Abstract
Background and objective
Faster and more accurate methods for registration of images are important for research involved in conducting population-based studies that utilize medical imaging, as well as improvements for use in clinical applications. We present a novel computation- and memory-efficient multi-level method on graphics processing units (GPU) for performing registration of two computed tomography (CT) volumetric lung images.
Methods
We developed a computation- and memory-efficient Diffeomorphic Multi-level B-Spline Transform Composite (DMTC) method to implement nonrigid mass-preserving registration of two CT lung images on GPU. The framework consists of a hierarchy of B-Spline control grids of increasing resolution. A similarity criterion known as the sum of squared tissue volume difference (SSTVD) was adopted to preserve lung tissue mass. The use of SSTVD consists of the calculation of the tissue volume, the Jacobian, and their derivatives, which makes its implementation on GPU challenging due to memory constraints. The use of the DMTC method enabled reduced computation and memory storage of variables with minimal communication between GPU and Central Processing Unit (CPU) due to ability to pre-compute values. The method was assessed on six healthy human subjects.
Results
Resultant GPU-generated displacement fields were compared against the previously validated CPU counterpart fields, showing good agreement with an average normalized root mean square error (nRMS) of 0.044 ± 0.015. Runtime and performance speedup are compared between single-threaded CPU, multi-threaded CPU, and GPU algorithms. Best performance speedup occurs at the highest resolution in the GPU implementation for the SSTVD cost and cost gradient computations, with a speedup of 112 times that of the single-threaded CPU version and 11 times over the twelve-threaded version when considering average time per iteration using a Nvidia Tesla K20X GPU.
Conclusions
The proposed GPU-based DMTC method outperforms its multi-threaded CPU version in terms of runtime. Total registration time reduced runtime to 2.9 min on the GPU version, compared to 12.8 min on twelve-threaded CPU version and 112.5 min on a single-threaded CPU. Furthermore, the GPU implementation discussed in this work can be adapted for use of other cost functions that require calculation of the first derivatives.
Keywords: GPU, Mass-preserving image registration, Multi-level, Lungs
1. Introduction
Image registration is a process used in medical image analysis to determine a spatial transformation that aligns image data in a common coordinate frame. It is used in a variety of applications, e.g., linking images across modalities [1], tracking the motion of lung tissues [2–4], and providing subject-specific boundary conditions for computational fluid dynamics analysis of airflow and particle deposition [5–7]. The process involves two or more images, a moving (floating) image, which is deformed by a transformation to align with a fixed (reference) image. A similarity measure (also called a cost function) quantifies the degree of alignment and is used to optimize the transformation’s parameters to achieve maximal alignment. Maintz and Viergever [8], Brown [9], Zitova and Flusser [10], and Sotiras et al. [11] provide extensive reviews on various image registration methods. Faster and more accurate methods for registration of images are important for conducting large population-based studies that make use of medical imaging, as well as improved potential future clinical applications such as image guided surgery. For example, Eggers et al. [12] demonstrated a method that makes image-guided surgery a usable application in clinical settings by developing a system using intraoperative CT imaging and automated registration. Heinrich et al. [13] developed a CPU method using a novel self-similarity descriptor that improves registration accuracy for multimodal fusion of images with realtime ultrasound images and taking less than 30 s, nearly fast enough for clinical use. Choi et al. [5] applied image registration to compare Asthmatic and normal patients to identify sensitive variables that can help provide improved and personalized diagnoses of compromised lung function. Shamonin et al. [14] used GPU for fast diagnostic classification of Alzheimer’s disease. Staring et al. [15] developed methods to better investigate emphysema and its progression over time. Progress made in accelerating registration algorithms has been demonstrated through various approaches [16–20], more recently through parallel computing using GPUs [21–28]. Previous works by multiple authors [29–31] have reviewed the use of GPU-based methods for image registration.
The objective of this paper is the development of an efficient multi-level method to effectively implement mass-preserving registration on GPU architectures. We employ a cubic B-Spline-based Free-Form Deformation (FFD) transformation model for its effectiveness at capturing nonrigid deformations [32] of objects such as the human lungs. Pratx and Xing [30] summarized the key strategies for GPU acceleration, such as aligning the B-Spline grid of control nodes with the image voxel grid for B-Spline methods and optimizing data parallel computation to fit the GPU programming model [33]. Previous works [21,23–26,29] in accelerating intensity-based B-Spline deformable registration methods using GPU have focused on sum of squared intensity differences (SSD), normalized cross correlation, and mutual information as similarity measures. For example, the work of [24] reports a speedup of 6–8 times using GPU-based methods with mutual information. Shackleford et al. [23] developed methods to exploit data parallelism within B-spline based deformable registration methods using a tiling approach with the mean squared error (MSE) similarity measure, a variation of the simplest and commonly used similarity measure, the sum of squared intensity difference (SSD), obtaining a speedup of 15 times using NVIDIA’s Tesla C1060 with 240 CUDA cores. A 55× speedup was reported in [25] using the Demons algorithm, when compared to single threaded implementations [34]. The work of [22] introduced a GPU framework for the optimal mass transport registration method, showing speed improvement of 4826% compared to CPU when using a Nvidia GeForce 8800 GPU and Intel Dual Xeon 1.6 GHz CPU.
We focus on mass-preserving registration and propose a composite transform necessary for its implementation in a multi-level framework on GPU in a memory- and computationally-efficient manner. We use mass-preserving methods due to their demonstrated ability to effectively match the intra-subject lung CT images acquired at different inflation levels [35] and to further develop physiologically consistent lung functional measurements [6,15,36,37]. In particular, the use of the sum of squared tissue volume difference (SSTVD) has demonstrated improved registration accuracy for large deformations compared to SSD and MI [35]. In the mass preserving method, SSTVD is minimized and a one-to-one transformation with positive Jacobian values must be enforced. The composite transform introduced in this work is essential to reduce the memory needs of SSTVD at high resolutions to fit limitations of high-end GPU.
