Abstract
Massively parallel sequencing is rapidly becoming a widely used method in genetic diagnostics. However, there is still no clear consensus as to which approach can most efficiently identify the pathogenic mutations carried by a given patient, while avoiding false negative and false positive results. We developed a targeted exome approach (MyoPanel2) in order to optimize genetic diagnosis of neuromuscular disorders. Using this approach, we were able to analyse 306 genes known to be mutated in myopathies as well as in related disorders, obtaining 98.8% target sequence coverage at 20 ×. Moreover, MyoPanel2 was able to detect 99.7% of 11,467 known mutations responsible for neuromuscular disorders. We have then used several quality control parameters to compare performance of the targeted exome approach with that of whole exome sequencing. The results of this pilot study of 140 DNA samples suggest that targeted exome sequencing approach is an efficient genetic diagnostic test for most neuromuscular diseases.
Keywords: Targeted exome, Gene panel, Whole exome, Massively parallel sequencing, Genetic diagnosis, Neuromuscular disorders
1. Introduction
Massively parallel sequencing has been widely used in genetic research since it has been developed more than a decade ago. This technology is also rapidly expanding into the genetic diagnostics field, expected to soon replace the gold-standard Sanger sequencing. However, a number of recent studies have suggested that adoption of massively parallel sequencing methods for genetic diagnosis of patients must be done with caution, as this technology can have false negative results due to locus-specific sequencing bias (Ross et al., 2013) or suboptimal variant calling by bioinformatics algorithms (O'Rawe et al., 2013, Park et al., 2014). Several studies have attempted to asses and compare the efficiency of different massively parallel sequencing methods (Lelieveld et al., 2015, Xue et al., 2014). However, because of rapid evolution of this technology as well as inherent differences in diagnosis between specific types of genetic disorders, further investigations are critical to determine the appropriate clinical sequencing approaches. We have developed an optimized targeted exome test for genetic diagnosis of neuromuscular diseases. We have then conducted a pilot study comparing this approach with whole exome sequencing, suggesting that optimized targeted exome approach is an efficient genetic diagnostic test for this group of disorders.
2. Materials and methods
2.1. Targeted exome approach design
Two different targeted exome designs are described in this study: the initial MyoPanel1 and the optimized MyoPanel2 designs. MyoPanel1 targeted exome approach was developed using HaloPlex target enrichment system (Agilent, CA, USA) adapted for Ion Torrent Next Generation Sequencing technology and has been recently described (Sevy et al., 2015). MyoPanel1 was composed of genes implicated in neuromuscular diseases and cardiomyopathies listed in the Gene Table of Neuromuscular Disorders (Kaplan and Hamroun, 2013) as well as differential diagnosis genes (298 genes total). The DNA capture probes for both MyoPanels were designed using the Agilent SureDesign web-based application (https://earray.chem.agilent.com/suredesign/home.htm, June 2015). The target regions used as an input for SureDesign tool included protein coding exons and 10 bp intron flanking regions. The characteristics of both designs are shown in Fig. 1D. The following modifications of MyoPanel1 were done to obtain the optimized MyoPanel2. Three genes were removed from the panel: SMN1 because of low target coverage by the capture probes proposed by SureDesign, DUX4 and KCNJ18 due to off-target read alignment. Eleven new genes were added: ALG14, BICD2, GMPPB, KLHL40, PTPLA, RBCK1, SLC5A7, SMCHD1, TIA1, TNPO3, and TRAPPC11. Capture probes for 436 regions with poor coverage by MyoPanel1 were redesigned. Shorter probes were selected for these regions using FFPE option in SureDesign and added to the total number of probes designed with the default parameters.
