Abstract
The application of multiphoton microscopy in the field of biomedical research and advanced diagnostics promises unique insights into the pathophysiology of inflammatory skin diseases. In the present study, we combined multiphoton-based intravital tomography (MPT) and fluorescence lifetime imaging (MPT-FLIM) within the scope of a clinical trial of atopic dermatitis with the aim of providing personalised data on the aetiopathology of inflammation in a non-invasive manner at patients’ bedsides. These ‘optical biopsies’ generated via MPT were morphologically analysed and aligned with classical skin histology. Because of its subcellular resolution, MPT provided evidence of a redistribution of mitochondria in keratinocytes, indicating an altered cellular metabolism. Two independent morphometric algorithms reliably showed an even distribution in healthy skin and a perinuclear accumulation in inflamed skin. Moreover, using MPT-FLIM, detection of the onset and progression of inflammatory processes could be achieved. In conclusion, the change in the distribution of mitochondria upon inflammation and the verification of an altered cellular metabolism facilitate a better understanding of inflammatory skin diseases and may permit early diagnosis and therapy.
Atopic dermatitis (AD) is a highly prevalent inflammatory skin disease with increasing incidence, mainly in developed countries. Manifestation of AD usually occurs in early childhood and is often followed by the development of allergies and asthma. According to current data, approximately 10–30% of newborns are affected by AD1. The underlying pathophysiological mechanisms of the disease are under debate, and a complex interplay between genetic, epigenetic and environmental factors is suggested2. However, further knowledge of the pathophysiology of AD is essential to improve the present therapeutic options.
To overcome the limitations of current mouse models and the restricted ability to analyse the skin of human patients via invasive techniques such as biopsies, we have applied multiphoton-based intravital tomography (MPT) equipped with a spectral fluorescence lifetime imaging module3 (MPT-FLIM) for the non-invasive in vivo analysis of human skin. Currently, the diagnosis of skin diseases is mainly based on the skills of the dermatologist or on the histological analysis of biopsies (the current gold standard). Physical examination of the patient is limited to the macroscopic surface level, and taking biopsies is an invasive approach, resulting in the formation of scars, thus excluding a longitudinal analysis of specific skin lesions. In contrast to alternative in vivo techniques such as ultrasound or confocal laser scanning microscopy4, MPT-FLIM allows subcellular in vivo imaging of thus far unknown parameters of inflammation. Therefore, the aim of the present study was to validate multiphoton-based tomography as a unique non-invasive tool for the morphological and biochemical characterisation of human skin.
Five-dimensional MPT-FLIM analysis comprises spatial (first to third dimension) and spectrally resolved fluorescence lifetime imaging (fourth and fifth dimension) by means of femtosecond laser pulses. This laser technology allows deep penetration of light into the skin and therefore the visualisation of the epidermis as well as the upper part of the dermis with subcellular resolution. In vivo investigations in humans exclude the application of fluorescence labelling of cells or proteins but are open to the use of naturally occurring fluorophores, such as melanin, keratin or NADH5,6. Multiphoton excitation of these endogenous fluorophores therefore enables the non-invasive high-resolution examination of human skin without damaging the surrounding tissue7.
Previous in vitro studies applying multiphoton microscopy showed a link between the pro-inflammatory activation of macrophages and the NADH signal intensity, suggesting an increased production of reactive oxygen species and a rearrangement of cellular metabolism8. To determine the cellular metabolism of epidermal cells in vivo, we utilised MPT-FLIM, an approach that enables an excitation energy independent readout of the NADH status9,10. In vivo FLIM measurement of NADH has been used in the context of skin cancer diagnoses in rodents11 and was recently successfully applied in the clinic12,13,14. The FLIM signal depends strongly on the microenvironment of the excited substance, enabling the discrimination of protein-bound and free NADH15. Free NADH was found to exhibit a fluorescence lifetime between 200 and 450 ps, whereas protein-bound NADH exhibits a prolonged lifetime in the range of 2,000 to 3,000 ps16,17. The ratio of free to protein-bound NADH is reflected by the mean fluorescence lifetime (taum)18, which serves as an intravital readout of the cellular metabolic state19.
Our clinical trial comprised 45 patients and followed their individual inflammatory progression over three months. For the first time, we established MPT-FLIM as a reliable and valuable tool for the clinical investigation of inflammatory skin diseases, facilitating a rapid, cost-efficient, non-invasive in vivo examination for diagnostic and therapeutic purposes in humans.
