Abstract
Autophagy is a lysosomal mediated degradation activity providing an essential mechanism for recycling cellular constituents, and clearance of excess or damaged lipids, proteins and organelles. Autophagy involves more than 30 proteins and is regulated by nutrient availability, and various stress sensing signaling pathways. This article provides an overview of the mechanisms and regulation of autophagy, its role in health and diseases, and methods for its measurement. Hopefully this teaching review together with the graphic illustrations will be helpful for instructors teaching graduate students who are interested in grasping the concepts and major research areas and introducing recent developments in the field.
Keywords: mTOR, Beclin, LC3, Mitochondria, Neurodegenerative diseases, Aging
Graphical abstract
Highlights
-
•
mTOR, Beclin–VPS34, LC3 homologs, and adaptor proteins in autophagy.
-
•
Autophagosomal membranes may derive from multiple sources.
-
•
Autophagosomal–lysosomal fusion contributes to the control of autophagic flux.
-
•
Assess autophagy by autophagosomal and protein turnover, and morphological alterations.
-
•
Autophagy adysfunction in cancer, aging, neurodegeneration and infection.
Introduction
Autophagy is defined as the processes that degrade intracellular materials in the lysosome/vacuole. It was first discovered from ultrastructural studies in the 1950s [1]. Vacuoles that contain amorphous materials were observed in kidneys from the newborn undergoing remodeling [2]. In the liver after glucagon perfusion or injection of a detergent, Triton WR-1339, there was a clear increase in vesicles containing partially digested cytoplasmic materials, in particular partially digested mitochondria [3], [4]. These vacuolar structures were then named “autophagosomes”, and examples surveyed in a review encompassing all major tissues [1].
Research in autophagy mechanisms and regulation was boosted by two key discoveries. One is the identification of AuTophaGy-related (ATG) genes in the 1990s, mutations of which resulted in dysregulated vacuole formation in yeast [5], [6]. The other is the finding that one key autophagy mammalian ATG gene homolog, ATG6/BECN1, is a tumor suppressor gene [7], [8].
Following that, autophagy has been demonstrated to be important not only in cancer biology, but also in the development, function and survival of essentially all cell types and tissues. It is subjected to regulation by a variety of signals, including nutrient availability, metabolic activities, and environmental changes [9]. Consequently, cellular lipids, proteins and organelles that exceed their “expiration date”, either due to structural or conformational changes or due to oxidative damages, are to be degraded by autophagy. Degradation by autophagy serves the purpose of decreasing the cellular energy cost for the maintenance of cellular structure and function, and attenuating further propagation of cellular damage. Autophagy is now considered the primary process that deals with damaged lipids, proteins, and organelles and can also contribute to the antioxidant defenses of the cells [9], [10], [11], [12].
Mechanisms and regulation of autophagy
Three types of autophagy
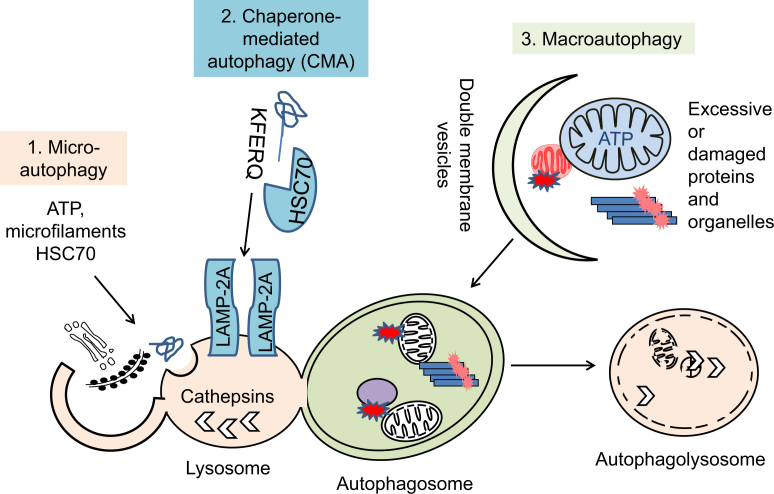
Three types of autophagy have been described: microautophagy, chaperone-mediated autophagy and macroautophagy (Fig. 1) [13]. Microautophagy describes extension and/or invagination of lysosomal membranes to take in intracellular contents [14], [15]. The signaling pathways have been shown in yeast to be dependent on Ca2+/Calmodulin, but independent of components involved in vesicle fusions [16], [17]. In response to ER stress, massive ER expansion triggers the formation of ER whorls, which are selectively taken up into the yeast vacuoles by invagination of the vacuolar membrane, in a process equivalent to microautophagy [18]. Electron microscopy studies have suggested that a decrease in microautophagic structures correlates with a decrease in protein turnover both during starvation and in response to re-feeding in hepatocytes [19]. The appearance of microautophagic structures has been shown to be dependent on ATP and microfilaments in mammalian cells [20], [21]. Recent studies have also indicated microautophagy-like mechanisms of direct trapping of proteins into late endosomes via a process facilitated by heat shock cognate 70 (HSC70)-endosomal acidic phospholipid interactions in mammalian systems [22].
Fig. 1.
The 3 types of autophagy. Lysosomal-mediated degradation of cellular contents, or autophagy, is divided into 3 general categories. 1. Microautophagy is defined by ultrastructural studies as performed by lysosomal/endosomal invagination of nearby cellular contents, or lysosomal extension to wrap around cellular contents, to engulf and degrade lipids, proteins and organelles. It has been shown to depend on ATP and microfilaments and be facilitated by HSC70-phospholipid interactions. 2. Chaperone-mediated autophagy is dependent on lysosomal LAMP-2A membrane receptor and chaperone HSC70 protein recognizing unfolded proteins that carry KFERQ consensus sequences. 3. Macroautophagy is characterized by formation of double membrane vesicles that recognize and encircle intracellular excessive or damaged lipids, proteins and organelles. The autophagic vesicles, or autophagosomes, then fuse with lysosomes, and their contents degraded by lysosomal enzymes. These 3 types of autophagy have shared as well as independent machineries, and their activities may compensate or regulate one another.
Chaperone-mediated autophagy involves chaperone protein HSC70 recognizing proteins with a consensus KFERQ sequence and bringing the proteins one by one, to the lysososome-associated membrane protein 2A (LAMP-2A) receptor to transport them into the lysosomes and be degraded. This is highly specific and is studied by incubating proteins with isolated lysosomes or genetic manipulations of chaperones or the lysosomal receptor [23].
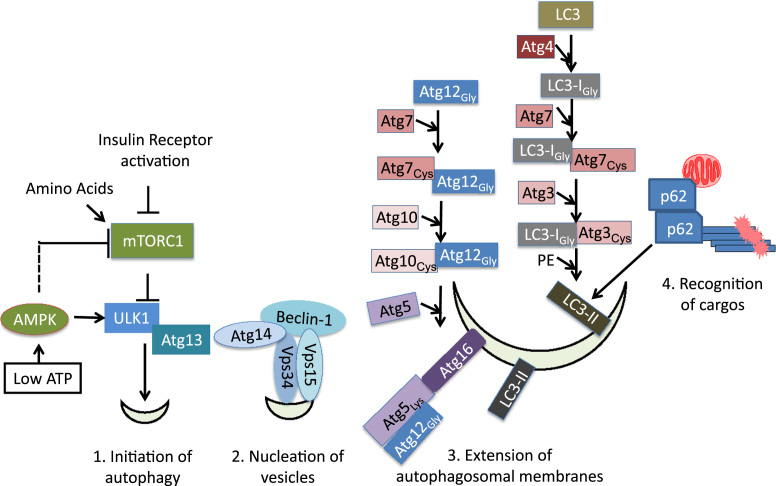
Macroautophagy is a high capacity process, initiated by membrane circling target proteins, organelles and to form autophagosomes. These autophagosomes then fuse with the lysosomes so that the contents can be degraded. This machinery consists of more than 30 proteins and regulators [8]. The main regulatory pathways involves sensing amino acid and nutrient availability by mammalian target of rapamycin (mTOR)-unc-51 like autophagy activating kinase 1 (ULK1)-Atg13 pathway, nucleation of autophagic vesicles by the Beclin-1/vacuolar protein sorting 34 (VPS34)/VPS15/Atg14 complex, expansion of autophagosomes by ubiquitin-like conjugation pathways ending with Atg5/Atg12/Atg16 as well as MAP1 light chain 3 (LC3)-II insertion into autophagosomal membranes, and cargo recognition, including p62/sequestosome 1 (SQSTM1) binding to both ubiquitinated proteins and LC3-II (Fig. 2) [10], [24], [25]. Transcriptional and post-transcriptional regulation of autophagy and lysosomal proteins also play important roles in macroautophagy [26], [27].
Fig. 2.
Proteins involved in autophagosome formation. 1. Initiation of autophagosomal formation is regulated by mTOR inhibition. In response to insulin receptor withdrawal, amino acid starvation, low ATP, mTOR is inhibited, resulting in ULK1 activation and initiation of autophagy. 2. Beclin-1/VPS34/VPS15/Atg14 complex formation also plays an important role in nucleation of autophagic vesicles. 3. Extension of autophagosomal membranes are regulated by 2 ubiquitin-like conjugation pathways, with both LC3 and Atg12 resembling ubiquitin structure. One involves Atg7 and Atg10 acting as E1 and E2 enzymes, sequentially conjugating with Atg12 via glycine–cysteine thioester bonds, eventually conjugating Atg12 to Atg5 at a lysine residue through an isopeptide bond, resulting in Atg5/Atg12/Atg16 complex association with autophagosomal membranes. The other involves Atg4 cleaving pro-LC3 exposing a glycine residue at the C-terminal, conjugation of LC3-I with Atg7, then Atg3 via thioester bonds, resulting in conjugation of LC3-I with phosphotidylethanolamine (PE) through an amide bond, forming LC3-II and insertion into autophagosomal membranes. 4. Recognition of cargo can be mediated by adaptor proteins such as p62 that has an ubiquitin binding domain as well as an LC3-II interacting domain.
mTOR and nutrient sensing
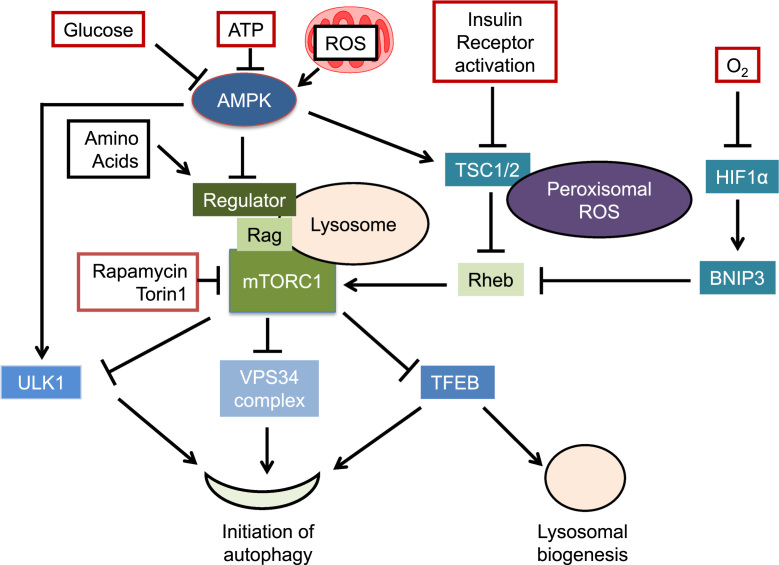
mTOR is a key autophagy regulator integrating amino acid starvation, growth factor deprivation, the drop of ATP or oxygen levels, and accumulation of reactive oxygen species to the autophagy pathway activities [28], [29], [30], [31] (Fig. 3). mTOR is associated with either complex 1 (mTORC1) or complex 2 (mTORC2) depending on cofactors in the complexes. In the presence of amino acids, mTORC1 is associated with the lysosome with the Regulator complex and Rag GTPase [32]. The lysosomal association facilitates the binding and activation of mTORC1 with Ras homolog enriched in brain (Rheb) GTPase. Amino acid deprivation leads to mTOR disassociation with lysosomes. Growth factor deprivation leads to inactivation of Akt protein kinase, activation of tuberous sclerosis 1/2 (TSC1/2), and inactivation of Rheb, thus attenuated Rheb activation of mTOR [33]. ATP depletion either due to glucose starvation or mitochondrial dysfunction activates AMP-activated protein kinase (AMPK) which can activate TSC1/2 and thereby inhibits mTOR, or activate ULK1 to activate autophagy [34], [35], [36]. Recent studies also identified an association of AMPK with the late endosomes in response to glucose starvation, leading to dissociation and inactivation of mTOR [37].
Fig. 3.
mTOR in sensing autophagy signals : mTOR inhibition integrates amino acid starvation, growth factor deprivation, decreased ATP or oxygen levels, enhanced reactive oxygen species (ROS) to activate autophagy. Amino acid deprivation leads to mTOR dissociation with lysosomes. An association of AMPK with the late endosomes in response to glucose starvation also leads to dissociation and inactivation of mTOR. ATP depletion activates AMPK which can activate TSC1/2 and thereby inhibits mTOR, or activates ULK1 to activate autophagy. In addition, mitochondrial generated ROS may also induce autophagy via an AMPK mediated pathway. Growth factor deprivation leads to activation of TSC1/2, inactivation of Rheb, and inactivation of mTOR. Peroxisomal TSC1/2 localizes to peroxisomes through binding to PEX19 and PEX5, and can be activated by peroxisomal ROS to inhibit mTOR activities and induce autophagy. Oxygen deprivation may activate autophagy by HIF1α-mediated transcription activation of BNIP3 which inhibits Rheb. mTOR inhibition activates autophagy by activation of ULK1, VPS34 and TFEB, a master transcription activator of genes encoding autophagy and lysosomal proteins. Conformational inhibitors of mTOR such as rapamycin and catalytic inhibitors such as Torin1 have been shown to induce autophagy.
Oxygen deprivation may activate autophagy by attenuating mitochondrial function or by hypoxia-inducible factor 1α (HIF1α) mediated transcription activation of Bcl-2/adenovirus E1B 19-kDa interacting protein 3 (BNIP3) which inhibits Rheb [38], [39]. Inhibition of mTOR activities has also been found to play a role in autophagy induction by reactive oxygen species (ROS) [29], [40]. It has been shown that TSC1/2 localizes to peroxisomes through binding to peroxisomal biogenesis factor (PEX) 19 and 5, and can be activated by peroxisomal ROS to inhibit mTOR activities and induce autophagy [29].
Downstream of mTOR inhibition, autophagy is activated by increased phosphorylation of ULK1 [34], [35], [36], VPS34 activation [41], as well as transcription factor EB (TFEB) activation [42], [43]. Notably, mTOR knockout mice are embryonically lethal [44], ULK1 has a homolog ULK2 and ULK1/2 double knockout mice exhibit respiratory deficits [45]. The soil bacterium Streptomyces hygroscopius produced macrolide rapamycin, and the mTOR ATP-competitive inhibitor Torin1, both have been shown to be potent inducers of autophagy in diverse cellular contexts [46].