The next section of the paper reviews cubic B-Spline deformable registration and introduces a composite transform operation for efficient multi-level registration. The third section focuses on developing multi-level framework for GPU using SSD and SSTVD as similarity measures, and discusses the advantages of this method. In the fourth section experimental results are reported. Discussion of results occupies the fifth section and the paper concludes with the sixth section.
2. An efficient diffeomorphic multi-level transform composite (DMTC) method
2.1. Preliminaries
Multi-level methods are commonly used in FFD image registration to help improve registration quality and efficiency, reduce computational complexity, as well as help prevent incorrect local minima solutions during optimization when dealing with high dimensional transformations. Competing needs, such as capturing global motion as well as local deformations can be addressed by constructing a hierarchy of control node meshes, referred to as control grids, and applying registration using each grid in a coarse to fine sequence. The control nodes serve as parameters to the FFD, where large control grid spacing allows for capturing large deformations and a smaller spacing between nodes captures local deformations. However, as the resolution of the control grid is increased, the dimension of the transformation and computational complexity also increases.
Another reason for employing a composite multi-level method is to enforce deformation constraints defined in terms of control node spacing to ensure a positive Jacobian. In the case of cubic B-Splines, the constraints of Choi and Lee [38] are employed to ensure diffeomorphic FFD transformations at each level. In their work, they use geometric argument to conclude that the transformation will be invertible if the x, y, and z directional displacements φx, φy, φz, along with B-spline grid spacing δx, δy, and δz, satisfy the constraints φx < δx/K, φy < δy/K, and φz < δz/K, where K = 2.479472335. Modeling large global deformations requires a coarse control grid with larger spacing (δx, δy, δz) to enforce the given constraints. However, local deformation modeling requires a fine control grid. Thus, a multi-level cubic B-spline technique is used to meet the conflicting needs of the control grid.
Let Φ0, Φ1, …, Φn define a hierarchy of control grids used to derive a sequence consisting of uniformly spaced FFD control nodes with displacement vectors φ = (φx, φy, φz) defined at each node to parameterize the corresponding transformations T0, T1, …, Tn. Denote by δx, δy, and δz the spacing of the control nodes in the x, y, and z directions, respectively, which vary for each grid refinement. Each control grid overlays a uniformly spaced voxel grid of intensity values, denoted as Ω, with a size of Nx × Ny × Nz voxels. The alignment of the control grid partitions the voxel grid into equally sized tiles. Fig. 1 demonstrates this with a 2D example consisting of an image grid of 32 × 32 uniformly spaced voxels, shown as the smallest squares, and a 5 × 5 grid of control nodes separated by 16 voxels per direction, represented as gray circles. This partitions the voxel grid into four tiles of 16 × 16 voxels, shown by bold lines.
Fig. 1.
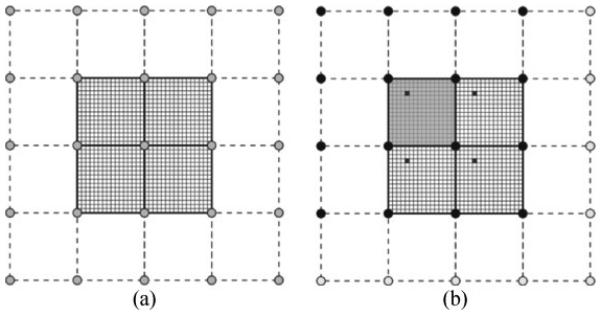
(a) 2D example image of 32 × 32 voxels (small squares) with aligned overlaying 5 × 5 control node (gray circles) grid creating partition of 4 tiles of 16 × 16 voxels each. (b) Emphasis on upper-left tile (shaded darker gray) and its 16 surrounding control nodes (solid black circles). In each tile, the voxels in solid black (5, 4), (21, 4), (5, 20), and (21, 20) share the same local index (5,4) within their respective tiles, as well as the same local normalized coordinates.
Define for each voxel coordinate x = (x, y, z) the cubic B-Spline transformation T,
| (1) |
where u = x/δx − x/δx, v = y/δy − y/δy, w = z/δz − z/δz denote the normalized local coordinates of the voxel within the tile it resides, and Bl, Bm, Bn denote the cubic B-Spline basis functions, referred to throughout as weights. Each voxel deformation is influenced by the φ values from 4 surrounding control nodes per direction, 64 in total for 3D. Two useful properties follow from applying Eq. (1) to a regularly spaced voxel grid with aligned control grid:
(1) The displacement field at each voxel within the same tile is determined by the same set of surrounding control nodes, and
(2) The normalized local coordinates (u, v, w) are identical for corresponding voxels within each tile. Thus, B-Spline weights Bl(u), Bm(v), Bn(w) are identical for voxels at the same offset within each tile. Fig. 1b provides an example for clarification.
Due to these properties, the B-Spline weights can be pre-computed for a single tile rather than the entire image and reused, reducing redundant computation when applying the transformation to all voxel positions. We take advantage of this feature by storing the pre-computed tile values in a lookup table (LUT) prior to cost-related computations, repeating for each control grid Φi.
2.2. Composite transformations and the DMTC
A smooth and one-to-one transformation can be obtained by a single-level manipulation employing the displacement constraints of Choi and Lee [38]. Given T0, T1, …, Ti as the sequence of transformations determined for i + 1 control grids, we define a composite transformation, Tc, by composing the sequence of transformations to represent the aggregate mapping after performing registration through the corresponding progression from coarse to fine control grids.
The most commonly used composite transform for multi-level cubic B-Spline based deformable registration [35,37,39,40] is
| (2) |
as proposed by Hagenlocker and Fujimura [41]. The composite transform is implemented by using the warping image, ω, which stores the resultant aggregate mapping from the previous levels, i.e. ω = Ti−1 ∘ ⋯ ∘ T0. This defines the composite transform in terms of the warping image at voxel position x as Tc [x, φ] = Ti [ω(x), φ]. The composite operation can be treated as a recursive process that manipulates the moving image physical space to better align corresponding intensity values with those of the fixed image. After level i + 1, the warping image is updated to store the displacement field following the i + 1 transformations. Fig. 2(a) illustrates the composite transform based on Eq. (2) using a 2D example with two transformation levels T0 and T1, thus with ω = T0 at the second level. Fig. 2(a) shows that original voxel positions are first transformed into new locations after applying ω, represented by the dotted arrow, leading to scattered positions for all voxels as denoted by the gray circles. T1 is applied to the displaced voxel positions following warping, which yields the correction to ω to complete the composite transformation as shown by the dashed arrow in Fig. 2(a). Note that application of ω results in a non-uniform voxel grid after the first level, and thus the B-Spline weights and derivative weights must be recomputed at each level for the full voxel grid, eliminating the use of LUTs and increasing memory storage requirements.
