Abstract
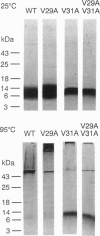
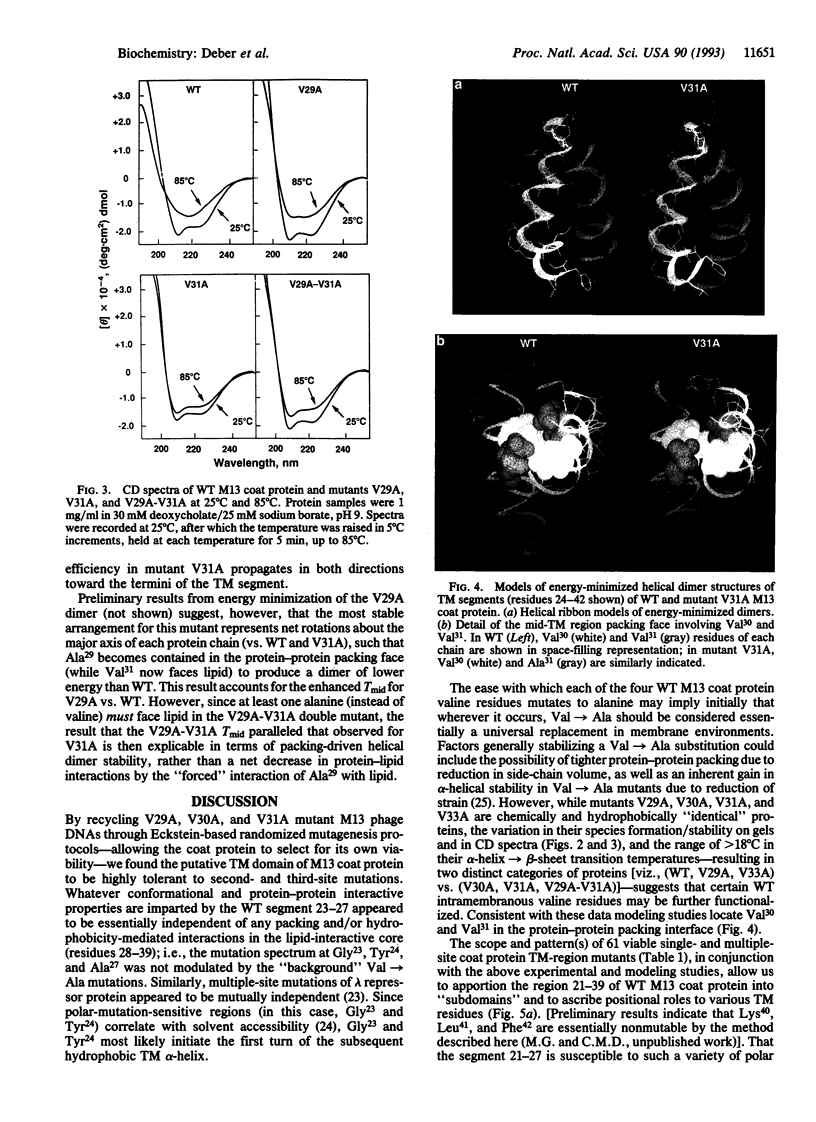
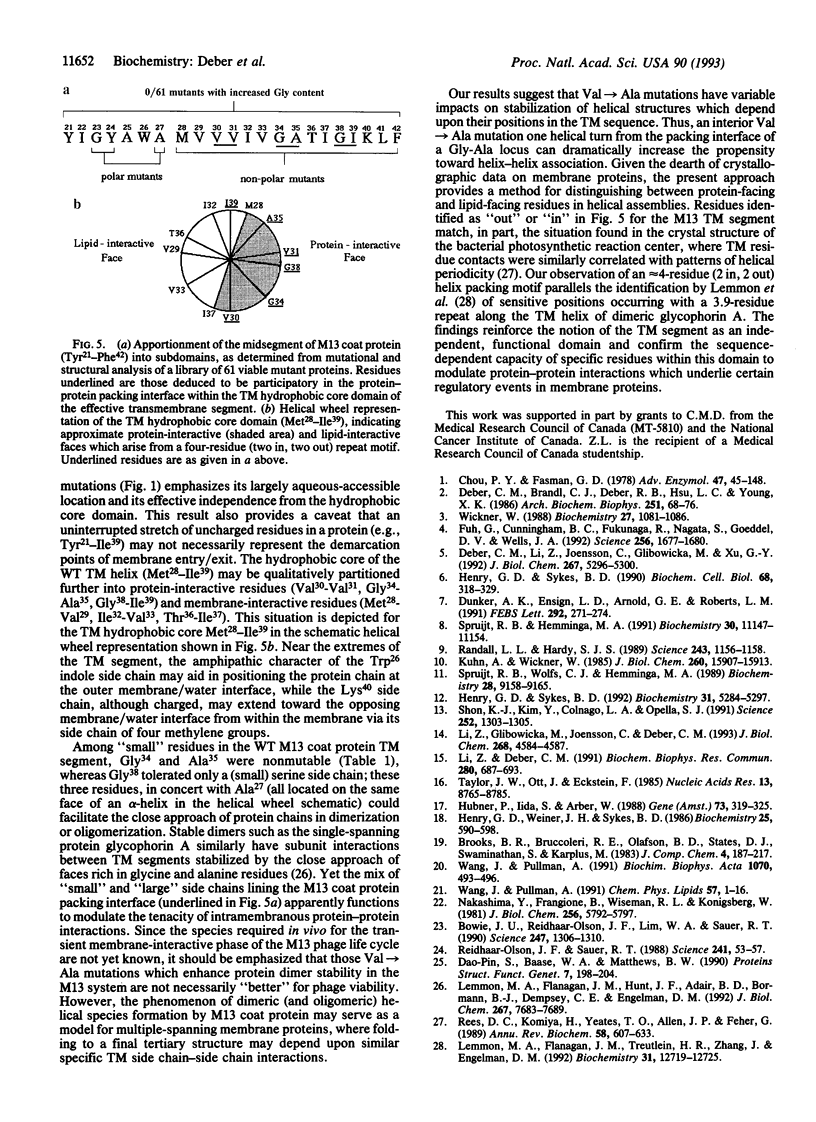
Val-->Ala mutations within the effective transmembrane segment of a model single-spanning membrane protein, the 50-residue major coat (gene VIII) protein of bacteriophage M13, are shown to have sequence-dependent impacts on stabilization of membrane-embedded helical dimeric structures. Randomized mutagenesis performed on the coat protein hydrophobic segment 21-39 (YIGYAWAMV-VVIVGATIGI) produced a library of viable mutants which included those in which each of the four valine residues was replaced by an alanine residue. Significant variations found among these Val-->Ala mutants in the relative populations and thermal stabilities of monomeric and dimeric helical species observed on SDS/PAGE, and in the range of their alpha-helix-->beta-sheet transition temperatures confirmed that intramembranous valine residues are not simply universal contributors to membrane anchoring. Additional analyses of (i) nonmutatable sites in the mutant protein library, (ii) the properties of the double mutant V29A-V31A obtained by recycling mutant V31A DNA through mutagenesis procedures, and (iii) energy-minimized helical dimer structures of wild-type and mutant V31A transmembrane regions indicated that the transmembrane hydrophobic core helix of the M13 coat protein can be partitioned into alternating pairs of potential protein-interactive residues (V30, V31; G34, A35; G38, I39) and membrane-interactive residues (M28, V29; I32, V33; T36, I37). The overall results consitute an experimental approach to categorizing the distinctive contributions to structure of the residues comprising a protein-protein packing interface vs. those facing lipid and confirm the sequence-dependent capacity of specific residues within the transmembrane domain to modulate protein-protein interactions which underlie regulatory events in membrane proteins.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bowie J. U., Reidhaar-Olson J. F., Lim W. A., Sauer R. T. Deciphering the message in protein sequences: tolerance to amino acid substitutions. Science. 1990 Mar 16;247(4948):1306–1310. doi: 10.1126/science.2315699. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Dao-Pin S., Baase W. A., Matthews B. W. A mutant T4 lysozyme (Val 131----Ala) designed to increase thermostability by the reduction of strain within an alpha-helix. Proteins. 1990;7(2):198–204. doi: 10.1002/prot.340070208. [DOI] [PubMed] [Google Scholar]
