Abstract
Crystal structure of a vicilin, SM80.1, was determined towards exploring its possible physiological functions. The protein was purified from Solanum melongena by combination of ammonium sulphate fractionation and size exclusion chromatography. Structure was determined ab initio at resolution of 1.5 Å by X-ray crystallography showing the three-dimensional topology of the trimeric protein. Each monomer of SM80.1 consists of two similar domains with hydrophobic binding pocket and each accommodating different ligands, i.e. acetate and pyroglutamate. The relatively high stability of these independent anionic ligands in similar pockets indicated a strict requirement of stabilization by hydrogen bonds with the charged residues, suggesting a degree of plasticity within the binding pocket. Comparison of SM80.1 structure with those of other 7S vicilins indicated conservation of putative binding pocket for anionic ligands. Here we propose the possibility of trapping of these ligands in the protein for their requirement in the metabolic processes.
Seed proteins play a vital role in cellular growth and development, nutrient accumulation1, thiamine storage2, plant defense3, regulation of indole acetic acid (IAA) levels4. They have been shown to exhibit antimicrobial and antifungal activity5,6, hemagglutination activity7, desiccation tolerance8, ribosome inhibitory activity9 and many more. This suggests the seed proteins are indispensible for germination and further growth and development. Seed proteins are classified into four groups on the basis of solubility- albumin, globulin, prolamins and glutelins10.
Among the abundant seed proteins, globulins are considered a major family of seed proteome. Members of this family are classified on the basis of their sedimentation coefficient as 7S or 11S11. 7S globulins are also called vicilins and perform various functions, including the role in sucrose binding12, desiccation13, defense against microbes14 and oxidative stress15. The vicilin peptides formed by trypsin or chymotrypsin digestion exhibit antihypertensive effects16. Vicilins are considered to be the most potent class of allergenic proteins from seed proteome17.
Structurally, vicilins are trimeric, formed predominantly by non-covalent interactions. Canavalin, Phaseolin, β-conglycinin and AraH1 are a few vicilin structures determined from jackbean, french bean, soybean and peanut, respectively18,19,20,21. It is apparent that there could be many more physiological functions of vicilins that are yet to be identified. Comparative structural proteomics studies will help identify other possible physiological roles of vicilins or vicilin-like proteins. The structural insights themselves can prove to be equally interesting, as we found in the case of SM80.1 from Solanum melongena (eggplant) seeds.
The vicilin, SM80.1, from S. melongena was purified and the crystal structure determined using ab initio phasing method. Crystal structure showed presence of acetate and pyroglutamate molecules bound in structurally quasi-equivalent cavities in each of the two domains of SM80.1. To the best of our knowledge, the anionic ligands bound to 7S vicilin have been identified for the first time. Association of such specific moieties indicates possible roles of this protein in critical metabolic pathways, in addition to helping protein’s architectural stability. We were able to visualize a highly flexible loop region, which is otherwise missing or found disordered in other homologous vicilin structures. Indeed, vicilin-like fold exists in a wide range of organisms and is involved in variety of different functions.
Results and Discussion
Comparative structural biology of seed proteins has not been adequately explored, in spite of these being most abundant and easily accessible. Therefore, the available structure-function data of seed proteins are limited. Systematic studies involving purification of individual proteins and comparative structural analyses would provide greater physiological insights concerning their functions15. S. melongena (eggplant) is among the most abundantly used plant fruits which are rich in seeds. The protein composition of seeds of this plant, therefore contributes significantly to human health. Systematic analysis of the seed proteome was therefore initiated.
Protein purification and crystallization
Whole proteome of S. melongena seeds was subjected to fractionation followed by purification and crystallization of a dominant protein SM80.1. Washed, delipidified and grounded S. melongena seeds were subjected to ammonium sulphate fractionation over the range of 0 to 95%, which resulted into selective precipitation of different proteins on the basis of solubility. All fractions were electrophoresed along with low molecular weight markers (Sigma) (Supplementary Figure S1a). Five major protein bands were selected and transferred on PVDF membrane for characterization. Transferred proteins were stained with Ponceau S and subjected to Edman degradation for identification of N-terminal amino acid sequence. Band 1 showed significant homology with an allergy-related protein from Solanum lycopersicum22. Bands 2 and 3 showed homology with hypothetical protein from Vitis vinifera whereas bands 4 and 5 belonged to 11S globulin family and have similarity with 11S legumin from Sesamum indicum (Table 1). Based on the limited N-terminal sequence and abundance, band 1 was selected for further purification and characterization which was discussed in our previous paper23. Purification profile of 45 kDa band 1 SM80.1 protein by size exclusion chromatography is shown in Supplementary Figure S1b.
Table 1. N-terminal sequence of different protein bands.
| Segregated Proteins | N-terminal Sequence | Homologous Proteins |
|---|---|---|
| Band 1 | PGREQQEENVPYLFKSQRSQSRSRASHMDF | Allergenic vicilin from Solanum lycopersicum |
| Band 2 | YKEYPGQHGQQGQTGI-P/I-LTXQARHQR/V | Hypothetical protein sorbidraft, Vitis vinifera |
| Band 3 | GLEENIQTTKIRTNMEEYYYADIYVI | Hypothetical protein Vitis vinifera, Arabidopsis lyrata |
| Band 4 | GLEETIRSAKLRENNDNPPAAADVYNPQGG | 11S globulin storage Sesamum indicum |
| Band 5 | GIEETYTMKLRENIGHPXXXDDVNNPRGR | 11S globulin storage Sesamum indicum |
To acquire the full length sequence of SM80.1, proteolytic digestion was carried out. Enzymes like trypsin, chymotrypsin, endoproteinase Glu V8 were used for protein digestion to generate short peptide fragments (Fig. 1a). These fragments were vacuum-dried and then passed through nano LC-mass spectrometer followed by data processing. Sequence alignment obtained from search engine MASCOT revealed that this protein has about 85% sequence homology with the vicilins of S. lycopersicum.
Figure 1. Sequence alignment of SM80.1 protein.
(a) The sequence alignments of peptides obtained by MASCOT database search utilizing MS-MS of proteolytically (Trypsin, Chymotrypsin, GluV8) digested protein fragments with protein sequence of 7S vicilin from Solanum lycopersicum. N-terminal is obtained by Edman degradation chemistry. Residues that are non-identified between the two proteins are marked in grey. (b) The alignment of amino acid sequence of SM80.1 identified on the basis of crystal structure with sequence obtained from mass spectrometry by MASCOT database search and N-terminal. Different residues among the sequences are highlighted with grey background.
