.6 | 0.005 |
| BSA, m2 | 2.0 ± 0.3 | 2.0 ± 0.3 | 2.0 ± 0.3 | 0.24 |
|
| ||||
| N (%) | N (%) | N (%) | ||
|
| ||||
| Female | 390 (49.7) | 176 (45.1) | 24 (37.5) | 0.08 |
| Racea | <0.001 | |||
| European Americans | 340 (43.4) | 305 (78.2) | 62 (96.9) | |
| African Americans | 444 (56.6) | 85 (21.8) | 2 (3.1) | |
| Indication for Warfarin therapy | ||||
| Venous thromboembolism | 359 (45.6) | 142 (36.4) | 23 (35.9) | 0.005 |
| Stroke /TIA | 50 (6.4) | 15 (3.9) | 3 (4.7) | 0.19 |
| Atrial Fibrillation | 300 (38.3) | 193 (49.5) | 32 (50.0) | <0.001 |
| Myocardial infarction | 12 (1.5) | 10 (2.6) | 0 (0.0) | 0.24 |
| Peripheral arterial disease | 9 (1.2) | 3 (0.8) | 1 (1.6) | 0.77 |
| Other | 53 (6.8) | 27 (6.9) | 5 (7.8) | 0.95 |
| Comorbid conditions | ||||
| Hypertension | 528 (68.0) | 244 (63.9) | 41 (64.1) | 0.34 |
| Hyperlipidemia | 359 (46.3) | 196 (51.3) | 31 (48.4) | 0.27 |
| Diabetes mellitus | 253 (32.6) | 117 (30.6) | 16 (25.0) | 0.40 |
| Chronic Kidney diseaseb | 0.03 | |||
| eGFR > 60ml/min/1.73m2 | 492 (62.8) | 240 (61.7) | 36 (56.3) | |
| eGFR >30-59ml/min/1.73m2 | 209 (26.7) | 123 (31.6) | 25 (39.1) | |
| eGFR < 30 ml/min/1.73m2 | 82 (10.5) | 26 (6.7) | 3 (4.7) | |
| Concurrent medications | ||||
| Statinsc | 411 (52.8) | 222 (56.9) | 33 (51.7) | 0.38 |
| Anti-plateletsd | 443 (56.9) | 240 (61.5) | 40 (62.5) | 0.26 |
| Amiodarone | 75 (9.6) | 45 (11.5) | 10 (15.6) | 0.24 |
| Percent patients possessing > 1 minor allele | ||||
| CYP2C9*2 | 109 (14.0) | 81 (20.8) | 16 (25.0) | 0.003 |
| CYP2C9*3 | 58 (7.5) | 37 (9.5) | 5 (7.8) | 0.47 |
| CYP2C9*5, *6, *11 | 13 (1.7) | 6 (1.5) | 0 (0.0) | 0.58 |
| VKORC1 | 283 (36.3) | 203 (52.5) | 39 (61.9) | <0.001 |
| rs12777823 | 292 (37.3) | 130 (33.3) | 17 (26.6) | 0.12 |
All patients enrolled were prescribed warfarin with a target INR range of 2-3.
Patients requiring a higher (INR 2.5 to 3.5; e.g. mechanical heart valves) or lower (INR 1.5-2.5; e.g. pulmonary arterial hypertension) intensity of anticoagulation were excluded.
SD: Standard Deviation, BSA: body surface area, TIA: transient ischemic attack, eGFR: estimated glomerular filtration rate
Asians (n=4; 0.3%) and Hispanics (n=4; 0.3%) were combined with the European Americans.
All eGFR are based on National Kidney Foundation staging using the Modification of Diet in Renal Disease study equation and categorized into 3 categories: eGFR > 60; eGFR >30-59 and eGFR < 30 ml/min/1.73m2
Statins included any of the 3-hydroxy-3-methyl-glutaryl-CoA (HMG-COA) reductase inhibitors
Antiplatelet agents included aspirin, clopidogrel, and, dipyridamole as mono or dual therapy
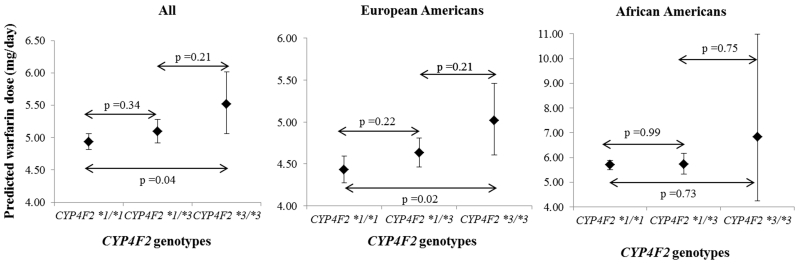
Patients with the CYP4F2*3 variant required higher warfarin dose compared to the wild-type genotype (Table 2) after adjusting for race, clinical and genetic factors. Among European Americans, compared to CYP4F2*1/*1, possession of *1/*3 genotype was associated with a statistically nonsignificant increase in dose by 4.5% and possession of *3/*3 genotype was associated with statistically significant increase in dose by 13.2% (Figure 1). Although African Americans possessing *3/*3 genotype required 19.9% higher warfarin dose, this finding was not statistically significant, likely due to the limited (n=2) prevalence of this genotype among African Americans (Figure 1). Gender (p=0.76), smoking status (p=0.54), and antiplatelet therapy (p=0.55) were not statistically significant in the model and were therefore removed from further analysis. Age, BSA, CYP2C9*3, and VKORC1 were significantly associated with dose among both European and African Americans (p<0.001, all). CYP2C9*2 was significantly associated only with dose among European Americans (p<0.001). Similarly, rs12777823 was significantly associated only with dose among African Americans (p<0.001).