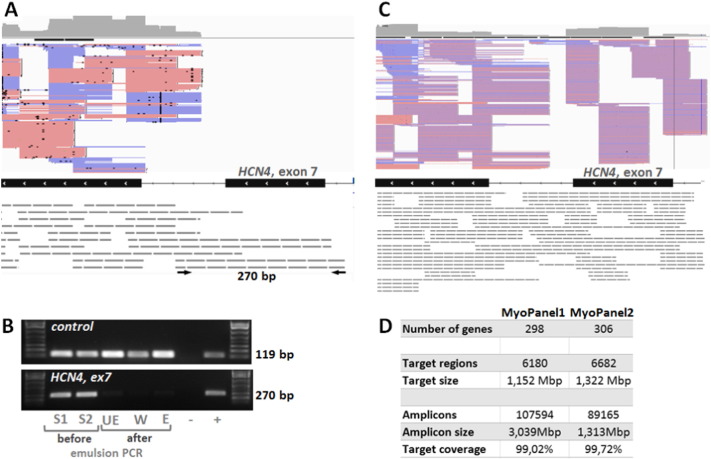
Fig. 1.
Optimization of the targeted exome approach.
A. IGV visualization of MyoPanel1 HaloPlex probes (grey dashed lines) covering the exons 7 and 8 of HCN4 gene. Pink and blue lines correspond to reads (forward and reverse respectively) obtained from the sequencing experiment. Both longer and shorter HaloPlex probes cover exon 8, generating a number of reads. Only longer (280 bp and more) probes cover exon 7, producing no reads. PCR primers specific to the 280 bp probe are shown as black arrows. B. Longer HaloPlex probes are lost during the emulsion PCR step. PCR using individual probe-specific primers was performed on aliquots from the following library preparation steps: two different samples before emulsion PCR (S1 and S2), before enrichment (UE), wash solution after elution (W), after enrichment (E). A PCR fragment (119 bp) is amplified using primers specific to a shorter 157 bp control probe, while no PCR fragment is produced from post-emulsion PCR steps library aliquots using primers specific to a 280 bp probe (HCN4, exon 7). Similar results were obtained for three other longer probes. C. IGV visualization of the same region as shown in A for the targeted exome MyoPanel2. Shorter HaloPlex probes cover exon 7, producing a number of reads. D. Characteristics of MyoPanel1 and 2 designs.
2.2. Identification of regions poorly covered by MyoPanel1
Coverage of the target exons was obtained for each sample with the help of VarAFT tool (http://varaft.eu, June 2015), which uses BedTools (Quinlan and Hall, 2010) to compute coverage statistics. For each exon, we verified whether it was 100% covered at 5 × in at least eight out of ten samples. 436 exons did not meet this requirement and served as an input for SureDesign tool to obtain better DNA capture probe design.
2.3. Detection of longer probes during library preparation steps
PCR using individual probe-specific primers was performed on aliquots from the following library preparation steps: two different samples before emulsion PCR, before enrichment, wash solution after elution and after enrichment. The following primers were used for detection of a 280 bp probe covering in HCN4 gene - 5'GACCTGGCTTAGGCATAAAGG and 5'TCCTGAGTCCTGATGCTCTG producing 270 bp fragment; for detection of a 400 bp probe in COL6A1 gene - 5'TGTCTGACCTGCATCTGACTC and 5'GGCCAATCAACTGTCAGACTT producing a 390 bp fragment; for detection of a 379 bp probe in PNPLA2 gene 5'TAGTGAAGGGAGGTGGCTGT and 5'CGAGTAATCCTCCGCTTGG producing 370 bp fragment; for detection of a 337 bp probe in LMNA gene 5'GAGATGCGGGCAAGGATG and 5'ACTCCAGTTTGCGCTTTTTG producing 321b. Primers to detect a shorter 157 bp control probe in FAT1 gene were 5'CAAGGACTTCGACTTCCCG and 5'CACTGGTGCCGTGAGTACG producing a 119 bp fragment.