Further, crystallization of the purified protein was attempted as reported earlier23. Another crystal form was also obtained which diffracted to a higher resolution. Two different crystal forms of SM80.1 thus obtained, belonged to P6322 and R32 space groups with the unit cell dimensions a = 117.9 Å, c = 123.5 Å and a = 119.4 Å, c = 158.0 Å, respectively. All subsequent crystallographic analyses were carried out using only the R32 crystal form that diffracted at higher resolution. The data collection and processing statistics are summarized in Table 2. The asymmetric unit of SM80.1 crystals contains one monomer in the asymmetric unit with 49.0% solvent content, as calculated on the basis of Matthews coefficient.
Table 2. Data collection and S-SAD phasing statistics.
| Dataset1 SAD 0 Kappa* | Dataset2 SAD 40 Kappa* | Dataset3 SAD 80 Kappa* | Merged SAD* | Dataset4 Native# | |
|---|---|---|---|---|---|
| Space group | R32 | R32 | R32 | R32 | R32 |
| Unit-cell parameters | |||||
| a (Å) | 119.49 | 119.47 | 119.45 | 119.48 | 119.36 |
| b (Å) | 119.49 | 119.47 | 119.45 | 119.48 | 119.36 |
| c (Å) | 158.09 | 157.95 | 157.93 | 158 | 158 |
| Wavelength (Å) | 1.771 | 1.771 | 1.771 | 1.771 | 0.95372 |
| Detector distance | 102.17 | 102.17 | 102.17 | – | 119.30 |
| Oscillation | 1 | 1 | 1 | – | 0.5 |
| No of images | 360 | 360 | 360 | 1080 | 400 |
| Total reflections | 494720 (45039) | 492603 (45736) | 490806 (42860) | 1479455 (134513) | 829874 (106878) |
| Unique reflections | 22886 (2237) | 22714 (2212) | 22847 (2230) | 22848 (2242) | 70495 (10235) |
| Resolution range (Å) | 50–2.18 | 50–2.18 | 50–2.18 | 50–2.18 | 22.71–1.49 |
| Completeness (%) | 100.0 (99.6) | 99.8 (98.4) | 99.9 (99.4) | 100 (99.1) | 99.9 (100) |
| Redundancy | 21.6 (20.1) | 21.7 (20.7) | 21.5 (19.2) | 64.8 (60.0) | 11.8 (10.4) |
| Mean I/(I) | 32.9 (7.8) | 36.0 (7.2) | 34.3 (6.2) | 59.8 (12.3) | 21.1 (4.3) |
| Rmerge† (%) | 8.5 (40.1) | 8.8 (44.9) | 9.1 (46.9) | 8.8 (44.7) | 6.3 (49.9) |
| Rmeas†† | 8.7 (41.1) | 9.0 (46.0) | 9.3 (48.1) | 8.9 (45.1) | 6.5 (52.4) |
| Rpim††† | 1.9 (9.1) | 1.9 (10.1) | 2.0 (10.9) | 1.1 (5.8) | 1.9 (15.9) |
| CC1/2 | 0.999 (0.984) | 0.999 (0.984) | 0.999 (0.978) | 1.000 (0.994) | 0.999 (0.934) |
†Rmerge = ΣhklΣi|Ii(hkl) − <I(hkl)> |/Σhkl ΣiIi(hkl) where Ii(hkl)is the ith measurement of the intensity of reflection hkl and <I(hkl)> is the mean intensity of reflection hkl.
††Rmeas (redundancy-independent Rmerge) = Σhkl[Nhkl/(Nhkl − 1)]1/2Σi|Ii(hkl) − <I(hkl)> |/Σhkl ΣiIi(hkl).
†††Rpim (precision-indicating Rmerge) = Σhkl[1/(Nhkl − 1)]1/2Σi|Ii(hkl) − <I(hkl)> |/Σhkl ΣiIi(hkl).
*Data processed using HKl2000 and merging statistics calculated using Phenix.
#Data processed using imosflm.
Structure determination
We tried ab initio S-SAD phasing method for determining the preliminary phases. Eight inherent sulphur atoms (six methionines and two cysteines) were used. PHENIX package24 was utilized to extract the initial phase information from experimental anomalous intensities. For the purpose of SAD phasing, three different kappa 0, 40, 80, and fine-slicing oscillations were taken into consideration which contributed to 64.8 fold redundancy (Table 2). A preliminary partial poly-Ala model was built automatically by PHENIX Autosol software and it was further enhanced using AutoBuild through iterative model building and refinement. A total of 225 residues were built and 30 chains were placed with Rwork and Rfree values of 46.0% and 50.0% respectively. Model completion, addition of waters and occupancy refinement were carried out using COOT25 and CNS26. The model was refined to Rwork of 19.94% and Rfree of 21.02% with 98.5% of the residues in the allowed region of the Ramachandran plot. Remaining residues are labeled as outlier and are present in loop region. This region shows high flexibility with relatively poor electron density. The final refinement parameters are listed in Table 3. Protein sequencing methods and interpretation of electron density map at a resolution of 1.5 Å identified the complete sequence of SM80.1 (Fig. 1b).
Table 3. Refinement statistics.
