Abstract
Regenerative medicine holds great potential to address many shortcomings in current medical therapies. An emerging avenue of regenerative medicine is the use of self-assembling peptides (SAP) in conjunction with stem cells to improve the repair of damaged tissues. The specific peptide sequence, mechanical properties, and nanotopographical cues vary widely between different SAPs, many of which have been used for the regeneration of similar tissues. To evaluate the potential of SAPs to guide stem cell fate, we extensively reviewed the literature for reports of SAPs and stem cell differentiation. To portray the most accurate summary of these studies, we deliberately discuss both the successes and pitfalls, allowing us to make conclusions that span the breadth of this exciting field. We also expand on these conclusions by relating these findings to the fields of nanotopography, mechanotransduction, and the native composition of the extracellular matrix in specific tissues to identify potential directions for future research.
Keywords: Self-assembling peptide, stem cell, nanotopography, extracellular matrix
Introduction
Current medical therapies fail to restore tissue damage due to degenerative conditions, normal aging processes, ischemia and traumatic injuries. Tissue regeneration and organ engineering have shown promise as novel treatment strategies for patients requiring tissue or whole organ replacement.1,2 Modern approaches to regenerative tissue therapies typically revolve around the use of stem cells. The development of pluripotent stem cells3 has helped resolve the challenge of acquiring a reliable, implantable stem cell source without the deleterious immune responses triggered by non-autologous cells. In spite of this breakthrough in stem cell technology, a great need still exists to optimize methods for implanting, organizing, differentiating, and integrating stem cells to reliably regenerate or replace damaged tissues.2
Lessons from stem cell biology have shed light on the numerous variables that affect stem cell differentiation and tissue formation.2,3 Among these are nanotopography, molecular extracellular matrix (ECM) composition, and mechanotransduction.2,3 A novel group of materials capable of manipulating these criteria are self-assembling peptides.
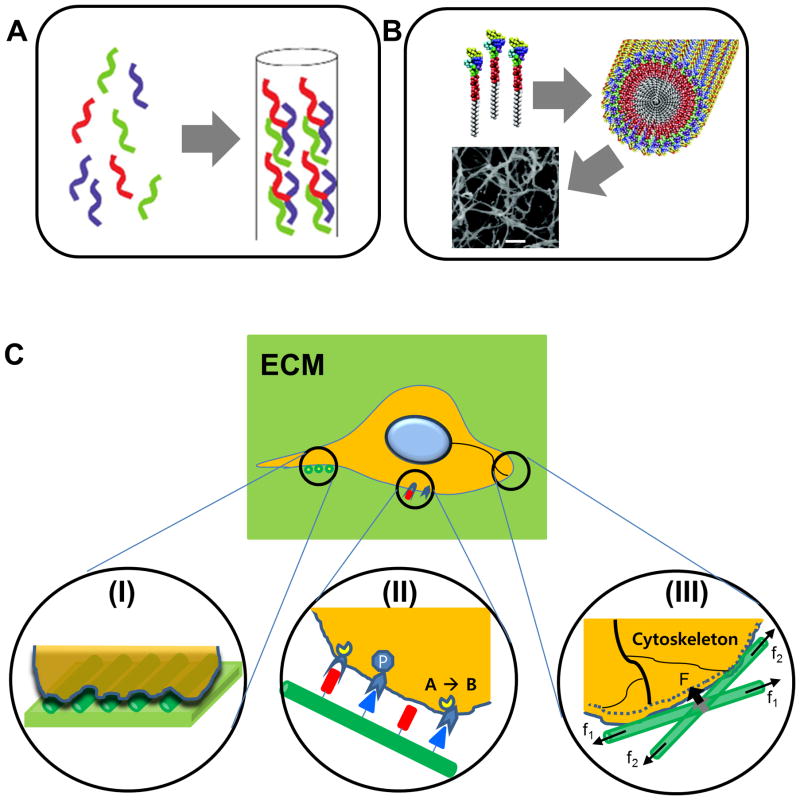
Self-assembling Peptides (SAPs) are defined as short peptides that self-organize into nanofiber structures (Figure 1A).4 The SAP family includes amino acids covalently modified with a lipophilic group that also self-assemble into nanofiber structures, called peptide amphiphiles (PAs) (Figure 1B). Both SAP and PA nanofibers can further assemble into macrostructures that resemble components of native extracellular tissue.5,6 Together, these properties enable scaffold architectural design on both a nano and macro scale. For most SAPs, assembly has been engineered to occur at a physiologic pH, allowing solutions of these peptides to be injected into damaged tissues before assembling into hydrogels.7 These scaffolds are inexpensive to produce, and their components are customizable to the resolution of a single amino acid, including the covalent addition of functional groups with integrin-binding prperties.8 These characteristics of SAPs are highly desirable for tissue engineering, and there has been a recent increase in the number of studies evaluating their potential to repair and regenerate tissue. This review summarizes these studies, focusing on the ability of SAPs to differentiate stem cells.
Figure 1.
Peptides self-assembled into nanostructures and their functional principles in the niche of stem cells. A. Graphical illustration of the formation of short peptides assembled into collagen-mimicking nanofiber.76 Reprinted with permission from ref. 76. B. Amphiphilic peptides for cylindrical nanostructures (scale bar in the SEM photo: 300 nm).28 Reprinted with permission from ref. 28. C. Action mechanisms of SAPs: (a) nanotopographical cues, (b) cell-ECM interactions, (c) transduction of mechanical forces.
Cell-fate altering mechanisms of SAPs
Self-assembling peptides are unique in that they can recapitulate niche cues in three distinct ways: nanotopography guidance, ECM binding, and transduction of mechanical forces (Figure 1C). Previous studies evaluating these characteristics with respect to stem cells have shown that altering these variables can either maintain stem cell populations or direct their differentiation.1–3
Nanotopographical cues have been shown to influence stem cell fate by mediating focal adhesion signaling and cytoskeletal organization.1,9 The ECM architecture of niches that maintain stem cells typically differs from the greater ECM architecture of surrounding tissue.10–12 One key mechanism by which nanotopography directs stem cell fate involves focal adhesion kinase (FAK).12,13 Nanotopography also alters cell fate through three-dimensional cytoskeletal re-organization.15,16 Our group has demonstrated this principle using nanogrooved substrates in conjunction with several cell lines, which results in a cellular architecture mimicking target tissues.17 The influence of nanotopography in guiding stem cell fate and organization aligns with idea that stem cells are influenced by three-dimensional changes in their respective niches.
The ECM composition and architecture for a given tissue changes in response to maturation, usage, and disease.13,14 Molecular ECM cues share many aspects of influence with nanotopographical cues. Both the molecular composition of the ECM, and the expression of focal adhesions that bind the ECM, play a role in determining the stem cell response.1 Specifically, the unique combination of integrin receptors and the presence (or absence) of their corresponding ligand will induce changes in signal transduction, which enforces the maintenance or differentiation of a given stem cell population.1
Mechanotransduction defines the physical interaction between stem cells and their surrounding niche via the transmission of physical forces.18 As cells bind and apply forces to their surrounding ECM, the ECM architecture will physically respond based on its mechanical characteristics. For example, stiffer materials will resist manipulation by cell forces to a greater degree than softer materials. The reverse of this principle is also true - the mechanical properties of the ECM determine the degree to which external forces are transferred on to a cell. The ability of cells to manipulate their environment via transduction of forces, and vice versa, has been shown to alter stem cell fate.19
While each of these three components can individually influence stem cell fate, physical interactions in-vivo between stem cells and their niche employ all three cooperatively. With respect to SAPs, mediating these interactions is dependent on peptide sequence. The ultrastructure of SAPs after assembly provides nanotopographical cues that can recapitulate aspects of ECM architecture. The specific peptide sequence can mimic native ECM ligands and interact with stem cells via physical binding. Finally, the mechanical properties of the SAPs determine the degree to which forces are directed onto and away from stem cells. In summary, by changing the peptide sequence, it is possible to alter the nanostructure, potential cell binding sites, and mechanotransduction of the resulting SAP, and therefore establish control over stem cell fate.