Fig. 2.
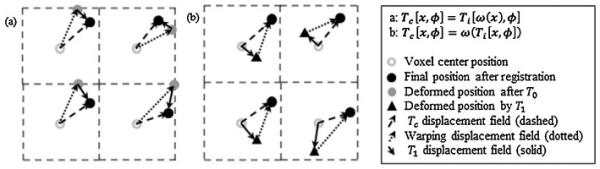
a and b Serve to illustrate the composite transform following registration after two levels. Please note that a and b have the same initial and final positions. (a) The initial positions (unfilled gray circles) are deformed to intermediate positions (filled gray circles) by using transform T0 (stored as ω), followed by deformation to final positions (filled dark circles) by further use of transform T1. The aggregate operation represents the composite transform (dashed line) of Eq. (2). (b) The initial positions (unfilled gray circles) are deformed to intermediate positions (filled black triangles) by using transform T1, followed by deformation to final positions (filled dark circles) by further use of transform T0, stored as the warping image ω at voxel centers. The aggregate operation represents the composite transform (dashed line) of Eq. (3).
Schnabel et al. [42] developed an efficient method for multi-level frameworks with non-uniform grids. However, we wish to preserve the B-Spline LUT properties in order to reduce memory storage needs to meet GPU limitations. To accomplish this, we must maintain the regular spacing of the voxel grid, which we accomplish by introducing the DMTC
| (3) |
The warping image stores all previous transforms as ω = T0 ∘ T1 ∘ ⋯ ∘ Ti−1 with the resultant composite transform at voxel position x defined as Tc [x, φ] = ω(Ti [x, φ]). Fig. 2(b) illustrates this method. We first seek the correction to ω in the non-deformed uniformly spaced grid, as determined by T1; thus, each tile has equivalent uniformly spaced input coordinates to T1, allowing use of LUTs for weights. We then add the warping image displacement vector, denoted by the dotted arrow, to complete the composite transformation as shown in Fig. 2(b) and denoted by the dashed arrow. Because ω was defined at the voxel center, the warping image displacement at the deformed position by T1 (denoted by black triangles in the figure) is obtained by linear interpolation of ω, which is less costly than computation of B-Spline weights and derivative weights in Eq. (2). Because the composition of diffeomorphisms is a diffeomorphism, a final one-to-one diffeomorphic mapping is guaranteed by imposing the displacement constraints for each control grid level in either Eq. (2) or Eq. (3). Besides preserving the regular spacing of the voxel grid for all iterations during optimization at each level to allow LUT use, several additional benefits of the DMTC will be exploited during cost and gradient computations, as discussed in the following section.
3. Multi-level GPU-based registration framework
3.1. Overview
The multi-level registration process is summarized in Fig. 3. At each level, a limited-memory quasi-Newton minimization method with bounds on the variables (L-BFGS-B) is adopted. This method allows bound constraints on the independent variables, a necessary characteristic to implement the aforementioned displacement constraints to enforce the one-to-one mapping condition (i.e. positive Jacobian). As previously mentioned, a key property for the multi-level framework using DMTC is the preservation of regular spacing of the voxel grid and thus the ability to utilize B-Spline weight LUTs. This is key for reducing memory needs and developing computational efficiencies necessary for GPU implementation.
Fig. 3.
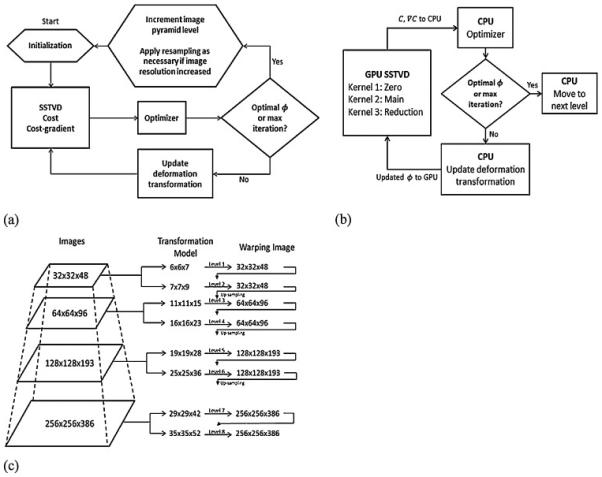
(a) Flow of the multi-level image registration framework and (b) division of work between CPU and GPU during the SSTVD computations. (c) Sample image pyramid used by multi-level framework.
We develop the GPU method for mass-preserving image registration using the SSTVD similarity measure, described in the next section. Use of SSD is also implemented in the same multi-level GPU framework to demonstrate fiexibility of the multi-level framework. A GPU tiling technique [43] is implemented that employs necessary principles of GPU computing to unlock the promised computational capability of the GPU. The GPU-based DMTC framework is implemented using Nvidia’s CUDA API, a parallel computing platform giving access to the GPU for general purpose computing.
3.2. Similarity measures
Let If and Im denote the intensity value of each image. We treat If(x) and Im(x) as continuous functions of intensity at voxel coordinate x. One of the simplest similarity measures is SSD, defined as
| (4) |
The gradient with respect to φ is computed at a control node via the chain rule to give:
| (5) |
where
However, SSD does not account for CT voxel intensity changes with lung inflation. For this reason the SSTVD similarity measure was introduced to minimize the local tissue volume differences between matched regions as follows [35].
| (6) |
where vf and vm are the local volumes of corresponding regions in the fixed and moving images, respectively, and vm(T [x, φ]) can be calculated from the Jacobian value JT as vm(T [x, φ]) = JT(x, φ) vf(x). In addition, is the tissue fraction estimated from the Hounsfield Unit (HU) by
| (7) |
where the intensities of air and tissue are set to HUair = −1000 HU and HUtissue = 55 HU [44]. Tissue volume V is computed as V = v.