| Resolution range (Å) | 22.71 −1.49 |
| No. of reflections, working set | 66896 |
| No. of reflections, test set | 3556 |
| Final Rwork (%) | 19.94 |
| Final Rfree (%) | 21.02 |
| No. of non-H atoms | |
| Protein | 2912 |
| Ion | 1 |
| Ligand | |
| ACETATE | 4 |
| PYROGLUTAMATE | 9 |
| Water | 256 |
| Total | 3182 |
| R.M.S. deviations | |
| Bonds (Å) | 0.009 |
| Angles (°) | 1.97 |
| Average B factors (Å2) | |
| Protein | 20.51 |
| Ion | 14.770 |
| Ligand | |
| ACETATE | 14.343 |
| PYROGLUTAMATE | 46.509 |
| Water | 32.08 |
| Ramachandran plot | |
| Most favoured (%) | 92.1 |
| Allowed (%) | 6.4 |
| Outlier (%) | 1.5 |
Overall Structure
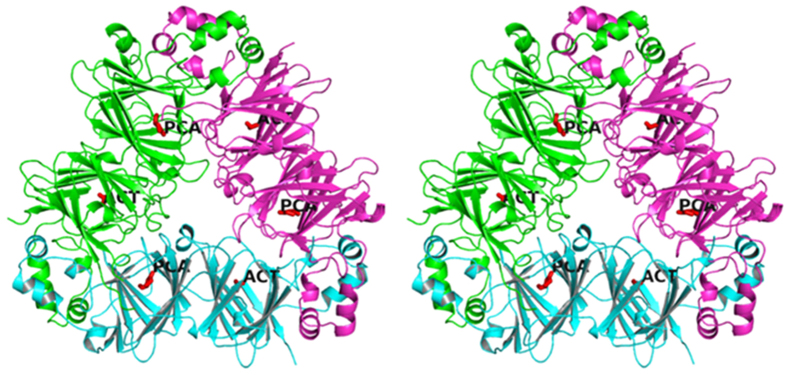
The overall crystal structure of SM80.1 consists of 392 residues of which the region 287–297 is structurally disordered and showed broken density whereas the core is clearly resolved. SM80.1 structure consists of α-helices, β-sheets and flexible loops. There is one monomer in the asymmetric unit. Further, examination of the region around the asymmetric unit in crystal structure revealed that the protein exists as a homotrimer. This trimer formation around the threefold crystallographic axis, as shown in ribbon diagram (Fig. 2), resembles that in the other vicilins. The trimerization helps in providing thermostability to the protein11. Each monomer also shows formation of core region by pair of β-barrels and helices protruding outward from each core to interact with the neighboring monomers, resulting in the formation of a trimer. It is observed that quaternary structure is stabilized by electrostatic interactions.
Figure 2. Stereo view of ribbon diagram of the trimeric protein along three fold axis of symmetry showing the overall structure of SM80.1 protein.
PCA and ACT are pyroglutamate and acetate identified in two domains of SM80.1 protein.
Monomer can further be divide across a pseudo-dyad axis into two similar halves, having a core region and extended arms, forming two domains. These two domains are named N- and C-terminal domains. Core region of each domain is formed by β-strands whereas the helices are involved in the formation of extended arms. Apart from these secondary structures, short β-turn is also present in the N-terminal of SM80.1 protein structure. Magnesium (Mg) ion is also present in the structure and exists at the interface of the monomers. The chemical environment of Mg ion is favoured by Gly28 and Ala25 with 3 water molecules present at a distance of 2.36, 2.34, 2.44, 2.43 and 2.39 Å, respectively. B-factor of the magnesium ion is 14.77 Å2 which is low in comparison to overall B-factor of protein or water molecules. This is because coordinating residues also show low B-factors (Bavg = 14.31 Å2). Anomalous difference Fourier map was calculated using “SAD” data to check the position of Mg ion and Sulphur atoms. The anomalous difference Fourier map identified peak height of 4.5 σ for magnesium ion. Also, all the sulphur atoms were identified at correct positions with an average peak height of 16.63 σ. This information is reflected in the supplementary material (Supplementary Table S1). The final model has 256 water molecules. More than 95% residues, i.e. around 359 out of 384 residues are surface exposed with overall surface area being around 17750.3 Å2.
Apart from electrostatic interactions, hydrophobic interactions at each monomer-monomer interface are critical in stabilization of the tertiary structure of SM80.1. Hydrophobic residues form hydrophobic patches which are involved in trimer formation by locking the extended arms. This suggested importance of these interactions for the formation of quaternary structure in this family of proteins.
Structural comparisons with related proteins
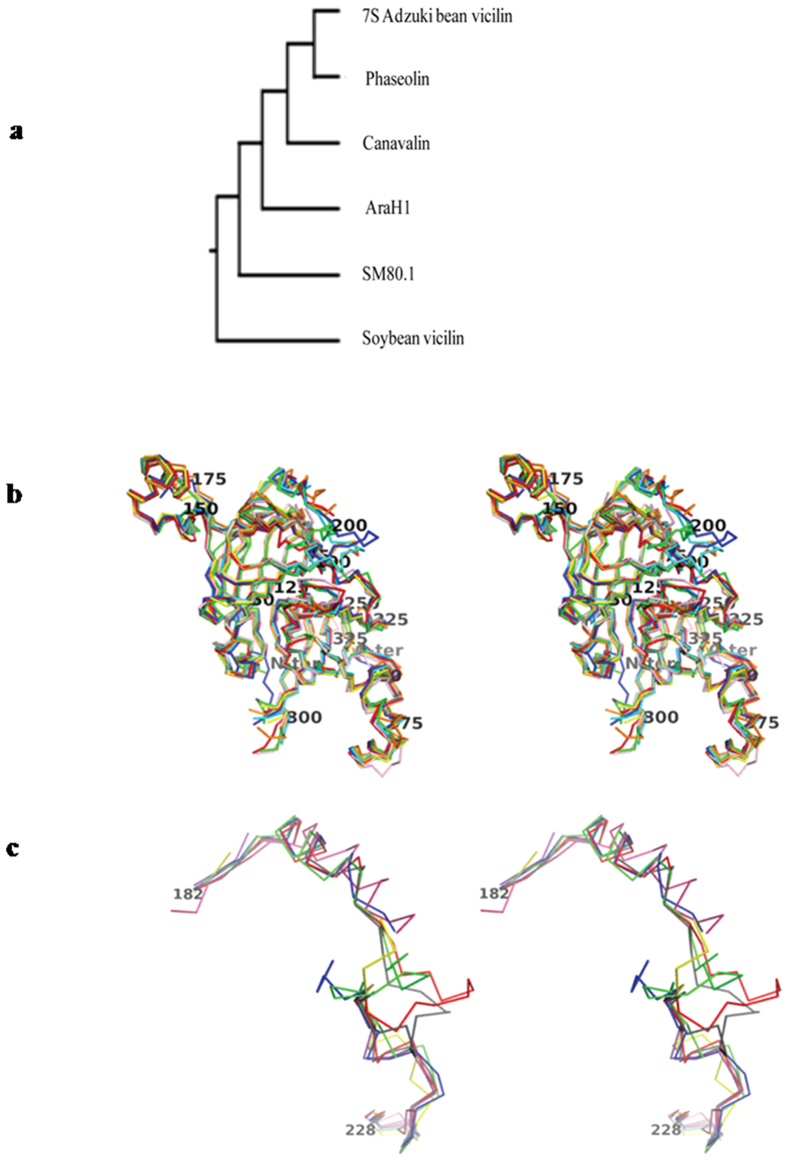