Table 2. Predicted warfarin dose in mg/day and 95% confidence intervals (95%CI) based on CYP4F2 genotypes.
| CYP4F2 *1/*1 | CYP4F2 *1/*3 | CYP4F2 *1/*3 | |||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Total | N | Dose (95%CI) | N | Dose (95%CI) | N | Dose (95%CI) | |
| All | 1238 | 784 | 4.9 (4.8 – 5.1) | 390 | 5.1 (4.9 – 5.3) | 64 | 5.5 (5.1 – 6.0) |
| European Americans | 707 | 340 | 4.4 (4.3 – 4.6) | 305 | 4.6 (4.5 – 4.8) | 62 | 5.0 (4.6 – 5.5) |
| African Americans | 531 | 444 | 5.7 (5.5 – 5.9) | 85 | 5.7 (5.3 – 6.2) | 2 | 6.8 (4.3 – 10.9) |
Adjusted for age (centered at 40 years); body surface area (BSA) centered at 2.01, chronic kidney disease (CKD), amiodarone therapy, CYP2C9*2, CYP2C9*3, CYP2C9*5, *6 and *11 together, VKORC1, and rs12777823 variants. CKD was based on the Modification of Diet in Renal Disease Study equation. Patients were categorized into 3 categories: eGFR ≥60 (no CKD or mild CKD stage 1 and 2), eGFR=30-59 (moderate CKD; stage 3) and eGFR < 30 (severe CKD; stage 4 and 5). CYP2C9*2, CYP2C9*3, and VKORC1 were included as additive; 0 if no variants; 1 if heterozygous and 2 if homozygous for the variant allele. CYP2C9*5, *6 and *11 together, and rs12777823 were categorized as 0 if no variants and 1 if heterozygous or homozygous for the variant allele.
Figure 1.
Warfarin dose by CYP4F2 genotypes for all, European Americans and African Americans patients.
Time to target INR (>2) did not differ by CYP4F2 genotype among European Americans (p=0.10) or African Americans (p=0.47). These associations remained unchanged after adjusting for clinical and genetic factors. Similarly, time to stable warfarin dose did not significantly differ by CYP4F2 genotype among European Americans (p = 0.98) and African Americans (p = 0.08).
Patients possessing CYP4F2*3 variant had higher PTTR compared to those with the wild-type genotype. However this association was not statistically significant after accounting for clinical and genetic factors (Table 3). As the CYP4F2- PTTR association did not differ significantly by race, the final model adjusted for race. As previously shown, age (p<0.001), African-American race (p<0.001), BSA (p=0.002) and chronic kidney disease (p<0.001) were significantly associated with PTTR whereas gender (p=0.62), amiodarone use (p=0.76), statin use (p=0.13), antiplatelet use (p=0.16), CYP2C9 (p=0.97), VKORC1 (p=0.94), and rs12777823 (p=0.34) had no significantly association with anticoagulation control.
Table 3. Anticoagulation control as assessed by percent time in target range (PTTR; INR 2-3) by CYP4F2 genotype.