Results
Alignment of intravital multiphoton tomographic data with classical skin biopsy analysis
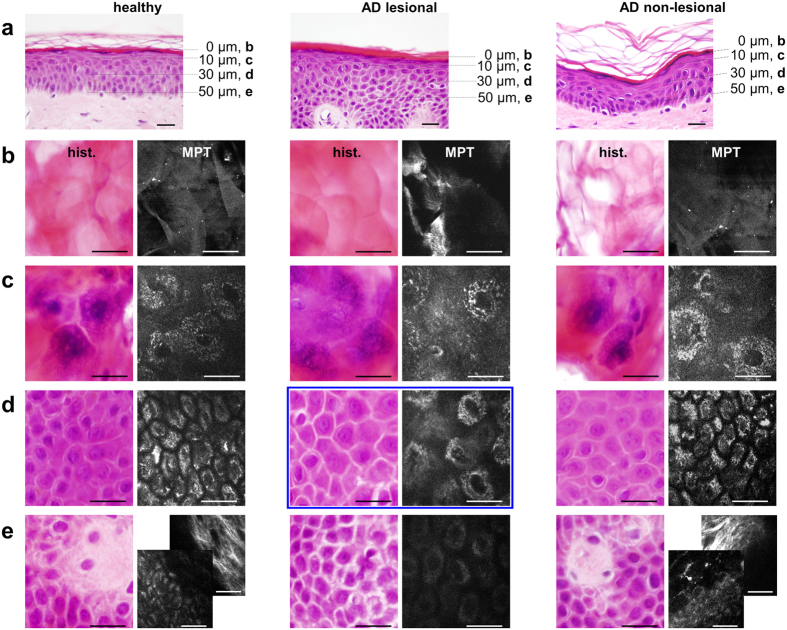
We used MPT as an artefact-free, painless approach that provides an ‘optical biopsy’ with subcellular resolution in human skin (Fig. 1a, see also Supplementary Movie S3). Previous studies of multiphoton imaging have suggested that NADH is the major natural fluorophore in human cells20,21,22. The bright spots observed in the cytoplasm of keratinocytes surrounding dark nuclei when using this technique (Fig. 1b) are mainly due to multiphoton-excited NADH, ensured by the combination of the utilised excitation wavelength and spectral emission filtering (see Methods section). These spots are located within the mitochondria as shown by immunohistochemical and fluorescence microscopic alignment (Fig. 1). Based on these settings, we compared the same skin region in healthy subjects and patients affected by AD via MPT and subsequently via histological analyses (Fig. 2). It must be emphasised that MPT allows horizontal sectioning of the skin, and we therefore also collected comparative biopsies cut in a horizontal manner. Each subject was clinically examined by a dermatologist and by MPT over a period of three months. During each session, we analysed one lesional (inflamed tissue) and one non-lesional (ostensibly healthy) skin region in comparison to the areas in age-correlated healthy subjects. As shown in Fig. 2, the analyses of histological biopsies and multiphoton-based intravital tomographic images of lesional skin areas were of identical pathognomonic validity (compare ‘hist.’-columns and skin depth correlated ‘MPT’-columns, Fig. 2b–e). Upon MPT, we were able to detect the characteristic skin morphologies of AD, such as intercellular oedema and an impaired architecture, accompanied by thickening of the epidermis in lesional skin. In particular, due to the absence of artefact-inducing embedding, dehydration and staining procedures, the MPT technique is markedly superior regarding assessment of the intensity of intercellular oedema (Fig. 2d, AD lesional, marked with a blue box).
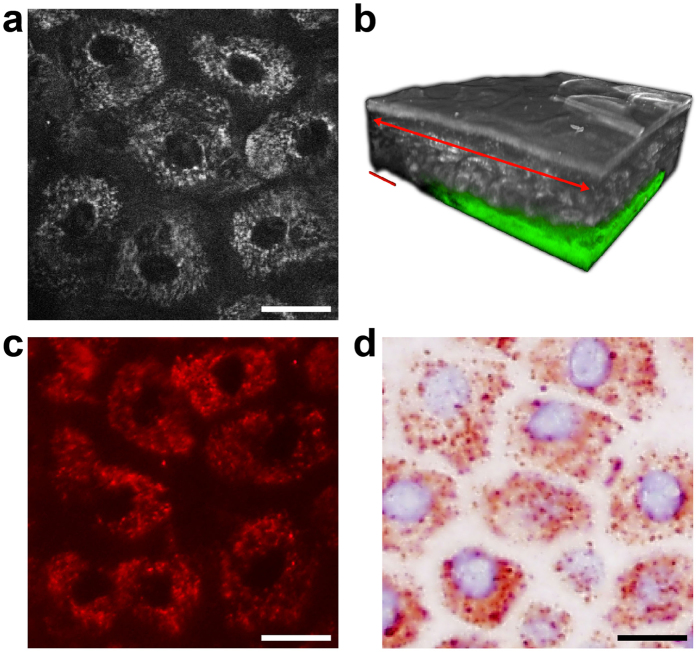
Figure 1. Multiphoton tomographic optical biopsy and mitochondria staining of healthy human skin.
(a) A representative multiphoton optical section of the Stratum granulosum of healthy skin in vivo. (b) A multiphoton tomographic three-dimensional reconstructed skin cube. The examined target section of (a) is indicated (red arrow), and dermal collagen in the Stratum papillare is pseudocolour-coded in green. Alignment with mitochondria-specific (c) immunofluorescence staining and (d) immunohistochemistry of a corresponding skin region in the Stratum granulosum performed ex vivo. Scale bars correspond to 20 μm.