Influence of SAPs on stem cell fate in different tissues
Neural Tissue
In adult neural tissues, the ECM plays an important nanotopographical role. The neural ECM (nECM) is rich with chondroitin sulfate proteoglycans (CSPGs) and hyaluronan, with low amounts of fibronectin, collagen, and laminin.17 Sulfated proteoglycans in the CNS myelin have been found to be inhibitory for axonal outgrowth, including Nogo-A, myelin associated glycoprotein, and oligodendrocyte myelin glycoprotein.16 Despite their relative scarcity, collagens and laminins are fundamental to neural cell fate decisions.20 Mechanically, neural tissue is very soft relative to other tissues, with an elastic modulus around 500 Pa.21 Structurally, aggrecan and hyaluronan are observed to form focal lattice-like perineural networks (PNN), which influence structural plasticity.15 Therefore, the survival, differentiation, and proliferation of neural cells can be modulated by the nanotopography, ECM composition, and mechanotransduction properties of the nECM, and some of these factors have been recapitulated by SAPs.
SAP structures contain pores of 5–200 nm in width.18 This pore size resembles the structures found in native nECM, allow diffusion of growth factors, and provide an excellent substrate for customizable lineage-specific neural stem cell (NSC) migration, differentiation, and proliferation.18 Comparative studies have shown that SAP nanofiber hydrogels facilitate a more realistic and replicable culture system than traditional approaches relying on heterogeneous animal-derived materials such as matrigel and collagen I.19 There are three key SAP’s and PAs that have seen utility in neural tissue regeneration; LDLK12, IKVAV-PA, and RADA.22,23
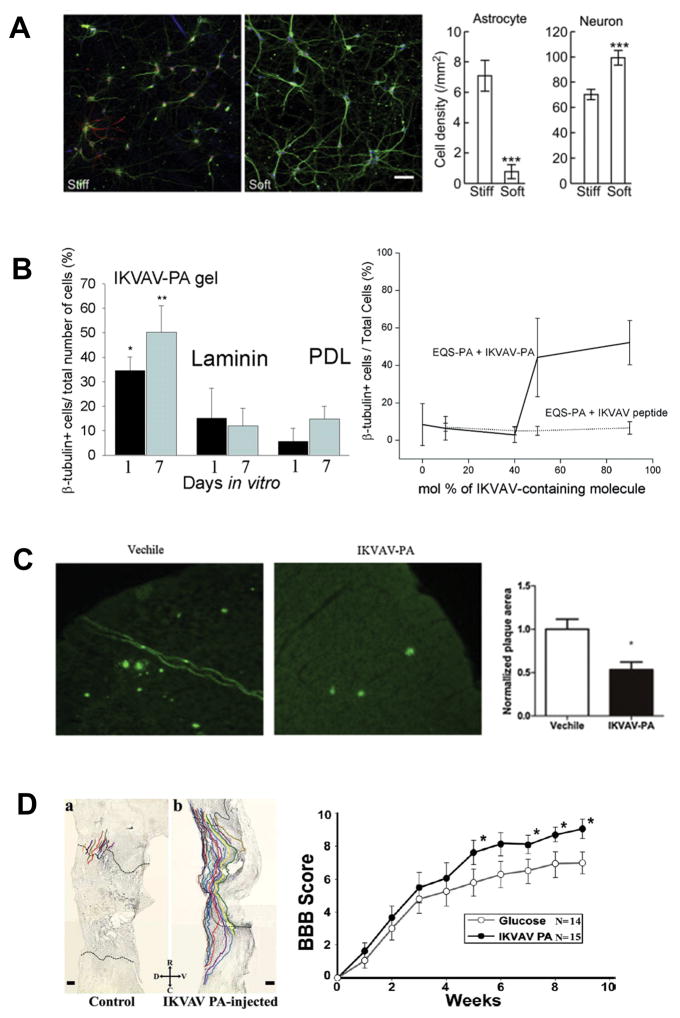
Notable progress has been made in achieving neuronal differentiation in vitro. Two key variables that have emerged for attaining SAP-mediated neuronal differentiation are mechanotransduction and bioactivity. Broadly speaking, stiff substrates like glass tend to bias NSC differentiation towards astrocytes, while softer substrates favor neuronal differentiation (Figure 2A).24,25 Several studies have shown this principle holds true for SAPs, such as a recent study by Caprini et al, in which an LDLK12 scaffold with a compression modulus of 100pa led to a greater degree of neuronal differentiation than stiffer substrates.26
Figure 2.
Effects of SAPs on in vitro differentiation of neural stem cells (A and B) and in vivo repair of neural tissues (C and D). A. (a) Neuronal density (green) is selectively increased relative to astrocytes (red) by soft (7.3 ± 0.9 kPa storage modulus) vs stiff (22.9 ± 5 kPa) PA substrate (DAPI stain) in hippocampal culture. (b) Comparison of cell densities (n = 3, p<0.001).25 Reprinted with permission from ref. 25. B. Left: IKVAV-PA gels enable statistically significant growth of neurons relative to laminin and poly-D-lysine (PDL) controls at one and seven days (P<0.5, P<0.01 in duplicate). Right: Increased concentrations of IKVAV-PA, but not IKVAV peptide, increases one-day neuronal differentiation. IKVAV-PA and EQS-PA (solid line) and EQS-PA combined with varied amounts of soluble IKVAV peptide (dashed line).28 Reprinted with permission from ref. 28. C. IKVAV-PA injection reduces the normalized cortical and hippocampal area of amyloid-beta plaques (Green, thioflavin S stain) in a murine model of Alzeheimer’s Disease relative to vehicle control (P<0.05).33 Reprinted with permission from ref. 33. D. Left: Neurolucida tracing of regenerating murine BDA-labeled corticospinal tract motor neurons 11 weeks post-SCI. IKVAV-PA presence results in greatly increased migration into the lesion. Right: This regrowth corresponds to statistically significant improvement of motor function after five weeks as quantified by BBB score.36 Reprinted with permission from ref. 36.