2.4. DNA samples and sequencing experiments
Results from five sequencing experiments were used for this study. Experiments 1 (10 DNA samples) and 2 (33 DNA samples) of MyoPanel1 were performed using HaloPlex (Agilent) capture method and sequenced on two different in-house PGM (Ion Torrent) sequencers. MyoPanel2 experiment was performed using HaloPlex enrichment method and NextSeq (Illumina) sequencing by Helixio (Biopôle Clermont-Limagne, France). A total of 46 DNA samples were sequenced. Of them, 17 DNA samples (batch1) were received from abroad and were most likely of lower quality, given clear differences in sequencing results for this batch of samples. No differences between the batch1 samples and the remaining 29 MyoPanel2 samples were observed at the time of library preparation. Results from two different whole exome sequencing experiments were included in this study, both performed using Agilent SureSelect V4 reagent kits and HiSeq (Illumina). Experiment 1 included 20 DNA samples and was sequenced by Integragen Genomic (Evry, France). Experiment 2 included 31 DNA samples and was sequenced by CNG (Centre National de Génotypage, Evry, France).
2.5. Library preparation, sequencing and variant calling
For MyoPanel1, DNA extraction and library preparation was performed as previously described (Sevy et al., 2015). Briefly, libraries of DNA samples were created using the Haloplex Target Enrichment System Kit (Agilent Technologies, CA, USA) according to manufacturer's instructions for Ion Torrent sequencing version D4. Emulsion clonal PCR amplification was performed using the Ion PGM (Personal Genome Machine) Template OT2 200 kit (Life Technologies, CA, USA) and Ion One Touch 2 instrument (Life Technologies, CA, USA), followed by an enrichment of the ion spheres particles (ISPs) using an Ion One Touch ES (Life Technologies, CA, USA) enrichment module according to the manufacturer's instructions. ISPs were loaded on a 318v2 chip and sequenced using an Ion PGM Sequencing 200 Kit v2 (Life Technologies, CA, USA) on a PGM sequencer system (Life Technologies, CA USA). Raw data generated by the PGM sequencer were processed by Torrent Suite Software v.4 and aligned using TMAPv.3 (https://github.com/iontorrent/TMAP, June 2015). Sequence variants were identified using the VariantCaller tool from the Ion Torrent package using default “germline high stringency” parameters.
For MyoPanel2, the initial DNA extraction and library preparation steps were similar to that of MyoPanel1, except that the adaptors used for creating the libraries of DNA samples with the Haloplex Target Enrichment System Kit (Agilent Technologies, CA, USA) were specific of Illumina sequencing technology. The libraries were then separated in three different pools for the step of clonal amplification by bridge PCR on the Next Seq 500 followed by paired end sequencing using the NextSeq 500 Mid Output kit (300 cycles) and the Illumina two-channel sequencing by synthesis (SBS) technology (Illumina, CA, USA). Sequencing reads were aligned using BWA-MEM 0.7.12 (Li and Durbin, 2009) with default parameters and Hg19 as the reference genome. Sambamba 0.51 (http://lomereiter.github.io/sambamba/index.html, June 2015) was used to convert SAM to BAM format. GATK 3.3 (McKenna et al., 2010) was used for local realignment around indels and for base quality score recalibration. Variant calling was performed using Haplotype Caller (GATK 3.3), followed by standard hard variant filtering using VariantFiltration module of GATK 3.3 with cut-offs depth < 10 and quality score < 50 according to GATK Best Practices recommendations (Van der Auwera et al., 2013, DePristo et al., 2011).
2.6. Coverage analysis comparison between target exome and whole exome approaches
In order to compare the coverage between MyoPanel1, MyoPanel2 and whole-exome results, a set of 295 genes was used. The bed file for a set of 5817 unique exons corresponding to the protein coding parts of these genes and including all the splice variants of RefSeq genes was downloaded from UCSC Genome Browser using Table Browser option (Rosenbloom et al., 2015). Coverage and depth statistics for this set of exons were obtained for each sample using VarAFT tool. Sample p54 was excluded from the analysis due to much lower quality of sequencing results obtained.