Beclin–VPS34 complex
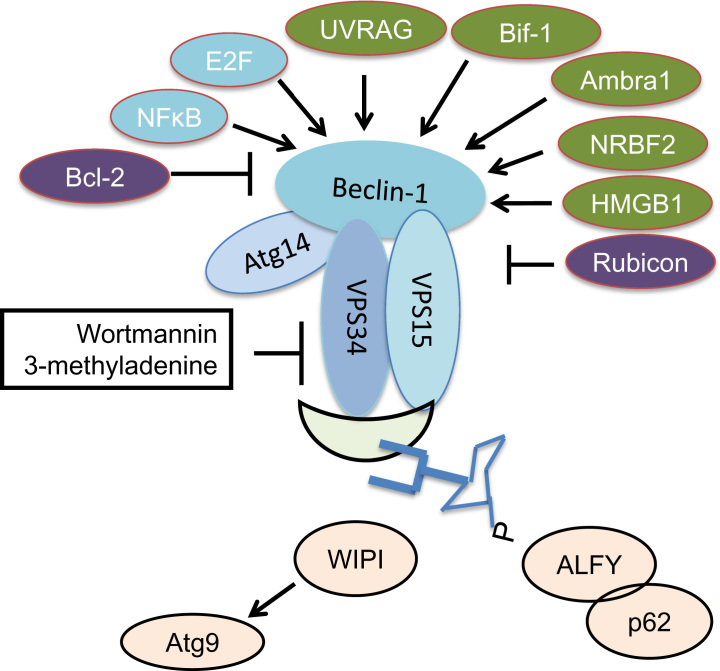
Beclin-1 is a Bcl-2-homology (BH)-3 domain only protein, which is under the transcription regulation of NF-kB, and E2F [47], [48]. Beclin-1 can be sequestered by the anti-apoptotic Bcl-2 protein, the interaction is also under the control of post-translational modification of both Bcl-2 and Beclin-1, or their binding to other regulatory proteins [47], [48], [49]. Beclin-1 association with class III PI3K, VPS34 plays an important role in nucleation of autophagosomes [47], [48] (Fig. 4). VPS34 can also be phosphorylated, which impacts its binding to Beclin-1 [47], [48]. Among others, UV radiation resistance-associated gene (UVRAG), Bax-interacting factor 1 (Bif-1)/Endophilin B1, Run domain Beclin-1 interacting and cysteine-rich containing protein (Rubicon), activating molecule in Beclin 1-regulated autophagy 1 (Ambra1), high-mobility group protein B1 (HMGB1), and nuclear receptor-binding factor 2 (NRBF2) have been shown to regulate Beclin-1–VPS34 binding or their participation in autophagic activities [47], [48], [50].
Fig. 4.
Beclin1–VPS34 complex in regulation of autophagy. Beclin-1/VPS34/VPS15/ATG14 complex plays an important role in regulating autophagy and is subjected to transcriptional (e.g., by transcription factor NFκB and E2F) and post-translational controls (phosphorylation or ubiquitination). Beclin-1 can also complex with the anti-apoptotic Bcl-2 protein, attenuating its involvement in autophagic activities. Beclin-1–VPS34 complex is subject to regulation by UVRAG, Bif-1, Rubicon, Ambra1, NRBF2, and HMGB1. PI3P production by VPS34 can be sensed by WIPIs which control the localization of Atg9 and its involvement in autophagy. ALFY can also bind PI3P and associate with p62 and cytoplasmic protein aggregates and mediates their autophagic degradation. PI3K inhibitors wortmannin and 3-methyladenine can inhibit autophagy, although they may have additional impact on other PI3K activities.
Downstream of Beclin-1/VPS34 complex activities that produce phosphatidylinositol 3-phosphate (PI3P), WD repeat domain phosphoinositide-interacting protein (WIPI)s sense PI3P and control the localization of ATG9 and its role in autophagy regulation [48], [51], [52], autophagy-linked FYVE protein (ALFY) binds PI3P and associates with cytoplasmic protein aggregates and mediates their autophagic degradation [53], [54], [55]. Beclin-1 and VPS34 gene knockout mice are embryonic lethal [56], [57], [58], [59], [60]. PI3K inhibitors wortmannin and 3-methyladenine have been shown to inhibit autophagy, however, they may have additional impact on other PI3 kinase activities depending on the specific cellular context [61], [62], [63].
Ubiquitin-like conjugation pathways
Extension of autophagosomal membranes are regulated by 2 ubiquitin-like conjugation pathways, with both LC3 and Atg12 resembling a ubiquitin structure [64] (Fig. 2). One involves Atg7 and Atg10 acting as E1 and E2 enzymes, sequentially conjugating with Atg12 via glycine–cysteine thioester bonds, eventually conjugating Atg12 to Atg5 at a lysine residue through an isopeptide bond, resulting in the Atg5/Atg12/Atg16 complex association with autophagosomal membranes. This complex associates with the autophagosomal membrane at an early step and dissociates from the autophagosomes upon completion of autophagy. The other involves a redox sensitive protease Atg4 cleaving pro-LC3 exposing a glycine residue at the C-terminal, conjugation of LC3-I with Atg7, then Atg3 via thioester bonds, resulting in conjugation of LC3-I with phosphatidylethanolamine (PE) through an amide bond, forming LC3-II and insertion into autophagosomal membranes. LC3-II association with the autophagosomes will be sustained until autophagosomes fuse with lysosomes. Atg4 can also delipidate LC3-II that is associated with the autophagosome outer membrane back to LC3-I.
MAP-1 LC3 homologs
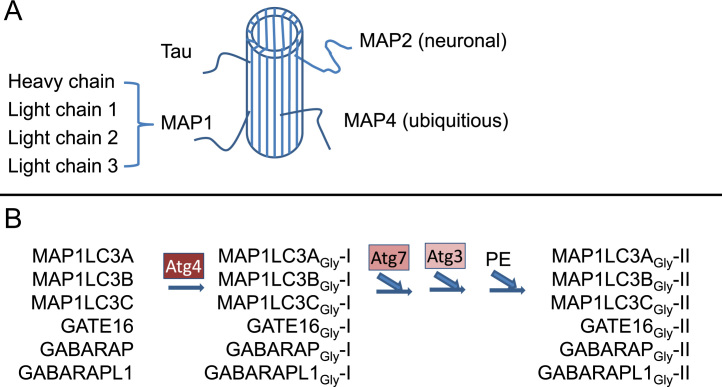
LC3 is one of the microtubule associated proteins (Fig. 5A). There are 4 groups of these microtubule associated proteins important for sustaining cell shape. MAP2 is specifically expressed in neurons, the others (Tau, MAP4 and MAP1) are enriched in neurons but are also in other cell types. The MAP1 family has one heavy chain and 3 light chains, LC3 is the smallest light chain [65]. In mammalian cells, LC3A, B, C, Golgi-associated ATPase enhancer of 16 kDa (GATE16), gamma-aminobutyric acid receptor-associated protein (GABARAP), gamma-aminobutyric acid receptor-associated protein like 1 (GABAPRAPL1) are homologs encoded by independent genes. All can be processed by Atg4 protease and ubiquitin-like conjugation by E1 and E2 like Atg7 and Atg3, ending with lipidation by phosphatidylethanolamine (PE) and insertion into autophagosomes (Fig. 5B) [66], [67], [68].
Fig. 5.
LC3 function and homologs. (A) LC3 is a microtubule-associated protein, MAP1 light chain 3. These microtubule associated proteins, including MAP2 (A, B, and C) that are restricted to the central nervous system, Tau that is enriched in the central nervous system but also expressed elsewhere, MAP4 that is widely expressed in mammalian tissues, and MAP1 (A, B, and C heavy chain, and light chain 1 and 2 that are enriched in the central nervous system, but also expressed in other tissues), are thought to be important for maintaining cell structure and shape. (B) LC3 has 6 mammalian homologs including LC3A, LC3B, LC3C, GATE16, GABARAP and GABARAPL1. All these homologs can be cleaved by Atg4 at the C-terminal which exposes the glycine residue generating LC3A-I, LC3B-I, LC3C-I, GATE16-I, GABARAP-I and GABARAPL1-I, allowing E1, E2, and E3-like conjugation by Atg7, Atg3 mediated reaction, ending with conjugation with phosphatidylethanolamine (PE), resulting in membrane associated form of LC3A-II, LC3B-II, LC3C-II, GATE16-II, GABARAP-II and GABARAPL1-II.
Adaptor proteins and selective autophagy
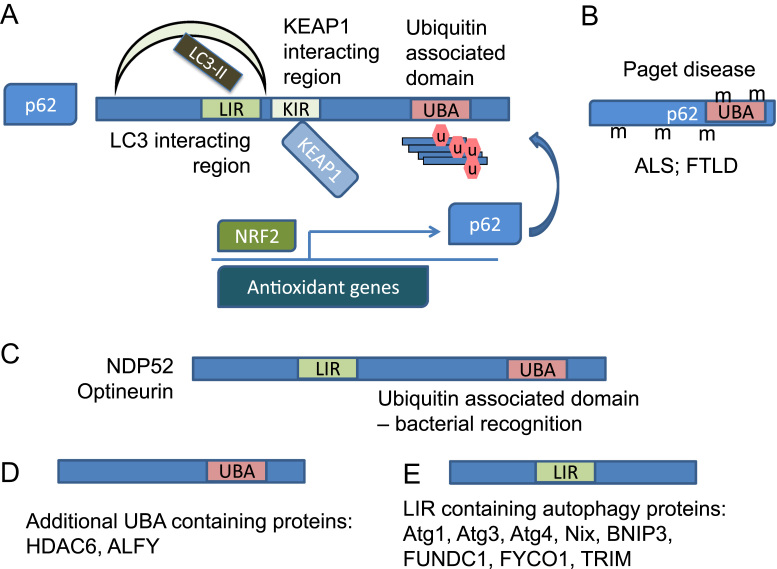
Certain adaptor proteins recognize selective autophagy substrates, although the full range of mechanisms of substrate selection is still largely unclear. Ubiquitinated proteins can be recognized by adaptor proteins, p62/SQSTM1 (sequestosome 1), NBR1 (neighbor of Brca1 gene 1) NDP52 (nuclear dot protein 52), and Optineurin, through an ubiquitin associated domain (UBA), and brought to autophagosomes through binding of these adaptor proteins to LC3 via their LC3-interaction regions (LIR) with XXXWXXLX consensus sequences, the core aromatic residue can be W, F, or Y [69], [70] (Fig. 6A). P62 also has a kelch-like ECH-associated protein 1 (KEAP1) interacting domain important for KEAP1 degradation by autophagy allowing nuclear factor (erythroid-derived 2)-like 2 (NRF2) transcription factor to translocate to the nucleus and activate antioxidant gene expression [11], [69], [71] (Fig. 6A). The increase of p62 protein may be either due to insufficient autophagic degradation of p62, or activation of p62 transcription, which has been shown to be under the control of the redox sensing transcription factor NRF2 [11], [72] (Fig. 6A). P62 has a PB1 domain which mediates its homopolymerization, and has been found to accumulate in ubiquitin-positive inclusions in neurodegenerative diseases [73]. When autophagy is blocked, for example due to Atg5, Atg7, or VPS34 deficiency, p62 as well as other autophagy substrates accumulate [57], [74], [75]. In addition to participating in autophagy and being an autophagy substrate, p62 can shuttle poly-ubiquitinated tau for proteasomal degradation [76]. Mutations in the UBA of p62 are associated with adult onset Paget disease of the bone, while mutations widespread in p62 coding regions have been associated with amyotrophic lateral sclerosis and frontotemporal lobar degeneration [77] (Fig. 6B).
Fig. 6.
Adaptor proteins that recognize autophagy substrates and/or bridge autophagy substrates with autophagy machineries. (A) P62/SQSTM1 is an important autophagy adaptor protein, with an LC3 interacting region (LIR) that can recognize autophagosomes through binding LC3/GABARAP family proteins, and an ubiquitin association domain (UBA) that can recognize ubiquitinated proteins that are autophagy substrates. Interestingly, p62 can also bind KEAP1 (via a KEAP1 interacting region, KIR), a redox sensor protein, to send KEAP1 for autophagy degradation. This is important for regulation of cellular redox status, since KEAP1 binding to NRF2 allows NRF2 degradation by the proteasomes, and KEAP1 modification or degradation frees NRF2 from the proteasomal degradation, therefore allowing NRF2 to activate transcription of antioxidants, as well as p62 itself. (B) P62/SQSTM1 mutations in the UBA have been found in Paget disease, in wide spread regions of the gene in amyloid lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD). (C) NDP52 and optineurin also contain LIR and UBA and are important for recognition of ubiquitinated bacteria and degrade bacteria via xenophagy. (D) Other UBA containing proteins involved in autophagy include HDAC6 and ALFY that are important for aggrephagy. (E) Atg1, 3, 4, Nix, BNIP3, FUNDC1, FYCO1 and TRIM also contain LIR and regulate autophagy.
Like p62, NBR1 has been found to be an autophagy substrate, able to bind LC3, and involved in aggrephagy and peroxophagy. Unlike p62, NBR1 PB1 domain cannot mediate NBR1 homopolymerization, but can bind to p62. Furthermore, compared to p62 which is found in flies and worms, NBR1 is found in plants and fungi, and NBR1 has a different UBA domain with differential affinity to different ubiquitin branches as p62. Ubiquitinated bacteria are recognized with differential affinity by NDP52, optineurin and p62, and degraded through xenophagy. NDP52 and optineurin also contain LIR motifs that enable LC3 binding (Fig. 6C) [78]. In addition to bringing autophagy substrate to the autophagosomes, the adaptor proteins may have other functions in cell signaling. For example, p62 also binds to kinases MEKK3, MEK5, and ERK1; as well as TNF receptor associated factor 6 (TRAF6) and capase-8. Proteins that have ubiquitin associate domains also include histone deacetylase 6 (HDAC6) and ALFY (autophagy linked FYVE protein), that bind PI3P, Atg5, and p62 (Fig. 6D). Furthermore, proteins that have LC3 interaction regions also include ULK1, ULK2, Atg3, Atg4, FYCO1 (FYVE and coiled-coil domain-containing protein 1), and others (Fig. 6E) [70], [78]. A subset of pattern recognition receptors tripartite motif-containing protein (TRIM)5α, 6, 16, 17, 20, 22, 49 and 55, binds LC3, p62, ULK1 and Beclin 1 and regulates autophagy, with TRIM5α shown to recognize viral capsid sequences via its SPRY (SPla and the RYanodine Receptor) domain [79] (Fig. 6E). Nix, Bcl-2/adenovirus E1B 19-kDa interacting protein 3 (BNIP3) and FUN14 domain containing 1 (FUNDC1), although do not have a UBA, can localize to the mitochondria thus bringing the organelles to the autophagosomes and promoting mitophagy (Fig. 6E) [80].
Mitophagy
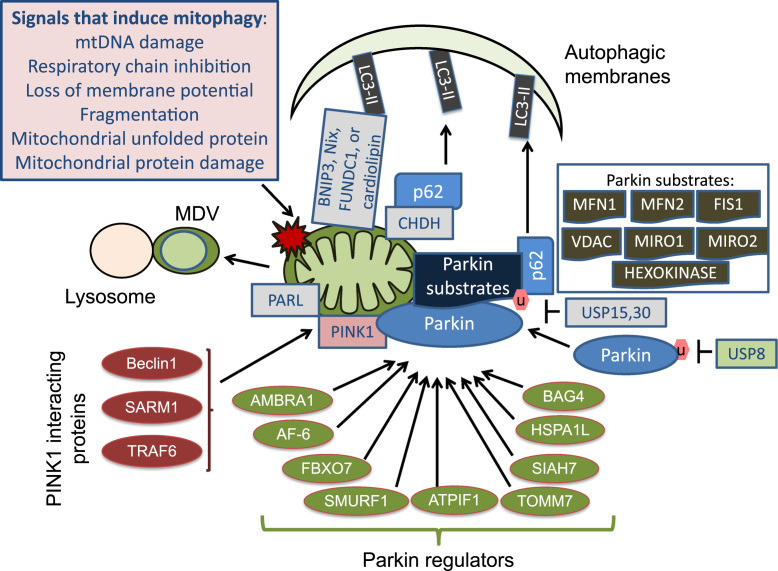
Mitophagy, or autophagy of the mitochondria, is important for mitochondrial quality control, thereby essential for providing cellular energy, calcium homeostasis, redox signaling and apoptotic signaling [81], [82] (Fig. 7). Apart from Nix, BNIP3 and FUNDC1 that target mitochondria to autophagosomes by binding to LC3 in response to hypoxia or during erythrocyte development, there are additional pathways regulating mitophagy. For example, mitochondrial dynamics, including fission and fusion, play a key role in signaling mitophagy [83], [84]. Mitochondrial DNA damage, inhibition of respiratory chain activity, loss of iron, and mitochondrial protein misfolding or damage, have all been shown to signal mitophagy [80], [85], [86], [87], [88], [89], [90], [91], [92], [93]. The mitochondrial membrane lipid cardiolipin also mediates mitophagy by binding to LC3 [88]. Recent studies demonstrated that choline dehydrogenase (CHDH) accumulates on the outer membrane in response to loss of membrane potential and recruits p62 and targets mitochondria for mitophagy [94]. Furthermore, mitochondrial spheroids or mitochondria derived vesicles (MDV) may directly bring part of the mitochondrion to the lysosomes for degradation [92], [95], [96], [97].
