Abstract
Latency-reversing agents (LRAs), including histone deacetylase inhibitors (HDACi), are being investigated as a strategy to eliminate latency in HIV-infected patients on suppressive antiretroviral therapy. The effectiveness of LRAs in activating latent infection in HIV strains derived from the central nervous system (CNS) is unknown. Here we show that CNS-derived HIV-1 strains possess polymorphisms within and surrounding the Sp transcription factor motifs in the long terminal repeat (LTR). These polymorphisms result in decreased ability of the transcription factor specificity protein 1 to bind CNS-derived LTRs, reducing the transcriptional activity of CNS-derived viruses. These mutations result in CNS-derived viruses being less responsive to activation by the HDACi panobinostat and romidepsin compared with lymphoid-derived viruses from the same subjects. Our findings suggest that HIV-1 strains residing in the CNS have unique transcriptional regulatory mechanisms, which impact the regulation of latency, the consideration of which is essential for the development of HIV-1 eradication strategies.
Introduction
Despite the enormous impact of combination antiretroviral therapy (cART) on HIV-related morbidity and mortality, treatment is lifelong, expensive and has side-effects. The major barrier to cure is the persistence of long-lived latently infected cells. One strategy being investigated to eliminate latency is to activate transcription from latently infected cells using latency-reversing agents (LRAs) such as histone deacetylase inhibitors (HDACi). The effect of LRAs on infected cells within the central nervous system (CNS) or on viral strains that are uniquely found in the CNS has not been examined. Determining the effect of LRAs on infected cells within compartments other than CD4+ T cells is essential for the implementation of strategies aimed either at complete elimination of all infected cells, or for the controlled activation and elimination of particular reservoirs.
Following the discovery of latent viral reservoirs in cART-treated HIV-1-positive patients, the need for a greater understanding of the regulatory mechanisms that control silencing of HIV-1 transcription has emerged. Numerous studies have concentrated on the use of potential therapeutic interventions to reverse HIV-1 latency. Initially, studies concentrated on the use of pan-HDACi (vorinostat, panobinostat; HDACi, which lack specificity to particular classes of HDAC) but these have had limited success in clinical trials. The more recent use of class-specific HDACi (romidepsin, entinostat, givinostat, belinostat) may achieve improved specificity and activity against the long terminal repeat (LTR).1 Non-HDACi LRAs are currently being pursued as alternatives and may potentially be more effective and specific activators of HIV-1 viral transcription. Disulfiram, a modulator of Akt signaling, HMBA, which is a Tat (viral transactivator of transcription) mimetic and has roles in chromatin remodeling, JQ1, which is a bromodomain inhibitor, and chaetocin, which is a histone methyltransferase inhibitor are all being investigated for their potential use in reversing HIV-1 latency.2, 3, 4, 5
The cellular targets of HIV-1 infection of the CNS are the relatively long-lived microglia, perivascular macrophages and astrocytes.6 HIV-1 infection in the CNS is characterized by the compartmentalization of unique viral variants demonstrated at the level of env sequence and the LTR,7, 8, 9, 10, 11, 12, 13, 14 with HIV-1 isolated from the CNS displaying reduced basal and Tat-activated transcriptional activity in CNS-derived cell types.15 The mechanism regulating the reduced transcriptional activity maps to elements within the LTR sequence, which may impact the effectiveness of LRAs.
HIV-1 transcription is regulated by the interaction of host and viral factors, with a variety of cis-acting sequences present in the HIV-1 LTR.16, 17 Following integration of HIV-1 into regions of active chromatin, transcription factor-binding sites in the LTR recruit activating and repressing host cell transcription factors.18, 19, 20, 21 Within the HIV-1 LTR, binding sites for nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB), specificity protein 1 (Sp1), Yin Yang 1, upstream binding protein 1, activator protein 1 and other transcription factors recruit chromatin-modifying complexes.22, 23, 24, 25, 26 Activating complexes recruit histone acetyltransferases that mediate chromatin remodeling and enhance transcriptional activation in either a Tat-dependent or independent manner.27, 28, 29, 30 Alternatively, repressing complexes recruit HDAC that stabilize the HIV-1 associated chromatin, resulting in a decrease in transcriptional activity and the establishment of viral latency.22, 23, 26 The relative recruitment of activating and repressing factors influences the balance between quiescence and active transcription, and the dynamics of switching between these two states.31 It follows that the regulation of HIV-1 latency is determined not only by the host cell environment, but also the sequences of LTRs within these cells. CNS-derived LTRs have tissue-specific sequences and CNS-restricted transcriptional activities when compared with lymphoid-derived LTRs,32, 33, 34, 35, 36, 37, 38, 39 suggesting that the relative recruitment of host transcription factors, in a tissue and LTR specific manner, may act to modulate HIV-1 transcription in the CNS.
The Sp motifs within the HIV-1 LTR are important for basal transcriptional activity, Tat transactivation and the establishment and control of viral latency.23, 31, 40, 41, 42 Recently we demonstrated that sequence changes within the LTR discriminated CNS- and lymphoid-derived LTRs with respect to sequence and transcriptional activity.15
Given these findings we hypothesized that polymorphisms within the Sp motifs in LTRs isolated from the CNS will impact their responsiveness to LRAs. The compartmentalization of HIV-1 LTR sequences between lymphoid cells and the CNS predicts that LRAs may function differently in the CNS with potentially important implications for the design and delivery of eradication strategies.
Materials and methods
Patient cohort
The patient cohort consisted of four HIV-1 dementia patients from which we had matched CNS and lymphoid autopsy samples. The primary CNS- and lymphoid-derived HIV-1 viruses isolated from subjects CB1, CB3, MACS2(M2) and MACS3(M3) have been described in detail previously.15, 43 For this study we obtained a minimum of six independent LTR clones per patient, per tissue sample (n=82) to ensure we could perform appropriate statistical tests. Sequencing revealed that 50 of these clones were unique, and among these we identified 17 unique Sp motifs from the cohort that were subsequently analyzed.
HIV-1 transcription assays
The human fetal astrocytic cell line (SVG)44 or Jurkat T-cell line45 were seeded at 2000 cells per well in antibiotic-free media into 96-well flat-bottom plates. The following day, cells were transfected with 150 ng per well of patient-derived or control LTR contained within the pGL3-basic expression vector, using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen, Waltham, MA, USA). For basal conditions, cells were co-transfected with 10 ng per well of pTarget-empty. For Tat-activated conditions, cells were co-transfected with 10 ng per well of pTargeT-HxB2-Tat. Four hours post transfection, the media was changed and cells were incubated for a further 20 h. For experiments performed with LRAs, pharmacologically relevant concentrations of panobinostat (60 nM), romidepsin (40 nM), vorinostat (1000 nM), chaetocin (100 nM), disulfiram (1000 nM) or JQ1 (500 nM) were added 24 h prior to harvesting (Supplementary Table 1). The viability of SVGs treated with LRAs was measured using the MTS assay and the 50% cytotoxicity concentration (CC50) was determined for all LRAs tested, which is summarized in Supplementary Table 1. Cells were then lysed 72 h post transfection in 1 × Cell Culture Lysis Reagent (Promega, Fitchburg, WI, USA) and the transcriptional activity of the LTR was measured by quantifying luciferase expression using the Luciferase Assay System (Promega) according to manufacturer's instructions. Luminescence was measured using a FLUOStar Optima microplate reader (BMG Labtech, Ortenberg, Germany).
Electrophoretic mobility shift assays (EMSAs)
Double-stranded DNA complementary to unique SpIII, SpII or SpI motifs from patient-derived CNS and lymphoid LTRs were used in EMSAs utilizing recombinant Sp1 protein as previously described.46 To demonstrate binding specificity, rabbit polyclonal anti-Sp1 (sc-59X, Santa-Cruz Biotech, Dallas, TX, USA) antibody was used to Supershift probe-Sp1 complexes. Sp1 complexes were quantitated by gel densitometry using ImageQuant software (GE Healthcare, Little Chalfont, UK). To account for variability in probe activity, Sp1 complex density was normalized to lane density and subsequently expressed relative to HxB2 control oligonucleotide. Alternatively, a 76 bp sequence spanning all three Sp motifs (corresponding to nucleotides 357–433 of HIV-1 HXB2) in the core promoter was generated using PCR. Sp1-binding affinity was determined using EMSAs as described above.