The gradient for the SSTVD cost function is then computed at a control node with parameter φ as
| (8) |
Regularization constraints are neglected in this work in order to isolate components accelerated by GPU and report computational improvements in the registration framework. However, segmented airway tree and lobar masks were used in this work for two purposes: (1) to eliminate the possibility of registering ‘background’ voxels during the registration process, as an incorrect registration of background voxel pairs would add to the overall cost total, inaccurately inflating the value, and (2) to improve accuracy of registration along the boundaries, including discontinuity preservation for sliding motion between lobes. Other works have used regularization for discontinuity preservation, such as [45] where they present a method that removes the need for accurate lung image segmentation.
3.3. GPU Implementation of SSTVD
GPUs possess a number of streaming multiprocessors that execute in parallel to one another. Each streaming multiprocessor consists of groups of streaming processors, or CUDA cores. For this work a Nvidia Tesla K20X was used. The K20X consists of 14 streaming multiprocessors of 192 CUDA cores each, for a total of 2688 cores. Each core executes a serial thread, or sequence of instructions on given data, and the architecture is designed so that cores execute in SIMT fashion (Single Instruction, Multiple Threads), meaning that all cores in the same group execute the same instruction simultaneously. A block is a group of threads sent to a streaming multiprocessor for parallel execution. Though individual threads cannot communicate, those within the same block can communicate through use of thread-block synchronization and shared memory. Effective use of GPU capabilities requires making use of the small amounts of available shared memory, which is faster to access than the larger global memory, and minimizing CPU–GPU communication.
As with the MSE implementation reported by Shackleford et al. [23], the majority of runtime using SSTVD (111/112 min on single thread CPU) is spent on B-Spline interpolation and cost gradient computation, and due to the data-parallelism present these components were targeted for GPU acceleration. However, SSTVD calculates local tissue volume values by introducing Jacobian terms. This leads to a more computationally complex cost function and cost gradient with additional dependencies between terms on the transformation parameters, requiring modifications of the procedure used by Shackleford.
This GPU implementation consists of three kernels (routines executed on the GPU): a zero kernel that initializes relevant values (cost, cost gradient, bins) to zero prior to optimization, the main kernel responsible for the bulk of the cost and cost gradient computations, and a reduction kernel that computes the final sums of the cost gradients. Final cost gradient results are transferred to CPU for optimization, and the process repeats with updated displacement parameters φ until optimal values are found. The division of work between CPU and GPU is shown in Fig. 3(b), and specific GPU kernel details and pseudo-code can be found in the supplementary materials.
3.4. Benefits
The use of DMTC with Eq. (3) leads to several benefits for both the CPU and GPU versions that reduce memory and computational needs. The reduction in memory needs is particularly important for the GPU implementation since the limited 6 GB of DRAM available on the Tesla K20X is fully utilized at the high-resolution levels of registration. The benefits are as follows:
(1) The use of the DMTC preserves the regular spacing of the voxel grid throughout registration for all resolution levels. This allows the use of weight and derivative weight LUTs that are pre-computed for a single tile, rather than all tiles, drastically reducing computation (48 kB for 1D x, y, and z weights in tile of 103 voxels, as opposed to 6 GB for the same 1D weights in a full 5123 voxel grid). Further, the memory required to store the LUTs for a single tile is far less than for all voxels in an image, easily fitting within a GPU’s limited resources. Since weights only need to be pre-computed once per level, due to the preservation of the regular spacing of the grid, CPU–GPU communication is minimized for this component.
(2) Several variables in Eq. (8), the composite transformation Tc, the Jacobian JTc, the Jacobian gradient ∇φJTc, and the moving tissue volume gradient ∇φm, can be mathematically manipulated to separate warping image components from components dependent on the current transformation Ti. This leads to the following forms for these variables:
| (9) |
where
| (10) |
where α is the x, y, or z directional component of φ, and is the spatial gradient computed at voxel center. Application of the product rule to ∇φJTc leads to
| (11) |
where, similar to Eq. (10),
| (12) |
By preserving the regular spacing of the image grid, along with the independence of the warping image terms from the control grid displacement parameters φ, the warping components Jω, ∇xmω, and ∇xJω can be pre-computed a single time during initialization of each registration level and reused throughout optimization. The ∂Ti/∂φα values are equivalent to product weights stored in the LUT. JTi consists of products of derivative weight LUT values, and ∇φJTi consists of determinant minors of values from the derivative weight LUT. This simplifies the original complex computations required for the composite transforms to only solving the components dependent on Ti for each iteration of optimization as the φ values are updated, as the remainder of the values can be accessed from pre-computed values of LUTs.
Thus, the DMTC framework minimized CPU–GPU communication by successfully reducing redundant and complex computations following application of the chain rule to split cost gradient terms into warping-related terms (ω) and current transformation (Ti) terms. The reduction in computation comes at the expense of linear interpolation, which is less costly than the original full computations. As the warping-related terms are independent of φ, they can be pre-computed a single time during initialization and transferred a single time to the GPU. The final cost and cost-gradient were the only terms requiring transfer from GPU to CPU every optimization iteration. These repeated transfers only required 0.2 s through 8 full levels of registration that totalled about 3.7 min, averaged for six subject data pairs and including roughly 500 iterations during the optimization process.
4. Evaluation of multi-level GPU framework
4.1. Data sets
Lung volumetric computed tomography (CT) image data for six healthy human subjects were used in this study. Two static images collected at 20% of vital capacity (VC) and 80% VC were used in this work. A Siemens Sensation 64-slice Multi-Detector-row CT scanner (Forchheim, Germany), with 120 kV, 75 mAs, 0.75 mm slice thickness, 500 mm field of view, was used to acquire the images. The scanning protocol to examine the patients was approved by the University Institutional Review Board. Each data set contains 550–760 image planes with a reconstruction matrix of 512 × 512 pixels. The software Apollo (VIDA Diagnostics, Coralville, Iowa) was used to segment the airway tree and lobes of the CT images.