2.7. Mutation coverage analysis
A set of 11,467 published mutations was obtained from Locus Specific Mutation Databases (http://grenada.lumc.nl/LSDB_list/lsdbs, June 2015) for target regions of 78 genes present in both MyoPanel1 and Myopanel2. This set was then used to calculate coverage of individual mutation positions for each sample using VarAFT tool. Number of mutations with no coverage, less than 6 × coverage and less that 20 × coverage was calculated.
3. Results
We performed a pilot study designed to test and optimize the targeted exome approach for genetic diagnosis of neuromuscular diseases. Our second goal was to compare the quality of sequencing data obtained from targeted exome as well as whole-exome experiments in order to determine which approach was more suitable for genetic diagnosis.
3.1. Optimization of targeted exome design
Our initial targeted exome design, MyoPanel1, was developed to simultaneously analyse 298 genes (5972 unique exons) implicated in neuromuscular disorders. It was tested in two sequencing experiments. The first one, MyoPanel1-exp1, included 10 samples from patients with known neuromuscular mutations. The second experiment, MyoPanel1-exp2, included 33 DNA samples from both undiagnosed and control patients with neuromuscular conditions (Sevy et al., 2015). Detailed coverage analysis from these experiments identified 436 exons that were less than 100% covered at 5 × in at least eight out of ten samples. We then examined these low-coverage regions in detail and noticed that many of them were designed to be enriched by longer HaloPlex probes. One such region, exon 7 of HCN4 gene, is shown in Fig. 1A. Indeed, this exon was covered by design but was not sequenced in the MyoPanel1 experiments. We hypothesized that the longer fragments were lost either during one of the library preparation steps or during sequencing. In order to answer this question, we designed PCR primers specific to longer probes covering regions with no corresponding sequencing reads. Results of PCR amplification using primers specific to a 280 bp HaloPlex probe on aliquots from several library preparation steps are shown in Fig. 1B. As seen from these results, the DNA fragments are initially present, but then lost during the emulsion PCR step. Similar results were obtained for three longer probes from other genomic regions. Amplification using primers specific to a shorter (157 bp) probe produced PCR products from all library preparation steps. Thus, longer (> 280 bp) HaloPlex probes were lost after the emulsion PCR step, leading to gaps in obtained sequence coverage. We therefore redesigned HaloPlex probes for 436 poorly covered regions using FFPE (formalin-fixed paraffin-embedded) option in SureDesign tool. When this option is selected, shorter probes are designed to compensate for DNA fragmentation observed in the formalin-processed samples. We have also added 11 new genes and removed three genes from the new MyoPanel2 design: SMN1 because of low target coverage by the probes proposed by SureDesign, DUX4 and KCNJ18 due to off-target read alignment. In order to diminish the possibility of losing probes during the emulsion PCR step and to test a different sequencing method, we have designed the MyoPanel2 probes for the Illumina platform. The summary of the optimized MyoPanel2 and the initial MyoPanel1 designs is shown in Fig. 1D.
3.2. Quality analysis of MyoPanel2 sequencing results
We have obtained sequencing results for 46 DNA samples using MyoPanel2 design for targeted exome approach. Many more reads were obtained for regions that were poorly covered in MyoPanel1 experiments. An example of such region, exon 7 of HCN4 gene, is shown in Fig. 1C. Interestingly, most of the coverage for this region still comes from the shorter capture probes, suggesting that improvement in the coverage is mostly due to probe re-design and not just due to the change in sequencing technology.