Results
Extensive polymorphisms exist within the core promoter of LTR sequences isolated from the CNS
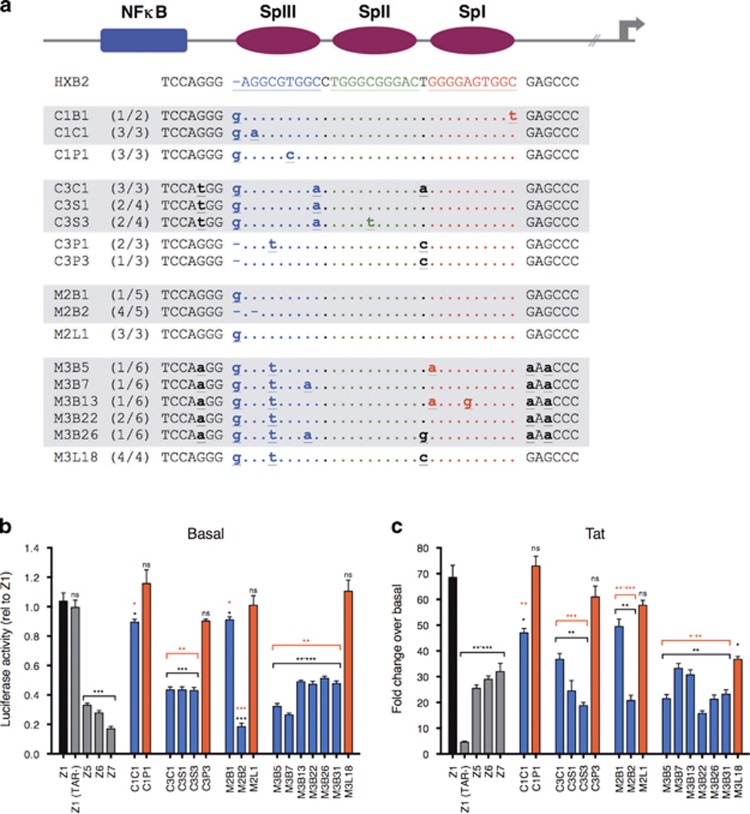
The Sp motifs in the core basal promoter region of the HIV-1 LTR of isolates obtained from the lymphoid compartment (peripheral blood mononuclear cell or lymph node) and the CNS (spinal cord, cerebrospinal fluid or brain), which have been described previously,15 were examined relative to the consensus T-cell tropic laboratory isolate HXB2 (Figure 1a). Extensive heterogeneity was observed across this region. We have previously shown that these polymorphisms compartmentalized between patients, and within patients between tissues.15 Mutations within the Sp motifs in the core promoter region of the LTR were observed within all LTRs isolated. In addition, several isolates contained sequence polymorphisms in the sequences intervening and adjacent to the Sp motifs. Notably, the functionally important SpIII (distal) site showed a high degree of heterogeneity in both the CNS and lymphoid compartment-derived isolates. However, polymorphisms in adjacent sequences were mainly observed in the CNS-derived isolates. These results suggest that alterations in sequences flanking the Sp-binding motifs of CNS-derived HIV-1 strains may reflect functional differences between CNS- and lymphoid-derived viruses.
Figure 1.
Unique Sp motifs isolated from the CNS compartments of HIV patients have reduced basal and Tat-activated transcription compared with lymphoid-derived LTRs, in CNS cells. (a) Nucleotide alignment of the HIV-1 LTR core promoter region relative to a consensus T-cell-derived LTR, HXB2, as indicated: Sp proximal (SpI) in red, Sp medial (SpII) in green and Sp distal (SpIII) in blue. Gray and white shading indicates CNS- and lymphoid-derived LTRs, respectively. Lower case bold lettering indicates mutations within the intervening and adjacent flanking sequences. Dots indicate conserved nucleotides and dashes indicate gaps. Gray arrow indicates the start site of transcription. Numbers in parentheses indicate the number of clones within that patient sample that had the same genetic makeup. (b) Basal and (c) Tat-activated transcriptional activity of LTRs isolated from the CNS and lymphoid compartments of patients with HIV. Lymphoid isolates are in red, CNS isolates in blue. SVG cells were transfected with LTR luciferase constructs containing LTRs from the CNS and lymphoid compartments of patients and subjected to HIV-1 transcription assays either in the presence or absence of Tat. Control promoters based on consensus T-cell isolate HXB2 are indicated by Z (Z1: wild-type sequence; Z1(TAR-): deletion of TAR region; Z5: mutation of SpI; Z6: mutation of SpII and SpIII; Z7: mutation of SpIII and NF-κB). Patient isolates are indicated. Luciferase activity is relative to a T-cell consensus sequence and Tat activation is shown as fold activation over basal for each LTR sequence. Data shown are representative of four independent experiments, each experiment performed in triplicate. Shown are the means and s.e.m. of these data. Significance values (calculated by student's t-test): *P=<0.05, **P=<0.01, ***P=<0.001, ns=not significant. Black values indicate significance of isolates relative to Z1 T-cell tropic consensus sequence (HXB2), red values represent significance of CNS-derived isolates relative to lymphoid-derived isolates from the same patient. CNS, central nervous system; LTR, long terminal repeat.
Compartment-specific polymorphisms in the core promoter region result in anatomical site-specific transcriptional activity
To determine whether polymorphisms in the core promoter region of the LTR sequences isolated from the CNS and lymphoid compartments have differential effects on transcriptional activity, LTRs with unique core promoters were analyzed for both basal and Tat-activated transcriptional activity in the human fetal astrocyte cell line SVG, as described previously15 (Figure 1b and c). SVG cells were used for this analysis as they contain similar levels of the Sp1 and Sp3 proteins to that observed in primary fetal astrocytes and in HIV-1 infected human brain tissue (Supplementary Figure 1). Control LTRs carrying mutations designed to inhibit binding of Sp1 alone (Z5 and Z6) and in combination with mutations in the NF-κB site (Z7) significantly reduced both basal and Tat-activated transcription as expected. Deletion of the TAR region had no effect on basal transcription, however significantly reduced Tat-activated transcription as expected.
All CNS-derived LTRs (C1C1, C3C1, C3S1, C3S3, M2B1, M2B2, M3B5, M3B7, M3B13, M3B26, M3B31) displayed basal transcription significantly lower than the consensus T-cell-derived control LTR (Z1). However, there was no significant difference in the transcriptional activity between LTR sequences isolated from the lymphoid compartment (C1P1, C3P3, M2L1, M3L18), and the consensus control LTR, Z1. Importantly, CNS-derived LTR sequences demonstrated significantly reduced transcriptional activity relative to lymphoid-derived LTRs from the same patients. Similar trends were observed for Tat-activated transcription, with the exception of M3L18 where there was a significant decrease in Tat-activated transcription in comparison with the consensus control LTR, Z1.
These data suggest that Sp motif polymorphisms result in a significant reduction in both basal and Tat-activated transcriptional activity of CNS-derived LTR sequences relative to lymphoid-derived LTRs from the same patients. Lymphoid-derived LTRs generally maintained levels of transcriptional activity similar to that of the consensus control LTR.
Reduced Sp1-binding capacity in the promoter proximal motif (SpI) does not significantly influence overall binding of Sp1 to the core promoter region in both CNS- and lymphoid-derived sequences
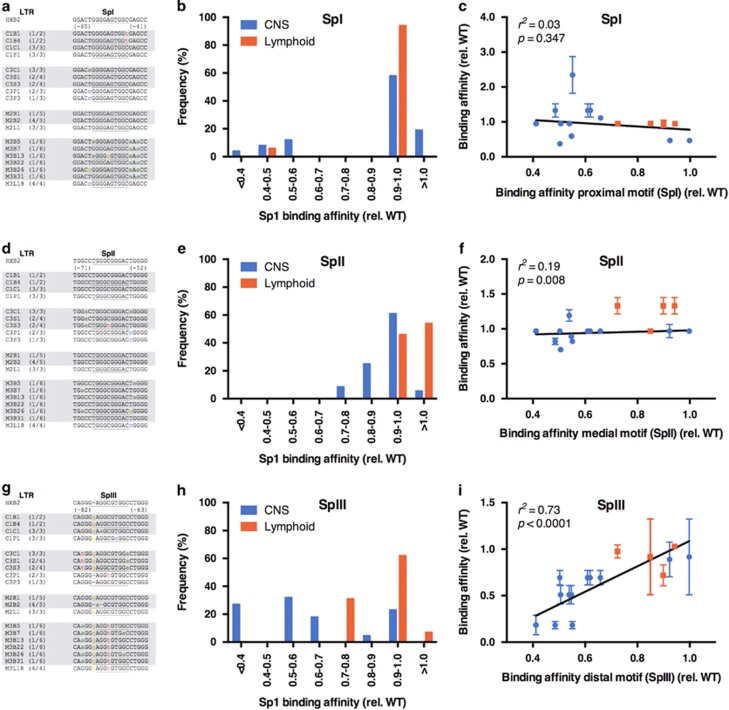
To investigate the role of mutations within the core basal promoter on reduced transcriptional activity, the binding affinities of Sp transcription factor motifs in LTRs isolated from the CNS and lymphoid compartments were determined using EMSAs. Radiolabeled probes encompassing the three Sp transcription factor motifs were generated for each unique sequence from the panel of CNS- and lymphoid-derived LTRs (Figure 1a).