4.2. Registration validation and performance comparison
4.2.1. Registration validation
Registration was performed on a system with an Intel Xeon E5-2620 6-core CPU clocked at 2.1 GHz and Tesla K20X GPU.
SSTVD was previously evaluated in [35], demonstrating better mean landmark error over SSD and MI, particularly in regions of large deformation. Landmarks generated by experts using a semi-automatic annotating system [46] at vessel bifurcations were used for evaluation by comparing distances before and after registration. Each data set contains between 102 and 210 landmarks, with approximately 20–40 in each lobe, and with initial distance differences between fixed and moving images ranging between 4 mm and 70 mm to check accuracy for small and large deformations.
The same data sets were used in this work to evaluate landmark error after registration when using the DMTC with SSTVD for method validation. The same framework and data sets were used with SSD and results were compared with SSTVD. In the appendix, Table 3 shows results averaged over all six subjects for landmark distance after registration categorized by initial landmark distance in groupings of 20 mm. Table 4 shows results for each subject. Table 3 shows that for the group of landmarks with initial separation distance of 20 mm, the distance after registration with SSTVD is 1.3 mm on average, while SSD is 7.14 mm. For landmarks initially separated between 20 and 40 mm, SSTVD results in average distance of 2.15 mm, and SSD with 10.88 mm. For initial separation distances of 40–60 mm, SSTVD yields results within 2.59 mm on average, and SSD yields 17.23 mm. Finally, for the largest deformations with distance greater than 60 mm, SSTVD results in average distance of 2.84 mm, compared to SSD which results in 28 mm distance after registration. The comparison of SSTVD to SSD was done by using a paired T-test with two tails to compare the relative error of SSTVD to the relative error of SSD, where relative error here means landmark distance after registration relative to landmark distance before. The low p values in each case demonstrate that the results are statistical significant.
Fig. 6 in the appendix shows a comparison of landmark distance results (horizontal axis), with initial landmark distance before registration (upper) and landmark distance after registration (lower) for SSTVD and SSD cases. Both cases are plotted against distance from the apex (trachea). Inspection clearly shows the improvement of SSTVD over SSD, as in [35].
The normalized root mean square error (nRMSE) was used to evaluate accuracy of the GPU resultant displacement field against the validated single-threaded CPU implemenation of Yin et al. [35].
| (13) |
where v is the resultant displacement field associate with a voxel at coordinate x.
The average nRMS error for the six human subjects is 0.044 ± 0.015. The RMS value was 0.26 ± 0.07.
4.2.2. Performance assessment
Performance of the multi-level GPU framework was compared to the multi-threaded CPU implementations under several measures, such as total runtime, average time per iteration per level, and speedup factor. Table 1 shows the total registration time results for GPU and CPU implementations. Also shown is the isolated cost and cost-gradient time, to directly compare the portions of the code modified for GPU implementation. SSTVD and SSD times are reported to show the success of the method for a non-mass-preserving and mass-preserving similarity criteria. The multi-level framework consisted of 8 levels of registration, beginning with the coarsest control grid and downsampled images, and increased in control grid resolution every level and image resolution every other level. See Fig. 3(c) for a sample image pyramid.
Table 1.
Totala registrationb and total cost and gradient computational time for GPU and CPUc implementations.
| Version | Total registration time (min) |
Total cost and gradient time (min) |
||
|---|---|---|---|---|
| SSTVDd | SSDd | SSTVDd | SSDd | |
| K20X GPU | 2.9 | 2.0 | 1.1 | 1.0 |
| 12 T CPU | 12.9 | 7.2 | 11.7 | 6.3 |
| 6 T CPU | 23.7 | 12.6 | 22.6 | 11.7 |
| 1 T CPU | 112.5 | 63.6 | 111.3 | 63.8 |
Time is totalled over all 8 levels of registration and averaged for 6 healthy subjects.
Registration time consists of SSTVD (cost and cost gradient) and optimization.
CPU implementations include single(1 T)-, six(6 T)-, and twelve(12 T)- threaded versions.
SSTVD and SSD time each consists of their respective cost and cost gradient calculations.
Table 1 demonstrates that total times substantially decrease with GPU compared to the multi-threaded implementations. For greater than 12 threads, the multi-threaded implemenation’s runtime performance does not improve much, only 30 s faster than 12 threads when using 24 threads. Registration using SSD performs faster than SSTVD, as expected since SSD requires far less computation i.e. no Jacobian calculations. However, with the GPU implementations the difference in timing between the two is much closer (1 min on K20X) than that of the multi-threaded CPU implementations (5.7 min with 12 threads).
To compare the components targeted for acceleration by GPU with the multi-threaded CPU counterparts, the cost and cost-gradient times per level were divided by the total number of iterations to create a normalized comparison from start to finish, a table of these results can be found in the supplementary section. A common metric used to compare the performance GPU to CPU is the speedup factor, speedup = timeCPU/timeGPU, where time refers to time per iteration. Fig. 4 shows the speedup factors by level for both GPU and CPU implementations, comparing time per iteration. At every level, the GPU performs better than each of the multithreaded CPU versions. Greatest speedup occurs for both GPUs over CPU at the highest resolution level, where the Tesla K20X demonstrates an average of 112 times and nearly 11 times speedup over the single-threaded and twelve-threaded versions, respectively. The twelve-threaded version demonstrates a 10.4 time speedup over the single-threaded CPU version at the highest level.
Fig. 4.
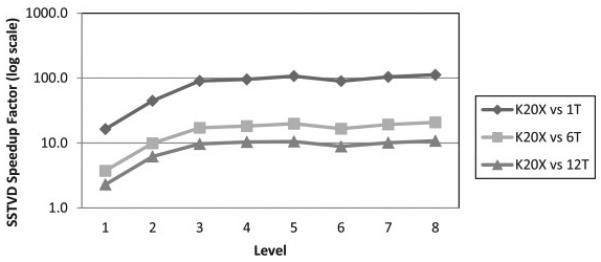
Speedup factors for SSTVD implementation of the cost and cost gradient components, comparing the GPU implementation to single-, six-, and twelve-threaded (1 T, 6 T, 12 T) CPU versions. Log scale used for speedup axis. Values shown are averaged for 6 healthy subjects at each of the 8 levels of the image pyramid.