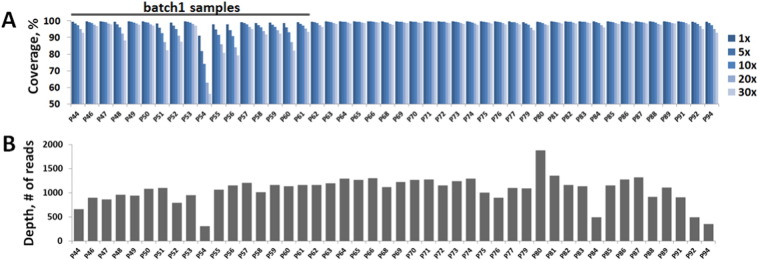
The sequence coverage statistics for each sample is shown in Fig. 2A. A set of 17 DNA samples (batch1) had clearly lower target sequence coverage comparing to the rest of the DNA samples. These samples all came from the same source abroad and most likely contained lower quality DNA. Interestingly, we observed no apparent differences between this batch and the rest of the samples during numerous quality checks at different library preparation steps. It is possible that minor DNA degradation was present in these samples, suggesting that an additional PCR-based DNA integrity verification step might be necessary before the library preparation procedure. Average target sequence coverage in MyoPanel2 experiment was 99.7% at 1 ×, 99.6% at 5 ×, 99.3% at 10 ×, 98.8% at 20 × and 98.3% at 30 × for high quality DNA samples. For lower quality (batch1) samples, the coverage was 99.3% at 1 ×, 98.1% at 5 ×, 96.8% at 10 ×, 94.1% at 20 × and 91.6% at 30 ×. Thus, even for the lower quality DNA samples, the target sequence coverage is comparable to that of a typical clinical exome sequencing experiment (Lelieveld et al., 2015, Biesecker and Green, 2014).
Fig. 2.
Coverage and depth statistics for MyoPanel2 sequencing experiment.
A. Coverage statistics per sample in MyoPanel2 sequencing experiment. Average coverage was 99.7% at 1 ×, 99.6% at 5 ×, 99.3% at 10 ×, 98.8% at 20 × and 98.3% at 30 × for high quality DNA samples. For lower quality (batch1) samples, the coverage was 99.3% at 1 ×, 98.1% at 5 ×, 96.8% at 10 ×, 94.1% at 20 × and 91.6% at 30 ×. B. Depth of coverage per sample in MyoPanel2 sequencing experiment. Average depth of coverage was 1106 reads for higher quality DNA samples and 989 reads for lower quality DNA samples.
The depth of coverage per sample in MyoPanel2 sequencing experiment is shown in Fig. 2B. Contrary to the sequence coverage statistics, we did not observe any clear differences in depth between batch1 and the rest of the samples. An average depth of coverage was 1106 reads for higher quality DNA samples and 989 reads for lower quality (batch1) DNA samples. Interestingly, sample quality had a drastic effect on sequence coverage independently of coverage depth. For example, samples P68 and P60 had similar depth of sequencing (average of 1092 and 1114 reads respectively), while coverage of target regions was much better for P68 comparing to that of P60 (99.8% and 88.2% respectively at 20 ×). These results show that percentage of target coverage is not directly related to the depth coverage. Thus, increasing the depth of coverage cannot compensate for lower sample DNA quality.