The consensus-binding sequence of the proximal Sp motif (SpI) was conserved in lymphoid-derived LTRs, but polymorphisms were observed in several CNS-derived LTRs with a number of polymorphisms located in the adjacent sequences (Figure 2a). The frequency of polymorphisms resulting in reduced Sp1 binding to the proximal Sp motif predominated in CNS-derived LTR sequences, with 26% of these sequences binding Sp1 with an affinity of <0.6 × that of consensus LTR, whereas 100% of lymphoid-derived LTR sequences bound Sp1 with an affinity approaching that of consensus LTR (Figure 2b). There was no significant correlation between binding to the proximal Sp motif (SpI) and overall binding to the core promoter (r2=0.03, P=0.347) (Figure 2c).
Figure 2.
Frequency distribution of Sp1-binding affinities relative to compartment and correlation of effect of individual Sp motif affinities to overall binding to the core basal promoter. Sp1-binding capacity of individual Sp motifs relative to compartment distribution was determined using EMSA: SpI (a and b), SpII (d and e) or SpIII (g and h). Gray and white shading in the sequence alignments indicates CNS- and lymphoid-derived LTRs, respectively. The correlation between binding to the individual motif and binding to the complete core promoter is shown (c, f and i). Blue symbols represent CNS-derived LTRs; red symbols represent lymphoid-derived LTRs. Shown are the means and s.e.m. of these data. Correlations were calculated using linear regression analyses in Prism 6.0e (GraphPad, La Jolla, CA, USA) with P and r2 values shown. CNS, central nervous system; EMSA, electrophoretic mobility shift assay; LTR, long terminal repeat.
These results show that the proximal Sp motif is generally well conserved in both CNS- and lymphoid-derived LTRs, with binding of Sp1 to this motif generally close to that of the consensus motif, and there was no significant functional impact on binding to this motif or binding to the overall core promoter region.
Sp1-binding capacity of the promoter medial motif (SpII) significantly influences overall binding of Sp1 to the core promoter region in both CNS- and lymphoid-derived sequences
The medial Sp-binding motif (SpII) is well conserved in both the CNS- and lymphoid-derived LTR sequences, however, heterogeneity was observed in the distal/medial and proximal/medial intervening sequences. Within each patient, polymorphisms in these regions were compartment specific. C3 carried a T to C change at nucleotide −56 in the proximal/medial intervening sequence only in the lymphoid-derived isolates, and the CNS-derived isolates from the same patients maintained the consensus T in this region or contained a T to A polymorphism (Figure 2d).
Polymorphisms in and surrounding the medial motif had only minor effects on the affinity of this site for Sp1 protein irrespective of compartment of origin, with all of the LTRs maintaining Sp1 binding to the medial motif with an affinity similar to the consensus motif (>0.7 ×) (Figure 2e). There was a strong correlation between binding to the medial motif and binding to the overall core promoter region (r2=0.19, P=0.008) (Figure 2f).
Thus, despite relatively few polymorphisms observed within the medial motif, the majority of sequences maintained a high affinity for binding Sp1 protein, and binding to this motif significantly correlates with binding to the overall core promoter region.
Sp1-binding capacity of the promoter distal motif (SpIII) significantly influences overall binding of Sp1 to the core promoter region in both CNS- and lymphoid-derived sequences
The promoter distal Sp motif (SpIII) contained several polymorphisms in the CNS-derived LTRs, but lymphoid-derived LTR sequences were more conserved (Figure 2g). Significantly, although nucleotide changes were patient specific, polymorphisms were also compartment specific. All CNS-derived sequences from patient C3 contained a C to A nucleotide change at position 68, a similar mutation was also observed in two of the M3 patient brain-derived sequences B7 and B26. Additional CNS-derived LTRs contained nucleotide changes at position 80 directly adjacent to the core SpIII motif (G to T for C3 CNS isolates and G to A for M3 CNS isolates). The binding activity of these CNS-derived isolates (C3C1/S1/S3; M3B5/7/13/22/26/31) was significantly reduced relative to the consensus LTR (ranging from 0.183±0.039 (M3B7/26) to 0.693±0.078 (M3B5/13/22/31)) with similar reductions in transcriptional activity (Figure 1b). Polymorphisms in the lymphoid-derived LTRs had minimal effect on binding of Sp1 to the distal motif, with 100% of the LTRs maintaining binding affinity of 0.7 × or greater relative to the consensus motif (Figure 2h). Conversely, >70% of CNS-derived LTRs demonstrated binding affinities of <0.7 × that of the consensus sequence, with approximately 60% having affinities <0.6 × that of the consensus sequence (Figure 2h). A significant correlation (r2=0.73, P<0.0001) was observed between binding to the distal motif and binding to the overall core promoter region (Figure 2i).
These results show that polymorphisms affecting Sp1 binding to the distal Sp motif predominate in CNS-derived sequences with the majority binding Sp protein poorly, and that there was a significant correlation between binding to this motif and overall binding to the core promoter region.
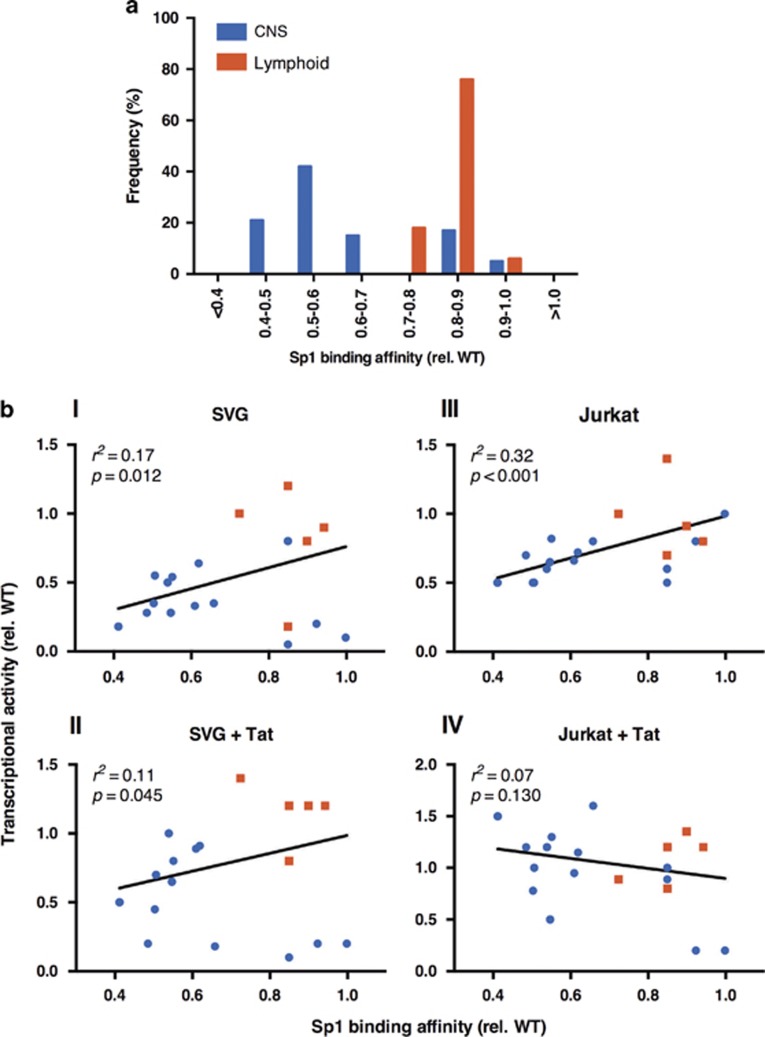
Decreased Sp1-binding affinities for the complete core promoter predominate in CNS-derived LTRs and strongly correlate with reduced transcriptional activity
DNA sequences encompassing the complete core promoter region (SpI-III) for each of the unique sequences isolated from the CNS and lymphoid compartments were next analyzed for their ability to bind Sp1 protein using EMSA. Core promoter regions with a reduced capacity to bind Sp1 protein predominated in sequences isolated from the CNS; >80% of CNS-derived LTRs bound Sp1 protein with an affinity of <0.7 × that of the consensus motif, and 60% bound with an affinity of <0.6 × consensus binding (Figure 3a). In contrast, 100% of LTRs isolated from the lymphoid compartment of the same patients maintained an affinity to bind Sp1 protein approaching that of the consensus motif.