| Parameter | CYP4F2 *1/*1 | CYP4F2 *1/*3 | CYP4F2 *3/*3 | P Value | |
|---|---|---|---|---|---|
| Number of patients | 784 | 390 | 64 | ||
| Number of visits | 21008 | 10420 | 1864 | ||
| Follow-up (months) | 13193 | 6643 | 1289 | ||
| Follow-up months/patient | 16.8 ± 10.6 | 17.0 ± 10.8 | 20.1 ± 10.4 | 0.06 | |
|
| |||||
| Distribution of treatment time spent below range, in range and above range | |||||
|
| |||||
| PTBR | 29.6 | 26.2 | 26.0 | 0.09 | |
| PTTR | 52.5 | 57.1 | 56.4 | 0.002 | |
| PTAR | 17.9 | 16.7 | 17.6 | 0.57 | |
|
| |||||
|
Distribution of treatment time spent below range, in range and above range after controlling
for confoundersa | |||||
|
| |||||
| PTBR | 28.6 | 27.5 | 29.4 | 0.77 | |
| PTTR | 53.5 | 55.5 | 53.8 | 0.34 | |
| PTAR | 17.9 | 17.0 | 16.8 | 0.73 | |
INR: International Normalized Ratio
PTBR denotes percent time spent below range (INR 2-3); PTTR denotes percent time spent in range (INR 2-3); PTAR denotes percent time spent above range (INR 2-3)
Adjusted analysis includes age, race, BSA, current smoking, chronic kidney disease, concurrent statin and antiplatelet use, CYP2C9, and VKORC1 variants
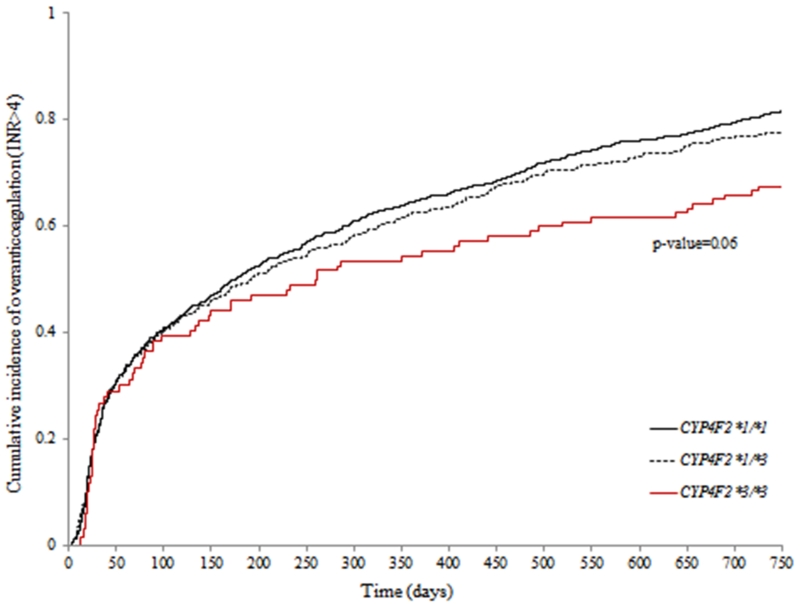
Compared to patients possessing the *1/*1 genotype, the risk of over anticoagulation (INR>4) was not significantly different among those with *1/*3 genotype (HR: 0.90, 95% CI: 0.76 – 1.07; p=0.22) and was lower among those with *3/*3 genotype (HR: 0.69, 95% CI: 0.47 – 1.01, p=0.06; Figure 2), although this was not statistically significantly different. As the CYP4F2-overanticoagulation association did not differ by race, the final model adjusted for race. African-American race (p<0.001), BSA (p=0.002), chronic kidney disease (p<0.001), antiplatelet use (p=0.008), CYP2C9 variants (p=0.006) and age (p=0.05) had a significant influence on over anticoagulation. Gender (p=0.81), amiodarone use (p=0.49), statin use (p=0.23), and rs12777823 (p=0.35) and VKORC1 (p=0.07) did not influence the risk of over anticoagulation.
Figure 2.
Cumulative incidence of overanticoagulation (INR>4) by CYP4F2 genotype
Over the accrued 1762 person-years of follow up, 140 major hemorrhagic events were observed. Major hemorrhages by site included gastrointestinal (n=84), genitourinary (n=18), retroperitoneal (n=6), intracranial bleeds (n=12), hemoptysis (n=4), and hematomas (n=16). On average, patients with major hemorrhage had significantly lower hematocrit values compared to those without major hemorrhages (35.5 ± 6.9 vs. 37.3 ± 8.7, p=0.007). The overall incidence of major hemorrhage was 7.9/100 person-years (95% CI: 6.7 – 9.3 per 100 p-yrs). The overall incidence of hemorrhage was 55% lower among patients with CYP4F2 *3/*3 genotype compared to *1/*3 or *1/*1 genotype, however this finding was not statistically significantly different (Table 4). European Americans with CYP4F2 *3/*3 had a 49% lower risk of hemorrhage compared to those with the *1/*3 or *1/*1 genotype;–although this finding was not statistically significant (Table 4). Due to the limited (n=2) prevalence of the *3/*3 genotype, we could not evaluate the gene-hemorrhage association among African Americans.