Fig. 7.
Mitophagy. Mitophagy can be induced by mtDNA damage, mitochondrial electron transport chain inhibition, loss of mitochondrial membrane potential, mitochondrial fission or fragmentation, mitochondrial unfolded or damaged proteins. Mitophagy is important for mitochondrial quality control and thus essential for maintaining cellular energy, calcium homeostasis, redox signaling, mitochondrial–cytosol or mitochondrial–nuclear signaling, and sequestration of apoptotic factors. One major signaling pathway is through inactivation of mitochondrial protease PARL due to loss of membrane potential, thus stabilization of PINK1. PINK1 recruits Parkin which ubiquitinates mitochondrial associated proteins. P62 recognizes ubiquitinated proteins and brings mitochondria to autophagosomes by binding to LC3. Parkin and PINK1 can be regulated by a variety of cellular proteins in many recent studies, including those listed in this diagram. Deubiquitinase USP8 plays an important role in Parkin recruitment to the mitochondria and mitophagy. In contrast USP15 and 30 remove ubiquitin from Parkin substrates and thus attenuate mitophagy. Parkin/PINK1-independent mitophagy also exist, including those mediated or signaled by BNIP3, Nix, FUNDC1 or cardiolipin, which can bind LC3 and directly bring mitochondria to the autophagosomes for mitophagy. In addition, mitochondrial spheroids and mitochondria derived vesicles (MDV) may bring mitochondria to the lysosomes for degradation. In response to loss of mitochondrial membrane potential, choline dehydrogenase (CHDH) can also recruit p62 directly and target mitochondria to mitophagy.
Parkin and PINK1 have been shown to play an important role in mitophagy [98]. PINK1 targets to the mitochondria but is normally degraded by presenilin associated rhomboid-like protease (PARL). In response to loss of mitochondrial membrane potential, PARL is inactivated, PINK1 is stabilized and recruits Parkin. Parkin ubiquitinates several mitochondrial associated proteins and they are then recognized by p62 and bring mitochondria to the autophagosomes. Thus mitochondria with membrane potential loss can be selectively degraded. Parkin substrates include voltage-dependent anion-selective channel protein (VDAC), components of the mitochondrial transport translocase of the outer membrane (TOM) complex, mitochondrial fission 1 (FIS1), mitochondrial Rho-GTPase (MIRO)1 and 2, and hexokinase, although whether ubiquitination of these proteins is either required or sufficient for mitophagy is unclear and highly dependent on the specific cellular context.
Parkin-mediated mitophagy may be regulated by additional factors including AF-6 (ALL-1 fusion partner from chromosome 6), FBXO7 (F-box only protein 7), SMURF1 (SMAD specific E3 ubiquitin protein ligase 1), ATPIF1 (ATPase inhibitory factor 1), AMBRA1 (activating molecule in Beclin 1-regulated autophagy 1), TOMM7 (translocase of outer mitochondrial membrane 7), SIAH7 (seven in absentia homolog), HSPA1L (Heat shock 70 kDa protein 1 L) and BAG4 (BCL2-Associated Athanogene 4) [80], [99], [100], [101], [102]. Deubiquitinase USP8 plays an important role in removing K6-linked ubiquitin from Parkin and subsequent recruitment of Parkin to the mitochondria [103]. In contrast, deubiquitinase USP30 and USP15 antagonize Parkin-mediated mitophagy by deubiquitinating Parkin substrates [104], [105]. Mitophagy can also be inhibited by caspase-1 mediated Parkin cleavage in response to the formation of NLRP3 (NOD-like receptor family, pyrin domain containing 3) inflammasomes [106]. BECN1, sterile α and TIR motif containing 1 (SARM1) and tumor necrosis factor receptor-associated factor 6 (TRAF6), may also play a role in mitophagy by interacting with PINK1 [107], [108].
Membrane source
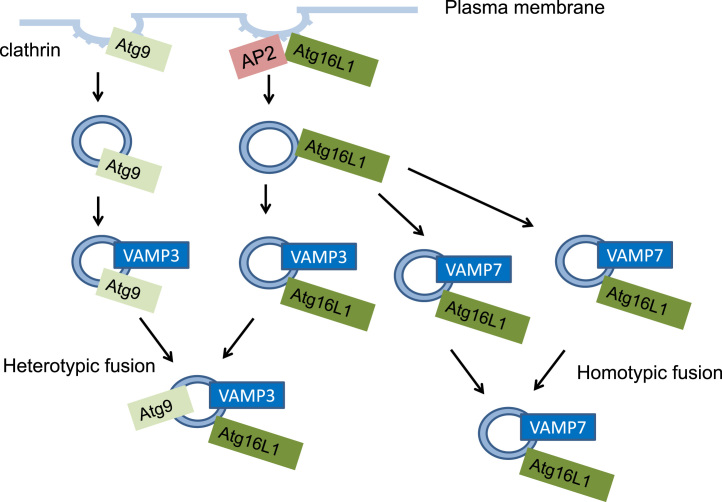
Where do autophagosomes come from? De novo synthesis of membrane materials has been proposed. However, recent studies also found that essentially all existing cellular membranes can contribute to the formation of autophagosomes, including plasma membrane, ER, Golgi, and mitochondrial membranes [109]. Localization of autophagosomal associated protein Atg16L1 to plasma membrane and interaction of Atg16L with assembly polypeptide 2 (AP2) and clathrin heavy chain have been found to occur prior to the formation of subsequent endosomal-like structures. Knockdown of clathrin heavy chain of AP2 decreased both basal and starvation-induced autophagosomal biogenesis [110]. The acquisition of LC3 by the plasma membrane-derived phagophore precursors requires homotypic fusion mediated by vesicle-associated membrane protein 7 (VAMP7) and partner soluble NSF attachment protein receptors (SNARES) [111]. Also internalized by clathrin-mediated endocytosis as Atg16L1, but in Atg16L1-free vesicles, is the transmembrane protein Atg9. Atg9- and Atg16L1-containing vesicles can undergo VAMP3-mediated heterotypic fusion, and this step also stimulates autophagosomal formation with plasma membrane as a contributing source [112] (Fig. 8).
Fig. 8.
Plasma membrane contributes to autophagosomal biogenesis. Localization of autophagosomal associated protein Atg16L1 to plasma membrane and interaction of Atg16L1 with assembly polypeptide 2 (AP2) and clathrin heavy chain have been found to occur prior to the formation of subsequent endosomal-like structures. Knockdown of clathrin heavy chain of AP2 decreased both basal and starvation-induced autophagosomal biogenesis. Atg9 is also internalized by clathrin-mediated endocytosis in Atg16L1-free vesicles. Atg9- and Atg16L1-containing vesicles can undergo VAMP3-mediated heterotypic fusion, and this step also stimulates autophagosomal formation with plasma membrane as a contributing source. Atg16L1-containing vesicles can undergo VAMP7-mediated homotypic fusion which precedes the acquisition of LC3 by the plasma membrane-derived phagophore precursors.
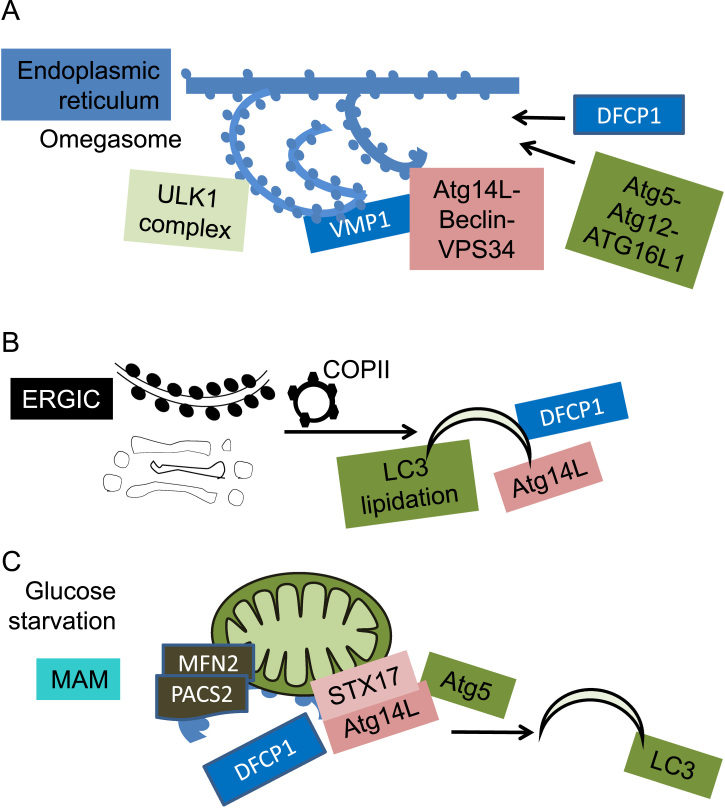
In addition to autophagy proteins Atg16L1 and Atg9 localizing to the plasma membrane and recruiting plasma membrane to formation of autophagosomes, double FYVE-containing protein 1 (DFCP1), a PI3P binding protein, has been found to localize to the ER and Golgi. In response to starvation, ULK complex and subsequently Atg14L moves to the ER at a site where vacuole membrane protein (VMP1) also transiently localizes. This is followed by DFCP1 moving to punctate structures called “omegasomes”, in a Beclin and VPS34 dependent manner, and colocalizes with LC3 and Atg5–Atg12–Atg16L1 complex. Then the formation of autophagosomes begins [113], [114], [115], [116]. Recent studies have also found that the VMP1–Beclin interaction plays an important role in autophagy induction [117] (Fig. 9A). Using a membrane fractionation approach, it has been demonstrated that the ER–Golgi intermediate compartment (ERGIC) which contains SEC22B (SEC22 vesicle trafficking protein homolog B), but not Atg9, RPN1 (ER marker), prohibitin1 (mitochondrial marker), PMP70 (peroxisome marker), LAMP2 (lysosomal membrane protein) or GM130 (cis-Golgi marker), is the most efficient membrane substrate for LC3 lipidation in an in vitro reconstitution experiment. Coat protein complex II (COPII) plays an important role in autophagosomal formation from ERGIC [118] (Fig. 9B). Glucose starvation also induces colocalization of LC3 and Atg5 with mitochondria, this localization has been shown to recruit mitochondrial lipids to autophagosomes. The mitochondria-associated ER membrane (MAM) fraction has been shown to contain Atg5 and DFCP1. MFN2 or phosphofurin acidic cluster sorting protein 2 (PACS2) are required for autophagosomal formation. Furthermore the SNARE protein syntaxin 17 (STX17) recruits Atg14L to the MAM sites [119], [120] (Fig. 9C).
Fig. 9.
ER and Golgi contribute to autophagosomal biogenesis. (A) ER donates membranes to autophagosomes. In response to starvation, ULK complex and subsequently Atg14L moves to the ER at a site where vacuole membrane protein (VMP1) also transiently localizes, followed by DFCP1 moving to punctate structures called “omegasomes”, in a Beclin and VPS34 dependent manner, and colocalizes with LC3 and Atg5–Atg12–Atg16L1 complex, this begins the formation of autophagosomes. Recent studies have also found that VMP1-Beclin interaction plays an important role in autophagy induction. (B) ER–Golgi intermediate complex compartment (ERGIC) promotes LC3 lipidation. Autophagosomal formation from ERGIC depends on coat protein complex II (COPII) vesicles. (C) The mitochondria-associated ER membrane (MAM) donates membranes to autophagosomes. Glucose starvation also induces colocalization of LC3 and Atg5 with mitochondria, this localization has been shown to recruit mitochondrial lipids to autophagosomes. Formation of autophagosomes from MAM requires MFN2 or phosphofurin acidic cluster sorting protein 2 (PACS2). The SNARE protein syntaxin 17 (STX17) recruits Atg14L to the MAM sites and further recruits DFCP1.
Autophagosome–lysosome fusion
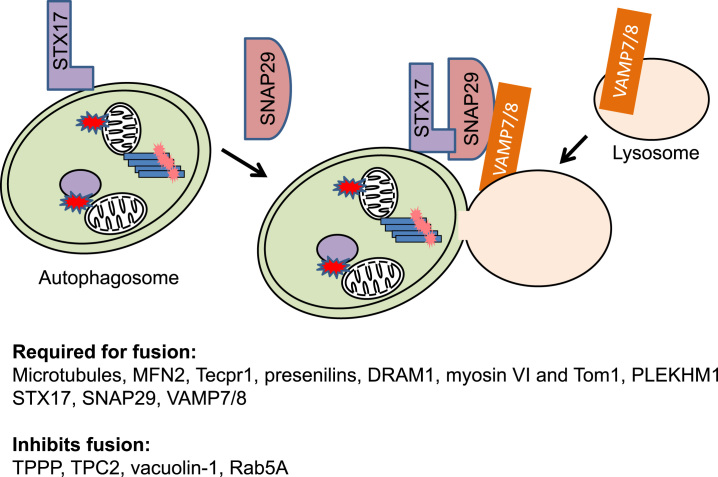
Degradation of autophagic cargo depends on lysosomal activities and therefore fusion of autophagosomes and lysosomes can be rate limiting in autophagic flux. It has been shown that mTOR localization to the lysosomes is an important regulator of autophagosomal–lysosomal fusion, and mTOR inhibition by Torin1 stimulates fusion [121]. It has been shown that microtubules [122], MFN2 [123], Tecpr1 [124], presenilins [125], DNA damage-regulated autophagy modulator 1 (DRAM1) [126], myosin VI and Tom1 [127], and pleckstrin homology domain containing protein family member 1 (PLEKHM1) [128], are required for or promote autophagosomal–lysosomal fusion. Tubulin polymerization-promoting protein (TPPP/p25α) [129], two pore channel 2 (TPC2) [130], and vacuolin-1 (by activating Rab5A) [131] inhibits autophagosomal–lysosomal fusion. Insertion of SNARE protein syntaxin 17 (STX17) to the autophagosomal membrane, binding to SNAP29 and complexing with lysosomal VAMP7/8, play important roles in the fusion step [132], [133] (Fig. 10). STX17 interaction with the homotypic fusion and protein sorting (HOPS)-tethering complex that consists of VPS33A, VPS16, VPS39 is required for this fusion [134].
Fig. 10.
Autophagosome–lysosome fusion. Fusion of autophagosomes and lysosomes depends on lysosomal VAMP7/8 and insertion of SNARE protein syntaxin 17 (STX17) to the autophagosomal membrane and binding SNAP29. Additional factors required or inhibit fusion are listed.