Figure 2. Gold standard alignment of intravital multiphoton tomography.
(a) Standard vertical histological sections of the skin from healthy subjects (healthy), the ostensibly healthy skin of a patient affected by AD (AD non-lesional) and lesional atopic skin (AD lesional) examined via multiphoton tomography. Alignment of horizontal histological sections (‘hist.’-column) with multiphoton tomographic sections (‘MPT’-column) of the same regions of healthy, AD non-lesional and AD lesional areas of skin at distinct skin depths of 0 μm (Stratum corneum (b)), 10 μm (upper Stratum granulosum (c)), 30 μm (Stratum spinosum (d)) and 50 μm (e), representing the interface between the epidermal Stratum basale and the Stratum papillare of the dermis. A comparison of ‘hist.’ with ‘MPT’ illustrates the identical validity of assessing the morphological skin architecture and detecting characteristic skin morphologies. In the ‘AD lesional’ patient, the dermis is not accessible at a depth of 50 μm due to typical thickening of the epidermis. The accessibility of the dermis was demonstrated through additional imaging of the collagen-specific generation of second harmonics, as shown in the backmost image of the composite ((e) ‘MPT healthy’ and ‘MPT AD non-lesional’). Furthermore, the keratinocytes appear to be dispersed by inflammatory intercellular oedema. At a depth of 30 μm ((d) ‘AD lesional’), MPT is conspicuously superiour to the histological preparation regarding the assessment of the intensity of intercellular oedema (marked with a blue box). Scale bars correspond to 20 μm.
Detailed morphological analysis of the cellular mitochondrial distribution
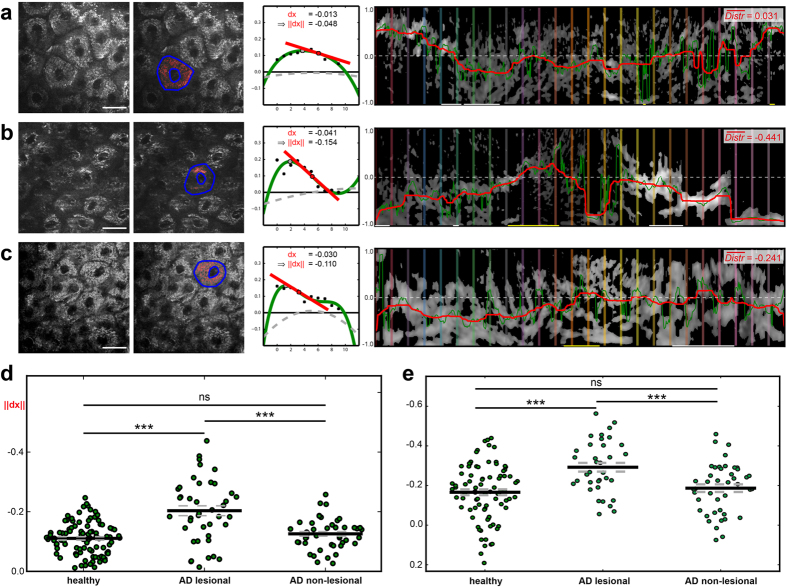
In addition to pathognomonic skin morphologies, we found a strikingly altered multiphoton tomographic pattern within single keratinocytes. Upon inflammation, the subcellular mitochondrial distribution (MD) appeared to be affected. In lesional skin, the mitochondrial signal was apparently stronger in the vicinity of the nuclei compared with an almost homogenous arrangement in healthy skin areas. For a detailed quantification of MD, we developed two independent computational algorithms: annuli filling analysis (AFA) and radial profiles analysis (RPA). Briefly, in AFA the signal intensity is plotted against the distance to an artificial nucleus from the centre to the periphery. As an appropriate indicator of MD characteristics, the normalised gradient (||dx||) at the dominant inflexion point ranges from −1.0 (maximal centralisation, Fig. 3a) across 0.0 (even signal distribution, Fig. 3b) to 1.0 (maximal peripheralisation, Fig. 3c). Owing to the potential limitations of AFA for irregularly shaped cells or eccentric nuclei, we introduced RPA, in which the mitochondrial signal is sampled on equiangular spoked lines (Fig. 4b,h) within the cytoplasm. Here, MD characteristics are quantified based on the RPA distribution value (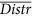 ): In contrast to the even signal distribution found in healthy cells (
): In contrast to the even signal distribution found in healthy cells (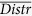 ≈ 0.0, Fig. 4a–e), lesional cells exhibit centralisation, resulting in a significant decrease of
≈ 0.0, Fig. 4a–e), lesional cells exhibit centralisation, resulting in a significant decrease of 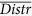 (Fig. 4f–j). For a detailed description of both approaches, please see also the Methods section.
(Fig. 4f–j). For a detailed description of both approaches, please see also the Methods section.