Covalently modifying SAP scaffolds through the inclusion of different functional motifs27 is another means of achieving neuronal differentiation. In 2004, a landmark paper by Silva et al. demonstrated that an IKVAV (beta-1 integrin ligand derived from laminin) epitope integrated with a peptide amphiphile and seeded with NSCs could significantly induce differentiation of NSCs to neurons, and down-regulate astroglia, better than both non-functionalized scaffold and 3D cultures mixed with dissociated IKVAV (Figure 2B).28 The key variable in differentiation efficacy was found to be density of biomotif exposure.28 Cheng et al. expanded these findings by placing the IKVAV motif onto a different scaffold (i.e. RADA16) and showed that neural progenitor cells (NPCs) were directed to a neuronal lineage by the RADA16-IKVAV scaffold, while the unaltered RADA16 SAP directed cells towards both neuronal and astroglia lineages.29 Li et al. extended these findings to pluripotent embryonic carcinoma stem cells, demonstrating that RADA-IKVAV drove cell fate towards a neuronal differentiation.30
Some of the most promising applications of SAPs have come from in-vitro models of neurodegenerative disease and nervous system trauma. Cui et al. found that implantation of NSCs in a RADA16-YIGSR scaffold into mouse neural tissue (1) decreased beta-amyloid mediated hippocampal apoptosis by alleviating down regulation of synapsin-1, (2) promoted a neuro-protective environment, and (3) facilitated greater cognitive rescue and restoration of learning and memory versus scaffold or NSC alone.31 Two studies by Yang et al. demonstrated that IKVAV-PA injection alone could lower amyloid plaque burden,32 ameliorate learning and memory losses, and increase endogenous hippocampal NPC proliferation and neural differentiation in mice (Figure 2C).33 Ni et al. applied RADA16 scaffolds with murine embryonic stem cells (ESCs) to an in vitro Parkinson’s disease model, which resulted in enhancement of dopaminergic neuronal differentiation.34 An emerging theme from these studies is that SAPs demonstrate potential to treat conditions of endogenous, pathological SAPs. Consistent with this idea, Hnasko et al. showed that RADA inoculation could disrupt prion accumulation and extend survival in a hamster model of scrapie.35
SAPs have also been used for treating spinal cord injury (SCI) and traumatic brain injury. Tysseling-Mattiace demonstrated in an acute compression model of SCI that injection of IKVAV-PA, but not IKVAV alone, reduced astrogliosis and promoted the growth of atypical motor and sensory neurons along with functional improvement (Figure 2D).36 Cigognini et al. showed that injection of RADA16 functionalized with BMPH1 and glycine repeats facilitated statistically significant functional improvement and increased axonal regeneration within the cyst of acute contusive rat SCI.37 Gelain et al. used a similar acute contusive model to show that injecting a LDLK12 SAP functionalized with Ac-FAQ achieved better locomotor recovery than LDLK12 or saline injection.38 In subacute SCI injury, Wasaki et al. discovered that K2(QL)6K2 (QL6) injection with NSC injection promoted functional improvement, higher NSC survival, and a statistically significant reduction in cystic cavity size.39 In brain injury, Cheng et al. showed that injecting RADA16-IKVAV with encapsulated NSCs led to improved cerebral neocortex/neopallidum regeneration in a model of TBI/tumor removal, with higher ratio of neurons and reduced formation of glial astrocytes.29 Finally, Ellis-Behnke et al. demonstrated that RADA16 injection alone could restore visual function in hamsters with a severed ophthalmic tract.40
Skeletal Tissue: Bone and Cartilage
The extracellular nanoarchitecture of skeletal tissues is highly organized and constantly remodeled by native cell populations to withstand changes in mechanical forces. For instance, in bone tissue the binding between collagen type I and individual hydroxyapatite crystals occurs in a periodic manner every 2 nm,41 and is responsible for giving bone its unique physical properties. This periodicity is carried into the ultrastructure of bone, yielding a 68 nm periodicity between collagen type I fibrils and hydroxyapatite deposits.42 As a result, bone achieves a Young’s modulus that surpasses 20 GPa.43 In contrast, the ECM of cartilage is defined by three distinct zones of collagen type II organization.44,45 While the ultrastructural organization of collagen is unique to each zone, all contain collagen bundled into parallel sheets with periodic banding patterns that occur approximately every 100 nm.44 This combination of nanoscale collagen organization within a stratified ultrastructural organization leads to a gradient in Young’s modulus across each zone ranging from 300 kPa up to 20 MPa.45 Nanofibrous scaffolds such as SAPs have the potential to recapitulate the periodic nature, and defined collagen architecture, of both bone and cartilage extracellular matrix. Applications of self-assembling peptides to skeletal tissue engineering have primarily revolved around the use of RADA and KLDL SAPs, and subsequent functionalized derivatives of RADA and KLDL.
A common component used in bone tissue engineering is hydroxyapatite. Garreta et al. performed one of the first studies with a hydroxyapatite-functionalized SAP, culturing human ESCs with or without osteogenic media on either RADA16/hydroxyapatite nanoparticles, RADA16, or hydroxyapatite alone.46 All ESCs cultured in an osteogenic media resulted in a greater degree of calcification than cultures without, and the combination of RADA16 and hydroxyapatite caused a greater degree of calcification than hydroxyapatite alone.46 Anderson et al. modified a peptide amphiphile with an RGDS group, and combined this with a hydroxyapatite-nanoparticle/gelatin solution to differentiate human mesenchymal stem cells (MSCs) in an osteogenic culture medium.47 RT-PCR revealed higher expression of osteogenic genes in cultures containing both the RGDS-PA and hydroxyapatite-nanoparticle/gelatin solution.47 Interestingly, Sargeant et al. demonstrated that peptide amphiphiles (PAs) functionalized with RGDS sequences could accumulate calcium mineralization without the addition of hydroxyapatite, which also induces the differentiation of human MSCs.48 These studies demonstrate the ability of functionalized SAPs or PAs to enhance osteogenic differentiation in both ESCs and MSCs.
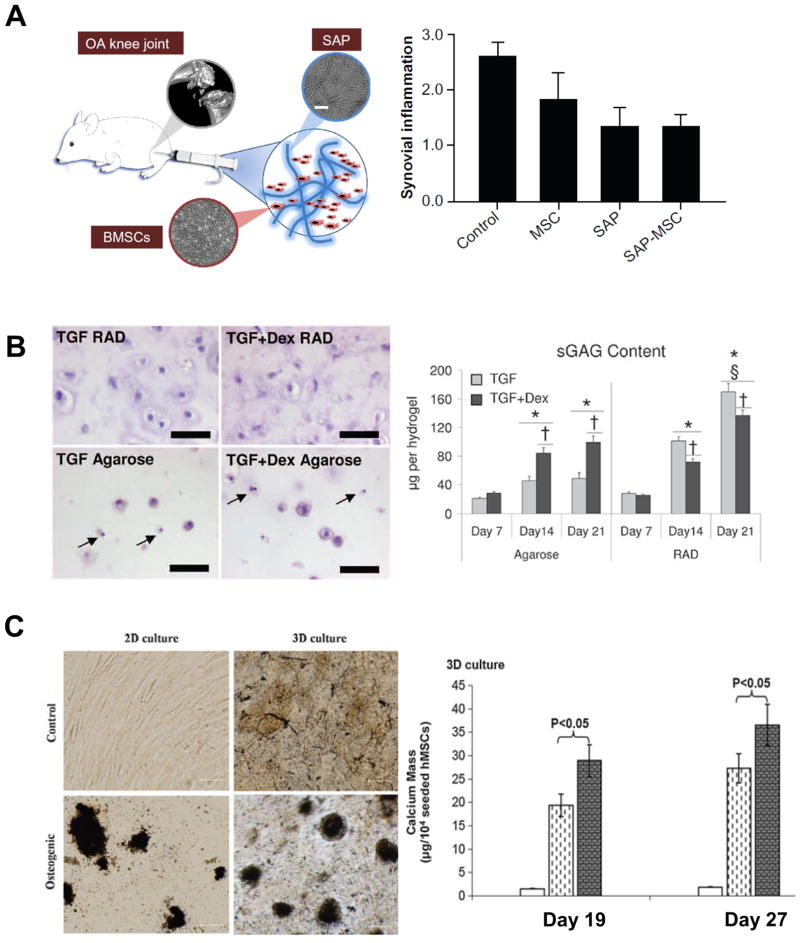
Other factors have also been utilized in combination with SAPs to improve osteogenic differentiation, including heat shock,49 neuropeptide substance P,50 and PRP.51 Human MSCs cultured in RADA16 hydrogels and exposed to 41 °C heat shock for 1 hour every three days demonstrated a greater degree of calcium mineralization and osteogenic gene expression than RADA16 cultures without heat shock (Figure 3C).49 The combination of neuropeptide substance P with KLD12 SAPs was shown to increase the rate of MSC migration when injected in vivo, resulting in a greater reduction of bone defect size in a rat model.50 An ultrastructural analysis revealed the formation of lacunae-like structures habited by migrating cells within the KLD12-substance P mixture, demonstrating the ability of KLD12 SAPs to recapitulate the nanoarchitecture of bone ECM.50 Yoshimi et al. injected MSCs and a combination of PRP and RADA16 into dog mandible bone defects, showing that combination of MSCs, PRP, and RADA16 reduced the defect to a greater degree than other groups.51 However, no significant decrease in defect size was noted for PRP and RADA16 injections without MSCs.51
Figure 3.