Methods for macroautophagy measurements
Macroautophagy can be assessed with a combination of methods including ultrastructural studies using electron microscopy (EM), degradation assays for measuring protein half-lives using radioisotope labels, assessment of fluorescence puncta with autophagy proteins fused to RFP or GFP, as well as western blot analyses of levels and modifications of autophagy proteins [135]. These methods are outlined briefly below:
Autophagic flux
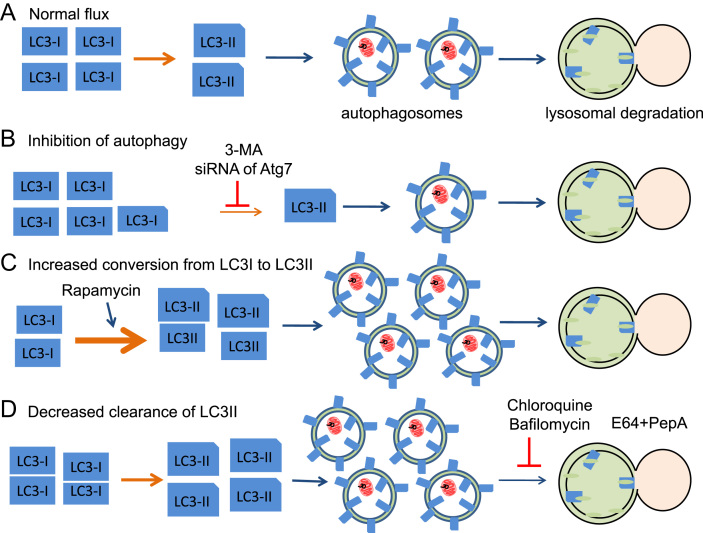
As discussed above, MAP1 LC3 is processed and lipidated by phosphatidylethanolamine (PE) and inserted into autophagosomal membranes [136]. It has been shown that the PE conjugated LC3, named LC3-II, migrates differently compared to the unconjugated LC3-I on SDS-PAGE and the levels of LC3-II is proportional to the amount of autophagosomes as assessed by electron microscopy studies [136]. Based on these properties, western blots to quantify LC3-II levels have been used to assess autophagosomal accumulation (Fig. 11A). Inhibition of autophagy prior to LC3-II generation, either by 3-methyladenine (3-MA) or by knockout/knockdown of Atg7 decreases LC3-II (Fig. 11B). Activation of autophagy either by starvation or by inhibitors of mTOR such as rapamycin increases LC3-II (Fig. 11C). However, if LC3-II levels are up in response to experimental conditions compared to controls, it can be due to increased conversion of LC3-I to LC3-II as in the case of starvation or inhibition of mTOR by rapamycin, or due to decreased clearance of LC3-II by the lysosomes. Chloroquine, bafilomycin, or ammonium chloride can change lysosomal pH and attenuate lysosomal degradation of autophagic cargo, inner membrane of autophagosomes, and LC3-II. E64 and Pepstatin A can inhibit lysosomal proteases and as a consequence, decrease clearance of autophagosomes (Fig. 11D). Therefore, to assess if a chemical or a genetic manipulation activates autophagy prior to LC3-I to LC3-II conversion, one needs to compare LC3-II in response to manipulation alone, lysosomal blockade alone (chloroquine, bafilomycin or E64+pepstatin A), and manipulation in the presence of lysosomal blockade. The relative steady-state ratio of LC3-I and LC3-II depends on cell type and experimental conditions. Different protein extraction methods and antibodies may affect the apparent ratio of LC3-I and LC3-II Western blot band intensities too.
Fig. 11.
Autophagy assessment by LC3-II levels. (A) During normal autophagic flux, LC3-I is conjugated with phosphatidylethanolamine to become LC3-II and inserts into autophagosomal membranes. LC3-II level has been found to be proportional to the amount of autophagosomes and thus has been used for autophagy assessment. (B) Inhibition of autophagy prior to LC3-II generation, either by 3-methyladenine (3-MA) or by knockout/knockdown of Atg7 decreases LC3-II. (C) Activation of autophagy either by starvation or by inhibitors of mTOR such as rapamycin increases LC3-II. (D) Inhibition of lysosomal clearance of autophagosomes, such as chloroquine, bafilomycin, E64+pepstatin A (PepA) also increases LC3-II accumulation. Therefore, to assess if a chemical or a genetic manipulation activates autophagy prior to LC3-I to LC3-II conversion, one approach is to compare LC3-II in response to manipulation alone, lysosomal blockade alone (chloroquine, bafilomycin or E64+pepstatin A), and manipulation in the presence of lysosomal blockade.
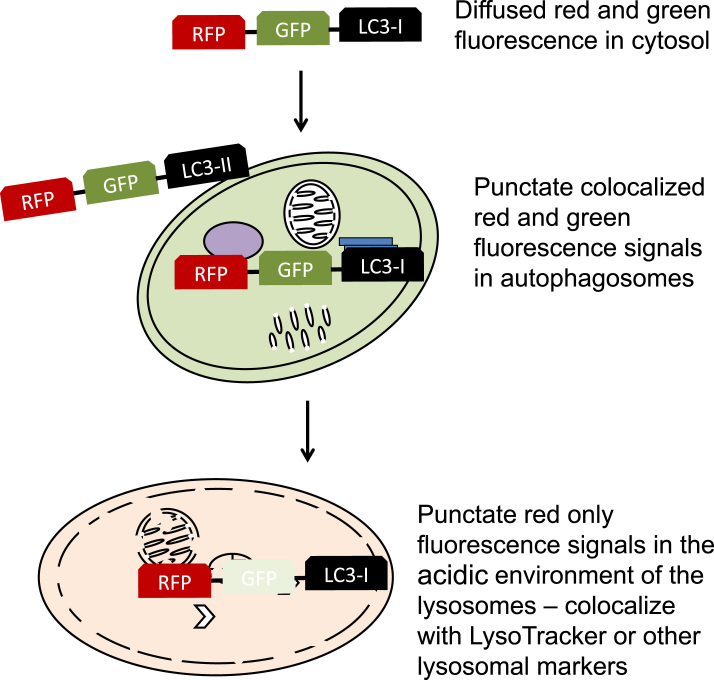
In addition to western blot analyses, LC3 localization to the autophagosomes can be visualized by immunostaining or by fluorescence microscopy of GFP-LC3 [136]. To provide assessment of autophagic flux from the autophagosomes to the lysosomes, the tfLC3 (tandem fluorescent tagged LC3, or RFP–GFP-LC3) method can be used [137]. The colocalized red and green fluorescence puncta represent the association of lipidated LC3-II with autophagosomes, with the number and intensity of the puncta correlating with the amount of autophagosomes present [136]. The red only puncta indicate the flux of RFP–GFP-LC3 protein into the acidic environment of the lysosomes, where the GFP but not the RFP fluorescence is quenched due to the differences in the pKa of GFP and RFP [137] (Fig. 12).
Fig. 12.
Autophagic flux assessment using RFP–GFP-LC3. To assess autophagic flux from the autophagosomes to the lysosomes, RFP–GFP-LC3 plasmids can be transfected into cells. Under conditions with minimum autophagosomal accumulation, RFP–GFP-LC3-I is diffused in the cytosol. When RFP–GFP-LC3-I is converted into RFP–GFP-LC3-II and inserted into autophagosomal membrane, RFP and GFP signals colocalize to punctate structures. As autophagosomes fuse with lysosomes, the outer membrane RFP–GFP-LC3-II is delipidated to RFP–GFP-LC3-I and diffused to the cytosolic space, the inner membrane RFP–GFP-LC3-II is quenched for the GFP signal but not the RFP signal. This is due to the lysosomal acidic environment that protonates the fluorophore of the GFP but not RFP because of differences in their pKa values. Therefore, the red only puncta indicate the flux of RFP–GFP-LC3-II protein into the acidic environment of the lysosomes.
Both GFP-LC3 and RFP–GFP-LC3 transgenic mice have been generated to facilitate assessment of autophagy in vivo [138], [139]. Using these mice, LC3 puncta can be observed in skeletal muscles and the heart in response to starvation [138]; a time dependent accumulation of RFP/GFP colocalization puncta followed by RFP only puncta have been observed in proximal tubular cells of the kidney in response to ischemia–reperfusion injury [139]. In vivo flux assays using lysosomal inhibitors in non-transgenic mice have also been used with systemic injection of chloroquine, bafilomycin, or leupeptin [140], [141]. Drawbacks to these approaches include the varied half-life and effective concentrations of the pharmacological probes in different tissues.
Ultrastructural studies
Electron microscopy is the first used visualization of autophagosomes. The advantage is that it can detect the different types and stages of autophagosomes, and the origin of the autophagosomal membranes [1], [142], [143]. The disadvantage is that it cannot easily sample large numbers of cells, or comparing multiple experimental conditions and time courses simultaneously.
Degradation of long lived protein
It was discovered that protein degradation rate in hepatocytes changes dramatically in response to insulin, glucagon, amino acid availability and lysosomotropic agents, and degradation is mediated largely by autophagy [61], [144], [145], [146], [147], [148], [149]. Since then, measurement of protein degradation rate using radiolabeled amino acid has been used to confirm and support many autophagy studies.
Mitophagy
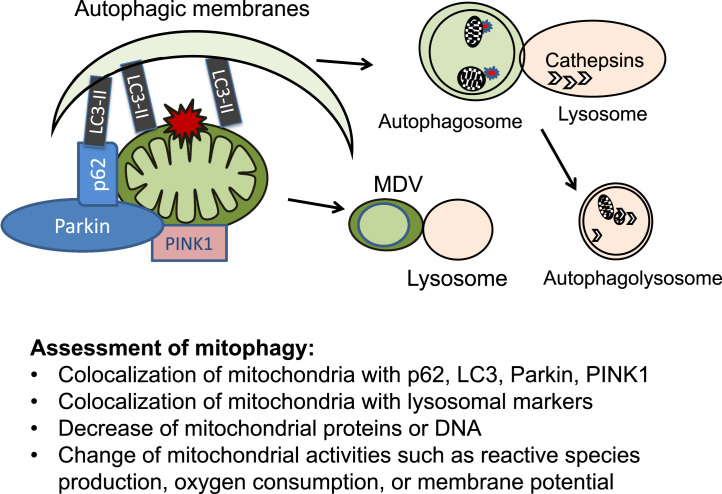
As macroautophagy, a variety of biochemical and cell biological methods can be used to assess mitophagy [85], [86], [87], [88], [93], [150], [151], [152], [153], [154], [155], [156], [157]. Mitophagy can be visualized by colocalization of mitochondria with mitophagic proteins. Depending on the steps and mechanisms of mitophagy, one can measure mitochondrial proteins (such as VDAC, MnSOD, or subunits of electron transport chain complexes) with LC3, p62, Parkin, or lysosomal protein LAMP1. Visualization of mitochondria and autophagosomes or mitochondria and lysosomes can also be facilitated by MitoTracker/GFP-LC3 or MitoTracker/LysoTracker colocalization provided neither mitochondria nor lysosomes lose their membrane potentials and the ability to take up the dyes under specific experimental conditions. Furthermore, loss of mitochondrial mass (such as loss of citrate synthase protein level and activities, loss of electron transport chain complexes, or loss of mitochondrial DNA) can also be used to assess mitochondrial clearance, provided biogenesis is unchanged. Finally, the consequence of mitophagy can be measured by mitochondrial activities including oxygen consumption, regulation of redox status, membrane potential or release of apoptosis signals [158], [159], [160] (Fig. 13).
Fig. 13.
Mitophagy assessment. Mitophagy is characterized by colocalization of autophagy proteins (such as p62, LC3, Parkin or PINK1) with the mitochondria; colocalization of mitochondria with lysosomal markers (either due to autophagosomal–lysosomal fusion or due to mitochondrial derived vesicles (MDV) fusing with the lysosomes). These characteristics can be used to assess mitophagy activities. In addition, the consequences of mitophagy can be assessed by either a decrease of mitochondrial proteins or DNA, or changes of mitochondrial activities such as reactive species production, oxygen consumption or changes of mitochondrial membrane potential.
Role of macroautophagy in health and diseases
Autophagy and mitophagy are highly regulated biological processes and their dysfunction have been found to contribute to all major diseases [10], [12], [24], [27], [80], [161], [162], [163], [164], [165], [166], [167], [168], [169], [170]. Representative evidence of their involvement in diseases are highlighted below focusing on cancer, aging, neurodegenerative diseases and infection (Fig. 14).
Fig. 14.
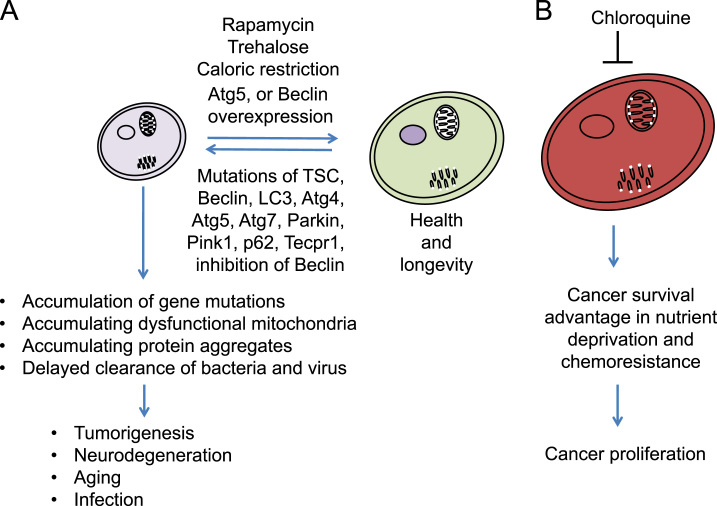
Role of autophagy and mitophagy in health and diseases. (A) Deficient autophagy due to inhibition of autophagic proteins or mutations of autophagy genes can lead to accumulation of gene mutations, dysfunctional mitochondria and protein aggregates, as well as inability to clear viral particles, as occurs in tumorigenesis, neurodegeneration, aging and infections. Autophagy induction, for example, by rapamycin, trehalose, caloric restriction, Atg5 or Beclin overexpression, has been shown to provide benefits to attenuate disease pathogenesis and extend lifespan. (B) Autophagy has also been shown to provide a detrimental role in established cancer by enhancing survival of cancer cells in nutrient deprived conditions or in response to chemotherapeutics. Inhibiting autophagy, for example by chloroquine treatment, has been investigated as a cancer therapeutic strategy.
Cancer
Cancer was the first disease connected to autophagy abnormalities. Beclin/Atg6 deficiency has been found to be associated with human breast, ovarian and prostate tumors [56], [171]. Beclin+/- mice develop tumors [56]. TSC1/2 that are mTOR inhibitors are known to be tumor suppressor proteins [172]. Atg4C, Atg5, and Atg7 genetic disruptions have been found to result in tumorigenesis in mice [173], [174]. Knockdown of GABARAPL1 in breast cancer cells has been found to increase cell proliferation [66]. It has been proposed that autophagy is required to guard against oxidative damage to the genome by clearance of dysfunctional mitochondria; however, once tumorigenic mutations have been established, autophagy provides a survival advantage to the tumor cells in nutrient deprived conditions and supports tumor cells chemoresistance. Thus, autophagy inhibitors, such as chloroquine and derivatives, has been tested in cancer therapy [175].
Aging
Impaired autophagy has been found to contribute to aging in model organisms [176], [177], [178]. Pharmacological or genetic manipulations that increase lifespan in model organisms often stimulate autophagy [177], [179], [180], [181], [182], [183], [184], [185], [186], [187]. Rapamycin, an mTOR inhibitor, as well as Atg5 overexpression in mice, have been shown to extend lifespan [179], [180], [181], [183]. It would be interesting to analyze autophagy in a library of cryopreserved “cell zoo” of fibroblasts from species of mammals and birds, which are notable because of their exceptionally long- or short-lives relative to their body size, such as shrews, bats, or a variety of rodents [188].
Neurodegenerative diseases
Accumulation of autophagosomes have been noted in aging and neurodegenerative diseases [189], [190]. Furthermore, accumulation of protein aggregates and dysfunctional mitochondria in neurodegenerative diseases corroborate with the idea that autophagy is insufficient in these pathologies. It has been found that Atg5 and Atg7 knockout mice accumulated ubiquitinated proteins and exhibit neurodegenerative pathologies in the brain [74], [75]. Beclin has been found to be decreased in Alzheimer's disease brains, its knockdown exacerbates and overexpression alleviates Alzheimer's disease-like pathology in mouse models [191]. Recessive gene mutations of Parkin or PINK1 cause a submit of familial Parkinson's disease [190]. Translating these ideas to potential treatment of neurodegenerative diseases, neuroprotection has been demonstrated by enhancers of autophagy by rapamycin [192] and trehalose [17] in animal models.