Recurring memory transfers between CPU and GPU represent a negligible amount of time per iteration, as intended by design and the use of the composite transform Eq. (3). This includes transfer of the cost, cost gradient, and parameters φ for the control grid at each iteration of optimization. The total time summed for all levels for these recurring transfers is 0.2 s. In addition, transfer of the quantities pre-computed during initialization (described in Section 3.4) to GPU occurs only once per registration level, prior to optimization start, and totaled 0.7 s for all levels, negligible compared to total registration runtime. Furthermore, memory allocation on the GPU totaled only 0.05 s. The negligible amount of time required for data transfers demonstrates the success of the algorithm in terms of minimizing CPU to GPU communication time. The majority of time is spent on cost and cost gradient computation, primarily handled by the main kernel described in Section 0 and the appendix. Of the three kernel executed on the GPU each iteration, the main kernel accounts for 99.8% of the time.
5. Discussion
We have presented a novel DMTC method for multi-level cubic B-Spline based mass preserving deformable registration developed to keep memory storage requirements within the GPU limitations. The method requires a uniformly spaced voxel grid and control grid for B-Spline weight computations to make use of the computational and memory efficiencies developed in this work. Constraints are placed on the maximum control node displacement in order to ensure a one-to-one transformation, and thus a multi-level framework is utilized in order to capture larger global deformations and smaller local deformation. In addition, the image size must be large enough to utilize the large number of threads available on the GPU to amortize the cost of parallelization and memory communication. SSTVD was chosen as similarity criteria due to its improvement of accuracy for registering two CT lung images when compared to SSD, and the composite multi-level framework was needed to capture both small and large deformations and ensure positive Jacobian values [35]. The proposed GPU method was validated against the previously established CPU version [35]. Corresponding landmarks (102–210) in 6 different subjects were used to compare the landmark error distance between the fixed image to moving image before registration and after registration, for both SSTVD and SSD. Results reported for DMTC (Section 4.2.1, Appendix Section 7.4), when compared to the results using the composite transform Eq. (2) of [35], show similar improvements of SSTVD over SSD, but lose some accuracy due to the required linear interpolation step when applying the warping image displacement to the transformed location (see Section 2, Fig. 2b). The final displacement fields show agreement with an average nRMS of 0.044 ± 0.015. Regularization constraints are currently being investigated for improved registration accuracy while fitting within the current computational framework to prevent loss of performance. The GPU implementation achieves a peak speedup at the highest resolution levels over the multi-threaded CPU version, with speedup of 112 times over the sequential CPU version and 11 times over the twelve-threaded version when run on a Tesla K20X GPU. Total registration time for the full eight resolution levels reduced runtime to 2.9 min on the K20X GPU version, compared to 12.8 min on twelve-threaded CPU version and 112.5 min on a single-threaded CPU, as averaged over six subjects. In addition, the GPU implementation of SSTVD performs considerably closer to SSD in terms of runtime than multi-threaded CPU versions, making it a practical replacement for improved accuracy and efficiency in image analysis.
The presence of the tissue volume and the Jacobian terms in the SSTVD cost function lead to a cost gradient with data dependencies between all term pairs. Unlike the GPU-based SSD algorithm by Shackleford et al. [23], splitting of terms of the cost gradient into separate kernels for independent computations is non-trivial. Attempting to duplicate their procedure would require additional storage of large data sets exceeding GPU limitations and would require recurring costly CPU-GPU memory transfers of a decomposed domain. Splitting the work between the three kernels discussed in Section 3.3 and the appendix prevents the issues just described. The Jacobian JTi, its matrix, and the transformed coordinates Ti [x, φ] were stored in registers throughout a voxel’s respective computations, the fastest memory to access on the GPU. Shared memory and thread-block synchronization enabled fast thread inter-communication within a thread-block during cost gradient computations and avoided the use of slower global memory. The use of the DMTC method with Eq. (3) prevented excessive CPU–GPU communication during optimization and enabled the use of B-Spline LUTs, pre-computed a single time per level for a single tile rather than all voxels in the image. DMTC minimized computations for the cost gradient by utilizing values pre-computed for the previous resolution levels. This provided a benefit for the GPU implementation by requiring only one memory transfer between CPU and GPU at initialization for the data variables introduced by SSTVD and its gradient at previous levels.
6. Conclusion
The proposed GPU-based DMTC method for mass preserving (SSTVD) multi-level registration of CT lung images outperforms its multi-threaded CPU version in terms of runtime. As a result, total registration time reduced runtime to 2.9 min on the K20X GPU version, compared to 12.8 min on twelve-threaded CPU version and 112.5 min on a single-threaded CPU. This is essential to process large data sets for population-based studies via clustering analysis to subpopulations of normal and diseased lungs for translational science [47]. Furthermore, the GPU implementation discussed in this work can be adapted for use of other cost functions that require calculation of the spatial first derivatives, as demonstrated with SSD. Future work to improve processing large data sets may include investigating regularization constraints and methods that enforce discontinuity preservation and avoid the need for accurate segmentation of images, as demonstrated in [45]. Other future work includes use of higher order interpolation, such as B-spline interpolation, to calculate the composite transformation Tc [x, φ] = w(Ti [x, φ]) as well as in the image resampling from the supporting neighbor voxels, where linear interpolation is currently used. However, as noted in [48] the accuracy is improved at the expense of some increase in computing time. Another development is of a symmetric SSTVD registration method that contains forward and backward (i.e. inverse) transformation information to further improve accuracy of the method.
Supplementary Material
Acknowledgements
This work supported in part by NIH grants R01 HL094315, U01 HL114494, and S10 RR022421. The authors would like to thank Dr. Sanghun Choi, Mr. Nariman Jahani, and Mr. Babak Haghighi for their assistance during the course of this study.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.cmpb.2015.12.018.