Influence of SAP on bone and cartilage. A. (Left) schematic representation of an SAP and MSC injection into a rat osteoarthritis model.18 (Right) our previously published data, which shows a repression of IL-2 secretion with the use of SAPs.60 Reprinted with permission from ref. 18 and ref. 60. B. The histology panel on the left highlights the abundance of apoptotic cells cultured in agarose gels compared to RADA SAP gels.54 The graph to the right shows an increase of GAG accumulation using RADA over agarose, supporting the use of RADA SAPs to direct chondrogenic stem cell fate. Reprinted with permission from ref. 54. C. The histology panel on the left shows an increase in calcification in a 3D culture over a 2D culture, and further enhancement of tissue calcification of both cultures in the presence of an osteogenic media.49 The graph on the right compares the effects of normal SAP cultures (white bar) to SAP cultures with an osteogenic media (dashed bar) and SAP cultures with an osteogenic media exposed to heat-shock (gray bar).49 These data highlight the enhanced osteogenic potential of SAPs for bone tissue engineering, and further optimization using novel stimuli like heat shock. Reprinted with permission from ref. 49.
One of the more promising applications of SAPs for bone tissue engineering is reconstituting large bone defects. In a two-study series, Hou et al.52 and Li et al.53 demonstrated the ability to apply a combination of RADA16 with demineralized bone matrix to differentiate MSCs for repairing small bone defects in a rat model,52 and then closing a large bone defect in a goat model.53 In the large defect model, bone marrow was used for the cell source instead of MSCs, and the combination of RADA16 with demineralized bone matrix and bone marrow was superior to demineralized bone and marrow without RADA16 for reduction of the bone defect.53
With respect to cartilage tissue engineering, Florine et al. showed the effect of SAPs on cartilage matrix production may be unique to stem cells (Figure 3B).54 In this study, both bovine MSCs and bovine chondrocytes were cultured in either agarose gels or RADA4 SAP hydrogel.54 In response to a chondrogenic media, bovine MSCs produced significantly higher amounts of glycosaminoglycans (GAGs) and collagen in the RADA4 SAP gel versus the agarose gel, while chondrocytes exhibited the opposite relationship.54 Erickson et al. performed a similar study that confirmed enhanced differentiation of MSCs in RADA hydrogels in comparison to alginate gels.55
Like bone tissue engineering, SAPs have been functionalized to enhance chondrogenic differentiation of stem cells. RADA16 functionalized with heparin was shown to improve chondrogenic differentiation at a ratio 190 parts RADA to one part heparin.56 At a higher mixing ratio (95:1, RADA:heparin) the mechanical properties of the resulting tissue were statistically indistinguishable from native chicken and calf cartilage, while lower ratios had significantly softer mechanical properties.56 TGF-beta is known to increase cartilage matrix production, and has also been used for this purpose in combination with SAPs. The addition of TGF-beta1 to a culture of bone marrow stromal cells in a KLDL3 SAP hydrogel resulted in a TGF-beta1 dose-dependent increase of GAG accumulation.57 Shah et al. combined a novel PA modified with a TGF-beta binding group to create a culture system that would release TGF-beta over time.58 The combination of PAs with the TGF-beta binding group and TGF-beta1 resulted in significantly higher expression of aggrecan compared to normal PAs and TGF-beta alone.58
In spite of promising in vitro results, the addition of growth factors does not appear to provide any additional chondrogenic benefit over SAPs or PAs alone in vivo. In the same study by Shah et al., PAs with the TGF-beta binding group had a significantly higher O’Driscoll score than PAs alone when injected in a rabbit cartilage defect, but the addition of TGF-beta1 to either group did not change the score.58 A study by Miller et al. found similar results using a rabbit cartilage defect model to evaluate KLDL3 in combination with chondrogenic growth factors and bone marrow stroma cells.59 Radiographic analysis showed significant improvement in all treatment groups compared to a sham control, but histologic scoring and analysis of newly formed tissue was only significantly improved over untreated animals in the KLDL group,59 suggesting the SAPs had greater therapeutic potential when administered alone than in conjunction with stem cells or growth factors.
Tatman et al. found inflammation to play a role in SAP based therapies for cartilage defects (Figure 3A).60 They injected either MSCs alone, KLD12 SAPs alone, or MSCs and KLD12 SAPs into rat cartilage defects. In comparison to MSCs alone, KLD12 and the combination of KLD12 with MSCs had lower levels of interleukins in the synovial fluid, suggesting SAPs may attenuate the immune response. The use of KLD12 also resulted in lower tissue mineral density, which indicates KLD12 may preferentially drive MSCs towards a chondrogenic lineage in vivo. Furthermore, they also found KLD12 SAPs significantly lowered a modified Mankin score, with or without MSCs, supporting their use to stymie the progression of osteoarthritis.
One potential pitfall of SAPs specific to bone and cartilage tissue engineering is a lack of mechanical integrity. Without chemical modification, the compression modulus of most SAPs is measured in units of kPA77, which is one order of magnitude –lower than native cartilage and two orders of magnitude –lower than native bone. The lack of resistance to compression may restrict the use of SAPs to non-weight bearing applications. However, the significance of this deficit remains unclear as SAPs have successfully closed large bone defects in weight bearing appendages of goats. 53 As osteochondral applications of SAPs are pursued in the future, an emphasis should be placed on mechanical integrity, in addition to tissue formation, to help define the potential clinical role of SAP derived therapies.
Cardiac Tissue
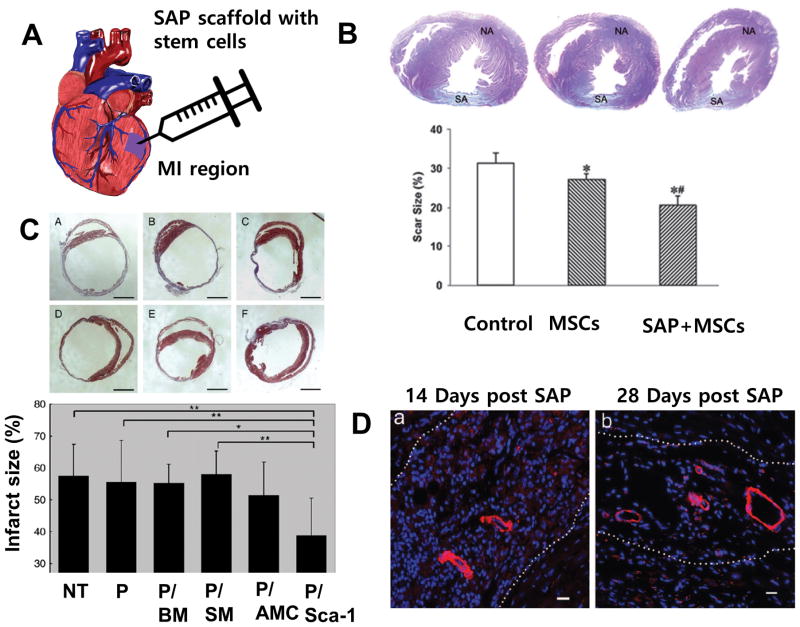
The cardiac extracellular matrix is a continuous macromolecular network extending from the pericardium to the endocardium.61 Collagen and GAGs comprise the majority of the cardiac ECM, which is organized into distinct zones. Collagen types I and III are most common in the pericardium, Collagen IV in the basement membrane, collagen type VI in the myocardium and pericellular area and fibronectin in smaller septal and pericapillary regions.62 The composition and ultrastructural organization of cardiac tissue directly translates to tissue function. We have previously shown that anisotropic force contraction can be significantly increased by aligning cardiomyocytes using nanotopographical cues.9 SAPs can also mimic cardiac ECM binding sites. RADA based SAPs closely mimic the RGD binding motif found on both alpha -1 beta-1 and alpha-5 beta-1 integrins, which are common cardiac integrins known to bind collagen VI and IV.63 Considering the importance of both nanoarchitecture and ECM composition to cardiac function, SAPs have the potential to improve stem cell differentiation into cardiac lineages by mimicking components of the native tissue (Figure 4A).
Figure 4.