Table 4. Incidence (per 100 person-years) of Major Hemorrhage by CYP4F2 genotype.
| CYP4F2 genotype | N | Events | FU years |
Incidence | Incidence rate ratio |
P Value |
|---|---|---|---|---|---|---|
| All | ||||||
| *1/*1 | 784 | 92 | 1099.5 | 8.37 (6.75 -10.26) | Reference | — |
| *1/*3 | 390 | 44 | 554.6 | 7.93 (5.76-10.65) | 0.95 (0.66 -13.53) | 0.85 |
| *3/*3 | 64 | 4 | 107.5 | 3.72 (1.00-9.53) | 0.44 (0.14-1.10) | 0.09 |
| European Americans | ||||||
| *1/*1 | 340 | 36 | 504.0 | 7.14 (5.00-9.89) | Reference | — |
| *1/*3 | 305 | 35 | 438.2 | 7.99 (5.56-11.11) | 1.12 (0.70-1.79) | 0.64 |
| *3/*3 | 62 | 4 | 103.8 | 3.85 (1.04-9.87) | 0.54 (0.16-1.40) | 0.24 |
| African Americans | ||||||
| *1/*1 | 444 | 56 | 595.5 | 9.40 (7.10-12.21) | Reference | |
| *1/*3 | 85 | 9 | 116.4 | 7.73 (3.53-14.68) | 0.82 (0.38-1.61) | 0.61 |
| *3/*3 | 2 | 0 | 3.7 | — | — | — |
|
| ||||||
| CYP4F2 genotype (*1/*1 or *1/*3 versus *3/*3) | ||||||
|
| ||||||
| All | ||||||
| *1/*1 or *1/*3 | 1174 | 136 | 1654.1 | 8.22 (6.89 -9.73) | Reference | — |
| *3/*3 | 64 | 4 | 107.5 | 3.72 (1.00-9.53) | 0.45 (0.14-1.11) | 0.09 |
| European Americans | ||||||
| *1/*1 or *1/*3 | 645 | 71 | 942.2 | 7.54 (5.89-9.51) | Reference | — |
| *3/*3 | 62 | 4 | 103.8 | 3.85 (1.04-9.87) | 0.51 (0.16-1.28) | 0.22 |
| African Americans | ||||||
| *1/*1 or *1/*3 | 529 | 65 | 711.9 | 9.13 (7.10-11.56) | — | — |
| *3/*3 | 2 | 0 | 3.7 | — | — | — |
|
| ||||||
| CYP4F2 genotype (*1/*1 versus *1/*3 or *3/*3) | ||||||
|
| ||||||
| All | ||||||
| *1/*1 | 784 | 92 | 1099.5 | 8.37 (6.75 -10.26) | Reference | — |
| *1/*3 or *3/*3 | 454 | 48 | 662.1 | 7.25 (5.35-9.61) | 0.87 (0.61-1.22) | 0.42 |
| European Americans | ||||||
| *1/*1 | 340 | 36 | 504.0 | 7.14 (5.00-9.89) | Reference | — |
| *1/*3 or *3/*3 | 367 | 39 | 542.0 | 7.20 (5.12-9.84) | 1.01 (0.64-1.59) | 0.98 |
| African Americans | ||||||
| *1/*1 | 444 | 56 | 595.5 | 9.40 (7.10-12.21) | Reference | — |
| *1/*3 or *3/*3 | 87 | 9 | 120.1 | 7.49 (3.42-14.23) | 0.80 (0.37-1.56) | 0.55 |
FU years: Follow-up years
As the CYP4F2 *3/*3 genotype appears to be protective for major hemorrhage, we assessed its influence further by accounting for other clinical and genetic factors, categorizing the CYP4F2 *1/*3 and *1/*1 as the referent genotype (recessive model). While possession of CYP4F2 *3/*3 genotype was associated with a 52% lower risk of major hemorrhage, this finding was not statistically significant (HR: 0.48, 95% CI: 0.14 – 1.66, p=0.24).
We categorized *1/*3 or *1/*1 genotype as the referent genotype based on observed incidence of hemorrhage in our cohort. However, a recent report 30 evaluated the CYP4F2*3-hemorrhage association using a dominant model categorizing *1/*1 as the referent genotype and *1/*3 and *3/*3 as the variant genotype. Using this categorization, CYP4F2 *3 was not significantly associated with risk of hemorrhage (Table 4). This finding remained unchanged after adjusting for other clinical and genetic factors (HR: 0.88; 95% CI: 0.59-1.33; p=0.54).
Discussion
To our knowledge, this is the first paper to comprehensively evaluate the association of CYP4F2*3 with warfarin dose, PTTR, risk of overanticoagulation, and risk of major hemorrhage among chronic warfarin users. We also assessed whether possession of the CYP4F2*3 variant has a consistent effect across race groups.