Dali server (DaliLite ver. 3) was used for identifying structural correlation with other canonical proteins. Protein structure based search engine27, indicated many structures similar to SM80.1 with very low sequence homology. Supplementary Table S2 shows that these proteins have below 30% sequence homology indicating that a common structural fold is being used, perhaps for different functions. A phylogenetic tree of all these proteins explains the relationship and diversification among the members (Fig. 3a). Structural comparisons showed that vicilins AraH1 (pdb: 3SMH), 7S globulin-1 adzuki bean (pdb: 2EA7), β-conglycinin (pdb: 1UIJ), 8S mungbean storage protein (pdb: 2CV6), Korean pine vicilin (pdb: 4LEJ), canavalin (pdb: 2CAV), phaseolin (pdb: PHL) have high structural homology (RMSD less than 2 Å) with SM80.118,20,21,28,29,30,31. Dali analysis also suggested that similar fold is spread across in all the species. It is evident that Bacillus subtilis (pdb: 1L3J), Pseudomonas aeruginosa (pdb: 1SQ4), Geobacillus kaustophilus (pdb: 2P17), Marinobacter aquaeolei (pdb: 3O14), Xanthomonas campestris (pdb: 3H50), Chlamydomonas reinhardtii (pdb: 2V4A) as well as Homo sapiens (pdb: 2W2I) exhibit similar fold although with weak homology. To have better understanding, carbon alpha (Cα) chains of monomeric unit of these proteins were superimposed (Fig. 3b), leading to identification of flexible loop with high B-factors. Superimposition of this particular loop is shown in Fig. 3c. High structural homology of SM80.1 with many other vicilins did not significantly reflect in sequence. AraH1 from peanut and 7S adzuki bean vicilin showed highest structural similarity with SM80.1 with RMSD value of 1.4 Å and 1.3 Å, respectively as analysed by Dali server for complete trimeric molecule.
Figure 3. Cα comparison of structures homologous to SM80.1 protein.
(a) Dendrogram of high fold similarity structure of SM80.1 with 7S vicilin of Adzuki bean, Phaseolin, Canavalin, AraH1 and Soybean. (b) Monomer superimposition of Cα chain of SM80.1 (green), Adzuki bean (blue), AraH1 (red), Phaseolin (orange), Canavalin (yellow), Soybean (cyan) and Korean pine vicilin (pink). N-terminal, C-terminal and every consecutive 25th amino acid of SM80.1 vicilin is labeled. (c) Superimposition of comparative loop of all the above structures.
As in the case of most vicilins, SM80.1 also showed presence of hydrophobic interactions playing critical role in trimer formation and stability, thus protecting epitopes from digestive enzymes21. Comparison of SM80.1 with adzuki bean 7S vicilin28 monomer using PyMol shows RMSD of 0.979 Å (Fig. 3b). SM80.1 shows difference in position of short β-turns which are present at the N-terminus and before first β-strand whereas in adzuki bean 7S vicilin a short β-turn is present after first β-strand. In the case of adzuki bean 7S vicilin, loops corresponding to residues Glu188 to Glu196 and Ser222 to Ser227 are disordered, which is not the case with SM80.1, although the corresponding residues show comparatively higher B-factors (Supplementary Figure S2).
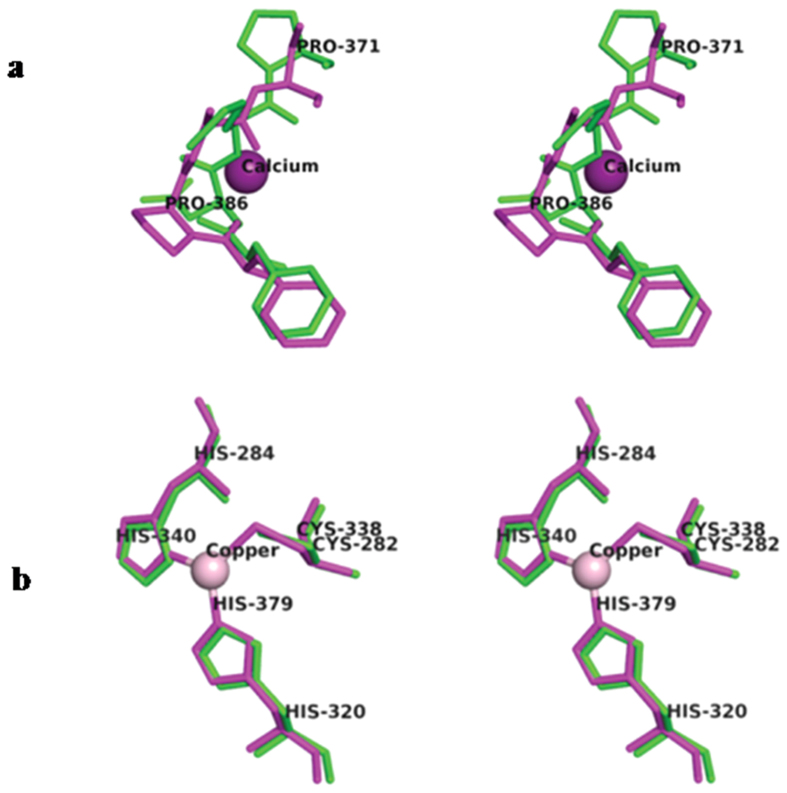
Metal binding
SM80.1 lacks bound calcium despite its structure being similar to that of 7S adzuki bean vicilin, a protein showing bound calcium28. This is probably due to a substantial difference in the geometrical arrangement of side chains at the canonical calcium binding site. Thus, even slight variation in this local sequence can disallow calcium binding, although binding motif of helix-loop-helix is conserved. Indeed, this topological arrangement of side chains is important for metal binding (Fig. 4a). On the contrary, sometimes even after conservation of geometrical arrangement of side chains, metal binding is not observed. For example, bound copper is not found in SM80.1 although residues required for coordinating copper are present. On the other hand, similar residues i.e two histidines and one cysteine are favouring existence of copper center in Korean pine vicilin (Fig. 4b)30. Careful analysis in SM80.1 structure around the positions where calcium or copper were present in other homologous proteins, 7S adzuki bean vicilin and Korean pine vicilin, indicating absence of electron density in difference maps implying absence of bound calcium or copper, in SM80.1. Above analyses indicate subtle but critical variations in 7S vicilins, a possible indication that it is associated with a variety of functions, which include assisting metabolites and other related molecules during germination and growth.