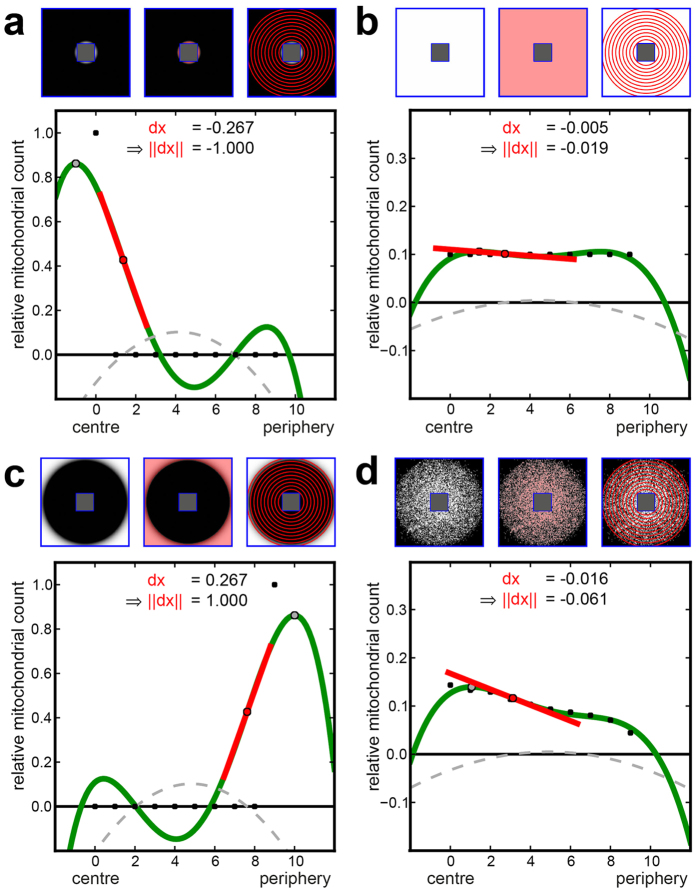
Figure 3. Annuli filling analysis (AFA) of generated dummy cells.
(a–d) Upper row: Quadratic dummy cells with nuclear (central rectangle) and cell membranes marked in blue; the cytoplasm lying in between is the used 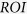 for AFA. Upper left: An intensity image of the mitochondrial signal within the
for AFA. Upper left: An intensity image of the mitochondrial signal within the 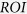 . Upper centre: Mitochondrial pixels marked with a light red overlay. Upper right: Circular regions (annuli) from the centre to the periphery (A0, …, A9) are visualised as red concentric circles. Graph: A scatterplot of the relative mitochondrial count m0, …, m9 per annulus (black dots) is shown. The interpolated polynomial is shown in green, the inflexion point as a red dot, the tangent at the inflexion point as a red line, the dominant maximum as a grey dot, and the second derivative of the polynomial as a dashed grey curve. (a) All mitochondrial pixels reside within A0, resulting in maximal centralisation and, thus, a minimal (normed) inflexion point gradient (dx = −0.267, ||dx|| = −1.0). (b) A homogeneous distribution over all annuli is shown. (c) All mitochondrial pixels are located within A9, resulting in maximal peripheralisation. (d) A cell-like even distribution results in slight centralisation.
. Upper centre: Mitochondrial pixels marked with a light red overlay. Upper right: Circular regions (annuli) from the centre to the periphery (A0, …, A9) are visualised as red concentric circles. Graph: A scatterplot of the relative mitochondrial count m0, …, m9 per annulus (black dots) is shown. The interpolated polynomial is shown in green, the inflexion point as a red dot, the tangent at the inflexion point as a red line, the dominant maximum as a grey dot, and the second derivative of the polynomial as a dashed grey curve. (a) All mitochondrial pixels reside within A0, resulting in maximal centralisation and, thus, a minimal (normed) inflexion point gradient (dx = −0.267, ||dx|| = −1.0). (b) A homogeneous distribution over all annuli is shown. (c) All mitochondrial pixels are located within A9, resulting in maximal peripheralisation. (d) A cell-like even distribution results in slight centralisation.
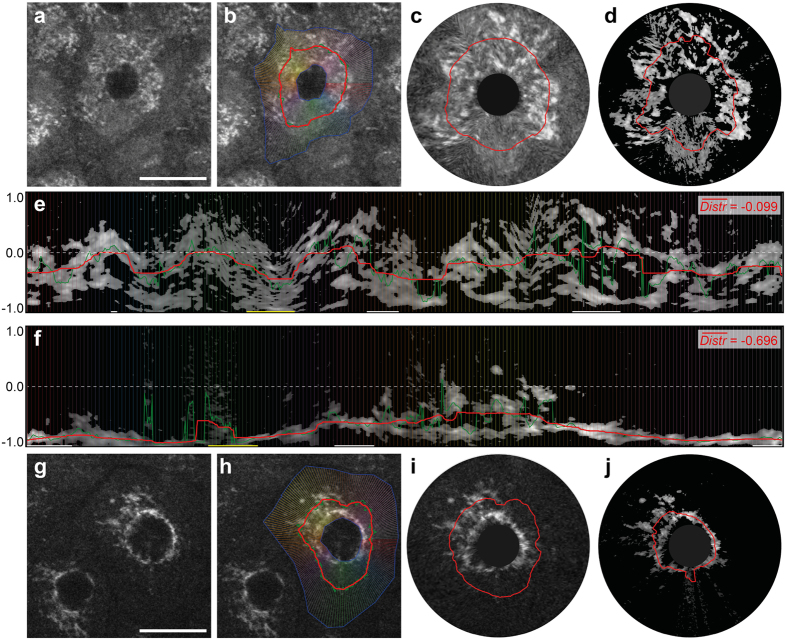
Figure 4. Radial profiles analysis (RPA) of human epidermal cells of the Stratum granulosum.