3.3. Known mutation coverage
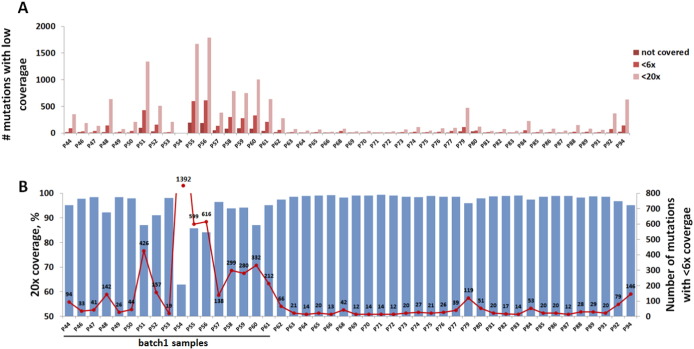
Percentage of target region coverage is now a widely accepted quality control parameter in massively parallel sequencing approaches. However, even the experiments with high average sequence coverage can still have a high rate of false negative results if pathogenic mutations are concentrated in the regions with gaps in sequencing. Thus, for genetic diagnostics, statistics about the coverage of known pathogenic mutation positions is a key quality control parameter. We analysed the performance of our targeted exome approach by using a set of 11,467 published mutations located in target regions of 78 neuromuscular genes. Numbers of known mutation positions missed or covered at less than 6 × and 20 × are shown in Fig. 3A. As expected, numbers of missed mutations are greater in samples with gaps in sequencing (i.e. samples with lower target sequence coverage). In order to better visualize this relationship, overlay of 20 × sequence coverage data (bars) and the number of known mutation positions covered at less than 6 × (red line) is shown in Fig. 3B. In several samples, as little as 12 out of 11,467 known mutation positions were missed (covered < 6 ×). Of these, six mutations cannot be detected by MyoPanel2 since they are not covered by the capture probes. Many more mutation positions are not covered in lower quality DNA samples (batch1). These results underline the importance of analysing both target sequence coverage and known mutation coverage.
Fig. 3.
Coverage of known mutations for MyoPanel2 design.
A. Number of known mutation positions with no or low sequence coverage per each MyoPanel2 sample. B. Number of mutations with < 6 × coverage (red line) in correlation with target sequence coverage (bars) per sample. As little as 12 out 11,467 published mutations are missed by our optimized targeted exome approach. Of these, six are not covered by capture probe design and therefore cannot be detected even if the sequencing conditions are optimal. Lower quality DNA samples from batch1 had many more missed known mutation positions.
3.4. Comparison between targeted and whole exome approaches
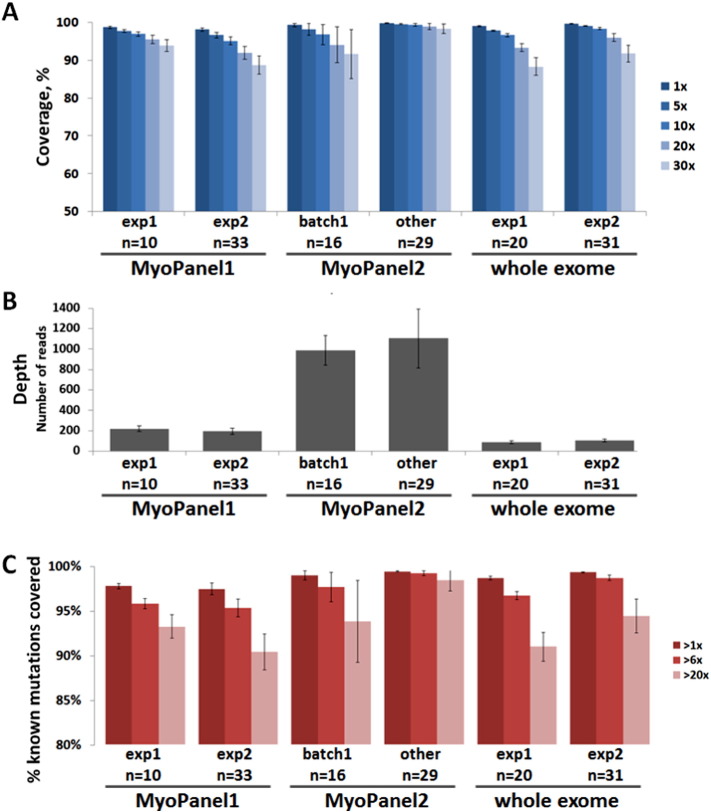
We then compared several quality control parameters between three different targeted exome and two whole exome experiments. Since target regions differed between MyoPanel1 and MyoPanel2, an overlap set of 5817 unique exons (295 genes) was used for coverage and depth analysis. The same set of exons was used to calculate the coverage and depth for exome data. Typical genome and exome sequencing experiments cover 85 to 95% of the targeted sequence (Biesecker and Green, 2014), with more recent studies reporting 95% at 20 × for whole exomes (Lelieveld et al., 2015). As seen from Fig. 4A, we observed similar coverage statistics in two different whole exome experiments: 93.4% and 96% coverage at 20 ×. Coverage of optimized MyoPanel2 was 98.8%, which is superior to exomes and to initial MyoPanel1 results. Depth of coverage was much higher for MyoPanel2 experiment (Fig. 4B). However, as discussed above, depth of coverage did not correlate with percentage of target sequence coverage, as seen from the results for batch1 samples.