Infectious disease
Autophagy plays an important role in immune surveillance by clearing bacterial and viral proteins or particles. As discussed before, ubiquitinated bacteria can be recognized by p62, NDP52, or optineurin [193], [194], [195] (Fig. 6). Autophagic clearance of Shigella can also be mediated by binding of Tecpr1 with Atg5 and WIPI-2 to target Shigella to autophagosomes [196]. Evasion of autophagic degradation by Mycobacterium tuberculosis, Shigella flexineri, Liseria monocytogenes, or Coxiella burnetii due to binding to proteins that inhibit autophagy protein binding to the bacteria, secreting protein that inhibits autophagy induction by bacterial protein, directly blocking autophagosome maturation, or hijacking autophagic machinery for replication, respectively, plays a role in infection [197], [198], [199], [200]. Enhancement of autophagy inhibits M. tuberculosis intracellular survival [197], and inhibition of autophagy delays clearance of Sindbis virus infection [201]. Herpes, influenza and HIV contain Bcl-2 family of proteins that interact with Beclin, and inhibit autophagosomal formation or maturation [202], [203].
Future directions
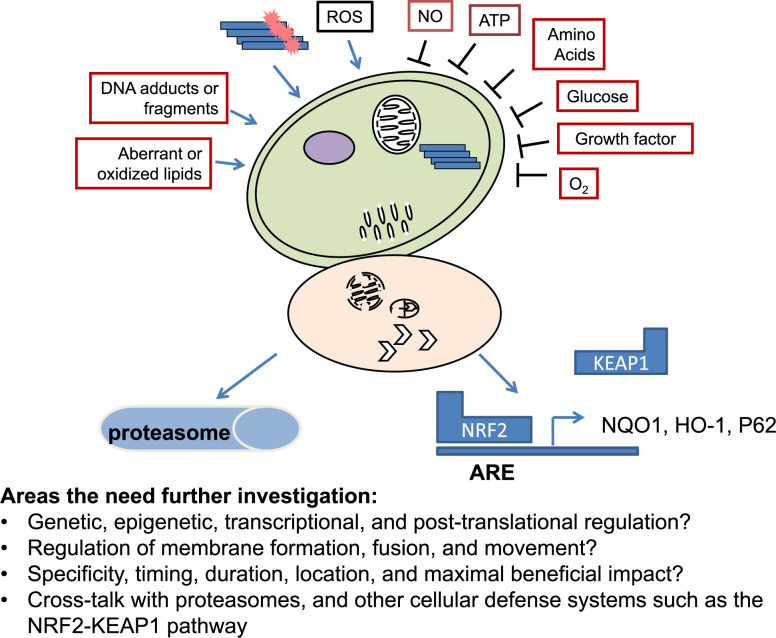
Autophagy is an essential cellular process involved in homeostasis and quality control of cellular constituents. Complex signaling, target recognition, vesicle formation, fusion and movement mechanisms interpose and influence the outcome of the process. A great deal is now known about the autophagy process and its contribution to normal and pathological conditions. Nonetheless there are still an infinite number of unanswered questions (Fig. 15). For example: How many different ways and exactly through which mechanisms do cellular damage induce autophagy, what are the signaling pathways sensing reactive species, old, excessive and toxic protein species, and what is the maximal capacity of autophagy that can be used to deal with perturbations of the cell? In addition, whether and how autophagy is regulated by epigenetic, transcription, and post-translational processes, and whether and how it is independent or inter-dependent on other cellular vesicles are unclear. Furthermore, how to enhance autophagosome–lysosomal fusion, what are rate-limiting steps in different cellular contexts and disease conditions, and how to enhance positive autophagy in these situations without off-target or undesirable negative outcomes are unknown. Last but not least, determining the extent and signaling mechanisms of the coordination and cross regulation of autophagy with the proteasomal and the NRF2-KEAP1 antioxidant defense pathways are important to our understanding of cellular and organismal homeostasis and adaptation to age, lifestyle and environmental exposures [11], [204], [205], [206], [207]. Research addressing these questions may therefore provide new insights into potential disease intervention.
Fig. 15.
Future directions. Although we now know that many different conditions (including growth factor, glucose, amino acids or ATP deprivation, deficient or excessive oxygen, reactive species, damage to DNA, lipid, proteins or organelles) may stimulate autophagy, the exactly mechanisms are unclear. Understanding the genetic, epigenetic, transcriptional, and post-translational regulation of autophagy, understanding the regulation of membrane formation, fusion and movement, and understanding the specificity, timing, duration, location of these regulations, understanding the cross regulation of autophagy with proteasomal and NRF2 transcription activities (in regulating expression of antioxidant proteins and p62), and understanding how to apply pharmacological or genetic interventions to achieve maximal beneficial impact without negative outcomes will be important in autophagy biology and its application to medicine.
Acknowledgment
This work was supported by NIH R01-NS064090 (to J.Z.).
References
- 1.De Duve C., Wattiaux R. Functions of lysosomes. Annual Review of Physiology. 1966;28:435–492. doi: 10.1146/annurev.ph.28.030166.002251. 5322983 [DOI] [PubMed] [Google Scholar]
- 2.Clark S.L., Jr. Cellular differentiation in the kidneys of newborn mice studies with the electron microscope. Journal of Biophysical and Biochemical Cytology. 1957;3(3):349–362. doi: 10.1083/jcb.3.3.349. 13438920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashford T.P., Porter K.R. Cytoplasmic components in hepatic cell lysosomes. Journal of Cell Biology. 1962;12:198–202. doi: 10.1083/jcb.12.1.198. 13862833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Novikoff A.B., Essner E. Cytolysomes and mitochondrial degeneration. Journal of Cell Biology. 1962;15:140–146. doi: 10.1083/jcb.15.1.140. 13939127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsukada M., Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Letters. 1993;333(1–2):169–174. doi: 10.1016/0014-5793(93)80398-e. 8224160 [DOI] [PubMed] [Google Scholar]
- 6.Takeshige K., Baba M., Tsuboi S., Noda T., Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. Journal of Cell Biology. 1992;119(2):301–311. doi: 10.1083/jcb.119.2.301. 1400575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang X.H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402(6762):672–676. doi: 10.1038/45257. 10604474 [DOI] [PubMed] [Google Scholar]
- 8.Klionsky D.J. Autophagy: from phenomenology to molecular understanding in less than a decade. Nature Reviews Molecular Cell Biology. 2007;8(11):931–937. doi: 10.1038/nrm2245. 17712358 [DOI] [PubMed] [Google Scholar]
- 9.Yang Z., Klionsky D.J. Eaten alive: a history of macroautophagy. Nature Cell Biology. 2010;12(9):814–822. doi: 10.1038/ncb0910-814. 20811353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dodson M., Darley-Usmar V., Zhang J. Cellular metabolic and autophagic pathways: traffic control by redox signaling. Free Radical Biology and Medicine. 2013;63:207–221. doi: 10.1016/j.freeradbiomed.2013.05.014. 23702245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levonen A.L., Hill B.G., Kansanen E., Zhang J., Darley-Usmar V.M. Redox regulation of antioxidants, autophagy, and the response to stress: implications for electrophile therapeutics. Free Radical Biology and Medicine. 2014;71:196–207. doi: 10.1016/j.freeradbiomed.2014.03.025. 24681256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giordano S., Darley-Usmar V., Zhang J. Autophagy as an essential cellular antioxidant pathway in neurodegenerative disease. Redox Biology. 2014;2:82–90. doi: 10.1016/j.redox.2013.12.013. 24494187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J. Autophagy and mitophagy in cellular damage control. Redox Biology. 2013;1(1):19–23. doi: 10.1016/j.redox.2012.11.008. 23946931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mijaljica D., Prescott M., Devenish R.J. Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy. 2011;7(7):673–682. doi: 10.4161/auto.7.7.14733. 21646866 [DOI] [PubMed] [Google Scholar]
- 15.Li W.W., Li J., Bao J.K. Microautophagy: lesser-known self-eating. Cellular and Molecular Life Sciences. 2012;69(7):1125–1136. doi: 10.1007/s00018-011-0865-5. 22080117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sattler T., Mayer A. Cell-free reconstitution of microautophagic vacuole invagination and vesicle formation. Journal of Cell Biology. 2000;151(3):529–538. doi: 10.1083/jcb.151.3.529. 11062255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uttenweiler A., Schwarz H., Mayer A. Microautophagic vacuole invagination requires calmodulin in a Ca2+-independent function. Journal of Biological Chemistry. 2005;280(39):33289–33297. doi: 10.1074/jbc.M506086200. 16055436 [DOI] [PubMed] [Google Scholar]
- 18.Schuck S., Gallagher C.M., Walter P. ER-phagy mediates selective degradation of endoplasmic reticulum independently of the core autophagy machinery. Journal of Cell Science. 2014;127(18):4078–4088. doi: 10.1242/jcs.154716. 25052096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mortimore G.E., Hutson N.J., Surmacz C.A. Quantitative correlation between proteolysis and macro- and microautophagy in mouse hepatocytes during starvation and refeeding. Proceedings of the National Academy of Sciences of the United States of America. 1983;80(8):2179–2183. doi: 10.1073/pnas.80.8.2179. 6340116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakai M., Ogawa K. Energy-dependent lysosomal wrapping mechanism (LWM) during autophagolysosome formation. Histochemistry. 1982;76(4):479–488. doi: 10.1007/BF00489903. 7166511 [DOI] [PubMed] [Google Scholar]
- 21.Sakai M., Araki N., Ogawa K. Lysosomal movements during heterophagy and autophagy: with special reference to nematolysosome and wrapping lysosome. Journal of Electron Microscopy Technique. 1989;12(2):101–131. doi: 10.1002/jemt.1060120206. 2668454 [DOI] [PubMed] [Google Scholar]
- 22.Sahu R., Kaushik S., Clement C.C., Cannizzo E.S., Scharf B., Follenzi A., Potolicchio I., Nieves E., Cuervo A.M., Santambrogio L. Microautophagy of cytosolic proteins by late endosomes. Developmental Cell. 2011;20(1):131–139. doi: 10.1016/j.devcel.2010.12.003. 21238931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaushik S., Bandyopadhyay U., Sridhar S., Kiffin R., Martinez-Vicente M., Kon M., Orenstein S.J., Wong E., Cuervo A.M. Chaperone-mediated autophagy at a glance. Journal of Cell Science. 2011;124(4):495–499. doi: 10.1242/jcs.073874. 21282471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee J., Giordano S., Zhang J. Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling. Biochemical Journal. 2012;441(2):523–540. doi: 10.1042/BJ20111451. 22187934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng Y., He D., Yao Z., Klionsky D.J. The machinery of macroautophagy. Cell Research. 2014;24(1):24–41. doi: 10.1038/cr.2013.168. 24366339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Settembre C., Di Malta C., Polito V.A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S.U., Huynh T., Medina D., Colella P., Sardiello M., Rubinsztein D.C., Ballabio A. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332(6036):1429–1433. doi: 10.1126/science.1204592. 21617040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wani W.Y., Boyer-Guittaut M., Dodson M., Chatham J., Darley-Usmar V., Zhang J. Regulation of autophagy by protein post-translational modification. Laboratory Investigation. 2015;95:14–25. doi: 10.1038/labinvest.2014.131. 25365205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dunlop E.A., Tee A.R. mTOR and autophagy: a dynamic relationship governed by nutrients and energy. Seminars in Cell and Developmental Biology. 2014;36C:121–129. doi: 10.1016/j.semcdb.2014.08.006. 25158238 [DOI] [PubMed] [Google Scholar]
- 29.Zhang J., Kim J., Alexander A., Cai S., Tripathi D.N., Dere R., Tee A.R., Tait-Mulder J., Di Nardo A., Han J.M., Kwiatkowski E., Dunlop E.A., Dodd K.M., Folkerth R.D., Faust P.L., Kastan M.B., Sahin M., Walker C.L. A tuberous sclerosis complex signalling node at the peroxisome regulates mTORC1 and autophagy in response to ROS. Nature Cell Biology. 2013;15(10):1186–1196. doi: 10.1038/ncb2822. 23955302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russell R.C., Yuan H.X., Guan K.L. Autophagy regulation by nutrient signaling. Cell Research. 2014;24(1):42–57. doi: 10.1038/cr.2013.166. 24343578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen H.M., Mizushima N. At the end of the autophagic road: an emerging understanding of lysosomal functions in autophagy. Trends in Biochemical Sciences. 2014;39(2):61–71. doi: 10.1016/j.tibs.2013.12.001. 24369758 [DOI] [PubMed] [Google Scholar]
- 32.Bar-Peled L., Schweitzer L.D., Zoncu R., Sabatini D.M. Ragulator Is a GEF for the rag GTPases that signal amino acid Levels to mTORC1. Cell. 2012;150(6):1196–1208. doi: 10.1016/j.cell.2012.07.032. 22980980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inoki K., Li Y., Zhu T., Wu J., Guan K.L. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nature Cell Biology. 2002;4(9):648–657. doi: 10.1038/ncb839. 12172553 [DOI] [PubMed] [Google Scholar]
- 34.Dunlop E.A., Tee A.R. The kinase triad, AMPK, mTORC1 and ULK1, maintains energy and nutrient homoeostasis. Biochemical Society Transactions. 2013;41(4):939–943. doi: 10.1042/BST20130030. 23863160 [DOI] [PubMed] [Google Scholar]
- 35.Zhao M., Klionsky D.J. AMPK-dependent phosphorylation of ULK1 induces autophagy. Cell Metabolism. 2011;13(2):119–120. doi: 10.1016/j.cmet.2011.01.009. 21284977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alers S., Löffler A.S., Wesselborg S., Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Molecular and Cellular Biology. 2012;32(1):2–11. doi: 10.1128/MCB.06159-11. 22025673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang C.S., Jiang B., Li M., Zhu M., Peng Y., Zhang Y.L., Wu Y.Q., Li T.Y., Liang Y., Lu Z., Lian G., Liu Q., Guo H., Yin Z., Ye Z., Han J., Wu J.W., Yin H., Lin S.Y., Lin S.C. The lysosomal V-ATPase-ragulator complex is a common activator for AMPK and mTORC1, acting as a switch between catabolism and anabolism. Cell Metabolism. 2014;20(3):526–540. doi: 10.1016/j.cmet.2014.06.014. 25002183 [DOI] [PubMed] [Google Scholar]
- 38.Semenza G.L. Oxygen sensing, hypoxia-inducible factors, and disease pathophysiology. Annual Review of Pathology. 2014;9:47–71. doi: 10.1146/annurev-pathol-012513-104720. 23937437 [DOI] [PubMed] [Google Scholar]
- 39.Li Y., Wang Y., Kim E., Beemiller P., Wang C.Y., Swanson J., You M., Guan K.L. Bnip3 mediates the hypoxia-induced inhibition on mammalian target of rapamycin by interacting with Rheb. Journal of Biological Chemistry. 2007;282(49):35803–35813. doi: 10.1074/jbc.M705231200. 17928295 [DOI] [PubMed] [Google Scholar]
- 40.Alexander A., Cai S.L., Kim J., Nanez A., Sahin M., MacLean K.H., Inoki K., Guan K.L., Shen J., Person M.D., Kusewitt D., Mills G.B., Kastan M.B., Walker C.L. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(9):4153–4158. doi: 10.1073/pnas.0913860107. 20160076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan H.X., Russell R.C., Guan K.L. Regulation of PIK3C3/VPS34 complexes by MTOR in nutrient stress-induced autophagy. Autophagy. 2013;9(12):1983–1995. doi: 10.4161/auto.26058. 24013218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Settembre C., Zoncu R., Medina D.L., Vetrini F., Erdin S., Erdin S., Huynh T., Ferron M., Karsenty G., Vellard M.C., Facchinetti V., Sabatini D.M., Ballabio A. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO Journal. 2012;31(5):1095–1108. doi: 10.1038/emboj.2012.32. 22343943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martina J.A., Chen Y., Gucek M., Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8(6):903–914. doi: 10.4161/auto.19653. 22576015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gangloff Y.G., Mueller M., Dann S.G., Svoboda P., Sticker M., Spetz J.F., Um S.H., Brown E.J., Cereghini S., Thomas G., Kozma S.C. Disruption of the mouse mTOR gene leads to early postimplantation lethality and prohibits embryonic stem cell development. Journal of Molecular Cell Biology. 2004;24(21):9508–9516. doi: 10.1128/MCB.24.21.9508-9516.2004. 15485918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheong H., Wu J., Gonzales L.K., Guttentag S.H., Thompson C.B., Lindsten T. Analysis of a lung defect in autophagy-deficient mouse strains. Autophagy. 2014;10(1):45–56. doi: 10.4161/auto.26505. 24275123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guertin D.A., Sabatini D.M. The pharmacology of mTOR inhibition. Science Signaling. 2009;2(67):e24. doi: 10.1126/scisignal.267pe24. 19383975 [DOI] [PubMed] [Google Scholar]