Figure 4. Stereo view comparison of the metal binding site in SM80.1 with other homologous vicilin.
(a) Helix loop helix motif of SM80.1 (green) and adzuki bean 7S vicilin (magenta). (b) Copper binding site in SM80.1 (green) and Korean pine vicilin (magenta).
Bound metabolites
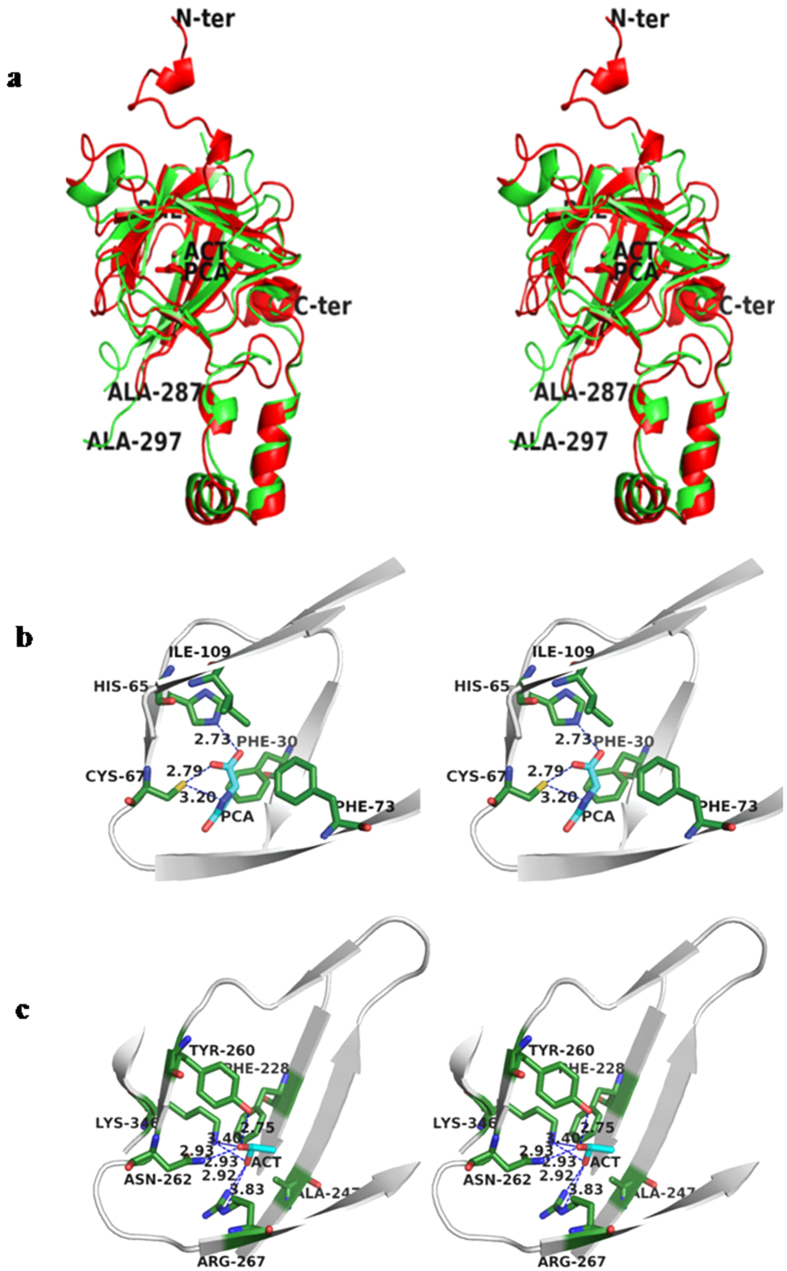
We were able to identify the bound ligands based on the high resolution electron density map at 1.5 Å resolution. Both the N- and the C-terminal domains of SM80.1 form a central cavity each. In these predominantly hydrophobic and structurally similar cavities, two different ligands were observed (Fig. 5a). The N-terminal domain has cyclic lactam of glutamate, called pyroglutamate. Acetate moiety was present in the corresponding pocket in the C-terminal domain (Supplementary Figure S3). It appears that the two ligands may be contributing to the structural stability of the protein. Presence of metabolite in the cavity is consistent with metabolic profiling data that identified pyroglutamate in the extract of tomato fruit and seeds32.
Figure 5. Stereo view of anionic ligand binding pockets in SM80.1.
(a) Superimposition of binding pocket of two domains showing the plasticity for different ligands. (b) Pyroglutamate binding pocket with pyroglutamate (PCA) present in center surrounded by interacting residues. (c) Acetate binding pocket with acetate present in the central cavity and interacting residues are labeled in black.
Pyroglutamate is present in the largely hydrophobic core of N-terminal β-barrel domain (Fig. 5b). Surface area occupied by pyroglutamate is around 249.8 Å2. Pyroglutamate is stabilized by three electrostatic interactions. Pyroglutamate (N), (O) and (OXT) are forming electrostatic bond with Cys67, His65, Cys67 at a distance of 3.20 Å, 2.73 Å, 2.79 Å (Supplementary Table S3). With buried surface area of 243.92 Å2 out of the total accessible surface area (ASA) of 249.77 Å2, pyroglutamate is completely buried. Phe30, His65, Cys67, Tyr73 and Ile109 are the residues in the close proximity and interacting with pyroglutamate in the N-terminal core region.
In the core cavity of the C-terminal β-barrel, clearly evident was the presence of an acetate moiety fully buried, occupying around 187.1 Å2 surface area (Fig. 5c). Acetate is making in total six electrostatic interactions, three each with [O] and [OXT]. Acetate [OXT] is forming bond with Asn262, Tyr260 and Lys346 at a distance of 2.93 Å, 2.75 Å and 3.40 Å respectively. Similarly, acetate [O] is forming bond with Lys 346 and two bonds with Arg267 at a distance of 2.93 Å, 2.92 Å and 3.83 Å respectively. Detailed statistics of ligand electrostatic interaction are enumerated in Supplementary Table S3. Superimposition of the ligand binding sites in the two domains is shown in Fig. 5a. It is evident that both the ligands exist at equivalent positions in the largely hydrophobic pockets and are stabilized by electrostatic interactions involving charged residues.