Fig. 4.
Comparison between targeted exome and whole exome approaches.
A. Percentage of target region coverage for all sequencing experiments. A set of 5817 unique exons, corresponding to 295 genes found in both MyoPanel1 and MyoPanel2, was used for coverage and depth analysis. Average sequence coverage at 20 × was 95.5% for MyoPanel1-exp1, 92% for MyoPanel1-exp2, 94.1% for lower-quality (batch1) MyoPanel2 samples, 98.8% for other MyoPanel2 samples, 93.4% and 96% for two different whole exome sequencing experiments. B. Depth of coverage for all sequencing experiments. Average depth of coverage was 221 for MyoPanel1-exp1, 195 for MyoPanel1-exp2, 989 for lower-quality (batch1) MyoPanel2 samples, 1106 for other MyoPanel2 samples, 88 and 104 for two different whole exome sequencing experiments. C. Percentage of known mutation positions detected by sequencing test. A set of 11,467 published mutations was used for calculations. Average number of mutation positions covered more than 6 × was 96.2% for MyoPanel1-exp1, 95.7% for MyoPanel1-exp2, 98.1% for lower-quality (batch1) MyoPanel2 samples, 99.7% for other MyoPanel2 samples, 97.1% and 99.2% for two different whole exome sequencing experiments. Averages ± SD are shown.
In order to assess the ability of each sequencing approach to detect potential disease-causing mutations, we compared the percentage of known mutation positions (11,467 total) identified with more than 1 × and 6 × coverage. MyoPanel2 was able to detect the highest number of published mutations. Indeed, 99.7% of known mutation positions was detected by MyoPanel2, comparing to 97.1% and 99.2% identified by two different whole exome sequencing experiments. That is, if a given patient carried a known pathogenic mutation responsible for the neuromuscular phenotype, the risk of missing this mutation would be lower if MyoPanel2 was used for diagnosis. Our results therefore suggest that, based on several quality control parameters, targeted exome approach is superior to whole exome sequencing for genetic diagnosis of most neuromuscular disorders.
4. Discussion
In this study we present an optimization process for targeted exome approach, leading to a more sensitive sequencing test, based on coverage statistics as well as on percentage of known mutation positions detected. We have also compared the performance of this optimized targeted exome approach with whole exome sequencing using a set of genes mutated in neuromuscular diseases. Using the optimized targeted exome approach we were able to analyse 306 genes with 98.8% target sequence coverage at 20 × and to detect 99.7% of 11,467 known mutations responsible for neuromuscular disorders. Based on the results of our pilot study, several quality control parameters for targeted exome approach were superior to that of whole exome sequencing (obtained using V4 reagent kits and HiSeq). It is important to note, however, that as sequencing technology continues to evolve rapidly, the newer versions of whole exome sequencing approach might be more efficient than the one used in this study.