- 47.Kang R., Zeh H.J., Lotze M.T., Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death & Differentiation. 2011;18(4):571–580. doi: 10.1038/cdd.2010.191. 21311563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wirth M., Joachim J., Tooze S.A. Autophagosome formation − the role of ULK1 and Beclin1–PI3KC3 complexes in setting the stage. Seminars in Cancer Biology. 2013;23(5):301–309. doi: 10.1016/j.semcancer.2013.05.007. 23727157 [DOI] [PubMed] [Google Scholar]
- 49.Marsh S.A., Powell P.C., Dell’Italia L.J., Chatham J.C. Cardiac O-GlcNAcylation blunts autophagic signaling in the diabetic heart. Life Sciences. 2013;92(11):648–656. doi: 10.1016/j.lfs.2012.06.011. 22728715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu J., He L., Behrends C., Araki M., Araki K., Jun W.Q., Catanzaro J.M., Friedman S.L., Zong W.X., Fiel M.I., Li M., Yue Z. NRBF2 regulates autophagy and prevents liver injury by modulating Atg14L-linked phosphatidylinositol-3 kinase III activity. Nature Communications. 2014;5:3920. doi: 10.1038/ncomms4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Polson H.E., de Lartigue J., Rigden D.J., Reedijk M., Urbé S., Clague M.J., Tooze S.A. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6(4):506–522. doi: 10.4161/auto.6.4.11863. 20505359 [DOI] [PubMed] [Google Scholar]
- 52.Tooze S.A., Jefferies H.B., Kalie E., Longatti A., McAlpine F.E., McKnight N.C., Orsi A., Polson H.E., Razi M., Robinson D.J., Webber J.L. Trafficking and signaling in mammalian autophagy. IUBMB Life. 2010;62(7):503–508. doi: 10.1002/iub.334. 20552641 [DOI] [PubMed] [Google Scholar]
- 53.Filimonenko M., Isakson P., Finley K.D., Anderson M., Jeong H., Melia T.J., Bartlett B.J., Myers K.M., Birkeland H.C., Lamark T., Krainc D., Brech A., Stenmark H., Simonsen A., Yamamoto A. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Molecular Cell. 2010;38(2):265–279. doi: 10.1016/j.molcel.2010.04.007. 20417604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clausen T.H., Lamark T., Isakson P., Finley K., Larsen K.B., Brech A., Øvervatn A., Stenmark H., Bjørkøy G., Simonsen A., Johansen T. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy. 2010;6(3):330–344. doi: 10.4161/auto.6.3.11226. 20168092 [DOI] [PubMed] [Google Scholar]
- 55.Simonsen A., Birkeland H.C., Gillooly D.J., Mizushima N., Kuma A., Yoshimori T., Slagsvold T., Brech A., Stenmark H. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. Journal of Cell Science. 2004;117(18):4239–4251. doi: 10.1242/jcs.01287. 15292400 [DOI] [PubMed] [Google Scholar]
- 56.Yue Z., Jin S., Yang C., Levine A.J., Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(25):15077–15082. doi: 10.1073/pnas.2436255100. 14657337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jaber N., Dou Z., Chen J.S., Catanzaro J., Jiang Y.P., Ballou L.M., Selinger E., Ouyang X., Lin R.Z., Zhang J., Zong W.X. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(6):2003–2008. doi: 10.1073/pnas.1112848109. 22308354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jaber N., Dou Z., Lin R.Z., Zhang J., Zong W.X. Mammalian PIK3C3/VPS34: the key to autophagic processing in liver and heart. Autophagy. 2012;8(4):707–708. doi: 10.4161/auto.19627. 22498475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou X., Wang L., Hasegawa H., Amin P., Han B.X., Kaneko S., He Y., Wang F. Deletion of PIK3C3/Vps34 in sensory neurons causes rapid neurodegeneration by disrupting the endosomal but not the autophagic pathway. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(20):9424–9429. doi: 10.1073/pnas.0914725107. 20439739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou X., Wang F. Effects of neuronal PIK3C3/Vps34 deletion on autophagy and beyond. Autophagy. 2010;6(6):798–799. doi: 10.4161/auto.6.6.12511. 20562532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seglen P.O., Gordon P.B. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proceedings of the National Academy of Sciences of the United States of America. 1982;79(6):1889–1892. doi: 10.1073/pnas.79.6.1889. 6952238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu Y.T., Tan H.L., Shui G., Bauvy C., Huang Q., Wenk M.R., Ong C.N., Codogno P., Shen H.M. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. Journal of Biological Chemistry. 2010;285(14):10850–10861. doi: 10.1074/jbc.M109.080796. 20123989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O’Farrell F., Rusten T.E., Stenmark H. Phosphoinositide 3-kinases as accelerators and brakes of autophagy. FEBS Journal. 2013;280(24):6322–6337. doi: 10.1111/febs.12486. 23953235 [DOI] [PubMed] [Google Scholar]
- 64.Geng J., Klionsky D.J. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. Protein modifications: beyond the usual suspects. EMBO Reports. 2008;9:859–864. doi: 10.1038/embor.2008.163. 18704115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maccioni R.B., Cambiazo V. Role of microtubule-associated proteins in the control of microtubule assembly. Physiological Reviews. 1995;75(4):835–864. doi: 10.1152/physrev.1995.75.4.835. 7480164 [DOI] [PubMed] [Google Scholar]
- 66.Boyer-Guittaut M., Poillet L., Liang Q., Bôle-Richard E., Ouyang X., Benavides G.A., Chakrama F.Z., Fraichard A., Darley-Usmar V.M., Despouy G., Jouvenot M., Delage-Mourroux R., Zhang J. The role of GABARAPL1/GEC1 in autophagic flux and mitochondrial quality control in MDA-MB-436 breast cancer cells. Autophagy. 2014;10(6):986–1003. doi: 10.4161/auto.28390. 24879149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Le Grand J.N., Chakrama F.Z., Seguin-Py S., Fraichard A., Age-Mourroux R., Jouvenot M., Boyer-Guittaut M. GABARAPL1 (GEC1): original or copycat? Autophagy. 2011;7:1098–1107. doi: 10.4161/auto.7.10.15904. [DOI] [PubMed] [Google Scholar]
- 68.Chakrama F.Z., Seguin-Py S., Le Grand J.N., Fraichard A., Age-Mourroux R., Despouy G., Perez V., Jouvenot M., Boyer-Guittaut M. GABARAPL1 (GEC1) associates with autophagic vesicles. Autophagy. 2010;6:495–505. doi: 10.4161/auto.6.4.11819. [DOI] [PubMed] [Google Scholar]
- 69.Johansen T., Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7(3):279–296. doi: 10.4161/auto.7.3.14487. 21189453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rogov V., Dötsch V., Johansen T., Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Molecular Cell. 2014;53(2):167–178. doi: 10.1016/j.molcel.2013.12.014. 24462201 [DOI] [PubMed] [Google Scholar]
- 71.Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y.S., Ueno I., Sakamoto A., Tong K.I., Kim M., Nishito Y., Iemura S., Natsume T., Ueno T., Kominami E., Motohashi H., Tanaka K., Yamamoto M. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nature Cell Biology. 2010;12(3):213–223. doi: 10.1038/ncb2021. 20173742 [DOI] [PubMed] [Google Scholar]
- 72.Fujita K., Maeda D., Xiao Q., Srinivasula S.M. Nrf2-mediated induction of p62 controls toll-like receptor-4-driven aggresome-like induced structure formation and autophagic degradation. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(4):1427–1432. doi: 10.1073/pnas.1014156108. 21220332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zatloukal K., Stumptner C., Fuchsbichler A., Heid H., Schnoelzer M., Kenner L., Kleinert R., Prinz M., Aguzzi A., Denk H. p62 is a common component of cytoplasmic inclusions in protein aggregation diseases. American Journal of Pathology. 2002;160(1):255–263. doi: 10.1016/S0002-9440(10)64369-6. 11786419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441(7095):880–884. doi: 10.1038/nature04723. 16625205 [DOI] [PubMed] [Google Scholar]
- 75.Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441(7095):885–889. doi: 10.1038/nature04724. 16625204 [DOI] [PubMed] [Google Scholar]
- 76.Babu J.R., Geetha T., Wooten M.W. Sequestosome 1/p62 shuttles polyubiquitinated tau for proteasomal degradation. Journal of Neurochemistry. 2005;94(1):192–203. doi: 10.1111/j.1471-4159.2005.03181.x. 15953362 [DOI] [PubMed] [Google Scholar]
- 77.Rea S.L., Majcher V., Searle M.S., Layfield R. SQSTM1 mutations – bridging Paget disease of bone and ALS/FTLD. Experimental Cell Research. 2014;325(1):27–37. doi: 10.1016/j.yexcr.2014.01.020. 24486447 [DOI] [PubMed] [Google Scholar]
- 78.Schreiber A., Peter M. Substrate recognition in selective autophagy and the ubiquitin-proteasome system. Biochimica et Biophysica Acta. 2014;1843(1):163–181. doi: 10.1016/j.bbamcr.2013.03.019. 23545414 [DOI] [PubMed] [Google Scholar]
- 79.Mandell M.A., Jain A., Arko-Mensah J., Chauhan S., Kimura T., Dinkins C., Silvestri G., Münch J., Kirchhoff F., Simonsen A., Wei Y., Levine B., Johansen T., Deretic V. TRIM proteins regulate autophagy and can target autophagic substrates by direct recognition. Developmental Cell. 2014;30(4):394–409. doi: 10.1016/j.devcel.2014.06.013. 25127057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Redmann M., Dodson M., Boyer-Guittaut M., Darley-Usmar V., Zhang J. Mitophagy mechanisms and role in human diseases. International Journal of Biochemistry & Cell Biology. 2014;53:127–133. doi: 10.1016/j.biocel.2014.05.010. 24842106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lemasters J.J. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Research. 2005;8(1):3–5. doi: 10.1089/rej.2005.8.3. 15798367 [DOI] [PubMed] [Google Scholar]
- 82.Mitchell T., Chacko B., Ballinger S.W., Bailey S.M., Zhang J., Darley-Usmar V. Convergent mechanisms for dysregulation of mitochondrial quality control in metabolic disease: implications for mitochondrial therapeutics. Biochemical Society Transactions. 2013;41(1):127–133. doi: 10.1042/BST20120231. 23356271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim I., Lemasters J.J. Mitochondrial degradation by autophagy (mitophagy) in GFP-LC3 transgenic hepatocytes during nutrient deprivation. American Journal of Physiology—Cell Physiology. 2011;300(2):C308–C317. doi: 10.1152/ajpcell.00056.2010. 21106691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Twig G., Shirihai O.S. The interplay between mitochondrial dynamics and mitophagy. Antioxidants & Redox Signaling. 2011;14(10):1939–1951. doi: 10.1089/ars.2010.3779. 21128700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mitchell T., Johnson M.S., Ouyang X., Chacko B.K., Mitra K., Lei X., Gai Y., Moore D.R., Barnes S., Zhang J., Koizumi A., Ramanadham S., Darley-Usmar V.M. Dysfunctional mitochondrial bioenergetics and oxidative stress in Akita(+/Ins2)-derived β-cells. American Journal of Physiology—Endocrinology and Metabolism. 2013;305(5):E585–E599. doi: 10.1152/ajpendo.00093.2013. 23820623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Higdon A.N., Benavides G.A., Chacko B.K., Ouyang X., Johnson M.S., Landar A., Zhang J., Darley-Usmar V.M. Hemin causes mitochondrial dysfunction in endothelial cells through promoting lipid peroxidation: the protective role of autophagy. American Journal of Physiology—Heart and Circulatory Physiology. 2012;302(7):H1394–H1409. doi: 10.1152/ajpheart.00584.2011. 22245770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Giordano S., Dodson M., Ravi S., Redmann M., Ouyang X., Darley Usmar V.M., Zhang J. Bioenergetic adaptation in response to autophagy regulators during rotenone exposure. Journal of Neurochemistry. 2014;131:625–633. doi: 10.1111/jnc.12844. 25081478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chu C.T., Ji J., Dagda R.K., Jiang J.F., Tyurina Y.Y., Kapralov A.A., Tyurin V.A., Yanamala N., Shrivastava I.H., Mohammadyani D., Qiang Wang K.Z., Zhu J., Klein-Seetharaman J., Balasubramanian K., Amoscato A.A., Borisenko G., Huang Z., Gusdon A.M., Cheikhi A., Steer E.K., Wang R., Baty C., Watkins S., Bahar I., Bayir H. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nature Cell Biology. 2013;15(10):1197–1205. doi: 10.1038/ncb2837. 24036476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gilkerson R.W., de Vries R.L., Lebot P., Wikstrom J.D., Torgyekes E., Shirihai O.S., Przedborski S., Schon E.A. Mitochondrial autophagy in cells with mtDNA mutations results from synergistic loss of transmembrane potential and mTORC1 inhibition. Human Molecular Genetics. 2012;21(5):978–990. doi: 10.1093/hmg/ddr529. 22080835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Allen G.F., Toth R., James J., Ganley I.G. Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Reports. 2013;14(12):1127–1135. doi: 10.1038/embor.2013.168. 24176932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jin S.M., Youle R.J. The accumulation of misfolded proteins in the mitochondrial matrix is sensed by PINK1 to induce PARK2/Parkin-mediated mitophagy of polarized mitochondria. Autophagy. 2013;9(11):1750–1757. doi: 10.4161/auto.26122. 24149988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McLelland G.L., Soubannier V., Chen C.X., McBride H.M., Fon E.A. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO Journal. 2014;33(4):282–295. doi: 10.1002/embj.201385902. 24446486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dagda R.K., Zhu J., Kulich S.M., Chu C.T. Mitochondrially localized ERK2 regulates mitophagy and autophagic cell stress: implications for Parkinson's disease. Autophagy. 2008;4(6):770–782. doi: 10.4161/auto.6458. 18594198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Park S., Choi S.G., Yoo S.M., Son J.H., Jung Y.K. Choline dehydrogenase interacts with SQSTM1/p62 to recruit LC3 and stimulate mitophagy. Autophagy. 2014;10(11):1906–1920. doi: 10.4161/auto.32177. 25483962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ding W.X., Li M., Biazik J.M., Morgan D.G., Guo F., Ni H.M., Goheen M., Eskelinen E.L., Yin X.M. Electron microscopic analysis of a spherical mitochondrial structure. Journal of Biological Chemistry. 2012;287(50):42373–42378. doi: 10.1074/jbc.M112.413674. 23093403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yin X.M., Ding W.X. The reciprocal roles of PARK2 and mitofusins in mitophagy and mitochondrial spheroid formation. Autophagy. 2013;9(11):1687–1692. doi: 10.4161/auto.24871. 24162069 [DOI] [PubMed] [Google Scholar]