- Deber C. M., Brandl C. J., Deber R. B., Hsu L. C., Young X. K. Amino acid composition of the membrane and aqueous domains of integral membrane proteins. Arch Biochem Biophys. 1986 Nov 15;251(1):68–76. doi: 10.1016/0003-9861(86)90052-4. [DOI] [PubMed] [Google Scholar]
- Deber C. M., Li Z., Joensson C., Glibowicka M., Xu G. Y. Transmembrane region of wild-type and mutant M13 coat proteins. Conformational role of beta-branched residues. J Biol Chem. 1992 Mar 15;267(8):5296–5300. [PubMed] [Google Scholar]
- Dunker A. K., Ensign L. D., Arnold G. E., Roberts L. M. A model for fd phage penetration and assembly. FEBS Lett. 1991 Nov 4;292(1-2):271–274. doi: 10.1016/0014-5793(91)80882-4. [DOI] [PubMed] [Google Scholar]
- Fuh G., Cunningham B. C., Fukunaga R., Nagata S., Goeddel D. V., Wells J. A. Rational design of potent antagonists to the human growth hormone receptor. Science. 1992 Jun 19;256(5064):1677–1680. doi: 10.1126/science.256.5064.1677. [DOI] [PubMed] [Google Scholar]
- Henry G. D., Sykes B. D. Structure and dynamics of detergent-solubilized M13 coat protein (an integral membrane protein) determined by 13C and 15N nuclear magnetic resonance spectroscopy. Biochem Cell Biol. 1990 Jan;68(1):318–329. doi: 10.1139/o90-044. [DOI] [PubMed] [Google Scholar]
- Henry G. D., Weiner J. H., Sykes B. D. Backbone dynamics of a model membrane protein: 13C NMR spectroscopy of alanine methyl groups in detergent-solubilized M13 coat protein. Biochemistry. 1986 Feb 11;25(3):590–598. doi: 10.1021/bi00351a012. [DOI] [PubMed] [Google Scholar]
- Lemmon M. A., Flanagan J. M., Hunt J. F., Adair B. D., Bormann B. J., Dempsey C. E., Engelman D. M. Glycophorin A dimerization is driven by specific interactions between transmembrane alpha-helices. J Biol Chem. 1992 Apr 15;267(11):7683–7689. [PubMed] [Google Scholar]
- Lemmon M. A., Flanagan J. M., Treutlein H. R., Zhang J., Engelman D. M. Sequence specificity in the dimerization of transmembrane alpha-helices. Biochemistry. 1992 Dec 29;31(51):12719–12725. doi: 10.1021/bi00166a002. [DOI] [PubMed] [Google Scholar]
- Li Z., Deber C. M. Viable transmembrane region mutants of bacteriophage M13 coat protein prepared by site-directed mutagenesis. Biochem Biophys Res Commun. 1991 Oct 31;180(2):687–693. [PubMed] [Google Scholar]