On comparison of bound ligand with other vicilins, it is observed that glycerol is found in Korean pine vicilin at the position where pyroglutamate is present in SM80.1. However, this pocket is empty in AraH1, Canavalin, Phaseolin and in β-conglycinin. But, when the electron density was evaluated in the corresponding pocket in the molecule of adzuki bean 7S vicilin, an unexplained density was seen around the same position, suggesting that it was not analysed in the light of such a molecule. Therefore, possibility of presence of pyroglutamate cannot be avoided in adzuki bean 7S vicilin on the basis on unexplained density. Thus, both SM80.1 and probably adzuki bean 7S vicilin have pyroglutamate or similar metabolite in the N-terminal domain and acetate molecule within same region in the C-terminal β-barrel domain.
Physiological implication
Structural analysis identified putative binding pocket for anionic ligands in the predominantly hydrophobic core. In addition to playing a role in maintaining structural integrity of the protein and probably serving as energy source, they are likely to be involved in metabolic pathways in temporal fashion.
Pyroglutamate, an intermediate metabolite has not been discovered earlier in any other vicilin structure. As such, pyroglutamate is ubiquitously present in living cells ranging from archaebacteria to humans and is involved in all glutamate linked processes33. Apart from N-terminal modification, pyroglutamate also exists as free cellular metabolite. Free cellular pyroglutamate may act as an analogue or precursor of glutamate. On the other hand, acetate the other metabolite present in SM80.1 probably meets the requirement of oil as precursor moiety34,35.
In SM80.1, pyroglutamate and acetate could have role in Calvin cycle and further, pyroglutamate may be involved in glutathione metabolism. Calvin cycle is one of the cardinal metabolic pathways in plants36. Acetate is required in Calvin cycle and is produced by protein catabolism37. Oxidation of acetate leads to the formation of central metabolite of Calvin cycle i.e. acetyl-CoA38. This undergoes series of chemical reactions and forms an intermediate product 2–oxoglutarate/α-ketoglutarate which acts as a precursor for the biosynthesis of pyroglutamate39. Pyroglutamate could be further involved in glutathione metabolism and play a critical role in oxidative stress40.
The fact that these two closely linked metabolites are found in bound form with SM80.1 suggests attractive possibility of this protein being important in stress management. Also due to the inert nature and high water binding capacity, pyroglutamate is an excellent candidate to serve as an osmoprotectant33, a property appropriate for stress management. Acetate and pyroglutamate both act as transitional partners and play a role in metabolic pathways thereby suggesting that they assist in growth and germination of seeds.
Materials and Methods
Protein purification
Eggplant (Solanum melongena) seeds were procured from the National Seeds Corporation, Indian Agricultural Research Institute (IARI), New Delhi, India. Seeds (50 g) were grounded to fine powder and defatted using petroleum ether. Defatted powder was then homogenized with 50 mM Tris-HCl, pH 8 containing 150 mM NaCl by continuous stirring for 4 hrs at 277 K in the presence of a protease inhibitor cocktail (Sigma). The crude extract was prepared by centrifugation at 7000 rpm for 3 hrs and then subjected to salt fractionation over a concentration range of 0–95% (w/v) ammonium sulphate. Pellets thus obtained at each step were resuspended in 50 mM Tris-HCl buffer pH 8 and dialyzed to remove the traces of salt. Resuspended pellets were further electrophoresed on 15% SDS PAGE to identify the proteins present in each fraction. The isolation and purification of proteins was carried out from 80% ammonium sulphate fraction by gel-filtration chromatographic technique, using manually packed sephacryl-200 column, pre-equilibrated with 50 mM Tris-HCl, pH 8. Protein concentration at different levels of purification was determined by BCA protein estimation assay.
Protein sequencing
To identify the complete sequence of the purified protein by internal sequencing trifluoroethanol (TFE) protein digestion protocol was followed with slight modification (Agilent Technologies). 250 μg of protein was dissolved in 100 mM ammonium bicarbonate buffer and denatured by TFE. It was then reduced and denatured at 333 K by addition of 200 mM DTT for 45 min. Subsequently, alkylation was done in the dark by addition of 200 mM iodoacetamide for an hour at room temperature. DTT was again added to destroy the excess of iodoacetamide in dark for one hour. Further, milliQ water was added to dilute the denaturant, and ammonium bicarbonate solution was used to raise the pH. Different enzymes were used to digest the protein at 1:50 enzyme:substrate concentration and all solutions were incubated overnight at 298 K. Absolute formic acid was used to lower the pH and stop the enzyme activity. The resulting solutions were dried in a vacuum centrifuge and resuspended in 50% acetonitrile +0.1% formic acid. The digest was diluted and analyzed by mass spectrometry.
Mass Spectrometry
Samples were subjected to an Eksigent Nano LC ultra 2D plus system (AB Sciex, Waldbronn, Germany) connected to Hybrid Quadrupole-TOF LC/MS/MS Mass Spectrometer (AB Sciex, Waldbronn, Germany). The calibration of system was done using beta-galactosidase and combined with C18 trap column and C18 RP analytical column for the analysis of digested peptide fragments. Sample trapping and washing was carried out at a flow of 5 μl in 12 min run time with 100% solvent A (water +0.1% formic acid), and elution was accomplished during a 45 min. gradient from 13% to 32% Solvent B (ACN +0.1% formic acid) at 550 nL/min. The positive ion mode was selected for running the sample, with MS window from 350 to 1250 m/z. Parent ions were fragmented by Atmospheric Pressure Ionization. The flow rate was split less and the column flow through was directly introduced into the Nano spray III ion source. MS-MS data were extracted for database searches.
Database searches
MS-MS data analysis and generation of peak list were performed with the protein pilot software (version 4.5, Applied Biosystems). Highly specific rigorous searches (MASCOT) were useful to identify either known proteins (i.e. in database), or unknown proteins (identical peptides). For the analysis, peptide precursor tolerance was set to 100 ppm for 2 and 3 peptide charges (ESI), and MS/MS tolerance was 0.9 Da (ESI) within MASCOT parameters. Also carbamidomethyl (C) was marked as fixed modification whereas oxidation (M) and deamidation (NQ) as variable modifications. Sequence alignment was carried out using Clustal W41. Sequence of vicilin from Solanum lycopersicum was used as a guide to align the sequences of proteolytic fragments obtained from MASCOT. The part of sequence which was not sequenced biochemically was identified crystallographically using high resolution electron density map.