Figure 3.
(a) Frequency distribution of Sp1-binding affinities to the overall core promoter (SpI-III) relative to compartment of origin. (b) Correlation of Sp1 binding to the complete core promoter and basal and Tat-activated transcriptional activity in SVG astrocyte cells (bI and bII) and Jurkat T cells (bIII and bIV). Blue symbols represent CNS-derived LTRs; red symbols represent lymphoid-derived LTRs. Shown are the means. Correlations were calculated using linear regression analysis in Prism 6.0e (GraphPad) with P and r2 values shown. CNS, central nervous system; LTR, long terminal repeat.
There was a strong correlation between Sp1-binding affinity and basal transcriptional activity of both CNS- and lymphoid-derived LTRs in both SVG astrocytes (r2=0.17, P=0.012) and Jurkat T cells (r2=0.32, P<0.001) (Figure 3 bI and bIII), and Tat-activated transcription in SVG astrocytes (r2=0.11, P=0.045) but not in Jurkat T cells (r2=0.07, P=0.130) (Figure 3 bII and bIV). These data suggest that the Sp motifs have an important role in the regulation of HIV-1 transcriptional activity irrespective of the compartment from which they are derived, and that the predominance of Sp motif polymorphisms observed in CNS-derived LTRs reduce Sp1 binding and result in the reduced transcriptional activity of these LTRs in CNS cells.
Reduced transcriptional activity of CNS-derived LTRs maps to mutations within the Sp-binding motifs within the core basal promoter region
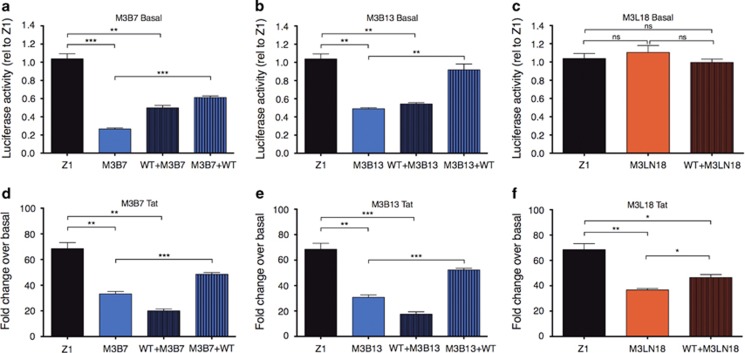
To determine whether polymorphisms in the core promoter region Sp motifs were directly responsible for the decreased transcriptional activity observed by CNS-derived LTRs, the Sp motifs from CNS isolates M3B7 and M3B13 and from the lymphoid-derived M3L18 LTR were introduced into the consensus T-cell-derived LTR Z1. Conversely, the Sp motifs from the consensus T-cell-derived LTR Z1 was introduced into the CNS-derived LTRs M3B7 and M3B13. Transcriptional activity of the mutated LTRs was determined in SVG astrocyte cells in the presence and absence of Tat.
A significant difference in transcriptional activity was observed between Z1 and M3B7 under basal conditions (M3B7 0.3 × Z1 P=0.005) (Figure 4a). Introduction of the M3B7 Sp motifs into Z1 (WT+M3B7) resulted in a significant decrease in the activity of Z1 (0.5 × Z1, P=0.006) and replacement of the M3B7 motifs with consensus Sp motifs (M3B7+WT) resulted in an increase in M3B7 transcriptional activity from 0.3 × to 0.65 × that of Z1 (P=0.003). Similar results were observed under Tat-activated transcription with a significant difference observed in Tat responsiveness between Z1 and M3B7 (Z1 70-fold over basal, M3B7 32-fold over basal, P=0.0024) (Figure 4d). Introduction of the M3B7 Sp motifs into Z1 (WT+M3B7) resulted in a significant decrease in Tat activation of Z1 (20-fold over basal, P=0.001) and replacement of the M3B7 motifs with consensus Sp motifs (M3B7+WT) resulted in an enhanced ability of Tat to activate M3B7 from 32-fold to 55-fold over basal (P=0.009).
Figure 4.
Effect of CNS-derived Sp motif polymorphisms on consensus T-cell-derived promoter activity. The Sp motifs from the CNS-derived isolates M3B7 and M3B13, and the lymphoid-derived isolate M3L18 were introduced into the Z1 T-cell-derived promoter Sp core region (WT+M3B7, WT+M3B13, WT+ML18), and the transcriptional activity determined. Conversely, the consensus Z1 T-cell Sp motifs were introduced into the CNS-derived isolates M3B7 and M3B13 (M3B7+WT; M3B13+WT) and the transcriptional activity determined. Basal transcription is shown in a, b and c, whereas Tat-activated transcription is shown in d, e and f. Blue bars represent CNS-derived LTR backbones; red bars represent lymphoid-derived LTR backbones; black bars represent consensus T-cell LTR backbones. Data shown are representative of four independent experiments, each experiment performed in triplicate. Shown are the means and s.e.m. of these data. Significance values (calculated using student's t-test): *P=<0.05, **P=<0.01, ***P=<0.001, ns=not significant. CNS, central nervous system; LTR, long terminal repeat.
A significant difference in transcriptional activity was also observed between Z1 and M3B13 under basal conditions (M3B13 0.45 × Z1 P=0.0017) (Figure 4b). Replacement of the consensus Sp motifs in Z1 with M3B13 Sp motifs (WT+M3B13) resulted in a significant decrease in the activity of Z1 (0.5 × Z1, P=0.0021) and introduction of the consensus Sp motifs into the M3B13 backbone (M3B13+WT) resulted in an increase in M3B13 transcriptional activity from 0.45 × to 0.9 × that of Z1 (P=0.0061). Similar results were observed under Tat-activated conditions, with a significant difference observed in Tat responsiveness between Z1 and M3B13 (Z1 70-fold over basal, M3B13 30-fold over basal, P=0.0018) (Figure 4e). Introduction of the M3B13 Sp motifs into Z1 (WT+M3B13) resulted in a significant decrease in Tat responsiveness of Z1 (18-fold over basal, P=0.0006) and replacement of the M3B13 motifs with consensus Sp motifs (M3B13+WT) resulted in an enhanced ability of Tat to activate M3B13 transcriptional activity from 30-fold to 57-fold over basal (P=0.0002).
There was no significant difference in basal transcriptional activity of the lymphoid-derived isolate from the same patient (M3L18) and Z1 (Figure 4c). Introduction of the M3L18 Sp motifs into the Z1 consensus backbone did not significantly alter basal transcription. Interestingly, a significant decrease in Tat responsiveness of M3L18 LTR was observed relative to Z1, along with a significant decrease in Tat responsiveness of the mutated construct WT+M3L18, suggesting regions outside the Sp motifs of primary isolates may contribute to Tat responsiveness of lymphoid-derived LTRs.
Together, these data strongly suggest that LTRs isolated from the CNS have a reduced basal transcriptional activity compared with lymphoid-derived isolates from the same patients and to the consensus T-cell-derived LTR, and that this phenotype can be mapped to polymorphisms within the Sp motifs that show reduced Sp1-binding affinity.