- Li Z., Glibowicka M., Joensson C., Deber C. M. Conformational states of mutant M13 coat proteins are regulated by transmembrane residues. J Biol Chem. 1993 Mar 5;268(7):4584–4587. [PubMed] [Google Scholar]
- Nakashima Y., Frangione B., Wiseman R. L., Konigsberg W. H. Primary structure of the major coat protein of the filamentous bacterial viruses, If1 and Ike. J Biol Chem. 1981 Jun 10;256(11):5792–5797. [PubMed] [Google Scholar]
- Randall L. L., Hardy S. J. Unity in function in the absence of consensus in sequence: role of leader peptides in export. Science. 1989 Mar 3;243(4895):1156–1159. doi: 10.1126/science.2646712. [DOI] [PubMed] [Google Scholar]
- Rees D. C., Komiya H., Yeates T. O., Allen J. P., Feher G. The bacterial photosynthetic reaction center as a model for membrane proteins. Annu Rev Biochem. 1989;58:607–633. doi: 10.1146/annurev.bi.58.070189.003135. [DOI] [PubMed] [Google Scholar]
- Shon K. J., Kim Y., Colnago L. A., Opella S. J. NMR studies of the structure and dynamics of membrane-bound bacteriophage Pf1 coat protein. Science. 1991 May 31;252(5010):1303–1305. doi: 10.1126/science.1925542. [DOI] [PubMed] [Google Scholar]
- Spruijt R. B., Hemminga M. A. The in situ aggregational and conformational state of the major coat protein of bacteriophage M13 in phospholipid bilayers mimicking the inner membrane of host Escherichia coli. Biochemistry. 1991 Nov 19;30(46):11147–11154. doi: 10.1021/bi00110a018. [DOI] [PubMed] [Google Scholar]
- Spruijt R. B., Wolfs C. J., Hemminga M. A. Aggregation-related conformational change of the membrane-associated coat protein of bacteriophage M13. Biochemistry. 1989 Nov 14;28(23):9158–9165. doi: 10.1021/bi00449a030. [DOI] [PubMed] [Google Scholar]
- Taylor J. W., Ott J., Eckstein F. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphorothioate-modified DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8765–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Pullman A. Do helices in membranes prefer to form bundles or stay dispersed in the lipid phase? Biochim Biophys Acta. 1991 Dec 9;1070(2):493–496. doi: 10.1016/0005-2736(91)90091-l. [DOI] [PubMed] [Google Scholar]
- Wickner W. Mechanisms of membrane assembly: general lessons from the study of M13 coat protein and Escherichia coli leader peptidase. Biochemistry. 1988 Feb 23;27(4):1081–1086. doi: 10.1021/bi00404a001. [DOI] [PubMed] [Google Scholar]