Crystallization
Initial crystallization trials for the purified protein (10 mg/ml) involved the exploration of various precipitants, detergents, concentrations and pH values. The final diffraction data were collected from crystals grown in a drop composed of 2 μl protein solution with 2 μl reservoir solution containing 10% (w/v) PEG 3350, 1.5 M–2 M Sodium malonate by hanging drop vapour diffusion at 25°C. Surprisingly, two different crystal forms were obtained in above conditions. Since Crystallization condition contains sodium malonate, no cryoprotectant was used for data collection.
Data Collection and processing
For ab initio structure solution, inherent sulphur atoms present in the protein were utilized for single anomalous diffraction (SAD) phasing42. Data for both native and SAD were collected at beamline BM14, European Synchrotron Radiation Facility (ESRF), Grenoble, France at wavelengths of 0.953 Å and 1.771 Å respectively. The data were collected at 120 K with crystal-to-detector distance of 120 mm, an oscillation range of 0.5° and an exposure time of 2 sec per image. Data were integrated and scaled with imosflm or HKL2000. Rmeas, Rpim and CC1/2 were extracted by using merging statistics tool of PHENIX software suite (Phenix version 3.0.1).
Ab initio structure solution
Although molecular replacement could be done using the protein structures having weak homology with 7S vicilins as discussed in our earlier paper23, we exploited the initial phase information by S-SAD phasing method to solve the structure. Two data sets at 1.5 Å and 2.18 Å resolution were collected from the same crystal, at 120 K. To increase the accuracy, high multiplicity of the data at the 2.18 Å resolution (64.8-fold) and fine slicing rotations (0.5°) and three different kappa angles were used. Single-wavelength anomalous diffraction (SAD) using sulphur was carried out for the determination of phases and structure was refined. Data were indexed, integrated and final scaling for the native data was carried out by SCALEPACK. For merging, SAD datasets were also processed with same settings as the native data set. The PHENIX software suite24 module, AutoSol was used to locate the anomalous scatterers from the S-SAD intensity data, calculating the initial experimental phases, density modification and preliminary model building. High resolution data were used by AutoBuild for improving the phases and model building. COOT25 was used to build the missing residues and side chains of the model. CNS26 was used for B-factor and minimization. The theoretical Bijvoet pairs were calculated using the modified Hendrickson formula in Autosol program of PHENIX software42,43. The quality of atomic model was assessed with PROCHECK44. The figures were generated using PyMOL45. The final model and structure factors have been deposited in protein data bank (PDB ID: 5CAD).
Additional Information
How to cite this article: Jain, A. et al. Crystal structure of the vicilin from Solanum melongena reveals existence of different anionic ligands in structurally similar pockets. Sci. Rep. 6, 23600; doi: 10.1038/srep23600 (2016).
Supplementary Material
Acknowledgments
We thank the EMBL staff Dr. Hassan Belrhali and Dr. Babu A. Manjasetty for providing support on the beamline and EMBL-DBT for providing access to the BM14 beamline at the ESRF. We thank Dr. Deepti Jain for critically reading the manuscript. We also acknowledge financial support from the Departments of Science & Technology and Biotechnology, Govt of India.
Footnotes
Author Contributions A.J. and D.M.S. conceived and designed the experiments, A.J. and A.K. performed the experiments, A.J., A.K. and D.M.S. analyzed the data, A.J. and D.M.S. wrote the manuscript. Manuscript is reviewed, discussed and approved by all the authors.
References
- Agrawal L. et al. Comparative proteomics reveals a role for seed storage protein AmA1 in cellular growth, development, and nutrient accumulation. J Proteome Res. 12, 4904–4930 (2013). [DOI] [PubMed] [Google Scholar]
- Gołda A., Szyniarowski P., Ostrowska K., Kozik A. & Rąpała-Kozik M. Thiamine binding and metabolism in germinating seeds of selected cereals and legumes. Plant Physiol. Biochem. 42, 187–195 (2004). [DOI] [PubMed] [Google Scholar]
- Borad V. & Sriram S. Pathogenesis-related proteins for the plant protection. Asian J. Exp. Sci. 22, 189–169 (2008). [Google Scholar]
- Delatorre P. et al. Interactions between indole-3-acetic acid (IAA) with a lectin from Canavalia maritima seeds reveal a new function for lectins in plant physiology. Biochimie 95, 1697–703 (2013). [DOI] [PubMed] [Google Scholar]
- de Souza Cândido E. et al. Plant storage proteins with antimicrobial activity: novel insights into plant defense mechanisms. FASEB J. 25, 3290–3305 (2011). [DOI] [PubMed] [Google Scholar]
- Ribeiro S. F. et al. Antifungal and other biological activities of two 2S albumin-homologous proteins against pathogenic fungi. Protein J. 31, 59–67 (2012). [DOI] [PubMed] [Google Scholar]
- Nair D. N., Singh V., Yamaguchi Y. & Singh D. D. Jatropha curcas hemagglutinin is similar to a 2S albumin allergen from the same source and has unique sugar affinities. Planta 236, 1499–1505 (2012). [DOI] [PubMed] [Google Scholar]
- Businge E., Bygdell J., Wingsle G., Moritz T. & Egertsdotter U. The effect of carbohydrates and osmoticum on storage reserve accumulation and germination of Norway spruce somatic embryos. Physiologia plantarum 149, 273–285 (2013). [DOI] [PubMed] [Google Scholar]
- Peumans W. J., Hao Q. & van Damme E. J. Ribosome-inactivating proteins from plants: more than RNA N-glycosidases? FASEB J. 15, 1493–1506 (2001). [DOI] [PubMed] [Google Scholar]
- Osborne T. The vegetable proteins. 1924. Brown, WL: J. biol. Chem. 154, 57 (1944). [Google Scholar]
- Hauser M. et al. Molecular properties of plant food allergens: a current classification into protein families. The Open Immunology Journal 1, 1–12 (2008). [Google Scholar]
- Rose T. L., Gomes V. M., Da Cunha M., Fernandes K. V. & Xavier-Filho J. Effect of sugars on the association between cowpea vicilin (7S storage proteins) and fungal cells. Biocell. 27, 173–9 (2003). [PubMed] [Google Scholar]