CNS-derived LTRs have reduced responsiveness to select LRAs in CNS cells
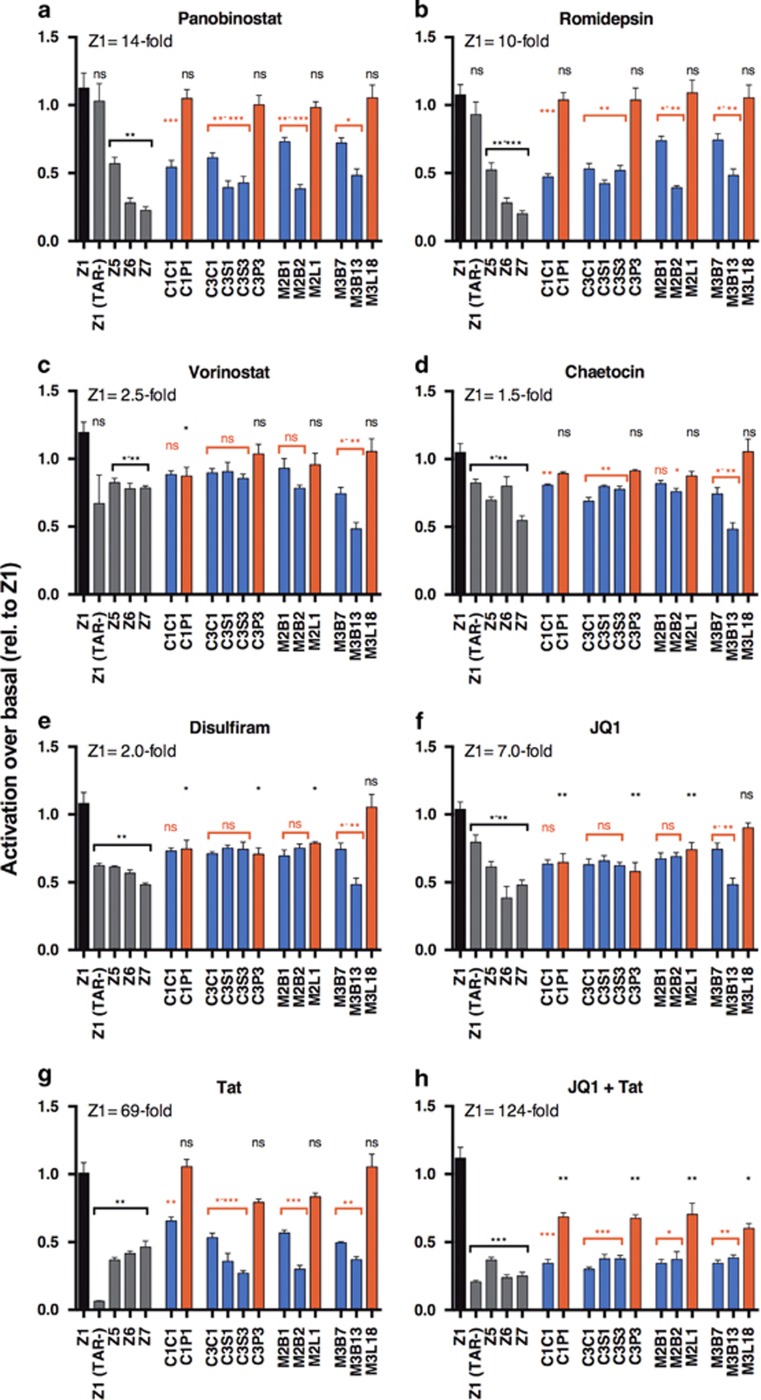
To determine whether CNS-derived LTRs have an altered responsiveness to LRAs, we next performed experiments in SVG cells in the presence and absence of a panel of LRAs. For the LRAs vorinostat, chaetocin, disulfiram and JQ1 alone, a modest increase in transcriptional activity of the control Z1 LTR was observed (up to sevenfold) (Figure 5c–f). For these LRAs, there was generally no significant difference between the responsiveness of the primary isolates and the consensus T-cell sequence LTR Z1 (with the exception of Disulfiram and JQ1, where the primary LTRs were generally significantly lower), or between CNS- and lymphoid-derived sequences, with the exception of M3B where an altered response was noted with vorinostat, chaetocin, JQ1 and disulfiram.
Figure 5.
Activity of LRAs to induce viral transcription of CNS- and lymphoid-derived LTRs in CNS cells. CNS cells were transfected with LTR luciferase constructs containing LTRs from the CNS and lymphoid compartments of patients and subjected to HIV-1 transcription assays either in the presence or absence of LRAs and/or Tat. Treatments included Panobinostat (a), Romidepsin (b), Vorinostat (c), Chaetocin (d), Disulfiram (e), JQ1 (f), Tat (g) and JQ1 + Tat (h). Lymphoid isolates are in red, CNS isolates in blue. The activity of the LRAs was plotted as fold change over basal activity for each construct with results made relative to Z1 (set as 1, with Z1 fold change values quoted in each panel). Control promoters (shown in black and gray) based on consensus T-cell isolate HXB2 are indicated by Z (Z1: wild-type sequence; Z1(TAR-): deletion of TAR region; Z5: mutation of SpI; Z6: mutation of SpII and III; Z7:mutation of SpIII and NF-κB). Patient isolates are indicated. Data shown are representative of four independent experiments, each experiment performed in triplicate. Shown are the means and s.e.m. of these data. Significance values (calculated using student's t-test): *P=<0.05, **P=<0.01, ***P=<0.001, ns=not significant. Black values indicate significance of isolates relative to Z1 T-cell-derived consensus sequence (HXB2), red values represent significance of CNS-derived isolates relative to lymphoid-derived isolates from the same patient. CNS, central nervous system; LRA, latency-reversing agent; LTR, long terminal repeat.
For the LRAs panobinostat, romidepsin, Tat and JQ1+Tat, there was a strong increase in the activity of Z1 LTR with a fold increase in activation of 14, 10, 69 and 124, respectively (Figure 5a, b, g and h). A significant decrease in the response to these LRAs compared with Z1 was observed for all CNS-derived LTRs and for primary LTRs generally when treated with a combination of JQ1+Tat, but not with Tat alone. Importantly, within each patient there was a significant reduction in the fold activation of the CNS-derived LTRs compared with the lymphoid-derived LTRs isolated from the same patients.
These data suggest that LTRs from primary isolates may respond differently compared with consensus T-cell-derived LTRs in the CNS, and importantly that LTRs derived from different compartments (CNS and lymphoid) from the same patient may have significantly different responsiveness to LRAs in CNS cells.
Discussion
We have previously shown that HIV-1 is compartmentalized between CNS and lymphoid compartments, and also between different cells within the CNS at the level of env and the LTR.9, 47, 48, 49 Furthermore, we have shown that HIV-1 LTR sequences isolated from the CNS have a significantly lower transcriptional activity than LTRs isolated from the lymphoid compartments of the same patients.15 We now demonstrate that these differences in transcriptional activity can be mapped to polymorphisms in the core promoter region of the LTR, specifically the three Sp motifs. We demonstrate that the presence of polymorphisms in the core promoter region of HIV-1 LTRs isolated from the CNS significantly correlate with reduced binding of Sp1 protein to the core promoter of these HIV-1 isolates, and is significantly correlated to a reduced transcriptional activity of these promoters. Importantly, these polymorphisms result in an altered response of CNS-derived LTRs to the LRAs panobinostat and romedepsin compared with lymphoid-derived LTRs from the same patients, suggesting the possibility of compartment specific effects within a patient in response to these LRAs, which are both under clinical investigation.
Our results demonstrate that naturally occurring polymorphisms within the Sp motifs of primary HIV-1 isolates have a significant effect on the binding of Sp1 protein to these core promoters (both as individual sites and as entire core promoter regions). Importantly, LTRs with a significantly reduced ability to bind Sp1 protein predominate in sequences isolated from the CNS, whereas lymphoid-derived isolates from the same patients maintain high-affinity binding to Sp1 protein irrespective of polymorphisms. Interestingly, the greatest numbers of polymorphisms were observed within the SpIII motif. While a previously reported polymorphism (5 T: C to T polymorphism in SpIII motif)50 was present in some of the CNS LTRs, this polymorphism was also present in lymphoid-derived sequences from the same patient where binding of Sp1 and transcriptional activity was approaching that of the wild-type consensus T-cell-derived promoter. Transcriptional activity was likely restored by compensatory nucleotide changes in the binding motif and the core promoter.
Previous studies have reported that the three Sp motifs have varying affinities for Sp1 protein and that the affinity of binding to the motif is directly related to promoter activity and correlates strongly with disease progression. The SpIII (NF-κB proximal) motif is reported to be the most functionally significant of the three motifs.24, 51, 52 We observed multiple polymorphisms in the NF-κB proximal Sp motif (SpIII) in both lymphoid- and CNS-derived LTRs and showed that binding to this site had a very significant (P=<0.0001) correlation with capacity of the entire core promoter region (SpI–SpIII) to bind Sp1 protein. Importantly, we demonstrated that despite polymorphisms being observed in both the CNS- and lymphoid-derived promoters from the same patients, polymorphisms resulting in a reduced capacity to bind Sp1 protein predominated in CNS-derived sequences. Furthermore, we demonstrated that the reduced binding capacity of the CNS-derived LTRs to bind Sp1 protein correlated with reduced transcriptional activity in both CNS cells and T cells under basal conditions, and in CNS cells but not T cells under Tat-activated transcription. Confirmation that the reduced Sp1-binding affinity was responsible for the lower transcriptional activity phenotype of the CNS-derived isolates was obtained by replacing the consensus Sp motifs of a T-cell LTR (Z1) with mutated core promoter regions from the CNS-derived isolates M3B7 and M3B13. These data confirm that the reduction in CNS-derived promoter transcriptional activity directly correlates with a reduced ability of the core promoter to bind Sp1 protein. Although the use of EMSA assays clearly describes differences in relative affinities for Sp1 protein between the consensus T-cell motif and the CNS-derived Sp motif-containing specific polymorphisms, they may provide relatively limited information on the mechanism of latency in the context of chromatin of CNS-derived LTRs. Future studies with chromatin immunoprecipitation assays may help clarify the mechanisms that regulate latency in the context of chromatin, and in the presence of low-affinity SpIII motifs described here.