The MAF for the CYP4F2*3 variant in our cohort was 30.3% among European Americans and 8.4% among African Americans, which is similar to previous reports for both European Americans and African Americans.8, 13, 17, 18, 27 Given that the bulk of the evidence regarding the influence of CYP4F2 among warfarin users is from patients of European descent, we compared our findings with previous reports in this race group. And given the limited reports in the literature among African Americans, we presented our findings acknowledging that our ability to interpret CYP4F2*3-outcome associations among African Americans was limited because of the lower MAF of the variant in this race group. Moreover, as only two patients possessed the *3/*3 genotype, we could not assess the CYP4F2*3-response association using the recessive model.
Possession of CYP4F2*3 variant was associated with higher warfarin dose requirements. Inclusion of CYP4F2*3 in the dose prediction algorithm improved the proportion of variability explained by 0.6% which is concordant with previous reports.13, 17, 40 Consistent with previous reports, the influence of CYP4F2 on warfarin dose varied by race in our study. Among European Americans, possession of CYP4F2*3 variant was associated with higher dose requirements with the CYP4F2*3/*3 genotype associated with significantly higher dose requirements (recessive effect).
The limited African-American participation has previously prevented the evaluation of the CYP4F2-dose association separately in this race group,41, 42 and has been unable to test the association after accounting for other clinical and genetic factors.8, 18 Our findings are consistent with those reported by others27 who demonstrated the lack of influence of CYP4F2*3 variant on warfarin dose requirement among 260 African Americans using an additive model. Moreover, as only 2 out of the 531 African Americans in our cohort possessed the CYP4F2*3/*3 genotype, we could not evaluate whether the CYP4F2*3 variant exerted a recessive effect on dose in this race group.
Concordant with previous reports among patients of European descent,17, 20, 28 CYP4F2*3 did not influence time to attainment of first target INR≥2 or time to stable dose among European Americans. Researchers evaluating the influence of 59 CYP4F2 SNPs on time to target INR and stable dose reported that the CYP4F2*3-time to therapeutic INR association (p=0.03) did not remain significant after correction for multiple testing. 28 Possession of the CYP4F2*3 variant did not influence PTTR after controlling for clinical and genetic factors in either race group in our study. The findings among European Americans are consistent with results of a previous study reporting that CYP4F2*3 was not significantly associated with PTTR (*1/*1 vs. *3/*3, p=0.39; *1/*3 vs. *3/*3, p=0.14).40 We are the first to report the lack of effect of the CYP4F2*3 variant on time to target INR, time to stable dose, and PTTR among African Americans.
Previous evaluations have not shown a significant association between CYP4F2 and overanticoagulation among patients of European descent.17, 20, 28, 40 We found no effect of CYP4F2 on risk of overanticoagulation among European or African Americans. Our results suggest a recessive effect of the *3 variant, with the CYP4F2*3/*3 genotype associated with a lower risk of over anticoagulation.
To our knowledge, this is the first report on the CYP4F2-hemorrhage association among African Americans. There are limited data on the CYP4F2-hemorrhage association among European Americans.17, 43 In a recent study of community-dwelling warfarin users of mainly European descent, possession of the *3 variant was associated with a 38% lower odds of major hemorrhage.30 The authors assessed the CYP4F2-hemorrhage association using a dominant model categorizing CYP4F2*1/*3 and *3/*3 versus *1/*1. However, our results do not support these findings. We recognize that categorization of genotypes as variant (*1/*3 and *3/*3) versus wild-type (*1/*1) is accepted. Therefore, we report the CYP4F2-hemorrhage association using both categorizations. However, our data regarding the incidence of hemorrhage demonstrates that the protective effect (if any) associated with CYP4F2 is related to possession of the *3/*3 genotype. Possession of the *3/*3 genotype is protective as supported by the absolute risk of hemorrhage observed among European Americans in our cohort. However, we could not evaluate the protective association of the CYP4F2*3 variant among African Americans given the lower minor allele frequency of the CYP4F2*3 in this group and the absence of major hemorrhage among the two African Americans with the CYP4F2*3/*3 genotype.
While we assessed the differential effect of CYP4F2*3 among African and European Americans, we recognize that these results may not be generalizable to Hispanics and Asians.23, 24, 27, 44, 45 Additional studies are needed to identify predictors that may differentially influence warfarin response in these race groups. As we assessed only the CYP4F2*3 polymorphism, we could not investigate for haplotype block structures in the two race groups. Assessment of additional SNPs in the CYP4F2 gene and investigating their association with warfarin response may reveal as yet undiscovered causal variants among African Americans. Moreover, it is likely that incorporation of other genetic (e.g. GGCX, CALU, CYP2C9*8)27, 46, 47 and clinical factors (including drug-drug interactions) not assessed in this study may further improve dose prediction and alter the effect-sizes of the predictors evaluated herein. We recognize this limitation.