Application of SAPs to cardiac tissue repair in the animal model. A. Conceptual illustration of injection of SAP and stem cells into the damaged myocardium. B. (above) Masson trichrome staining of heart sections at four weeks after myocardial infarction, observed under a light microscope. NA: normal area, SA: scar area. (below) Infarct area as determined by percent of total LV area for the no treatment control, stem cells only and cells with scaffold (p<0.001 vs. control, p<0.001 vs. cell alone).64 Reprinted with permission from ref. 64. C. (above) Treating with SAP and cSca-1 cells diminishes infarct expansion as seen in Masson trichrome stained heart slices and corresponding infarct size graph (below) 14 days after transplantation. The slices and graph depict; non-treated MI (A), SAP only (B), bone marrow stem cell with SAP (C), skeletal myoblasts with SAP (D), adipose-tissue derived mesenchymal cells with SAP (E) and cardiac progenitor cells with SAP (F). Data shows mean +/− SD. P < 0.05 cSca-1 with SAP vs. BM with SAP and p<0.01 for cSca-1 with SAP vs. non-treated MI, SAP only and skeletal myoblasts with SAP.69 Reprinted with permission from ref. 69. D. Mature vessel-like structures are detected in the SAP scaffold 14 days post injection (a) and after 28 days (b) several arteriole-like structures were seen within the microenvironment in all samples. Cells (blue, DAPI) are in the microenvironment positively stained for alpha - smooth muscle actin antibody.67 Reprinted with permission from ref. 67.
RADA peptides have been the primary focus of SAP research for cardiac regeneration. Cui et al. cultured MSCs expressing cardiac transcription markers Nkx2.5 and c-kit in RADA hydrogels, and found significantly increased cell survival compared to MSCs cultured on tissue culture plastic (Figure 4B).64 Additional immunofluorescence staining showed a higher level of cardiac specific marker cTnT, and a more organized sarcomere structure in comparison to the control.64 Functionalized SAPs have also been used in vitro to further control stem cell fate. Boopathy et al. combined RADA SAPs with Jagged1 (a ligand of Notch-1) and cultured cardiac progenitor cells.65 A significant increase in the cardiac specific transcription factors Nkx2.5, Hey1, MEF2C and GATA4 expression was observed in the RADA-Jagged1 culture over RADA alone and RADA with scrambled Jagged1.65 Guo et al. evaluated the combination of an RGDSP cell adhesion motif with a RADA SAP, and found an increases of cell survival over RADA alone.66
The first in vivo application of SAPs in cardiac tissue was performed by Davis et al.67 In this study a RADA hydrogel without cells was injected into the left ventricle of a healthy mouse.67 28 days after the injection, capillary-like structures were observed in the SAP environment, suggesting RADA SAPs may enhance vascularization in the heart (Figure 4D).67 Soler-Botija et al. progressed these findings by applying RADA SAPs to infarcted cardiac tissue in a mouse model and demonstrated the regeneration of functional blood vessels in the infarcted tissue.68 Cui et al. validated this phenomenon in a rat myocardial infarction (MI) model using MSCs and vWF immunostaining, finding significant increases in capillary density in transplanted RADA SAPs with MSCs expressing c-kit and Nkx2.5 over cells transplanted alone or mice without transplant.64 Collectively, these studies highlight the ability of RADA SAPs to induce vascularization in cardiac tissue, while also successfully sustaining stem cell populations in vivo.
The combination of stem cells and SAPs in vivo was further characterized by Cui et al and Tokunaga et al.69 Cui et al. demonstrated a reduction of infarct size with the addition of hydrogel and mesenchymal stem cells.64 Tokunaga et al went further, investigating four cell lines in conjunction with RADA SAPs: clonal stem cell antigen-1 positive cardiac progenitor cells (sSca-1), bone marrow mononuclear cells, skeletal myoblasts, and adipose tissue-derived mesenchymal cells. RADA SAPs and cells were injected around the border of the infarcted zone in a mouse MI model, showing that cSca-1s with RADA reduced the infarct size and increased vascularization significantly over RADA alone or no treatment (Figure 4C).69 However, no differences were noted between RADA alone and the treatment control.69
Functionally-modified RADA SAPs have also been used in vivo. Dubois et al. combined RADA16-I and II with Insulin-like growth factor 1 (IGF1).70 In spite of promising results in vitro, transplanting skeletal myoblasts into a rat cardiac MI model with IGF1 functionalized RADA SAPs did not significantly change the ejection fraction.70 Guo et al. functionalized a RADA16-1 SAP with a hairpin binding domain (sequence LRKKLGKA) to allow for the slow release of vascular endothelial growth factor (VEGF).71 The combination of VEGF with RADA modified SAPs resulted in a significant increase in cardiac ejection fraction and fractional shortening, while the myocardial scar size and fibrosis were significantly reduced.71 Boopathy et al. applied their Jagged1-RADA combination to a coronary artery ligation model, showing that Jagged1 functionalized RADA16 was able to completely rescue the ejection fraction and significantly decrease cardiac fibrosis when injected during reperfusion.65 Recently, Yoon and his colleagues utilized RGD modified PAs as a means to improve the survival of implanted mouse embryonic stem cells, and to enhance ejection fraction after a MI.78 Given their ability to promote vascularization and improve cardiac function after injury in conjunction with stem cells, SAPs provide a novel approach to further cardiac regenerative therapies.
Pitfalls of stem cell differentiation by SAPs
The use of SAPs and PAs for the differentiation of stem cells has yielded some very promising applications that have potential clinical implications. However, the use of SAPs for stem cell differentiation is not without limitations. One of the more obvious limitations brought to light in this review is the inability of SAPs to differentiate stem cells alone. With very few exceptions, the studies reviewed required the use of differentiation media or additional functionalization to achieve successful differentiation. In accordance with this finding, many studies have shown SAPs to actually maintain stem-ness for prolonged periods in culture using RGD and IKVAV peptides or similar derivatives.71 These findings are in contrast to much of the in vivo data.
In vivo, the studies reviewed support the ability of SAPs to aid in stem cell differentiation, induce tissue vascularization, stimulate the migration of native stem cell populations, and significantly improve tissue function for many organ-systems. SAPs also augment the immune response at sites of injury. These findings appear to contradict the effect of SAPs in vitro, but the presence of native cytokines and interaction with native ECM following in vivo SAP and stem cell injection could explain these differences. Many of these factors alter the stem cell niche and cause the observed changes in stem cell behavior, much like functionalizing SAPs in vitro. However, these in vivo studies do have limitations.