Sp factors are a family of zinc finger-binding transcriptional factors comprising Sp1-4, and although Sp1, 3 and 4 have similar affinities for the GC-rich consensus T-cell motif found in the HIV-1 LTR, Sp2 preferentially binds to GT-rich sequences. Although Sp1 and 4 are considered transcriptional activators, Sp3, by virtue of its inability to form the complex protein–protein interactions necessary for facilitating transcriptional activation, is considered a repressor of transcription. In addition, although considered an activator of transcription, Sp4 fails to activate transcription synergistically from adjacent multiple Sp motifs as found in the HIV-1 LTR. In tissues with high levels of Sp3 and Sp4 expression such as the CNS, competition with Sp1 for motifs in the HIV-1 LTR may be expected to regulate transcriptional activity distinctly from other tissues.
Sp factors and the Sp motifs have been demonstrated to have essential roles in the regulation of transcription, establishment and activation from latency.53 Sp factors bound to the HIV-1 core promoter cooperatively interact and recruit other transcription factors (Tat, NF-κB, TATA-binding protein, positive transcriptional elongation factor b) as well as the cellular transcriptional machinery (TFIID, RNA Pol II).54, 55, 56 Sp factors also have an important role in regulating transcription by remodeling chromatin either via the recruitment of histone acetyltransferases to promote transcription or recruitment of HDAC to inhibit transcription.57, 58 It follows that alterations in the ability of these motifs to bind Sp proteins, as observed in the CNS-derived isolates, will strongly influence their responsiveness to LRAs.
The Sp motifs have also been shown to confer a negative regulatory role on HIV-1 transcription in microglia via the action of the COUP-TF-interacting protein 2 (CTIP2).59 CTIP2 is expressed in the brain,60 where it has a key role in the development of corticospinal motor neuron axonal projections to the spinal cord.61 CTIP2 represses HIV-1 transcription by interacting with the core promoter of HIV-1 following interactions with the COUP-TF/Sp1 complex and by binding to the Sp motif.59, 62 Bound CTIP2 recruits HDAC1 and HDAC2 to promote local histone H3 deacetylation at the HIV-1 promoter region. In addition, DNA-bound CTIP2 also associates with the histone methyltransferase SUV39H1, which increases local histone H3 lysine 9 methylation and the simultaneous recruitment of heterochromatin proteins (HP1) to the viral promoter and formation of local heterochromatin, leading to HIV-1 silencing.59 The function of CTIP2 initially appears at odds with our data, as mutations within the Sp motif would be expected to relieve the repression observed in CNS cells. However, Marban et al.59 clearly demonstrates that the repressive effects of CTIP2 are exclusively directed through the proximal Sp sites (SpII and SpI). The mutations influencing basal and activated transcriptional activity presented here are predominantly located within the distal Sp site (SpIII), the site long established to be essential for the activation of HIV-1 transcription.
Finally, we clearly demonstrate that HIV-1 isolated from distinct compartments (CNS and lymphoid) within the same patient can have significantly different responses to LRAs, some of which are currently in clinical trials. Importantly, for the HDACi Panobinostat, and Romidepsin, and for JQ1 in combination with Tat, where a strong response was observed for activation of the T-cell consensus promoter, there was a significant reduction in the response of the CNS-derived LTRs following activation with the LRAs as compared with lymphoid-derived LTRs isolated from the same patients. For the LRAs that failed to activate transcription in CNS cells (disulfiram, JQ1 alone, vorinostat and chaetocin) there was no significant difference in the response of the CNS- and lymphoid-derived LTRs isolated from the same patients. The reduced capacity of CNS-derived Sp motif to bind Sp1 protein will likely impact the activation of viral transcription by LRAs. The recruitment of both repressing and activating factors to the LTR is dependent on the binding of Sp1 protein to its site in the LTR or by direct interaction with the Sp motif itself through a complex array of interactions dependent on cell type and cellular activation state (for example, HDAC1/2;23 CTIP2;59, 63 Histone acetylases;64 Tat;65 c-Myc66). The polymorphisms within the Sp motif described here, which result in a reduced function of the Sp motif, indicate that CNS-derived LTRs have developed unique mechanisms of transcriptional latency, which are likely to influence responses to LRAs.
It is important to note that these experiments were conducted in a non-integrating system with replication being extra-chromosomal. Although an integrating system is more representative of the ‘natural' situation and will be very important in the further characterization of the effects of these polymorphisms, the inclusion of stringent controls attests to the validity of our results. Importantly, it is clear that the control T-cell LTRs demonstrate activation levels in response to treatment with LRAs and that this activation is equivalent to that reported in the literature67 as determined in the ACH2 cell line and/or primary CD4+ T cells. Our data clearly indicate a differential response to LRAs examined between CNS-derived and non-CNS-derived HIV-1 LTRs.
Clinical trials of vorinostat, panobinostat and romedepsin in HIV-infected patients on cART have all recently been reported.1, 68, 69 Only one of these studies, panobinostat, included an assessment of the effects on the CNS1 and found no change in HIV RNA in cerebrospinal fluid collected prior to and following four treatment cycles.
There has recently been heightened interest in identifying strategies to eliminate latently infected cells by activating latent infection, commonly called ‘shock and kill'.70 Much of this work, in vitro, ex vivo and in vivo has focussed on the effects of LRAs on resting memory T cells that undoubtedly compose the major component of the viral reservoir.71 However, integrated proviral genomes residing in non-T-cell reservoirs and compartments outside the lymphoid system represent a major barrier to eradication. The CNS is one possible viral reservoir.6 We have clearly shown that patients may harbor integrated proviral genomes with distinct regulatory systems reflective of the compartment/tissue, and that these proviral genomes have distinct responses to activation by LRAs. These findings have important implications for the implementation of HIV ‘cure' strategies. First, in the negative, treatment of patients with specific LRAs while activating HIV in peripheral reservoirs will not broadly activate virus in the CNS and thus clearance of virus from this compartment will not be achieved. A more positive consequence of this differential regulation is that a lack of activation of latent virus from reservoirs within the CNS may be desirable as clearance of the CNS activated virus may be confounded by the immune privileged nature of the CNS and a restricted penetrance of cART. Strategies may be developed to promote a ‘functional cure' by which virus in the CNS remains dormant while eradication of virus from other reservoirs can be achieved.
In summary, we have identified that CNS-derived LTRs frequently harbor Sp motif mutations, which reduce their ability to bind Sp1. Furthermore, the lower Sp1 recruitment by CNS-derived LTRs resulted in reduced responsiveness to LRAs. These data suggest that the unique properties of non-T-cell reservoirs should be considered in the clinical development of LRAs, as a strategy to eliminate all latently infected cells in HIV-infected patients on cART.
Acknowledgments
This study was supported by grants from (i) the National Health and Medical Research Council of Australia (NHMRC) to MJC, PRG and SLW (APP1051093), (ii) and the National Institutes of Health (NIH) to MJC, PRG and SRL (R21 MH1022066) and (iii) the Delaney AIDS Research Enterprise (DARE) to MJC and SRL (U19 AI096109). PRG is supported by an Australian Research Council (ARC) Future Fellowship (FT2) and SRL is a NHMRC Practitioner Fellow. LRG was supported by an Australian NHMRC Early Career Fellowship. We gratefully acknowledge the contribution to this work of the Victorian Operational Infrastructure Support Program received by the Burnet Institute.