Conclusion
Our findings confirm that possession of CYP4F2*3 variant influences warfarin dose among European Americans but not African Americans. The influence of CYP4F2*3 on warfarin dose variability among European Americans is smaller than that accounted for by CYP2C9 and VKORC1. The CYP4F2-dose association follows a recessive model. Similarly, CYP4F2-overanticoagulation and CYP4F2-hemorrhage association follow a recessive pattern with possession of CYP4F2*3/*3 genotype likely demonstrating a protective effect. These findings need to be confirmed in larger studies that can evaluate the consistency of the effects in different race groups.
Acknowledgements
This work was supported in part by a grant from the National Heart Lung and Blood Institute (RO1HL092173), National Institute of Neurological Disorders and Stroke (K23NS45598) and the National Centre for Advancing Translational Sciences (UL1 TR000165).
Footnotes
Conflict of Interests: None reported
Financial interests: None reported
References
- 1.Absher RK, Moore ME, Parker MH. Patient-specific factors predictive of warfarin dosage requirements. Ann Pharmacother. 2002;10:1512–7. doi: 10.1345/aph.1C025. [DOI] [PubMed] [Google Scholar]
- 2.Kamali F, Khan TI, King BP, et al. Contribution of age, body size, and CYP2C9 genotype to anticoagulant response to warfarin. Clin Pharmacol Ther. 2004;3:204–12. doi: 10.1016/j.clpt.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Limdi NA, Arnett DK, Goldstein JA, et al. Influence of CYP2C9 and VKORC1 on warfarin dose, anticoagulation attainment and maintenance among European-Americans and African-Americans. Pharmacogenomics. 2008;5:511–26. doi: 10.2217/14622416.9.5.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Limdi NA, Beasley TM, Crowley MR, et al. VKORC1 polymorphisms, haplotypes and haplotype groups on warfarin dose among African-Americans and European-Americans. Pharmacogenomics. 2008;10:1445–58. doi: 10.2217/14622416.9.10.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Limdi NA, Wadelius M, Cavallari L, et al. Warfarin pharmacogenetics: a single VKORC1 polymorphism is predictive of dose across 3 racial groups. Blood. 2010;18:3827–34. doi: 10.1182/blood-2009-12-255992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schelleman H, Chen Z, Kealey C, et al. Warfarin response and vitamin K epoxide reductase complex 1 in African Americans and Caucasians. Clin Pharmacol Ther. 2007;5:742–7. doi: 10.1038/sj.clpt.6100144. [DOI] [PubMed] [Google Scholar]
- 7.Schelleman H, Chen J, Chen Z, et al. Dosing algorithms to predict warfarin maintenance dose in Caucasians and African Americans. Clin Pharmacol Ther. 2008;3:332–9. doi: 10.1038/clpt.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavallari LH, Langaee TY, Momary KM, et al. Genetic and clinical predictors of warfarin dose requirements in African Americans. Clin Pharmacol Ther. 2010;4:459–64. doi: 10.1038/clpt.2009.223. [DOI] [PubMed] [Google Scholar]
- 9.Drozda K, Labinov Y, Jiang R, et al. A pharmacogenetics service experience for pharmacy students, residents, and fellows. Am J Pharm Educ. 2013;8:175. doi: 10.5688/ajpe778175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perera MA, Cavallari LH, Limdi NA, et al. Genetic variants associated with warfarin dose in African-American individuals: a genome-wide association study. Lancet. 2013;9894:790–6. doi: 10.1016/S0140-6736(13)60681-9. PMCID:3759580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hernandez W, Gamazon ER, Aquino-Michaels K, et al. Ethnicity-specific pharmacogenetics: the case of warfarin in African Americans. Pharmacogenomics J. 2014;3:223–8. doi: 10.1038/tpj.2013.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagai R, Ohara M, Cavallari LH, et al. Factors influencing pharmacokinetics of warfarin in African-Americans: implications for pharmacogenetic dosing algorithms. Pharmacogenomics. 2015;3:217–25. doi: 10.2217/pgs.14.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caldwell MD, Awad T, Johnson JA, et al. CYP4F2 genetic variant alters required warfarin dose. Blood. 2008;8:4106–12. doi: 10.1182/blood-2007-11-122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stec DE, Roman RJ, Flasch A, Rieder MJ. Functional polymorphism in human CYP4F2 decreases 20-HETE production. Physiological genomics. 2007;1:74–81. doi: 10.1152/physiolgenomics.00003.2007. [DOI] [PubMed] [Google Scholar]