Archetypical keratinocytes of the Stratum granulosum in healthy (a–e) and lesional (f–j) skin areas have been selected. (a,g) Representative multiphoton tomographic cell images. (b,h) The cytoplasm bounded by the blue polygons is used as 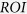 for RPA. Radial profile lines (coloured) and plots of mitochondrial distribution values (green: raw; red: smoothed) are shown. Virtual cell reconstructions based on radial profiles with smoothed distribution values are depicted before (c,i) and after thresholding (d,j). Consequently, the thresholded radial profiles for healthy (e) and lesional (f) cells are displayed with the calculated mean distribution values (
for RPA. Radial profile lines (coloured) and plots of mitochondrial distribution values (green: raw; red: smoothed) are shown. Virtual cell reconstructions based on radial profiles with smoothed distribution values are depicted before (c,i) and after thresholding (d,j). Consequently, the thresholded radial profiles for healthy (e) and lesional (f) cells are displayed with the calculated mean distribution values (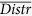 ). Outliers are marked with horizontal bars on the x-axis (yellow: short, white: long). Scale bars correspond to 20 μm.
). Outliers are marked with horizontal bars on the x-axis (yellow: short, white: long). Scale bars correspond to 20 μm.
In applying these models to the entire MPT dataset from the clinical trial, we focused on the outermost layer of living epidermal cells (Stratum granulosum) and observed even partitioning of the mitochondria in healthy skin, as expected (Fig. 5a). By contrast, the MD within inflamed (AD lesional) skin showed perinuclear accumulation, as quantified by a significant negative shift of both ||dx|| and 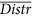 (Fig. 5b), whereas the MD in non-lesional skin (Fig. 5c) was comparable to that of healthy skin. The statistical analysis of MD in the Stratum granulosum of all subjects included in the clinical trial is shown in Fig. 5d,e.
(Fig. 5b), whereas the MD in non-lesional skin (Fig. 5c) was comparable to that of healthy skin. The statistical analysis of MD in the Stratum granulosum of all subjects included in the clinical trial is shown in Fig. 5d,e.
Figure 5. Mitochondrial distribution of patients’ epidermal cells calculated via AFA and RPA.
(a) Within a typical Stratum granulosum cell layer in healthy skin (left image), the selected cell marked in blue (second image) shows a homogenous mitochondrial distribution based on AFA, corresponding to a small gradient at the inflexion point (third image). Accordingly, RPA indicates a 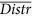 value near zero (right image). (b) The selected cell from inflamed skin (AD lesional) shows mitochondrial centralisation around the nucleus, resulting in a decreased inflexion point gradient and
value near zero (right image). (b) The selected cell from inflamed skin (AD lesional) shows mitochondrial centralisation around the nucleus, resulting in a decreased inflexion point gradient and 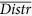 value compared with (a).(c) The mitochondria of the non-lesional cell show slight centralisation. (d) An AFA comparison of relative gradients (||dx||) of cells from healthy, lesional and non-lesional subjects. (e) An RPA
value compared with (a).(c) The mitochondria of the non-lesional cell show slight centralisation. (d) An AFA comparison of relative gradients (||dx||) of cells from healthy, lesional and non-lesional subjects. (e) An RPA 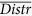 plot of cells from healthy, lesional and non-lesional patients. Each green point represents one segmented cell. Scale bars correspond to 20 μm.
plot of cells from healthy, lesional and non-lesional patients. Each green point represents one segmented cell. Scale bars correspond to 20 μm.