- 97.Soubannier V., Rippstein P., Kaufman B.A., Shoubridge E.A., McBride H.M. Reconstitution of mitochondria derived vesicle formation demonstrates selective enrichment of oxidized cargo. PLOS ONE. 2012;7(12):e52830. doi: 10.1371/journal.pone.0052830. 23300790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jin S.M., Youle R.J. PINK1- and Parkin-mediated mitophagy at a glance. Journal of Cell Science. 2012;125(4):795–799. doi: 10.1242/jcs.093849. 22448035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hasson S.A., Kane L.A., Yamano K., Huang C.H., Sliter D.A., Buehler E., Wang C., Heman-Ackah S.M., Hessa T., Guha R., Martin S.E., Youle R.J. High-content genome-wide RNAi screens identify regulators of Parkin upstream of mitophagy. Nature. 2013;504(7479):291–295. doi: 10.1038/nature12748. 24270810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lefebvre V., Du Q., Baird S., Ng A.C., Nascimento M., Campanella M., McBride H.M., Screaton R.A. Genome-wide RNAi screen identifies ATPase inhibitory factor 1 (ATPIF1) as essential for PARK2 recruitment and mitophagy. Autophagy. 2013;9(11):1770–1779. doi: 10.4161/auto.25413. 24005319 [DOI] [PubMed] [Google Scholar]
- 101.Orvedahl A., Sumpter R., Jr., Xiao G., Ng A., Zou Z., Tang Y., Narimatsu M., Gilpin C., Sun Q., Roth M., Forst C.V., Wrana J.L., Zhang Y.E., Luby-Phelps K., Xavier R.J., Xie Y., Levine B. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 2011;480(7375):113–117. doi: 10.1038/nature10546. 22020285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Van Humbeeck C., Cornelissen T., Hofkens H., Mandemakers W., Gevaert K., De Strooper B., Vandenberghe W. Parkin interacts with Ambra1 to induce mitophagy. Journal of Neuroscience. 2011;31(28):10249–10261. doi: 10.1523/JNEUROSCI.1917-11.2011. 21753002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Durcan T.M., Tang M.Y., Pérusse J.R., Dashti E.A., Aguileta M.A., McLelland G.L., Gros P., Shaler T.A., Faubert D., Coulombe B., Fon E.A. USP8 regulates mitophagy by removing K6-linked ubiquitin conjugates from Parkin. EMBO Journal. 2014;33:2473–2491. doi: 10.15252/embj.201489729. 25216678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bingol B., Tea J.S., Phu L., Reichelt M., Bakalarski C.E., Song Q., Foreman O., Kirkpatrick D.S., Sheng M. The mitochondrial deubiquitinase USP30 opposes Parkin-mediated mitophagy. Nature. 2014;510:370–375. doi: 10.1038/nature13418. 24896179 [DOI] [PubMed] [Google Scholar]
- 105.Cornelissen T., Haddad D., Wauters F., Van Humbeeck C., Mandemakers W., Koentjoro B., Sue C., Gevaert K., De Strooper B., Verstreken P., Vandenberghe W. The deubiquitinase USP15 antagonizes Parkin-mediated mitochondrial ubiquitination and mitophagy. Human Molecular Genetics. 2014;23(19):5227–5242. doi: 10.1093/hmg/ddu244. 24852371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yu J., Nagasu H., Murakami T., Hoang H., Broderick L., Hoffman H.M., Horng T. Inflammasome activation leads to caspase-1-dependent mitochondrial damage and block of mitophagy. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(43):15514–15519. doi: 10.1073/pnas.1414859111. 25313054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Michiorri S., Gelmetti V., Giarda E., Lombardi F., Romano F., Marongiu R., Nerini-Molteni S., Sale P., Vago R., Arena G., Torosantucci L., Cassina L., Russo M.A., Dallapiccola B., Valente E.M., Casari G. The Parkinson-associated protein PINK1 interacts with Beclin1 and promotes autophagy. Cell Death & Differentiation. 2010;17(6):962–974. doi: 10.1038/cdd.2009.200. 20057503 [DOI] [PubMed] [Google Scholar]
- 108.Murata H., Sakaguchi M., Kataoka K., Huh N.H. SARM1 and TRAF6 bind to and stabilize PINK1 on depolarized mitochondria. Molecular Biology of the Cell. 2013;24(18):2772–2784. doi: 10.1091/mbc.E13-01-0016. 23885119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rubinsztein D.C., Shpilka T., Elazar Z. Mechanisms of autophagosome biogenesis. Current Biology. 2012;22(1):R29–R34. doi: 10.1016/j.cub.2011.11.034. 22240478 [DOI] [PubMed] [Google Scholar]
- 110.Ravikumar B., Moreau K., Jahreiss L., Puri C., Rubinsztein D.C. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nature Cell Biology. 2010;12(8):747–757. doi: 10.1038/ncb2078. 20639872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Moreau K., Ravikumar B., Renna M., Puri C., Rubinsztein D.C. Autophagosome precursor maturation requires homotypic fusion. Cell. 2011;146(2):303–317. doi: 10.1016/j.cell.2011.06.023. 21784250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Puri C., Renna M., Bento C.F., Moreau K., Rubinsztein D.C. ATG16L1 meets ATG9 in recycling endosomes: additional roles for the plasma membrane and endocytosis in autophagosome biogenesis. Autophagy. 2014;10(1):182–184. doi: 10.4161/auto.27174. 24257061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Matsunaga K., Morita E., Saitoh T., Akira S., Ktistakis N.T., Izumi T., Noda T., Yoshimori T. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. Journal of Cell Biology. 2010;190(4):511–521. doi: 10.1083/jcb.200911141. 20713597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Axe E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A., Griffiths G., Ktistakis N.T. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. Journal of Cell Biology. 2008;182(4):685–701. doi: 10.1083/jcb.200803137. 18725538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hayashi-Nishino M., Fujita N., Noda T., Yamaguchi A., Yoshimori T., Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nature Cell Biology. 2009;11(12):1433–1437. doi: 10.1038/ncb1991. 19898463 [DOI] [PubMed] [Google Scholar]
- 116.Itakura E., Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6(6):764–776. doi: 10.4161/auto.6.6.12709. 20639694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Molejon M.I., Ropolo A., Re A.L., Boggio V., Vaccaro M.I. The VMP1–Beclin 1 interaction regulates autophagy induction. Scientific Reports. 2013;3:1055. doi: 10.1038/srep01055. 23316280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ge L., Melville D., Zhang M., Schekman R. The ER–Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. Elife. 2013;2:e00947. doi: 10.7554/eLife.00947. 23930225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hailey D.W., Rambold A.S., Satpute-Krishnan P., Mitra K., Sougrat R., Kim P.K., Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141(4):656–667. doi: 10.1016/j.cell.2010.04.009. 20478256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hamasaki M., Furuta N., Matsuda A., Nezu A., Yamamoto A., Fujita N., Oomori H., Noda T., Haraguchi T., Hiraoka Y., Amano A., Yoshimori T. Autophagosomes form at ER–mitochondria contact sites. Nature. 2013;495(7441):389–393. doi: 10.1038/nature11910. 23455425 [DOI] [PubMed] [Google Scholar]
- 121.Zhou J., Tan S.H., Nicolas V., Bauvy C., Yang N.D., Zhang J., Xue Y., Codogno P., Shen H.M. Activation of lysosomal function in the course of autophagy via mTORC1 suppression and autophagosome–lysosome fusion. Cell Research. 2013;23(4):508–523. doi: 10.1038/cr.2013.11. 23337583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Webb J.L., Ravikumar B., Rubinsztein D.C. Microtubule disruption inhibits autophagosome–lysosome fusion: implications for studying the roles of aggresomes in polyglutamine diseases. International Journal of Biochemistry & Cell Biology. 2004;36(12):2541–2550. doi: 10.1016/j.biocel.2004.02.003. 15325591 [DOI] [PubMed] [Google Scholar]
- 123.Zhao T., Huang X., Han L., Wang X., Cheng H., Zhao Y., Chen Q., Chen J., Cheng H., Xiao R., Zheng M. Central role of mitofusin 2 in autophagosome–lysosome fusion in cardiomyocytes. J. Biol. Chem. 2012;287(28):23615–23625. doi: 10.1074/jbc.M112.379164. 22619176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chen D., Fan W., Lu Y., Ding X., Chen S., Zhong Q. A mammalian autophagosome maturation mechanism mediated by TECPR1 and the Atg12–Atg5 conjugate. Mol. Cell. 2012;45(5):629–641. doi: 10.1016/j.molcel.2011.12.036. 22342342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Neely K.M., Green K.N. Presenilins mediate efficient proteolysis via the autophagosome-lysosome system. Autophagy. 2011;7(6):664–665. doi: 10.4161/auto.7.6.15448. 21460614 [DOI] [PubMed] [Google Scholar]
- 126.Zhang X.D., Qi L., Wu J.C., Qin Z.H. DRAM1 regulates autophagy flux through lysosomes. PLOS ONE. 2013;8(5):e63245. doi: 10.1371/journal.pone.0063245. 23696801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tumbarello D.A., Waxse B.J., Arden S.D., Bright N.A., Kendrick-Jones J., Buss F. Autophagy receptors link myosin VI to autophagosomes to mediate Tom1-dependent autophagosome maturation and fusion with the lysosome. Nature Cell Biology. 2012;14(10):1024–1035. doi: 10.1038/ncb2589. 23023224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.McEwan D., Popovic D., Gubas A., Terawaki S., Suzuki H., Stadel D., Coxon F., Miranda de Stegmann D., Bhogaraju S., Maddi K., Kirchof A., Gatti E., Helfrich M., Wakatsuki S., Behrends C., Pierre P., Dikic I. PLEKHM1 regulates autophagosome–lysosome fusion through HOPS complex and LC3/GABARAP proteins. Molecular Cell. 2014 doi: 10.1016/j.molcel.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 129.Ejlerskov P., Rasmussen I., Nielsen T.T., Bergström A.L., Tohyama Y., Jensen P.H., Vilhardt F. Tubulin polymerization-promoting protein (TPPP/p25α) promotes unconventional secretion of α-synuclein through exophagy by impairing autophagosome–lysosome fusion. Journal of Biological Chemistry. 2013;288(24):17313–17335. doi: 10.1074/jbc.M112.401174. 23629650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lu Y., Hao B.X., Graeff R., Wong C.W., Wu W.T., Yue J. Two pore channel 2 (TPC2) inhibits autophagosomal–lysosomal fusion by alkalinizing lysosomal pH. Journal of Biological Chemistry. 2013;288(33):24247–24263. doi: 10.1074/jbc.M113.484253. 23836916 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 131.Lu Y., Dong S., Hao B., Li C., Zhu K., Guo W., Wang Q., Cheung K.H., Wong C.W., Wu W.T., Markus H., Yue J. Vacuolin-1 potently and reversibly inhibits autophagosome–lysosome fusion by activating RAB5A. Autophagy. 2014;10(11):1895–1905. doi: 10.4161/auto.32200. 25483964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Itakura E., Kishi-Itakura C., Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151(6):1256–1269. doi: 10.1016/j.cell.2012.11.001. 23217709 [DOI] [PubMed] [Google Scholar]
- 133.Furuta N., Fujita N., Noda T., Yoshimori T., Amano A. Combinational soluble N-ethylmaleimide-sensitive factor attachment protein receptor proteins VAMP8 and Vti1b mediate fusion of antimicrobial and canonical autophagosomes with lysosomes. Molecular Biology of the Cell. 2010;21(6):1001–1010. doi: 10.1091/mbc.E09-08-0693. 20089838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jiang P., Nishimura T., Sakamaki Y., Itakura E., Hatta T., Natsume T., Mizushima N. The HOPS complex mediates autophagosome–lysosome fusion through interaction with syntaxin 17. Molecular Biology of the Cell. 2014;25(8):1327–1337. doi: 10.1091/mbc.E13-08-0447. 24554770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Klionsky D.J., Abdalla F.C., Abeliovich H., Abraham R.T., Acevedo-Arozena A., Adeli K., Agholme L., Agnello M., Agostinis P., Aguirre-Ghiso J.A., Ahn H.J. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8(4):445–544. doi: 10.4161/auto.19496. 22966490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO Journal. 2000;19(21):5720–5728. doi: 10.1093/emboj/19.21.5720. 11060023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kimura S., Noda T., Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3(5):452–460. doi: 10.4161/auto.4451. 17534139 [DOI] [PubMed] [Google Scholar]
- 138.Mizushima N. Methods for monitoring autophagy using GFP-LC3 transgenic mice. Methods in Enzymology. 2009;452:13–23. doi: 10.1016/S0076-6879(08)03602-1. 19200873 [DOI] [PubMed] [Google Scholar]
- 139.Li L., Wang Z.V., Hill J.A., Lin F. New autophagy reporter mice reveal dynamics of proximal tubular autophagy. Journal of the American Society of Nephrology. 2014;25(2):305–315. doi: 10.1681/ASN.2013040374. 24179166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zois C.E., Giatromanolaki A., Sivridis E., Papaiakovou M., Kainulainen H., Koukourakis M.I. “Autophagic flux” in normal mouse tissues: focus on endogenous LC3A processing. Autophagy. 2011;7(11):1371–1378. doi: 10.4161/auto.7.11.16664. 21997374 [DOI] [PubMed] [Google Scholar]
- 141.Haspel J., Shaik R.S., Ifedigbo E., Nakahira K., Dolinay T., Englert J.A., Choi A.M. Characterization of macroautophagic flux in vivo using a leupeptin-based assay. Autophagy. 2011;7(6):629–642. doi: 10.4161/auto.7.6.15100. 21460622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Eskelinen E.L., Reggiori F., Baba M., Kovács A.L., Seglen P.O. Seeing is believing: the impact of electron microscopy on autophagy research. Autophagy. 2011;7(9):935–956. doi: 10.4161/auto.7.9.15760. 21566462 [DOI] [PubMed] [Google Scholar]
- 143.Zakeri Z., Melendez A., Lockshin R.A. Detection of autophagy in cell death. Methods in Enzymology. 2008;442:289–306. doi: 10.1016/S0076-6879(08)01415-8. 18662576 [DOI] [PubMed] [Google Scholar]
- 144.Seglen P.O., Gordon P.B. Effects of lysosomotropic monoamines, diamines, amino alcohols, and other amino compounds on protein degradation and protein synthesis in isolated rat hepatocytes. Molecular Pharmacology. 1980;18(3):468–475. 7464813 [PubMed] [Google Scholar]
- 145.Seglen P.O., Gordon P.B., Poli A. Amino acid inhibition of the autophagic/lysosomal pathway of protein degradation in isolated rat hepatocytes. Biochimica et Biophysica Acta. 1980;630(1):103–118. doi: 10.1016/0304-4165(80)90141-5. 7388042 [DOI] [PubMed] [Google Scholar]
- 146.Gordon P.B., Seglen P.O. Autophagic sequestration of [14C]sucrose, introduced into rat hepatocytes by reversible electro-permeabilization. Experimental Cell Research. 1982;142(1):1–14. doi: 10.1016/0014-4827(82)90402-5. 7140848 [DOI] [PubMed] [Google Scholar]
- 147.Seglen P.O., Gordon P.B. Amino acid control of autophagic sequestration and protein degradation in isolated rat hepatocytes. Journal of Cell Biology. 1984;99(2):435–444. doi: 10.1083/jcb.99.2.435. 6746735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Seglen P.O., Gordon P.B., Tolleshaug H., Høyvik H. Autophagy and protein degradation in isolated rat hepatocytes. Biochemical Society Transactions. 1985;13(6):1007–1010. doi: 10.1042/bst0131007. 4092819 [DOI] [PubMed] [Google Scholar]
- 149.Seglen P.O., Gordon P.B., Tolleshaug H., Høyvik H. Pathways of intracellular sequestration and protein degradation in isolated rat hepatocytes. Progress in Clinical Biological Research. 1985;180:437–446. 4034550 [PubMed] [Google Scholar]