- 15.McDonald MG, Rieder MJ, Nakano M, Hsia CK, Rettie AE. CYP4F2 is a vitamin K1 oxidase: An explanation for altered warfarin dose in carriers of the V433M variant. Molecular pharmacology. 2009;6:1337–46. doi: 10.1124/mol.109.054833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takeuchi F, McGinnis R, Bourgeois S, et al. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;3:e1000433. doi: 10.1371/journal.pgen.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pautas E, Moreau C, Gouin-Thibault I, et al. Genetic factors (VKORC1, CYP2C9, EPHX1, and CYP4F2) are predictor variables for warfarin response in very elderly, frail inpatients. Clin Pharmacol Ther. 2010;1:57–64. doi: 10.1038/clpt.2009.178. [DOI] [PubMed] [Google Scholar]
- 18.Perini JA, Struchiner CJ, Silva-Assuncao E, Suarez-Kurtz G. Impact of CYP4F2 rs2108622 on the stable warfarin dose in an admixed patient cohort. Clin Pharmacol Ther. 2010;4:417–20. doi: 10.1038/clpt.2009.307. [DOI] [PubMed] [Google Scholar]
- 19.Wells PS, Majeed H, Kassem S, et al. A regression model to predict warfarin dose from clinical variables and polymorphisms in CYP2C9, CYP4F2, and VKORC1: Derivation in a sample with predominantly a history of venous thromboembolism. Thromb Res. 2010;6:e259–64. doi: 10.1016/j.thromres.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 20.Burmester JK, Berg RL, Yale SH, et al. A randomized controlled trial of genotype-based Coumadin initiation. Genet Med. 2011;6:509–18. doi: 10.1097/GIM.0b013e31820ad77d. [DOI] [PubMed] [Google Scholar]
- 21.Danese E, Montagnana M, Johnson JA, et al. Impact of the CYP4F2 p.V433M Polymorphism on Coumarin Dose Requirement: Systematic Review and Meta-Analysis. Clin Pharmacol Ther. 2012;6:746–56. doi: 10.1038/clpt.2012.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang R, Wang C, Zhao H, Huang J, Hu D, Sun Y. Influence of CYP4F2 genotype on warfarin dose requirement-a systematic review and meta-analysis. Thrombosis research. 2012;1:38–44. doi: 10.1016/j.thromres.2011.11.043. [DOI] [PubMed] [Google Scholar]
- 23.Cen HJ, Zeng WT, Leng XY, et al. CYP4F2 rs2108622: a minor significant genetic factor of warfarin dose in Han Chinese patients with mechanical heart valve replacement. Br J Clin Pharmacol. 2010;2:234–40. doi: 10.1111/j.1365-2125.2010.03698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh O, Sandanaraj E, Subramanian K, Lee LH, Chowbay B. Influence of CYP4F2 rs2108622 (V433M) on Warfarin Dose Requirement in Asian Patients. Drug Metabolism and Pharmacokinetics. 2011;2:130–36. doi: 10.2133/dmpk.dmpk-10-rg-080. [DOI] [PubMed] [Google Scholar]
- 25.Nakamura K, Obayashi K, Araki T, et al. CYP4F2 gene polymorphism as a contributor to warfarin maintenance dose in Japanese subjects. J Clin Pharm Ther. 2011 doi: 10.1111/j.1365-2710.2011.01317.x. [DOI] [PubMed] [Google Scholar]
- 26.Krishna Kumar D, Shewade DG, Loriot MA, et al. Effect of CYP2C9, VKORC1, CYP4F2 and GGCX genetic variants on warfarin maintenance dose and explicating a new pharmacogenetic algorithm in South Indian population. European journal of clinical pharmacology. 2014;1:47–56. doi: 10.1007/s00228-013-1581-x. [DOI] [PubMed] [Google Scholar]
- 27.Bress A, Patel SR, Perera MA, Campbell RT, Kittles RA, Cavallari LH. Effect of NQO1 and CYP4F2 genotypes on warfarin dose requirements in Hispanic-Americans and African-Americans. Pharmacogenomics. 2012;16:1925–35. doi: 10.2217/pgs.12.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang JE, Jorgensen AL, Alfirevic A, et al. Effects of CYP4F2 genetic polymorphisms and haplotypes on clinical outcomes in patients initiated on warfarin therapy. Pharmacogenetics and genomics. 2009;10:781–9. doi: 10.1097/FPC.0b013e3283311347. [DOI] [PubMed] [Google Scholar]
- 29.Nahar R, Saxena R, Deb R, et al. CYP2C9, VKORC1, CYP4F2, ABCB1 and F5 variants: influence on quality of long-term anticoagulation. Pharmacological reports : PR. 2014;2:243–9. doi: 10.1016/j.pharep.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 30.Roth JA, Boudreau D, Fujii MM, et al. Genetic risk factors for major bleeding in patients treated with warfarin in a community setting. Clin Pharmacol Ther. 2014;6:636–43. doi: 10.1038/clpt.2014.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kupstyte N, Zaliunas R, Tatarunas V, et al. Effect of clinical factors and gene polymorphism of CYP2C19*2, *17 and CYP4F2*3 on early stent thrombosis. Pharmacogenomics. 2015;3:181–9. doi: 10.2217/pgs.14.165. [DOI] [PubMed] [Google Scholar]