REFERENCES
- [1].Mattes D, Haynor DR, Vesselle H, Lewellen TK, Eubank W. PET-CT image registration in the chest using free-form deformations. IEEE Trans. Med. Imaging. 2003;22(1):120–128. doi: 10.1109/TMI.2003.809072. [DOI] [PubMed] [Google Scholar]
- [2].Christensen GE, Song JH, Lu W, El Naqa I, Low DA. Tracking lung tissue motion and expansion/compression with inverse consistent image registration and spirometry. Med. Phys. 2007;34(6):2155–2163. doi: 10.1118/1.2731029. [DOI] [PubMed] [Google Scholar]
- [3].Reinhardt JM, Ding K, Cao K, Christensen GE, Hoffman EA, Bodas SV. Registration-based estimates of local lung tissue expansion compared to xenon CT measures of specific ventilation. Med. Image Anal. 2008;12(6):752–763. doi: 10.1016/j.media.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jahani N, Yin Y, Hoffman EA, Lin C-L. Assessment of regional non-linear tissue deformation and air volume change of human lungs via image registration. J. Biomech. 2014;47(7):1626–1633. doi: 10.1016/j.jbiomech.2014.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Choi S, Hoffman EA, Wenzel SE, Tawhai MH, Yin Y, Castro M, Lin C-L. Registration-based assessment of regional lung function via volumetric CT images of normal subjects vs. severe asthmatics. J. Appl. Physiol. 2013;115(5):730–742. doi: 10.1152/japplphysiol.00113.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yin Y, Choi J, Hoffman EA, Tawhai MH, Lin C-L. Simulation of pulmonary air flow with a subject-specific boundary condition. J. Biomech. 2010;43(11):2159–2163. doi: 10.1016/j.jbiomech.2010.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Miyawaki S, Tawhai MH, Hoffman EA, Lin C-L. Effect of carrier gas properties on aerosol distribution in a CT-based human airway numerical model. Ann. Biomed. Eng. 2012;40(7):1495–1507. doi: 10.1007/s10439-011-0503-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Maintz JB, Viergever MA. A survey of medical image registration. Med. Image Anal. 1998;2(1):1–36. doi: 10.1016/s1361-8415(01)80026-8. [DOI] [PubMed] [Google Scholar]
- [9].Brown LG. A survey of image registration techniques. Comput. Surv. 1992;24(4):325–376. [Google Scholar]
- [10].Zitova B, Flusser J. Image registration methods: a survey. Image Vis. Comput. 2003;21(11):977–1000. [Google Scholar]
- [11].Sotiras A, Davatzikos C, Paragios N. Deformable medical image registration: a survey. IEEE Trans. Med. Imaging. 2013;32(7):1153–1190. doi: 10.1109/TMI.2013.2265603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Eggers G, Kress B, Rohde S, Muehling J. Intraoperative computed tomography and automated registration for image-guided cranial surgery. Dentomaxillofacial Radiol. 2009;38(1):28–33. doi: 10.1259/dmfr/26098099. [DOI] [PubMed] [Google Scholar]
- [13].Heinrich MP, Jenkinson M, Papieẓ BW, Brady M, Schnabel JA. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2013. Springer; 2013. Towards realtime multimodal fusion for image-guided interventions using self-similarities. [DOI] [PubMed] [Google Scholar]
- [14].Shamonin DP, Bron EE, Lelieveldt BPF, Smits M, Klein S, Staring M. I. Alzheimer’s Disease Neuroimaging, Fast parallel image registration on CPU and GPU for diagnostic classification of Alzheimer’s disease. Front. Neuroinf. 2013;7:50. doi: 10.3389/fninf.2013.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Staring M, Bakker ME, Stolk J, Shamonin DP, Reiber JHC, Stoel BC. Towards local progression estimation of pulmonary emphysema using CT. Med. Phys. 2014;41(2) doi: 10.1118/1.4851535. [DOI] [PubMed] [Google Scholar]
- [16].Yim Y, Hong H, Shin YG. Deformable lung registration between exhale and inhale CT scans using active cells in a combined gradient force approach. Med. Phys. 2010;37(8):4307–4317. doi: 10.1118/1.3460316. [DOI] [PubMed] [Google Scholar]
- [17].Yang D, Li H, Low DA, Deasy JO, El Naqa I. A fast inverse consistent deformable image registration method based on symmetric optical flow computation. Phys. Med. Biol. 2008;53(21):6143–6165. doi: 10.1088/0031-9155/53/21/017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lu WG, Chen ML, Olivera GH, Ruchala KJ, Mackie TR. Fast free-form deformable registration via calculus of variations. Phys. Med. Biol. 2004;49(14):3067–3087. doi: 10.1088/0031-9155/49/14/003. [DOI] [PubMed] [Google Scholar]
- [19].Ashburner J. A fast diffeomorphic image registration algorithm. Neuroimage. 2007;38(1):95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- [20].Kybic J, Unser M. Fast parametric elastic image registration. IEEE Trans. Image Process. 2003;12(11):1427–1442. doi: 10.1109/TIP.2003.813139. [DOI] [PubMed] [Google Scholar]
- [21].Shams R, Sadeghi P, Kennedy R, Hartley R. Parallel computation of mutual information on the GPU with application to real-time registration of 3D medical images. Comput. Methods Progr. Biomed. 2010;99(2):133–146. doi: 10.1016/j.cmpb.2009.11.004. [DOI] [PubMed] [Google Scholar]
- [22].Rehman TU, Haber E, Pryor G, Melonakos J, Tannenbaum A. 3D nonrigid registration via optimal mass transport on the GPU. Med. Image Anal. 2009;13(6):931–940. doi: 10.1016/j.media.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Shackleford JA, Kandasamy N, Sharp GC. On developing B-spline registration algorithms for multi-core processors. Phys. Med. Biol. 2010;55(21):6329–6351. doi: 10.1088/0031-9155/55/21/001. [DOI] [PubMed] [Google Scholar]