Author contributions
MJC, LRG, SLW and PRG designed and performed experiments, analyzed data and wrote the manuscript; DC and CW performed experiments and provided intellectual input; SRL and HKL provided intellectual input on experimental design, techniques and interpretation of results; BJB provided intellectual input, key reagents and assistance with experimental design and interpretation; all authors reviewed and edited the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
Supplementary Material
References
- Rasmussen TA, Tolstrup M, Brinkmann CR, Olesen R, Erikstrup C, Solomon A et al. Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: a phase 1/2, single group, clinical trial. Lancet HIV 2014; 1: e12–e21. [DOI] [PubMed] [Google Scholar]
- Spivak AM, Andrade A, Eisele E, Hoh R, Bacchetti P, Bumpus NN et al. A pilot study assessing the safety and latency-reversing activity of disulfiram in HIV-1-infected adults on antiretroviral therapy. Clin Infect Dis 2014; 58: 883–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary SK, Archin NM, Margolis DM. Hexamethylbisacetamide and disruption of human immunodeficiency virus type 1 latency in CD4(+) T cells. J Infect Dis 2008; 197: 1162–1170. [DOI] [PubMed] [Google Scholar]
- Li Z, Guo J, Wu Y, Zhou Q. The BET bromodomain inhibitor JQ1 activates HIV latency through antagonizing Brd4 inhibition of Tat-transactivation. Nucleic Acids Res 2013; 41: 277–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhard W, Barreto K, Saunders A, Dahabieh MS, Johnson P, Sadowski I. The Suv39H1 methyltransferase inhibitor chaetocin causes induction of integrated HIV-1 without producing a T cell response. FEBS Lett 2011; 585: 3549–3554. [DOI] [PubMed] [Google Scholar]
- Churchill M, Nath A. Where does HIV hide? A focus on the central nervous system. Curr Opinion HIV AIDS 2013; 8: 165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray L, Roche M, Churchill MJ, Sterjovski J, Ellett A, Poumbourios P et al. Tissue-specific sequence alterations in the human immunodeficiency virus type 1 envelope favoring CCR5 usage contribute to persistence of dual-tropic virus in the brain. J Virol 2009; 83: 5430–5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber BT, Kunstman KJ, Patterson BK, Furtado M, McEvilly MM, Levy R et al. Genetic differences between blood- and brain-derived viral sequences from human immunodeficiency virus type 1-infected patients: evidence of conserved elements in the V3 region of the envelope protein of brain-derived sequences. J Virol 1994; 68: 7467–7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohagen A, Devitt A, Kunstman KJ, Gorry PR, Rose PP, Korber B et al. Genetic and functional analysis of full-length human immunodeficiency virus type 1 env genes derived from brain and blood of patients with AIDS. J Virol 2003; 77: 12336–12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas ER, Dunfee RL, Stanton J, Bogdan D, Kunstman K, Wolinsky SM et al. High frequency of defective vpu compared with tat and rev genes in brain from patients with HIV type 1-associated dementia. AIDS Res Hum Retroviruses 2007; 23: 575–580. [DOI] [PubMed] [Google Scholar]
- Thomas ER, Dunfee RL, Stanton J, Bogdan D, Taylor J, Kunstman K et al. Macrophage entry mediated by HIV Envs from brain and lymphoid tissues is determined by the capacity to use low CD4 levels and overall efficiency of fusion. Virology 2007; 360: 105–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornatore C, Nath A, Amemiya K, Major EO. Persistent human immunodeficiency virus type 1 infection in human fetal glial cells reactivated by T-cell factor(s) or by the cytokines tumor necrosis factor alpha and interleukin-1 beta. J Virol 1991; 65: 6094–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins BA, Dorn HH, Kelly WB, Armstrong RC, Potts BJ, Michaels F et al. Specific tropism of HIV-1 for microglial cells in primary human brain cultures. Science 1990; 249: 549–553. [DOI] [PubMed] [Google Scholar]
- Wong JK, Ignacio CC, Torriani F, Havlir D, Fitch NJ, Richman DD. In vivo compartmentalization of human immunodeficiency virus: evidence from the examination of pol sequences from autopsy tissues. J Virol 1997; 71: 2059–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray LR, Cowley D, Crespan E, Welsh C, Mackenzie C, Wesselingh SL et al. Reduced basal transcriptional activity of central nervous system-derived HIV type 1 long terminal repeats. AIDS Res Hum Retroviruses 2013; 29: 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen CA, Sodroski JG, Haseltine WA. The location of cis-acting regulatory sequences in the human T cell lymphotropic virus type III (HTLV-III/LAV) long terminal repeat. Cell 1985; 41: 813–823. [DOI] [PubMed] [Google Scholar]
- Siekevitz M, Josephs SF, Dukovich M, Peffer N, Wong-Staal F, Greene WC. Activation of the HIV-1 LTR by T cell mitogens and the trans-activator protein of HTLV-I. Science 1987; 238: 1575–1578. [DOI] [PubMed] [Google Scholar]
- Brady T, Agosto LM, Malani N, Berry CC, O'Doherty U, Bushman F. HIV integration site distributions in resting and activated CD4+ T cells infected in culture. AIDS 2009; 23: 1461–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan A, Defechereux P, Verdin E. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J 2001; 20: 1726–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol 2004; 2: E234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 2002; 110: 521–529. [DOI] [PubMed] [Google Scholar]
- He G, Margolis DM. Counterregulation of chromatin deacetylation and histone deacetylase occupancy at the integrated promoter of human immunodeficiency virus type 1 (HIV-1) by the HIV-1 repressor YY1 and HIV-1 activator Tat. Mol Cell Bioly 2002; 22: 2965–2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G, Espeseth A, Hazuda DJ, Margolis DM. c-Myc and Sp1 contribute to proviral latency by recruiting histone deacetylase 1 to the human immunodeficiency virus type 1 promoter. J Virology 2007; 81: 10914–10923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Mak G, Franza BR Jr. In vitro study of functional involvement of Sp1, NF-kappa B/Rel, and AP1 in phorbol 12-myristate 13-acetate-mediated HIV-1 long terminal repeat activation. J Biol Chem 1994; 269: 30616–30619. [PubMed] [Google Scholar]
- Margolis DM, Somasundaran M, Green MR. Human transcription factor YY1 represses human immunodeficiency virus type 1 transcription and virion production. J Virol 1994; 68: 905–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SA, Chen LF, Kwon H, Ruiz-Jarabo CM, Verdin E, Greene WC. NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J 2006; 25: 139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaehlcke K, Dorr A, Hetzer-Egger C, Kiermer V, Henklein P, Schnoelzer M et al. Acetylation of Tat defines a cyclinT1-independent step in HIV transactivation. Mol Cell 2003; 12: 167–176. [DOI] [PubMed] [Google Scholar]
- Mahmoudi T, Parra M, Vries RG, Kauder SE, Verrijzer CP, Ott M et al. The SWI/SNF chromatin-remodeling complex is a cofactor for Tat transactivation of the HIV promoter. J Biol Chem 2006; 281: 19960–19968. [DOI] [PubMed] [Google Scholar]
- Marzio G, Tyagi M, Gutierrez MI, Giacca M. HIV-1 tat transactivator recruits p300 and CREB-binding protein histone acetyltransferases to the viral promoter. Proc Natl Acad Sci USA 1998; 95: 13519–13524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger DJ, Eberharter A, John S, Grant PA, Workman JL. Purified histone acetyltransferase complexes stimulate HIV-1 transcription from preassembled nucleosomal arrays. Proc Natl Acad Sci USA 1998; 95: 12924–12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett JC, Miller-Jensen K, Shah PS, Arkin AP, Schaffer DV. Control of stochastic gene expression by host factors at the HIV promoter. PLoS Pathog 2009; 5: e1000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ait-Khaled M, McLaughlin JE, Johnson MA, Emery VC. Distinct HIV-1 long terminal repeat quasispecies present in nervous tissues compared to that in lung, blood and lymphoid tissues of an AIDS patient. AIDS 1995; 9: 675–683. [DOI] [PubMed] [Google Scholar]
- Buzy JM, Lindstrom LM, Zink MC, Clements JE. HIV-1 in the developing CNS: developmental differences in gene expression. Virology 1995; 210: 361–371. [DOI] [PubMed] [Google Scholar]