- 32.Johnson JA, Gong L, Whirl-Carrillo M, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clinical pharmacology and therapeutics. 2011;4:625–9. doi: 10.1038/clpt.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kimmel SE, French B, Kasner SE, et al. A pharmacogenetic versus a clinical algorithm for warfarin dosing. The New England journal of medicine. 2013;24:2283–93. doi: 10.1056/NEJMoa1310669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pirmohamed M, Burnside G, Eriksson N, et al. A randomized trial of genotype-guided dosing of warfarin. The New England journal of medicine. 2013;24:2294–303. doi: 10.1056/NEJMoa1311386. [DOI] [PubMed] [Google Scholar]
- 35.Limdi NA, Beasley TM, Baird MF, et al. Kidney Function Influences Warfarin Responsiveness and Hemorrhagic Complications. J Am Soc Nephrol. 2009:912–21. doi: 10.1681/ASN.2008070802. PMC2663833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Limdi NA, Brown TM, Yan Q, et al. Race influences warfarin dose changes associated with genetic factors. Blood. 2015;4:539–45. doi: 10.1182/blood-2015-02-627042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosendaal FR, Cannegieter SC, van der Meer FJ, Briet E. A method to determine the optimal intensity of oral anticoagulant therapy. Thromb Haemost. 1993;3:236–9. [PubMed] [Google Scholar]
- 38.Schulman S, Kearon C, Subcommittee on Control of Anticoagulation of the S, Standardization Committee of the International Society on T, Haemostasis Definition of major bleeding in clinical investigations of antihemostatic medicinal products in non-surgical patients. Journal of thrombosis and haemostasis : JTH. 2005;4:692–4. doi: 10.1111/j.1538-7836.2005.01204.x. [DOI] [PubMed] [Google Scholar]
- 39.Levey A, Coresh J, Greene T, et al. Using standardized serum creatinine values in the modification of diet in renal disease study equation for estimating glomerular filtration rate. Ann Intern Med. 2006;4:247–54. doi: 10.7326/0003-4819-145-4-200608150-00004. [DOI] [PubMed] [Google Scholar]
- 40.Bejarano-Achache I, Levy L, Mlynarsky L, Bialer M, Muszkat M, Caraco Y. Effects of CYP4F2 polymorphism on response to warfarin during induction phase: a prospective, open-label, observational cohort study. Clinical therapeutics. 2012;4:811–23. doi: 10.1016/j.clinthera.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 41.Lubitz SA, Scott SA, Rothlauf EB, et al. Comparative performance of gene-based warfarin dosing algorithms in a multiethnic population. J Thromb Haemost. 2010 doi: 10.1111/j.1538-7836.2010.03792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sagreiya H, Berube C, Wen A, et al. Extending and evaluating a warfarin dosing algorithm that includes CYP4F2 and pooled rare variants of CYP2C9. Pharmacogenetics and genomics. 2010;7:407–13. doi: 10.1097/FPC.0b013e328338bac2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma C, Zhang Y, Xu Q, et al. Influence of warfarin dose-associated genotypes on the risk of hemorrhagic complications in Chinese patients on warfarin. International journal of hematology. 2012;6:719–28. doi: 10.1007/s12185-012-1205-8. [DOI] [PubMed] [Google Scholar]
- 44.Cavallari LH, Momary KM, Patel SR, Shapiro NL, Nutescu E, Viana MA. Pharmacogenomics of warfarin dose requirements in Hispanics. Blood Cells Mol Dis. 2011;2:147–50. doi: 10.1016/j.bcmd.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 45.Nakamura K, Obayashi K, Araki T, et al. CYP4F2 gene polymorphism as a contributor to warfarin maintenance dose in Japanese subjects. Journal of clinical pharmacy and therapeutics. 2012;4:481–5. doi: 10.1111/j.1365-2710.2011.01317.x. [DOI] [PubMed] [Google Scholar]
- 46.Voora D, Koboldt DC, King CR, et al. A polymorphism in the VKORC1 regulator calumenin predicts higher warfarin dose requirements in African Americans. Clinical pharmacology and therapeutics. 2010;4:445–51. doi: 10.1038/clpt.2009.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cavallari LH, Vaynshteyn D, Freeman KM, et al. CYP2C9 promoter region single-nucleotide polymorphisms linked to the R150H polymorphism are functional suggesting their role in CYP2C9*8-mediated effects. Pharmacogenet Genomics. 2013;4:228–31. doi: 10.1097/FPC.0b013e32835e95c7. [DOI] [PMC free article] [PubMed] [Google Scholar]