The in vivo injection of SAPs with cells or cytokines often did not exceed the benefit of injecting SAPs alone. Several factors may play a role in this observation, including stem cell choice/source, cytokine concentration, and functional group used. Allogenic stem cell sources are known to elicit an immune response, which may play a role in hindering the efficacy of in vivo SAP and stem cell injection. The cytokine profiles of an in vivo environment are known to be more complex than the single cytokine additions typically accompanying SAP injections, which could render these additions insignificant. Finally, the choice of functional group has a major role in recapitulating components of native ECM. RADA and KLD SAPs have sequences that resemble integrin binding motifs, but they do not stimulate integrin signaling in a tissue-specific manner, and are known to actually maintain a stem cell-like state.72
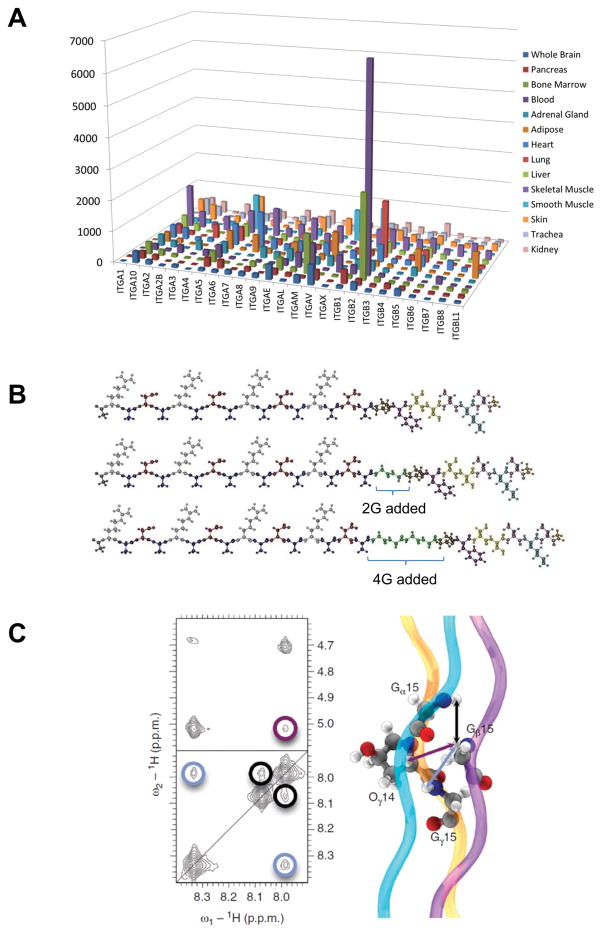
The lack of integrin specific signaling is a potential pitfall of the current SAPs, but the inclusion of such ligands has tremendous potential. To demonstrate this point, we have reanalyzed microarray data from GSE1133 and isolated the various integrins expressed across many tissues (Figure 5A).73 By comparing the tissue-specific integrin expression profiles, a picture of tissue-specific integrin signaling becomes clear. This reveals a potential avenue for future SAP research: by more deliberately selecting functional groups that resemble the ECM of a specific tissue, it may be possible to improve the interaction between SAPs and stem cells.
Figure 5.
Strategy to fine control over stem cell fate using SAPS. A. Normalized and processed microarray data were downloaded from GSE1133 from the NIH GEO database. The integrin gene family was isolated and the expression profiles for each tissue in the study are displayed graphically to visualize the integrin expression profiles unique to each tissue type.73 Reprinted with permission from ref. 73. B. By adding GLY spacers between the a standard RADA repeat sequence in the SAP amino acid sequence, better expose of the integrin binding motifs was achieved.74 Reprinted with permission from ref. 74. C. Computational and modeling of the self-assembling potential of peptide sequences has resulted in the development of an extensive library of theoretical self-assembling peptides that have yet to be investigated.76 Reprinted with permission from ref. 76.
Several studies in this review took advantage of tissue-specific functional groups to improve SAP mediated stem cell differentiation. Gelain et al. used phage-display to capture novel peptides on the surface of murine NPCs derived from NSCs. They were then able to functionalize RADA-FAQ scaffolds with these peptides, screen them for efficacy, and ultimately increase stem cell differentiation and survival in-vitro, while also promoting nervous tissue regrowth and functional recovery in-vivo using a rat contusive SCI model.38 With respect to skeletal tissue, the use of demineralized bone matrix in combination with SAPs had a similar effect on the reduction of large bone defects,53 which further advocates for the efficacy of SAP functionalization in a tissue-specific manor. These studies demonstrate the potential SAPs could have if further engineered to mimic tissue-specific ECM components, thus accentuating the need for additional development.
Advances in SAP synthesis to improve control over stem cell fate
The advantages of SAP functionalization have been described in this review as a means to control growth factor release, cell survival, cell fate, and reducing the immune response to implanted constructs. While further efforts utilizing these approaches are vital to the progression of SAP research, the addition of functional groups may disrupt the assembly of SAPs or the final SAP ultrastructure; two qualities that make SAPs desirable for tissue engineering. Another approach would be to incorporate tissue-specific ECM motifs into the SAP sequences themselves. This approach has seen some use in neuro engineering studies, where the addition of glycine repeats into a SAP sequence improved exposure of incorporated integrin receptor binding sites (Figure 5B).74 Further development of similar approaches could allow SAPs to assemble and self-organize into a final ultrastructure without interference from functional groups, while also providing additional tissue-specific ECM signaling.
Along this line of development, some groups have begun to catalog different permutations of self-assembling peptide sequences. Lesley et al. discovered a synthetic peptide sequence that self-assembled into a collagen-like triple helix, and then further organized into structures that resemble various native tissue ECMs.75 Building on this work, Jorge et al. computationally defined the relationships necessary to determine self-assembling collagen heterotrimers in silico (Figure 5C),76 making it possible to develop predictive models to screen peptides for the potential to self-assemble. Going one step further, Pim et al. devised a computational method to analyze the propensity of collagen-like peptides to self-assemble,8 which produced a library of nearly 8,000 potential peptides. Over 100 of these peptides were strongly predicted to self-assemble into a collagen-like triple helix, providing future SAP researchers with a vast array of novel self-assembling peptides to help achieve tissue specificity.
Novel functionalization and synthesis of SAPs is but one of many potential future directions for research. Conjugation of SAPs to metallic surfaces and metallic particles has yielded many intriguing results which warrant further investigation such as the ability to disrupt biofilm formation and other antimicrobial properties.79, 80 With respect to altering the fate of native eukaryotic cell populations, SAP coated titanium implants have demonstrated a greater degree of biocompatibility over traditional titanium implants denoted by increased implant mineralization and vascularization.81, 82 The addition of SAPs to metallic implants also provides a means to deliver growth factors and other bioactive substrates.81, 82 Further research into the use of SAPs as a means to improve existing medical interventions could enhance the integration of synthetic heart valves, dental implants, and other surgically implanted materials by controlling cell fate.
Conclusion
SAPs represent a novel and promising tool with emerging efficacy for stem cell differentiation. One of the most intriguing aspects of SAPs is their ability to attenuate the pathology of many diseases that plague national health, such as alzheimer’s, parkinson’s, heart failure, and arthritis. Further optimization of these scaffolds to more closely mimic target tissues could lead to the development of novel treatments, and relive the current burden of these diseases. Due to the highly tunable nature of SAPs, researchers will have the ability to control nanotopography, ligand binding interactions, and mechanotransduction in a single scaffold. Through elucidation of the native stem cell niche nanoarchitecture, ECM, and principles of mechanotransduction, SAPs could be designed to recapitulate these elements to advance tissue regeneration and organ engineering applications.
Supplementary Material
Acknowledgments
This work was supported by a National Institutes of Health R21 Grant (R21AR064395 to DHK) and a Muscular Dystrophy Association Research Grant (MDA255907 to DHK). DHK thanks the Department of Bioengineering at the University of Washington for the new faculty startup fund. This work was also supported by an Orthopaedic Research and Education Foundation New Investigator grant awarded to A.O.G. and an International Collaborative R&D Program Grant with KIAT funded by the MOTIE (N0000894) awarded to D.H.K.