- Canonne-Hergaux F, Aunis D, Schaeffer E. Interactions of the transcription factor AP-1 with the long terminal repeat of different human immunodeficiency virus type 1 strains in Jurkat, glial, and neuronal cells. J Virol 1995; 69: 6634–6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corboy JR, Buzy JM, Zink MC, Clements JE. Expression directed from HIV long terminal repeats in the central nervous system of transgenic mice. Science 1992; 258: 1804–1808. [DOI] [PubMed] [Google Scholar]
- Corboy JR, Garl PJ. HIV-1 LTR DNA sequence variation in brain-derived isolates. J Neurovirol 1997; 3: 331–341. [DOI] [PubMed] [Google Scholar]
- Koyanagi Y, Miles S, Mitsuyasu RT, Merrill JE, Vinters HV, Chen IS. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science 1987; 236: 819–822. [DOI] [PubMed] [Google Scholar]
- Kurth J, Buzy JM, Lindstrom L, Clements JE. In vivo transcriptional regulation of the human immunodeficiency virus in the central nervous system in transgenic mice. J Virol 1996; 70: 7686–7694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeichner SL, Hirka G, Andrews PW, Alwine JC. Differentiation-dependent human immunodeficiency virus long terminal repeat regulatory elements active in human teratocarcinoma cells. J Virol 1992; 66: 2268–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrich D, Garcia J, Wu F, Mitsuyasu R, Gonazalez J, Gaynor R. Role of SP1-binding domains in in vivo transcriptional regulation of the human immunodeficiency virus type 1 long terminal repeat. J Virol 1989; 63: 2585–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross EK, Buckler-White AJ, Rabson AB, Englund G, Martin MA. Contribution of NF-kappa B and Sp1 binding motifs to the replicative capacity of human immunodeficiency virus type 1: distinct patterns of viral growth are determined by T-cell types. J Virol 1991; 65: 4350–4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sune C, Garcia-Blanco MA. Sp1 transcription factor is required for in vitro basal and Tat-activated transcription from the human immunodeficiency virus type 1 long terminal repeat. J Virol 1995; 69: 6572–6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorry PR, Bristol G, Zack JA, Ritola K, Swanstrom R, Birch CJ et al. Macrophage tropism of human immunodeficiency virus type 1 isolates from brain and lymphoid tissues predicts neurotropism independent of coreceptor specificity. J Virol 2001; 75: 10073–10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major EO, Miller AE, Mourrain P, Traub RG, de Widt E, Sever J. Establishment of a line of human fetal glial cells that supports JC virus multiplication. Proc Natl Acad Sci USA 1985; 82: 1257–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider U, Schwenk HU, Bornkamm G. Characterization of EBV-genome negative "null" and "T" cell lines derived from children with acute lymphoblastic leukemia and leukemic transformed non-Hodgkin lymphoma. Int J Cancer 1977; 19: 621–626. [DOI] [PubMed] [Google Scholar]
- Churchill MJ, Ramsay RG, Rhodes DI, Deacon NJ. c-Myb influences HIV type 1 gene expression and virus production. AIDS Res Hum Retroviruses 2001; 17: 1481–1488. [DOI] [PubMed] [Google Scholar]
- Churchill MJ, Gorry PR, Cowley D, Lal L, Sonza S, Purcell DF et al. Use of laser capture microdissection to detect integrated HIV-1 DNA in macrophages and astrocytes from autopsy brain tissues. J Neurovirol 2006; 12: 146–152. [DOI] [PubMed] [Google Scholar]
- Smit TK, Wang B, Ng T, Osborne R, Brew B, Saksena NK. Varied tropism of HIV-1 isolates derived from different regions of adult brain cortex discriminate between patients with and without AIDS dementia complex (ADC): evidence for neurotropic HIV variants. Virology 2001; 279: 509–526. [DOI] [PubMed] [Google Scholar]
- Thompson KA, Churchill MJ, Gorry PR, Sterjovski J, Oelrichs RB, Wesselingh SL et al. Astrocyte specific viral strains in HIV dementia. Ann Neurol 2004; 56: 873–877. [DOI] [PubMed] [Google Scholar]
- Shah S, Alexaki A, Pirrone V, Dahiya S, Nonnemacher MR, Wigdahl B. Functional properties of the HIV-1 long terminal repeat containing single-nucleotide polymorphisms in Sp site III and CCAAT/enhancer binding protein site I. Virol J 2014; 11: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KA, Kadonaga JT, Luciw PA, Tjian R. Activation of the AIDS retrovirus promoter by the cellular transcription factor, Sp1. Science 1986; 232: 755–759. [DOI] [PubMed] [Google Scholar]
- McAllister JJ, Phillips D, Millhouse S, Conner J, Hogan T, Ross HL et al. Analysis of the HIV-1 LTR NF-kappaB-proximal Sp site III: evidence for cell type-specific gene regulation and viral replication. Virology 2000; 274: 262–277. [DOI] [PubMed] [Google Scholar]
- Kilareski EM, Shah S, Nonnemacher MR, Wigdahl B. Regulation of HIV-1 transcription in cells of the monocyte-macrophage lineage. Retrovirology 2009; 6: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang CM, Roeder RG. Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science 1995; 267: 531–536. [DOI] [PubMed] [Google Scholar]
- Emili A, Greenblatt J, Ingles CJ. Species-specific interaction of the glutamine-rich activation domains of Sp1 with the TATA box-binding protein. Mol Cell Biol 1994; 14: 1582–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yedavalli VS, Benkirane M, Jeang KT. Tat and trans-activation-responsive (TAR) RNA-independent induction of HIV-1 long terminal repeat by human and murine cyclin T1 requires Sp1. J Biol Chem 2003; 278: 6404–6410. [DOI] [PubMed] [Google Scholar]
- Doetzlhofer A, Rotheneder H, Lagger G, Koranda M, Kurtev V, Brosch G et al. Histone deacetylase 1 can repress transcription by binding to Sp1. Mol Cell Biol 1999; 19: 5504–5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widlak P, Gaynor RB, Garrard WT. In vitro chromatin assembly of the HIV-1 promoter. ATP-dependent polar repositioning of nucleosomes by Sp1 and NF kappaB. J Biol Chem 1997; 272: 17654–17661. [DOI] [PubMed] [Google Scholar]
- Marban C, Suzanne S, Dequiedt F, de Walque S, Redel L, Van Lint C et al. Recruitment of chromatin-modifying enzymes by CTIP2 promotes HIV-1 transcriptional silencing. EMBO J 2007; 26: 412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leid M, Ishmael JE, Avram D, Shepherd D, Fraulob V, Dolle P. CTIP1 and CTIP2 are differentially expressed during mouse embryogenesis. Gene Expr Patterns 2004; 4: 733–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron 2005; 45: 207–221. [DOI] [PubMed] [Google Scholar]
- Marban C, Redel L, Suzanne S, Van Lint C, Lecestre D, Chasserot-Golaz S et al. COUP-TF interacting protein 2 represses the initial phase of HIV-1 gene transcription in human microglial cells. Nucleic Acids Res 2005; 33: 2318–2331. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- du Chene I, Basyuk E, Lin YL, Triboulet R, Knezevich A, Chable-Bessia C et al. Suv39H1 and HP1gamma are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J 2007; 26: 424–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsili G, Remoli AL, Sgarbanti M, Battistini A. Role of acetylases and deacetylase inhibitors in IRF-1-mediated HIV-1 long terminal repeat transcription. Ann N Y Acad Sci 2004; 1030: 636–643. [DOI] [PubMed] [Google Scholar]
- Brady J, Kashanchi F. Tat gets the "green" light on transcription initiation. Retrovirology 2005; 2: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanova A, Caro C, Jarjour RJ, Oster SK, Penn LZ, Germinario RJ. Repression of the human immunodeficiency virus type-1 long terminal repeat by the c-Myc oncoprotein. J Cell Biochem 2004; 92: 400–413. [DOI] [PubMed] [Google Scholar]
- Churchill MJ, Cowley DJ, Wesselingh SL, Gorry PR, Gray LR. HIV-1 transcriptional regulation in the central nervous system and implications for HIV cure research. J Neurovirol 2015; 21: 290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott JH, Wightman F, Solomon A, Ghneim K, Ahlers J, Cameron MJ et al. Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS Pathog 2014; 10: e1004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Søgaard OS, Graversen ME, Leth S, Brinkmann CR, Kjær A-S, Olesen R et al. The HDAC inhibitor romidepsin is safe and effectively reverses HIV-1 latency in vivo as measured by standard clinical assays. 20th International AIDS Conference (AIDS 2014) 2014; 1, TUAA0106LB. [Google Scholar]
- Deeks SG. HIV: shock and kill. Nature 2012; 487: 439–440. [DOI] [PubMed] [Google Scholar]
- International ASSWGoHIVC, Deeks SG, Autran B, Berkhout B, Benkirane M, Cairns S et al. Towards an HIV cure: a global scientific strategy. Nat Rev Immunol 2012; 12: 607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.