In vitro characterisation of the energy status of human keratinocytes using MPT-FLIM
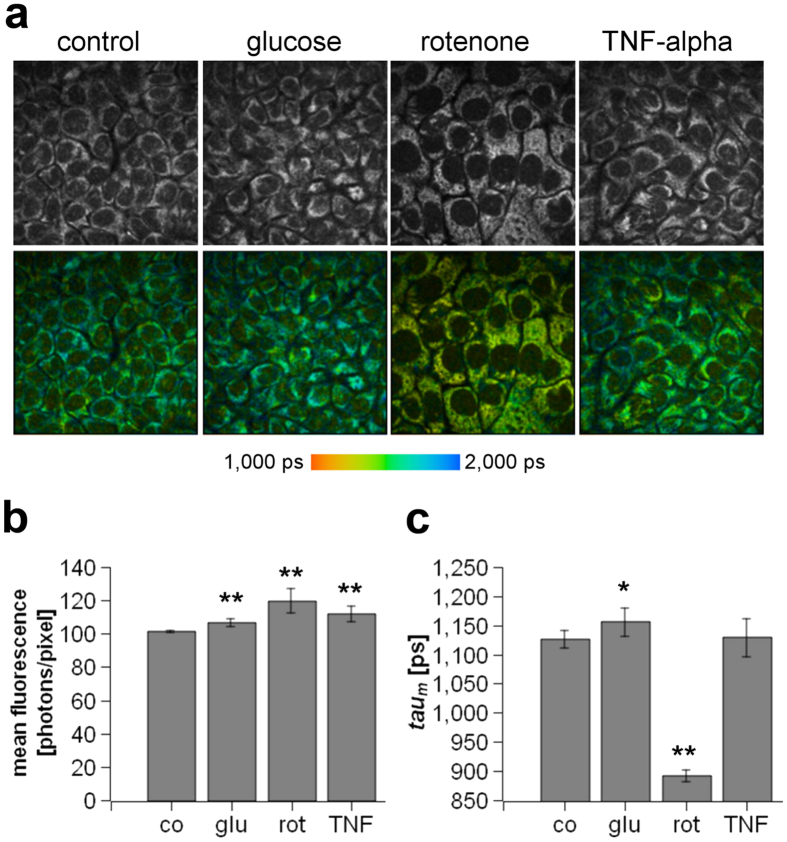
Because of the inflammation-correlated morphological alterations and shift of MD we observed in vivo (see Fig. 5d,e), we additionally aimed to demonstrate that MPT-FLIM is a suitable tool for the interpretation of cellular metabolism in human keratinocytes exposed to distinct stimuli that are known to alter their metabolic state in vitro. The molecular basis of skin inflammation is neither limited to keratinocytes nor to single stimuli, thus preventing the establishment of an accurate in vitro system. However, to allow a general understanding of the NADH metabolism of keratinocytes to be obtained under inflammatory conditions, we treated human keratinocytes in vitro with the pro-inflammatory cytokine TNF-alpha. For comparison, we manipulated the cellular metabolism of keratinocytes by adding glucose or rotenone. Representative images of the fluorescence intensity images (first row) and the corresponding fluorescence lifetime images (second row) are presented in Fig. 6a. Most of the fluorescence signals were localised in the cytoplasm and were related to mitochondrial NADH (see also Fig. 1)23. An increase of the fluorescence intensity was measured in glucose-, TNF-alpha- and rotenone-treated keratinocytes (Fig. 6b). The obtained taum values are depicted in Fig. 6c. A massively increased amount of free NADH was apparent in rotenone-treated keratinocytes, as indicated by a reduction of the taum value. In glucose-treated keratinocytes, an increase of taum was found, suggesting a greater amount of protein-bound NADH.
Figure 6. Evaluation of the energy status of human keratinocytes in vitro.
(a) Fluorescence intensity images reflecting the total amount of cellular NADH (first row) and fluorescence lifetime images (FLIM) (second row) of non-treated keratinocytes (control) or keratinocytes treated either with 20 mM glucose, 5 μM rotenone or 10 nM TNF-alpha. The colour-coded bar reflects the fluorescence lifetime, ranging from 1,000 ps (red) to 2,000 ps (blue). (b) Quantification of the mean fluorescence intensity of non-treated keratinocytes (co) or keratinocytes treated with glucose (glu), rotenone (rot), or TNF-alpha (TNF). (c) Quantification of the mean fluorescence lifetime (taum) of non-treated keratinocytes (co) or keratinocytes treated with glucose (glu), rotenone (rot), or TNF-alpha (TNF). *P < 0.05, **P < 0.01.
MPT-FLIM provides evidence of inflammation-related alteration of the cellular metabolism
To investigate whether the altered subcellular localisation of mitochondria is associated with an altered cellular metabolism in vivo, we applied MPT-FLIM in the setting of the clinical trial. We calculated the mean taum, reflecting the ratio of free-to-protein-bound NADH, for all images of the Stratum granulosum obtained for the entire study population. This FLIM analysis, shown in Fig. 7, revealed a significant decrease of taum in inflamed skin compared with healthy skin (1,270 +/− 28 ps to 1,452 +/− 13 ps, Fig. 7d). In contrast to the histological and MPT morphological analyses, the MPT-FLIM analysis allowed distinction between the healthy skin and non-lesional skin of AD-affected patients (1,452 +/− 13 ps to 1,377 +/− 22 ps, Fig. 7d), indicating a shift of the metabolic state.
Figure 7. Comparison of the fluorescence lifetime (taum) of healthy, lesional and non-lesional Stratum granulosum cells.
Overlay images of MPT morphology and colour-coded taum values of representative healthy (a), lesional (b) and non-lesional (c) cell layers, with one segmented cell contoured in red. A low taum (red = 100 ps) is visualised with warm colours and a high taum (blue = 2,000 ps) with cool colours. The subsequent graphs show the FLIM curve of one selected pixel (yellow crosshairs); its taum value is denoted in red in the colour bar. (d) The distribution of taum in healthy, lesional and non-lesional cell layers of patients is displayed. Each green point represents the mean taum value of one image. Scale bars correspond to 20 μm.