References
- 1.Dalby MJ, Gadegaard N, Oreffo ROC. Nat Mater. 2014;13:558–569. doi: 10.1038/nmat3980. [DOI] [PubMed] [Google Scholar]
- 2.Dvir T, Timko BP, Kohane DS, Langer R. Nature Nanotech. 2010;6:13–22. doi: 10.1038/nnano.2010.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim HN, Jiao A, Hwang NS, Kim MS, Kang DH, Kim DH, Suh KY. Advanced Drug Delivery Reviews. 2013;65:536–558. doi: 10.1016/j.addr.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scanlon S, Aggeli A. Nano Today. 2008;3:22–30. [Google Scholar]
- 5.Cui H, Muraoka T, Cheetham AG, Stupp SI. Nano Lett. 2009;9:945–951. doi: 10.1021/nl802813f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu K, Guo L, Mehta AK, Childers WS, Dublin SN, Skanthakumar S, Conticello VP, Thiyagarajan P, Apkarian RP, Lynn DG. Chem Commun. 2007:2729–2731. doi: 10.1039/b701029j. [DOI] [PubMed] [Google Scholar]
- 7.Hartgerink JD, Hartgerink Jeffrey D, Granja Juan R, Granja JR, Ronald A, Milligan A, Milligan RA, Ghadiri MR. J Am Chem Soc. 1996;118:43–50. [Google Scholar]
- 8.Frederix P, Scott GG, Abul-Haija YM. Nature. 2014;7:30–37. doi: 10.1038/nchem.2122. [DOI] [PubMed] [Google Scholar]
- 9.Kim DH, Lipke EA, Kim P, Cheong R, Thompson S, Delannoy M, Suh KY, Tung L, Levchenko A. Proc Natl Acad Sci USA. 2010;107:565–570. doi: 10.1073/pnas.0906504107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papagiannouli F, Lohmann I. Computational and Structural Biotechnology Journal. 2015;13:122–130. doi: 10.1016/j.csbj.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scadden DT. Nature. 2006;441:1075–1079. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- 12.Marthiens V, Kazanis I, Moss L, Long K, ffrench-Constant C. Journal of Cell Science. 2010;123:1613–1622. doi: 10.1242/jcs.054312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Risau W, Lemmon V. Developmental Biology. 1988;125:441–450. doi: 10.1016/0012-1606(88)90225-4. [DOI] [PubMed] [Google Scholar]
- 14.Daley WP, Peters SB, Larsen M. Journal of Cell Science. 2008;121:255–264. doi: 10.1242/jcs.006064. [DOI] [PubMed] [Google Scholar]
- 15.Giamanco KA, Matthews RT. Neuroscience. 2012;218:367–384. doi: 10.1016/j.neuroscience.2012.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hou S, Tian W, Xu Q, Cui F, Zhang J, Lu Q, Zhao C. Neuroscience. 2006;137:519–529. doi: 10.1016/j.neuroscience.2005.09.029. [DOI] [PubMed] [Google Scholar]
- 17.Bandtlow CE, Zimmermann DR. Physiol Rev. 2000;80:1267–1290. doi: 10.1152/physrev.2000.80.4.1267. [DOI] [PubMed] [Google Scholar]
- 18.Cunha C, Panseri S, Villa O, Silva D, Gelain F. Int J Nanomedicine. 2011;6:943–955. doi: 10.2147/IJN.S17292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koutsopoulos S, Zhang S. Acta Biomater. 2013;9:5162–5169. doi: 10.1016/j.actbio.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Liu M, Yan Y, Yang ST. World J Stem Cells. 2014;6:11–23. doi: 10.4252/wjsc.v6.i1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X, Katsanevakis E, Liu X, Zhang N, Wen X. Progress in Polymer Science. 2012;37:1105–1129. [Google Scholar]
- 22.Holmes TC, de Lacalle S, Su X, Liu G, Rich A, Zhang S. Proc Natl Acad Sci USA. 2000;97:6728–6733. doi: 10.1073/pnas.97.12.6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yokoi H, Kinoshita T, Zhang S. Proc Natl Acad Sci USA. 2005;102:8414–8419. doi: 10.1073/pnas.0407843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saha K, Keung AJ, Irwin EF, Li Y, Little L, Schaffer DV, Healy KE. Biophysical Journal. 2008;95:4426–4438. doi: 10.1529/biophysj.108.132217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sur S, Newcomb CJ, Webber MJ, Stupp SI. Biomaterials. 2013;34:4749–4757. doi: 10.1016/j.biomaterials.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caprini A, Silva D, Zanoni I, Cunha C, Volontè C, Vescovi A, Gelain F. N Biotechnol. 2013;30:552–562. doi: 10.1016/j.nbt.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 27.Gelain F, Bottai D, Vescovi A, Zhang S. PLoS ONE. 2006;1:e119. doi: 10.1371/journal.pone.0000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI. Science. 2004;303:1352–1355. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 29.Cheng TY, Chen MH, Chang WH, Huang MY, Wang TW. Biomaterials. 2013;34:2005–2016. doi: 10.1016/j.biomaterials.2012.11.043. [DOI] [PubMed] [Google Scholar]
- 30.Li Q, Cheung WH, Chow KL, Ellis-Behnke RG, Chau Y. Nanomedicine. 2012;8:748–756. doi: 10.1016/j.nano.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 31.Cui G-H, Shao S-J, Yang J-J, Liu J-R, Guo H-D. Mol Neurobiol. 2015 doi: 10.1007/s12035-014-9069-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang H, Yang H, Xie Z, Wang P, Bi J. Neurol Res. 2015;37:84–91. doi: 10.1179/1743132814Y.0000000417. [DOI] [PubMed] [Google Scholar]
- 33.Yang H, Qu T, Yang H, Wei L, Xie Z, Wang P, Bi J. Neurosci Lett. 2013;556:63–68. doi: 10.1016/j.neulet.2013.09.063. [DOI] [PubMed] [Google Scholar]
- 34.Ni N, Hu Y, Ren H, Luo C, Li P, Wan JB, Su H. PLoS ONE. 2013;8:e84504. doi: 10.1371/journal.pone.0084504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hnasko R, Bruederle CE. PLoS ONE. 2009;4:e4440. doi: 10.1371/journal.pone.0004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tysseling-Mattiace VM, Sahni V, Niece KL, Birch D, Czeisler C, Fehlings MG, Stupp SI, Kessler JA. J Neurosci. 2008;28:3814–3823. doi: 10.1523/JNEUROSCI.0143-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cigognini D, Satta A, Colleoni B, Silva D, Donegà M, Antonini S, Gelain F. PLoS ONE. 2011;6:e19782. doi: 10.1371/journal.pone.0019782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gelain F, Cigognini D, Caprini A, Silva D, Colleoni B, Donega M, Antonini S, Cohen BE, Vescovi A. Nanoscale. 2012;4:2946–2957. doi: 10.1039/c2nr30220a. [DOI] [PubMed] [Google Scholar]
- 39.Iwasaki M, Wilcox JT, Nishimura Y, Zweckberger K, Suzuki H, Wang J, Liu Y, Karadimas SK, Fehlings MG. Biomaterials. 2014;35:2617–2629. doi: 10.1016/j.biomaterials.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 40.Ellis-Behnke RG, Liang YX, You SW, Tay DKC, Zhang S, So KF, Schneider GE. Proc Natl Acad Sci USA. 2006;103:5054–5059. doi: 10.1073/pnas.0600559103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tao J, Battle KC, Pan H, Salter EA, Chien YC, Wierzbicki A, De Yoreo JJ. Proc Natl Acad Sci USA. 2015;112:326–331. doi: 10.1073/pnas.1404481112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McNally EA, Schwarcz HP, Botton GA, Arsenault AL. PLoS ONE. 2012;7:e29258. doi: 10.1371/journal.pone.0029258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rho JY, Kuhn-Spearing L, Zioupos P. Med Eng Phys. 1998;20:92–102. doi: 10.1016/s1350-4533(98)00007-1. [DOI] [PubMed] [Google Scholar]
- 44.Hughes LC, Archer CW, ap Gwynn I. Eur Cell Mater. 2005;9:68–84. doi: 10.22203/ecm.v009a09. [DOI] [PubMed] [Google Scholar]