46% diagnostic rate was observed with our initial targeted exome approach (MyoPanel1) applied to a cohort of patients affected with distal myopathies (Sevy et al., 2015). Most patients included in this study had previously undergone genetic testing for a number of myopathy-causing candidate genes. Thus, if our targeted exome approach is applied to a previously unexplored cohort, the diagnostic rate is expected to be even higher. Moreover, improvement in coverage and increase in gene number in MyoPanel2 will further advance the efficiency of genetic diagnosis. One obvious limitation of the targeted exome approach is that this test only detects mutations in genes previously implicated in the studied disease. If a mutation responsible for patient's phenotype is located in a novel gene, genetic diagnosis by this approach will not be possible. In this case, whole exome sequencing might provide diagnosis since it is designed to explore all protein coding genes, including little studied or uncharacterized candidate genes. However, both targeted exome or whole exome approaches are not the optimal diagnostic sequencing test choices for diseases that are often caused by structural rearrangements or large copy number variations in genomic sequence. Whole genome sequencing would be more appropriate in these cases. Given recent results from different studies applying massively parallel sequencing to diagnosis of myopathies (Gorokhova et al., 2015), exome based approach is likely to capture a large proportion of disease causing mutations in this group of disorders. The current pilot study now suggests that an optimized targeted exome approach is a sensitive and cost effective way to diagnose neuromuscular genetic disorders.
Acknowledgements
We thank Pr. Jean Pouget, Pr. Shahram Attarian, Dr. Emmanuelle Campana-Salort, Dr. Amandine Sevy, Pr. Jorge Bevilacqua and Pr. Meriem Tazir for providing clinical samples. We also thank Amira Cherrallah and Gaby Luciani for their help with library preparation as well as Arnaud Lagarde for his help with PGM sequencing. The research leading to these results has received funding from the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement n° 2012-305121 “Integrated European –omics research project for diagnosis and therapy in rare neuromuscular and neurodegenerative diseases (NEUROMICS)”.
References
- Ross M.G. Characterizing and measuring bias in sequence data. Genome Biol. 2013;14:R51. doi: 10.1186/gb-2013-14-5-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rawe J. Low concordance of multiple variant-calling pipelines: practical implications for exome and genome sequencing. Genome Med. 2013;5:28. doi: 10.1186/gm432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M.-H. Comprehensive analysis to improve the validation rate for single nucleotide variants detected by next-generation sequencing. PLoS One. 2014;9 doi: 10.1371/journal.pone.0086664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelieveld S.H., Spielmann M., Mundlos S., Veltman J.A., Gilissen C. Comparison of Exome and Genome Sequencing Technologies for the Complete Capture of Protein-Coding Regions. Hum. Mutat. 2015 doi: 10.1002/humu.22813. n/a–n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Ankala A., Wilcox W.R., Hegde M.R. Solving the molecular diagnostic testing conundrum for Mendelian disorders in the era of next-generation sequencing: single-gene, gene panel, or exome/genome sequencing. Genet. Med. 2014 doi: 10.1038/gim.2014.122. [DOI] [PubMed] [Google Scholar]
- Sevy A. Improving molecular diagnosis of distal myopathies by targeted next-generation sequencing. J. Neurol. Neurosurg. Psychiatry. 2015 doi: 10.1136/jnnp-2014-309663. jnnp–2014–309663. [DOI] [PubMed] [Google Scholar]
- Kaplan J.-C., Hamroun D. The 2014 version of the gene table of monogenic neuromuscular disorders (nuclear Genome) Neuromuscul. Disord. 2013;23:1081–1111. doi: 10.1016/j.nmd.2013.10.006. [DOI] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera G.A. From FastQ data to high confidence variant calls: the genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinform. Ed. Board Andreas Baxevanis Al. 2013;11:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo M.A. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom K.R. The UCSC genome browser database: 2015 update. Nucleic Acids Res. 2015;43:D670–D681. doi: 10.1093/nar/gku1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesecker L.G., Green R.C. Diagnostic clinical genome and exome sequencing. N. Engl. J. Med. 2014;370:2418–2425. doi: 10.1056/NEJMra1312543. [DOI] [PubMed] [Google Scholar]
- Gorokhova S. Clinical massively parallel sequencing for the diagnosis of myopathies. Rev. Neurol. (Paris) 2015;171:558–571. doi: 10.1016/j.neurol.2015.02.019. [DOI] [PubMed] [Google Scholar]