- Wang W. Q., Moller I. M. & Song S. Q. Proteomic analysis of embryonic axis of Pisum sativum seeds during germination and identification of proteins associated with loss of desiccation tolerance. J. Proteomics 77, 68–86 (2012). [DOI] [PubMed] [Google Scholar]
- Rose T. L. et al. Defense proteins from Vigna unguiculata seed exudates: characterization and inhibitory activity against Fusarium oxysporum. Plant and soil 286, 181–191 (2006). [Google Scholar]
- Gaur V., Qureshi I. A., Singh A., Chanana V. & Salunke D. M. Crystal structure and functional insights of hemopexin fold protein from grass pea. Plant Physiol. 152, 1842–50 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viernes L., Garcia R., Torio M. & Angelia M. Antihypertensive Peptides from Vicilin, the Major Storage Protein of Mung Bean (Vigna radiata (L.) R. Wilczek). Journal of Biological Sciences 12, 393 (2012). [Google Scholar]
- Dooper M. M., Plassen C., Holden L., Lindvik H. & Faeste C. K. Immunoglobulin E cross-reactivity between lupine conglutins and peanut allergens in serum of lupine-allergic individuals. J. Investig. Allergol. Clin. Immunol. 19, 283–91 (2009). [PubMed] [Google Scholar]
- Ko T. P., Day J. & McPherson A. The refined structure of canavalin from jack bean in two crystal forms at 2.1 and 2.0 A resolution. Acta Crystallogr. D Biol. Crystallogr. 56, 411–20 (2000). [DOI] [PubMed] [Google Scholar]
- Lawrence M. C. et al. The three-dimensional structure of the seed storage protein phaseolin at 3 A resolution. EMBO J. 9, 9–15 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama N. et al. Creation of soybean beta-conglycinin beta with strong phagocytosis-stimulating activity. Biochim. Biophys. Acta. 1648, 99–104 (2003). [DOI] [PubMed] [Google Scholar]
- Maleki S. J. et al. Structure of the major peanut allergen Ara h 1 may protect IgE-binding epitopes from degradation. J. Immunol. 164, 5844–9 (2000). [DOI] [PubMed] [Google Scholar]
- Bassler O. Y. et al. Evidence for novel tomato seed allergens: IgE-reactive legumin and vicilin proteins identified by multidimensional protein fractionation-mass spectrometry and in silico epitope modeling. J. Proteome Res. 8, 1111–22 (2009). [DOI] [PubMed] [Google Scholar]
- Jain A. & Salunke D. M. Purification, identification and preliminary crystallographic studies of an allergenic protein from Solanum melongena. Acta Crystallogr. F Struct. Biol. Commun. 71, 221–5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P. & Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–32 (2004). [DOI] [PubMed] [Google Scholar]
- Brunger A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–21 (1998). [DOI] [PubMed] [Google Scholar]
- Holm L. & Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T., Maruyama N., Salleh M. R., Mikami B. & Utsumi S. Characterization and crystallography of recombinant 7S globulins of Adzuki bean and structure-function relationships with 7S globulins of various crops. J. Agric. Food Chem. 56, 4145–53 (2008). [DOI] [PubMed] [Google Scholar]
- Itoh T. et al. Structure of 8Salpha globulin, the major seed storage protein of mung bean. Acta Crystallogr D Biol Crystallogr. 62, 824–32 (2006). [DOI] [PubMed] [Google Scholar]
- Jin T. et al. Crystal structure of Korean pine (Pinus koraiensis) 7S seed storage protein with copper ligands. J. Agric. Food Chem. 62, 222–8 (2014). [DOI] [PubMed] [Google Scholar]
- Lawrence M. C., Izard T., Beuchat M., Blagrove R. J. & Colman P. M. Structure of Phaseolin at 2· 2 Å Resolution: Implications for a Common Vicilin/Legumin Structure and the Genetic Engineering of Seed Storage Proteins. J Mol Biol. 238, 748–776 (1994). [DOI] [PubMed] [Google Scholar]
- Oms-Oliu G. et al. Metabolic characterization of tomato fruit during preharvest development, ripening, and postharvest shelf-life. Postharvest Biology and Technology 62, 7–16 (2011). [Google Scholar]
- Kumar A. & Bachhawat A. K. Pyroglutamic acid: throwing light on a lightly studied metabolite. Curr. Sci. 102, 288 (2012). [Google Scholar]
- Vigeolas H. & Geigenberger P. Increased levels of glycerol-3-phosphate lead to a stimulation of flux into triacylglycerol synthesis after supplying glycerol to developing seeds of Brassica napus L. in planta. Planta 219, 827–35 (2004). [DOI] [PubMed] [Google Scholar]
- Vigeolas H., van Dongen J. T., Waldeck P., Huhn D. & Geigenberger P. Lipid storage metabolism is limited by the prevailing low oxygen concentrations within developing seeds of oilseed rape. Plant Physiol. 133, 2048–60 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raines C. A. The Calvin cycle revisited. Photosynthesis research 75, 1–10 (2003). [DOI] [PubMed] [Google Scholar]
- Meléndez-Hevia E., Waddell T. G. & Cascante M. The puzzle of the Krebs citric acid cycle: assembling the pieces of chemically feasible reactions, and opportunism in the design of metabolic pathways during evolution. J. Mol. Evol. 43, 293–303 (1996). [DOI] [PubMed] [Google Scholar]
- Sheen J. Metabolic repression of transcription in higher plants. Plant Cell 2, 1027–1038 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounet F. et al. Quantitative metabolic profiles of tomato flesh and seeds during fruit development: complementary analysis with ANN and PCA. Metabolomics 3, 273–288 (2007). [Google Scholar]
- Noctor G. et al. Glutathione in plants: an integrated overview. Plant Cell Environ. 35, 454–484 (2012). [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G. & Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–80 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson W. A. & Teeter M. M. Structure of the hydrophobic protein crambin determined directly from the anomalous scattering of sulphur. Nature 290, 107–113 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauter Z., Dauter M. & Dodson E. Jolly SAD. Acta Crystallogr. D Biol. Crystallogr. 58, 494–506 (2002). [DOI] [PubMed] [Google Scholar]
- Laskowski R. A., MacArthur M. W., Moss D. S. & Thornton J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26, 283–291 (1993). [Google Scholar]
- DeLano W. L. The PyMOL Molecular Graphics System. DeLano Scientific LLC; Palo Alto, CA, USA. URL https://www.pymol.org/ (2008).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.