- 45.Tatman PD, Gerull W, Sweeney-Easter S, Davis JI, Kim DH, Gee A. Tissue Eng Part B Rev. 2015:150722062433005. doi: 10.1089/ten.TEB.2015.0142. [DOI] [PubMed] [Google Scholar]
- 46.Garreta E, Gasset D, Semino C, Borros S. Biomol Eng. 2007;24:75–80. doi: 10.1016/j.bioeng.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 47.Anderson JM, Patterson JL, Vines JB, Javed A, Gilbert SR, Jun HW. ACS Nano. 2011;5:9463–9479. doi: 10.1021/nn203247m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sargeant TD, Aparicio C, Goldberger JE, Cui H, Stupp SI. Acta Biomater. 2012;8:2456–2465. doi: 10.1016/j.actbio.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen J, Shi ZD, Ji X, Morales J, Zhang J, Kaur N, Wang S. Tissue Eng Part A. 2013;19:716–728. doi: 10.1089/ten.tea.2012.0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim SH, Hur W, Kim JE, Min HJ, Kim S, Min HS, Kim BK, Kim SH, Choi TH, Jung Y. Tissue Eng Part A. 2015;21:1237–1246. doi: 10.1089/ten.tea.2014.0472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshimi R, Yamada Y, Ito K, Nakamura S, Abe A, Nagasaka T, Okabe K, Kohgo T, Baba S, Ueda M. J Craniofac Surg. 2009;20:1523–1530. doi: 10.1097/SCS.0b013e3181b09b7e. [DOI] [PubMed] [Google Scholar]
- 52.Hou T, Li Z, Luo F, Xie Z, Wu X, Xing J, Dong S, Xu J. Biomaterials. 2014;35:5689–5699. doi: 10.1016/j.biomaterials.2014.03.079. [DOI] [PubMed] [Google Scholar]
- 53.Li Z, Hou T, Luo F, Chang Z, Wu X, Xing J, Deng M, Xu J. Int Orthop. 2014;38:2391–2398. doi: 10.1007/s00264-014-2388-9. [DOI] [PubMed] [Google Scholar]
- 54.Florine EM, Miller RE, Porter RM, Evans CH, Kurz B, Grodzinsky AJ. Cartilage. 2013;4:63–74. doi: 10.1177/1947603512455196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Erickson IE, Huang AH, Chung C, Li RT, Burdick JA, Mauck RL. Tissue Eng Part A. 2009;15:1041–1052. doi: 10.1089/ten.tea.2008.0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fernández-Muiños T, Recha-Sancho L, López-Chicón P, Castells-Sala C, Mata A, Semino CE. Acta Biomater. 2015;16:35–48. doi: 10.1016/j.actbio.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 57.Kopesky PW, Byun S, Vanderploeg EJ, Kisiday JD, Frisbie DD, Grodzinsky AJ. J Biomed Mater Res A. 2014;102:1275–1285. doi: 10.1002/jbm.a.34789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shah RN, Shah NA, Del Rosario Lim MM, Hsieh C, Nuber G, Stupp SI. Proc Natl Acad Sci USA. 2010;107:3293–3298. doi: 10.1073/pnas.0906501107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miller RE, Grodzinsky AJ, Vanderploeg EJ, Lee C, Ferris DJ, Barrett MF, Kisiday JD, Frisbie DD. Osteoarthr Cartil. 2010;18:1608–1619. doi: 10.1016/j.joca.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim JE, Lee SM, Kim SH, Tatman P, Gee AO, Kim DH, Lee KE, Jung Y, Kim SJ. Int J Nanomedicine. 2014;9(Suppl 1):141–157. doi: 10.2147/IJN.S54114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brown L. Am J Physiol Heart Circ Physiol. 2005;289:H973–4. doi: 10.1152/ajpheart.00443.2005. [DOI] [PubMed] [Google Scholar]
- 62.Bashey RI, Martinez-Hernandez A, Jimenez SA. Circ Res. 1992;70:1006–1017. doi: 10.1161/01.res.70.5.1006. [DOI] [PubMed] [Google Scholar]
- 63.Eble JA, Golbik R, Mann K, Kühn K. EMBO J. 1993;12:4795–4802. doi: 10.1002/j.1460-2075.1993.tb06168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cui XJ, Xie H, Wang HJ, Guo HD, Zhang JK, Wang C, Tan YZ. Tohoku J Exp Med. 2010;222:281–289. doi: 10.1620/tjem.222.281. [DOI] [PubMed] [Google Scholar]
- 65.Boopathy AV, Che PL, Somasuntharam I, Fiore VF, Cabigas EB, Ban K, Brown ME, Narui Y, Barker TH, Yoon YS, Salaita K, García AJ, Davis ME. Biomaterials. 2014;35:8103–8112. doi: 10.1016/j.biomaterials.2014.05.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guo HD, Cui GH, Wang HJ, Tan YZ. Biochem Biophys Res Commun. 2010;399:42–48. doi: 10.1016/j.bbrc.2010.07.031. [DOI] [PubMed] [Google Scholar]
- 67.Davis ME, Motion JPM, Narmoneva DA, Takahashi T, Hakuno D, Kamm RD, Zhang S, Lee RT. Circulation. 2005;111:442–450. doi: 10.1161/01.CIR.0000153847.47301.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Soler-Botija C, Bagó JR, Llucià-Valldeperas A, Vallés-Lluch A, Castells-Sala C, Martínez-Ramos C, Fernández-Muiños T, Chachques JC, Pradas MM, Semino CE, Bayes-Genis A. Am J Transl Res. 2014;6:291–301. [PMC free article] [PubMed] [Google Scholar]
- 69.Tokunaga M, Liu ML, Nagai T, Iwanaga K, Matsuura K, Takahashi T, Kanda M, Kondo N, Wang P, Naito AT, Komuro I. J Mol Cell Cardiol. 2010;49:972–983. doi: 10.1016/j.yjmcc.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 70.Dubois G, Segers VFM, Bellamy V, Sabbah L, Peyrard S, Bruneval P, Hagège AA, Lee RT, Menasché P. J Biomed Mater Res Part B Appl Biomater. 2008;87:222–228. doi: 10.1002/jbm.b.31099. [DOI] [PubMed] [Google Scholar]
- 71.Guo HD, Cui GH, Yang JJ, Wang C, Zhu J, Zhang LS, Jiang J, Shao SJ. Biochem Biophys Res Commun. 2012;424:105–111. doi: 10.1016/j.bbrc.2012.06.080. [DOI] [PubMed] [Google Scholar]
- 72.Dellatore SM, Garcia AS, Miller WM. Curr Opin Biotechnol. 2008;19:534–540. doi: 10.1016/j.copbio.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, Cooke MP, Walker JR, Hogenesch JB. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Taraballi F, Natalello A, Campione M, Villa O, Doglia SM, Paleari A, Gelain F. Front Neuroeng. 2010;3:1. doi: 10.3389/neuro.16.001.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.O’Leary LER, Fallas JA, Bakota EL, Kang MK, Hartgerink JD. Nat Chem. 2011;3:821–828. doi: 10.1038/nchem.1123. [DOI] [PubMed] [Google Scholar]
- 76.Fallas JA, Hartgerink JD. Nat Commun. 2012;3:1087. doi: 10.1038/ncomms2084. [DOI] [PubMed] [Google Scholar]
- 77.Hammond NA, Kamm RD. Biomaterials. 2008;29:3152–3160. doi: 10.1016/j.biomaterials.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ban K, Park HJ, Kim S, Andukuri A, Cho KW, Hwang JW, Cha HJ, Kim SY, Kim WS, Jun HW, Yoon YS. ACS Nano. 2014;8:10815–10825. doi: 10.1021/nn504617g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu L, Xu K, Wang H, Tan PKJ, Fan W, Venkatraman SS, Li L, Yang YY. Nature Nanotechnology. 2009;4:457–463. doi: 10.1038/nnano.2009.153. [DOI] [PubMed] [Google Scholar]
- 80.Kang JH, Super M, Yung CW, Cooper RM, Domansky K, Graveline AR, Mamotto T, Berthet JB, Tobin H, Cartwright MJ, Watters AL, Rottman M, Waterhouse A, Mamotto A, Gamini N, Rodas MJ, Kole A, Jiang A, Valentine T, Diaz A, Takahasi K, Ingber D. Nature Medicine. 2013;20:1211–1216. doi: 10.1038/nm.3640. [DOI] [PubMed] [Google Scholar]
- 81.Sargeant TD, Guler MO, Oppenheimer SM, Mata A, Satcher RL, Dunland DC, Stupp SI. Biomaterials. 2008;29:161–171. doi: 10.1016/j.biomaterials.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 82.Dettin M, Zamuner A, Lucci G, Messina GM, Battocchio C, Picariello G, Gallina G, Marletta G, Castagliuolo I, Brun P. Journal of Peptide Science. 2014;20:585–594. doi: 10.1002/psc.2652. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.