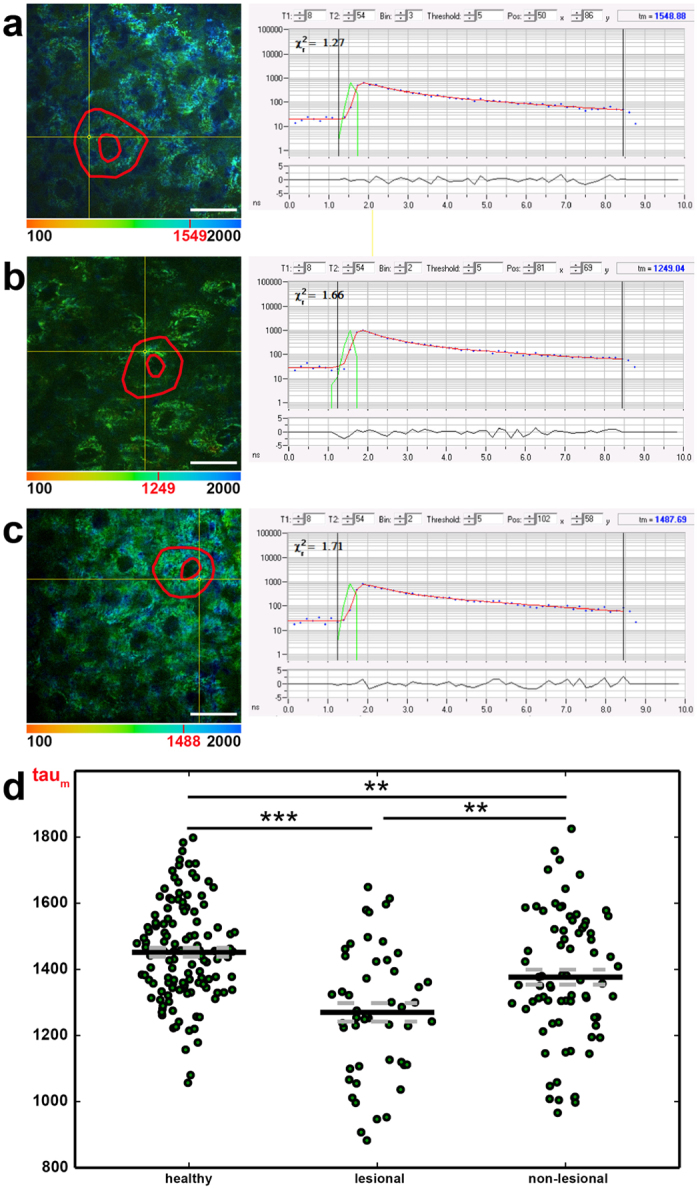
While the mean inflammatory state of patient skin slightly decreased during the course of the study (as indicated by a decreasing Severity Score of Atopic Dermatitis (SCORAD)), we could detect an increase of taum in lesional skin areas from 1,166 +/− 40 ps to 1,378 +/− 45 ps. Moreover, we observed a continuous significant difference in the measures throughout the study compared with the taum of healthy skin (Fig. 8a). Plotting the taum values of lesional skin against the corresponding inflammatory state measured via SCORAD, we found a significant linear correlation, in which taum decreased with an increasing SCORAD (PCC 0.65, p < 0.0001, Fig. 8b).
Figure 8. Overall fluorescence lifetime (taum) in the course of the clinical trial.
(a) Mean taum values of lesional skin areas in all patients suffering from AD continuously show a significant difference in the taum of healthy skin over the entire term. The bubble size represents the disease severity measured via SCORAD. (b) Plotting the taum of lesional skin against the corresponding inflammatory state measured via SCORAD, a significant linear correlation is revealed: taum decreases with increasing SCORAD.
To exclude systemic shifts influencing our MPT-FLIM measurements, we plotted taum as a function of the day and month of examination. As expected, in this nonsense correlation, the examination day did not show any impact on taum (see Supplementary Fig. S1).
Discussion
The present data demonstrate the translation of a medical assessment to in vivo MPT-FLIM and morphometric analyses in the context of a clinical setting for the first time. We investigated morphological alterations as well as the metabolic status of keratinocytes in the background of AD, an inflammatory skin disease with a complex pathophysiology and a high prevalence. In line with classical histological analyses, we were able to detect the formation of intercellular oedema and thickening of the epidermis that correlated with the severity of inflammation. Moreover, in the objective computational analysis of the morphological dataset, MPT provided information on the reorganisation of mitochondria. Two independent morphometric algorithms reliably showed an even MD in healthy skin and a perinuclear accumulation in inflamed skin (Fig. 5).