- [24].Vetter C, Guetter C, Xu C, Westermann R. Non-rigid multi-modal registration on the GPU. Medical Imaging 2007 Conference.2007. p. 6512. [Google Scholar]
- [25].Samant SS, Xia J, Muyan-Oezcelilk P, Owens JD. High performance computing for deformable image registration: towards a new paradigm in adaptive radiotherapy. Med. Phys. 2008;35(8):3546–3553. doi: 10.1118/1.2948318. [DOI] [PubMed] [Google Scholar]
- [26].Modat M, Ridgway GR, Taylor ZA, Lehmann M, Barnes J, Hawkes DJ, Fox NC, Ourselin S. Fast free-form deformation using graphics processing units. Comput. Methods Progr. Biomed. 2010;98(3):278–284. doi: 10.1016/j.cmpb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- [27].Gu X, Pan H, Liang Y, Castillo R, Yang D, Choi D, Castillo E, Majumdar A, Guerrero T, Jiang SB. Implementation and evaluation of various demons deformable image registration algorithms on a GPU. Phys. Med. Biol. 2010;55(1):207–219. doi: 10.1088/0031-9155/55/1/012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Saxena V, Rohrer J, Gong L. A parallel GPU algorithm for mutual information based 3D nonrigid image registration. Euro-Par 2010 – Parallel Processing Part Ii. 2010;6272:223–234. [Google Scholar]
- [29].Shams R, Sadeghi P, Kennedy RA, Hartley RI. A survey of medical image registration on multicore and the GPU. IEEE Signal Process. Mag. 2010;27(2):50–60. [Google Scholar]
- [30].Pratx G, Xing L. GPU computing in medical physics: a review. Med. Phys. 2011;38(5):2685–2697. doi: 10.1118/1.3578605. [DOI] [PubMed] [Google Scholar]
- [31].Fluck O, Vetter C, Wein W, Kamen A, Preim B, Westermann R. A survey of medical image registration on graphics hardware. Comput. Methods Progr. Biomed. 2011;104(3):E45–E57. doi: 10.1016/j.cmpb.2010.10.009. [DOI] [PubMed] [Google Scholar]
- [32].Rueckert D, Sonoda LI, Hayes C, Hill DLG, Leach MO, Hawkes DJ. Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans. Med. Imaging. 1999;18(8):712–721. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- [33].Nvidia C. Nvidia CUDA C Programming Guide. NVIDIA Corporation; 2011. p. 120. [Google Scholar]
- [34].Thirion JP. Image matching as a diffusion process: an analogy with Maxwell’s demons. Med. Image Anal. 1998;2(3):243–260. doi: 10.1016/s1361-8415(98)80022-4. [DOI] [PubMed] [Google Scholar]
- [35].Yin Y, Hoffman EA, Lin C-L. Mass preserving nonrigid registration of CT lung images using cubic B-spline. Med. Phys. 2009;36(9):4213–4222. doi: 10.1118/1.3193526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Yin Y, Choi J, Hoffman EA, Tawhai MH, Lin C-L. A multiscale MDCT image-based breathing lung model with time-varying regional ventilation. J. Comput. Phys. 2013;244:168–192. doi: 10.1016/j.jcp.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gorbunova V, Sporring J, Lo P, Loeve M, Tiddens HA, Nielsen M, Dirksen A, de Bruijne M. Mass preserving image registration for lung CT. Med. Image Anal. 2012;16(4):786–795. doi: 10.1016/j.media.2011.11.001. [DOI] [PubMed] [Google Scholar]
- [38].Choi Y, Lee S. Injectivity conditions of 2D and 3D uniform cubic B-spline functions. Graph. Models. 2000;62(6):411–427. [Google Scholar]
- [39].Rueckert D, Aljabar P, Heckemann RA, Hajnal JV, Hammers A. Diffeomorphic registration using B-splines. Medical Image Computing and Computer-Assisted Intervention – Miccai 2006. 2006;4191:702–709. doi: 10.1007/11866763_86. Pt 2. [DOI] [PubMed] [Google Scholar]
- [40].Yin Y, Hoffman EA, Ding K, Reinhardt JM, Lin C-L. A cubic B-spline-based hybrid registration of lung CT images for a dynamic airway geometric model with large deformation. Phys. Med. Biol. 2011;56(1):203–218. doi: 10.1088/0031-9155/56/1/013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hagenlocker M, Fujimura K. CFFD: a tool for designing fiexible shapes. Vis. Comput. 1998;14(5–6):271–287. [Google Scholar]
- [42].Schnabel JA, Rueckert D, Quist M, Blackall JM, Castellano-Smith AD, Hartkens T, Penney GP, Hall WA, Liu H, Truwit CL. A generic framework for non-rigid registration based on non-uniform multi-level free-form deformations. Medical Image Computing and Computer-Assisted Intervention – MICCAI. 2001:573–581. [Google Scholar]
- [43].Kirk DB, Wen-mei WH. Programming Massively Parallel Processors: A Hands-on Approach. Elsevier Inc.; 2012. [Google Scholar]
- [44].Hoffman EA. Effect of body orientation on regional lung expansion – a computed tomographic approach. J. Appl. Physiol. 1985;59(2):468–480. doi: 10.1152/jappl.1985.59.2.468. [DOI] [PubMed] [Google Scholar]
- [45].Papieẓ BW, Heinrich MP, Fehrenbach J, Risser L, Schnabel JA. An implicit sliding-motion preserving regularisation via bilateral filtering for deformable image registration. Med. Image Anal. 2014;18(8):1299–1311. doi: 10.1016/j.media.2014.05.005. [DOI] [PubMed] [Google Scholar]
- [46].Murphy K, van Ginneken B, Pluim J, Klein S, Staring M. Semi-automatic reference standard construction for quantitative evaluation of lung CT registration. Medical Image Computing and Computer-Assisted Intervention – MICCAI. 2008:1006–1013. doi: 10.1007/978-3-540-85990-1_121. [DOI] [PubMed] [Google Scholar]
- [47].Lin C-L, Peng GCY, Karniadakis G. Multi-scale modeling and simulation of biological systems preface. J. Comput. Phys. 2013;244:1–3. [Google Scholar]
- [48].Parker JA, Kenyon RV, Troxel DE. Comparison of interpolating methods for image resampling. IEEE Trans. Med. Imaging. 1983;2(1):31–39. doi: 10.1109/TMI.1983.4307610. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