- 150.Rodriguez-Enriquez S., Kim I., Currin R.T., Lemasters J.J. Tracker dyes to probe mitochondrial autophagy (mitophagy) in rat hepatocytes. Autophagy. 2006;2(1):39–46. doi: 10.4161/auto.2229. 16874071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ding W.X., Yin X.M. Mitophagy: mechanisms, pathophysiological roles, and analysis. Biological Chemistry. 2012;393(7):547–564. doi: 10.1515/hsz-2012-0119. 22944659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chu C.T., Zhu J., Dagda R. Beclin 1-independent pathway of damage-induced mitophagy and autophagic stress: implications for neurodegeneration and cell death. Autophagy. 2007;3(6):663–666. doi: 10.4161/auto.4625. 17622797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dagda R.K., Cherra S.J., III, Kulich S.M., Tandon A., Park D., Chu C.T. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. Journal of Biological Chemistry. 2009;284(20):13843–13855. doi: 10.1074/jbc.M808515200. 19279012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Cherra S.J., III, Dagda R.K., Tandon A., Chu C.T. Mitochondrial autophagy as a compensatory response to PINK1 deficiency. Autophagy. 2009;5(8):1213–1214. doi: 10.4161/auto.5.8.10050. 19786829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Narendra D., Tanaka A., Suen D.F., Youle R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. Journal of Cell Biology. 2008;183(5):795–803. doi: 10.1083/jcb.200809125. 19029340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Narendra D., Kane L.A., Hauser D.N., Fearnley I.M., Youle R.J. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy. 2010;6:1090–1106. doi: 10.4161/auto.6.8.13426. 20890124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Narendra D.P., Jin S.M., Tanaka A., Suen D.F., Gautier C.A., Shen J., Cookson M.R., Youle R.J. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLOS Biology. 2010;8(1) doi: 10.1371/journal.pbio.1000298. 20126261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hill B.G., Benavides G.A., Lancaster J.R., Jr., Ballinger S., Dell’Italia L., Jianhua Z., Darley-Usmar V.M. Integration of cellular bioenergetics with mitochondrial quality control and autophagy. Biological Chemistry. 2012;393(12):1485–1512. doi: 10.1515/hsz-2012-0198. 23092819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chacko B.K., Kramer P.A., Ravi S., Johnson M.S., Hardy R.W., Ballinger S.W., Darley-Usmar V.M. Methods for defining distinct bioenergetic profiles in platelets, lymphocytes, monocytes, and neutrophils, and the oxidative burst from human blood. Laboratory Investigation. 2013;93(6):690–700. doi: 10.1038/labinvest.2013.53. 23528848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chacko B.K., Kramer P.A., Ravi S., Benavides G.A., Mitchell T., Dranka B.P., Ferrick D., Singal A.K., Ballinger S.W., Bailey S.M., Hardy R.W., Zhang J., Zhi D., Darley-Usmar V.M. The Bioenergetic Health Index: a new concept in mitochondrial translational research. Clinical Science (London) 2014;127(6):367–373. doi: 10.1042/CS20140101. 24895057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Rubinsztein D.C., Codogno P., Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nature Reviews Drug Discovery. 2012;11(9):709–730. doi: 10.1038/nrd3802. 22935804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Choi A.M., Ryter S.W., Levine B. Autophagy in human health and disease. New England Journal of Medicine. 2013;368(19):1845–1846. doi: 10.1056/NEJMc1303158. 23656658 [DOI] [PubMed] [Google Scholar]
- 163.Murrow L., Debnath J. Autophagy as a stress-response and quality-control mechanism: implications for cell injury and human disease. Annual Review of Pathology. 2013;8:105–137. doi: 10.1146/annurev-pathol-020712-163918. 23072311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Allison D.B., Antoine L.H., Ballinger S.W., Bamman M.M., Biga P., Darley-Usmar V.M., Fisher G., Gohlke J.M., Halade G.V., Hartman J.L., Hunter G.R., Messina J.L., Nagy T.R., Plaisance E.P., Powell M.L., Roth K.A., Sandel M.W., Schwartz T.S., Smith D.L., Sweatt J.D., Tollefsbol T.O., Watts S.A., Yang Y., Zhang J., Austad S.N. Aging and energetics’ ‘top 40’ future research opportunities 2010–2013. F1000 Research. 2014;3:219. doi: 10.12688/f1000research.5212.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lavandero S., Troncoso R., Rothermel B.A., Martinet W., Sadoshima J., Hill J.A. Cardiovascular autophagy: concepts, controversies, and perspectives. Autophagy. 2013;9(10):1455–1466. doi: 10.4161/auto.25969. 23959233 [DOI] [PubMed] [Google Scholar]
- 166.Ni H.M., Williams J.A., Jaeschke H., Ding W.X. Zonated induction of autophagy and mitochondrial spheroids limits acetaminophen-induced necrosis in the liver. Redox Biology. 2013;1(1):427–432. doi: 10.1016/j.redox.2013.08.005. 24191236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lavallard V.J., Gual P. Autophagy and non-alcoholic fatty liver disease. BioMed Research International. 2014;2014:120179. doi: 10.1155/2014/120179. 25295245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Mallat A., Lodder J., Teixeira-Clerc F., Moreau R., Codogno P., Lotersztajn S. Autophagy: a multifaceted partner in liver fibrosis. BioMed Research International. 2014;2014:869390. doi: 10.1155/2014/869390. 25254217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lee M. Role of islet beta cell autophagy in the pathogenesis of diabetes. Trends in Endocrinology & Metabolism. 2014;25:620–627. doi: 10.1016/j.tem.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 170.Heath J.M., Sun Y., Yuan K., Bradley W.E., Litovsky S., Dell’Italia L.J., Chatham J.C., Wu H., Chen Y. Activation of AKT by O-Linked N-acetylglucosamine induces vascular calcification in diabetes mellitus. Circulation Research. 2014;114(7):1094–1102. doi: 10.1161/CIRCRESAHA.114.302968. 24526702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Qu X., Yu J., Bhagat G., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E.L., Mizushima N., Ohsumi Y., Cattoretti G., Levine B. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. Journal of Clinical Investigation. 2003;112(12):1809–1820. doi: 10.1172/JCI20039. 14638851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Inoki K., Corradetti M.N., Guan K.L. Dysregulation of the TSC-mTOR pathway in human disease. Nature Genetics. 2005;37(1):19–24. doi: 10.1038/ng1494. 15624019 [DOI] [PubMed] [Google Scholar]
- 173.Mariño G., Salvador-Montoliu N., Fueyo A., Knecht E., Mizushima N., López-Otín C. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in ATG4C/autophagin-3. Journal of Biological Chemistry. 2007;282:18573–18583. doi: 10.1074/jbc.M701194200. 17442669 [DOI] [PubMed] [Google Scholar]
- 174.Takamura A., Komatsu M., Hara T., Sakamoto A., Kishi C., Waguri S., Eishi Y., Hino O., Tanaka K., Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes & Development. 2011;25(8):795–800. doi: 10.1101/gad.2016211. 21498569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Janku F., McConkey D.J., Hong D.S., Kurzrock R. Autophagy as a target for anticancer therapy. Nature Reviews Clinical Oncology. 2011;8:528–539. doi: 10.1038/nrclinonc.2011.71. 21587219 [DOI] [PubMed] [Google Scholar]
- 176.Takacs-Vellai K., Vellai T., Puoti A., Passannante M., Wicky C., Streit A., Kovacs A.L., Müller F. Inactivation of the autophagy gene bec-1 triggers apoptotic cell death in C. elegans. Current Biology. 2005;15(16):1513–1517. doi: 10.1016/j.cub.2005.07.035. 16111945 [DOI] [PubMed] [Google Scholar]
- 177.Juhász G., Erdi B., Sass M., Neufeld T.P. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in drosophila. Genes & Development. 2007;21(23):3061–3066. doi: 10.1101/gad.1600707. 18056421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Tóth M.L., Sigmond T., Borsos E., Barna J., Erdélyi P., Takács-Vellai K., Orosz L., Kovács A.L., Csikós G., Sass M., Vellai T. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;4(3):330–338. doi: 10.4161/auto.5618. 18219227 [DOI] [PubMed] [Google Scholar]
- 179.Harrison D.E., Strong R., Sharp Z.D., Nelson J.F., Astle C.M., Flurkey K., Nadon N.L., Wilkinson J.E., Frenkel K., Carter C.S., Pahor M., Javors M.A., Fernandez E., Miller R.A. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460(7253):392–395. doi: 10.1038/nature08221. 19587680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bjedov I., Partridge L. A longer and healthier life with TOR down-regulation: genetics and drugs. Biochemical Society Transactions. 2011;39(2):460–465. doi: 10.1042/BST0390460. 21428920 [DOI] [PubMed] [Google Scholar]
- 181.Zhang Y., Bokov A., Gelfond J., Soto V., Ikeno Y., Hubbard G., Diaz V., Sloane L., Maslin K., Treaster S., Réndon S., van Remmen H., Ward W., Javors M., Richardson A., Austad S.N., Fischer K. Rapamycin extends life and health in C57BL/6 mice. Journals of Gerontology Series A: Biological Sciences and Medical Science. 2014;69(2):119–130. doi: 10.1093/gerona/glt056. 23682161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Vellai T., Takács-Vellai K., Sass M., Klionsky D.J. The regulation of aging: does autophagy underlie longevity? Trends in Cell Biology. 2009;19(10):487–494. doi: 10.1016/j.tcb.2009.07.007. 19726187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mai S., Muster B., Bereiter-Hahn J., Jendrach M. Autophagy proteins LC3B, ATG5 and ATG12 participate in quality control after mitochondrial damage and influence lifespan. Autophagy. 2012;8(1):47–62. doi: 10.4161/auto.8.1.18174. 22170153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Pyo J.O., Yoo S.M., Ahn H.H., Nah J., Hong S.H., Kam T.I., Jung S., Jung Y.K. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nature Communications. 2013;4:2300. doi: 10.1038/ncomms3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Madeo F., Tavernarakis N., Kroemer G. Can autophagy promote longevity? Nature Cell Biology. 2010;12(9):842–846. doi: 10.1038/ncb0910-842. 20811357 [DOI] [PubMed] [Google Scholar]
- 186.Vellai T. Autophagy genes and ageing. Cell Death & Differentiation. 2009;16(1):94–102. doi: 10.1038/cdd.2008.126. 19079287 [DOI] [PubMed] [Google Scholar]
- 187.Morselli E., Maiuri M.C., Markaki M., Megalou E., Pasparaki A., Palikaras K., Criollo A., Galluzzi L., Malik S.A., Vitale I., Michaud M., Madeo F., Tavernarakis N., Kroemer G. Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy. Cell Death & Disease. 2010;1:e10. doi: 10.1038/cddis.2009.8. 21364612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Austad S.N. Cats, “rats”, and bats: the comparative biology of aging in the 21st century. Integrative and Comparative Biology. 2010;50(5):783–792. doi: 10.1093/icb/icq131. 21558241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Anglade P., Vyas S., Javoy-Agid F., Herrero M.T., Michel P.P., Marquez J., Mouatt-Prigent A., Ruberg M., Hirsch E.C., Agid Y. Apoptosis and autophagy in nigral neurons of patients with Parkinson's disease. Histology and Histopathology. 1997;12(1):25–31. 9046040 [PubMed] [Google Scholar]
- 190.Nixon R.A. The role of autophagy in neurodegenerative disease. Nature Medicine. 2013;19(8):983–997. doi: 10.1038/nm.3232. 23921753 [DOI] [PubMed] [Google Scholar]
- 191.Pickford F., Masliah E., Britschgi M., Lucin K., Narasimhan R., Jaeger P.A., Small S., Spencer B., Rockenstein E., Levine B., Wyss-Coray T. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. Journal of Clinical Investigation. 2008;118(6):2190–2199. doi: 10.1172/JCI33585. 18497889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Hochfeld W.E., Lee S., Rubinsztein D.C. Therapeutic induction of autophagy to modulate neurodegenerative disease progression. Acta Pharmacologica Sinica. 2013;34(5):600–604. doi: 10.1038/aps.2012.189. 23377551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Zheng Y.T., Shahnazari S., Brech A., Lamark T., Johansen T., Brumell J.H. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. Journal of Immunology. 2009;183(9):5909–5916. doi: 10.4049/jimmunol.0900441. 19812211 [DOI] [PubMed] [Google Scholar]
- 194.Thurston T.L., Ryzhakov G., Bloor S., von Muhlinen N., Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nature Immunology. 2009;10(11):1215–1221. doi: 10.1038/ni.1800. 19820708 [DOI] [PubMed] [Google Scholar]
- 195.Wild P., Farhan H., McEwan D.G., Wagner S., Rogov V.V., Brady N.R., Richter B., Korac J., Waidmann O., Choudhary C., Dötsch V., Bumann D., Dikic I. Phosphorylation of the autophagy receptor optineurin restricts salmonella growth. Science. 2011;333(6039):228–233. doi: 10.1126/science.1205405. 21617041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ogawa M., Yoshikawa Y., Kobayashi T., Mimuro H., Fukumatsu M., Kiga K., Piao Z., Ashida H., Yoshida M., Kakuta S., Koyama T., Goto Y., Nagatake T., Nagai S., Kiyono H., Kawalec M., Reichhart J.M., Sasakawa C. A Tecpr1-dependent selective autophagy pathway targets bacterial pathogens. Cell Host & Microbe. 2011;9(5):376–389. doi: 10.1016/j.chom.2011.04.010. 21575909 [DOI] [PubMed] [Google Scholar]
- 197.Gutierrez M.G., Master S.S., Singh S.B., Taylor G.A., Colombo M.I., Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119(6):753–766. doi: 10.1016/j.cell.2004.11.038. 15607973 [DOI] [PubMed] [Google Scholar]
- 198.Ogawa M., Yoshimori T., Suzuki T., Sagara H., Mizushima N., Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307(5710):727–731. doi: 10.1126/science.1106036. 15576571 [DOI] [PubMed] [Google Scholar]
- 199.Yoshikawa Y., Ogawa M., Hain T., Yoshida M., Fukumatsu M., Kim M., Mimuro H., Nakagawa I., Yanagawa T., Ishii T., Kakizuka A., Sztul E., Chakraborty T., Sasakawa C. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nature Cell Biology. 2009;11(10):1233–1240. doi: 10.1038/ncb1967. 19749745 [DOI] [PubMed] [Google Scholar]
- 200.Gutierrez M.G., Vázquez C.L., Munafó D.B., Zoppino F.C., Berón W., Rabinovitch M., Colombo M.I. Autophagy induction favours the generation and maturation of the Coxiella-replicative vacuoles. Cellular Microbiology. 2005;7(7):981–993. doi: 10.1111/j.1462-5822.2005.00527.x. 15953030 [DOI] [PubMed] [Google Scholar]
- 201.Orvedahl A., MacPherson S., Sumpter R., Jr., Tallóczy Z., Zou Z., Levine B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host & Microbe. 2010;7(2):115–127. doi: 10.1016/j.chom.2010.01.007. 20159618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.He C., Levine B. The Beclin 1 interactome. Current Opinion in Cell Biology. 2010;22(2):140–149. doi: 10.1016/j.ceb.2010.01.001. 20097051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Shoji-Kawata S., Levine B. Autophagy, antiviral immunity, and viral countermeasures. Biochimica et Biophysica Acta. 2009;1793(9):1478–1484. doi: 10.1016/j.bbamcr.2009.02.008. 19264100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Höhn A., Jung T., Grune T. Pathophysiological importance of aggregated damaged proteins. Free Radical Biology and Medicine. 2014;71:70–89. doi: 10.1016/j.freeradbiomed.2014.02.028. 24632383 [DOI] [PubMed] [Google Scholar]
- 205.Höhn T.J., Grune T. The proteasome and the degradation of oxidized proteins: Part III – Redox regulation of the proteasomal system. Redox Biology. 2014;2:388–394. doi: 10.1016/j.redox.2013.12.029. 24563857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Jung T., Grune T. The proteasome and the degradation of oxidized proteins: Part I – Structure of proteasomes. Redox Biology. 2013;1(1):178–182. doi: 10.1016/j.redox.2013.01.004. 24024151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ngo J.K., Pomatto L.C., Davies K.J. Upregulation of the mitochondrial Lon protease allows adaptation to acute oxidative stress but dysregulation is associated with chronic stress, disease, and aging. Redox Biology. 2013;1(1):258–264. doi: 10.1016/j.redox.2013.01.015. 24024159 [DOI] [PMC free article] [PubMed] [Google Scholar]