As this observation suggested a direct metabolic impact, we first performed in vitro MPT-FLIM examinations of a human keratinocyte cell line upon stimulation with various agents known to alter keratinocyte metabolism (Fig. 6). Based on our data, a decrease of the mean fluorescence lifetime (taum) indicated an accumulation of free NADH in the mitochondria, suggesting an overload of the respiratory electron transport. This overload could be related to either a lack of oxygen or hyperproduction of NADH during glycolysis. Malfunction of the mitochondrial electron transport chain is a marker of cellular stress such as apoptosis and is accompanied by the production of reactive oxygen species24.
In line with these in vitro findings, we were able to measure a comparable change of cellular metabolism in the inflamed skin of patients using MPT-FLIM (Fig. 7). Additionally, the decrease of taum correlated with the severity of inflammation (Fig. 8b), pointing towards increased stress in keratinocytes within the affected skin regions.
Bridging the gap between the physiological basis of the cellular metabolism and its potential impact on a patient’s pathogenesis, this study demonstrates that multiphoton tomography equipped with a spectral FLIM module might be successfully used as a diagnostic tool in the field of skin inflammation research. In contrast to the exclusive morphological analysis of inflamed skin, MPT-FLIM is able to non-invasively demonstrate that even the apparently healthy (non-lesional) skin of patients affected by AD exhibits pathological features resembling those of eczematous skin lesions. Therefore, our data support previous genetic, histological and molecular biological findings from non-lesional skin regions25,26,27.
In contrast to classical examination tools, the non-invasive nature of MPT-FLIM might help to identify regions with nearly subclinical inflammation. Clear identification of these skin regions will be beneficial to initiate early proactive treatment, preventing inflammatory exacerbation. As a unique feature, MPT-FLIM combined with standardised computational analysis generates new in vivo parameters that can be used to detect and monitor pathophysiological alterations in the earliest state of inflammatory processes in human skin, prior to clinical manifestation. Thus, the in vivo application of MPT-FLIM offers the possibility of directly examining the pathogenesis of inflammatory skin diseases, in addition to consecutive observation of individual aetiopathology in a non-invasive manner.
Methods
Multiphoton tomography
In the present study, we applied a certified CE-marked five-dimensional intravital multiphoton tomographic system (MPT). The technical setup, based on a DermaInspect multiphoton microscope, was established within the framework of the BMBF project ‘5D-IVT’ by JenLab (JenLab GmbH, Jena, Germany). Near-infrared laser pulses with a pulse length of 100 fs provided by a Titanium:Sapphire tuneable laser system (Mai Tai, Newport Spectra-Physics, Santa Clara, CA, USA) were used for the excitation of endogenous fluorophores in human skin. The excitation laser beam was attenuated by the use of a Glan calcite polariser and scanned by two galvanometric mirrors. After passing a beam expander and collimator, the laser pulses are reflected by a dichroic beam splitter into a 40x oil immersion microscope objective with a numerical aperture of 1.3 (Carl Zeiss Jena GmbH, Jena, Germany).
The emitted fluorescence light was transmitted and cleared by an additional shortwave-pass filter (F37-490 BrightLine HC 490/LP and F75-680 Multiphoton-Emitter HC 680/SP, F39-461 BrightLine HC 460/60, F39-390 BrightLine HC 390/40; Semrock Inc., Rochester, NY, USA) and was split into three distinct spectral regions by a set of dichroic beam splitters (F43-031 425DCXR, F33-499 495DCXR; Chroma Technology Corp., Bellows Falls, VT, USA).
Three separate photomultipliers (PMTs) provided intensity images of the sample, and each PMT was readout via time-correlated single photon counting to calculate wavelength-range-specific fluorescence lifetime values (MPT-FLIM). The PMT data were processed by a high-resolution time-correlated single photon counting imaging module (SPC 830, Becker & Hickl GmbH, Berlin, Germany).
This setup allows us to non-invasively excite fluorophores in human skin in vivo to create high-resolution autofluorescence images and fluorescence lifetime images with a penetration depth of 150 μm. An excitation wavelength of 710 nm and an emission bandpass filter (460/60 nm) were used to match the spectrum of NADH.
Study design
Twenty healthy volunteers and 25 patients affected by AD who were undergoing basic therapy were enrolled in this study. For each subject, two different skin areas were analysed during four clinical visits over three months to obtain a survey of the diverse states of inflammation. In each visit, a physician diagnosed the state of inflammation according to the SCORAD28, and a local score of inflammatory manifestation in the investigated skin areas was assigned via clinical inspection. To offer the patients the most comfortable examination, we chose one lesional and one non-lesional skin area of the volar forearm. The targeted skin area was directly coupled to MPT using an in vivo adapter provided by JenLab. Sequences of confocal sections with 10 μm penetration steps, an edge length of 100 μm and an exposure time of 25 s per image were recorded, starting at the outmost epidermal layer (Stratum corneum), defined as a depth of 0 μm, and ending at the papillary dermis, at approximately 150 μm. The study was conducted conforming to the guidelines of the Declaration of Helsinki29 and The International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use (ICH)30. It was approved by the Bundesinstitut für Arzneimittel und Medizinprodukte (BfArM) of Germany and the Ethics Committee of the Medical Association Westfalen-Lippe (Münster, Germany). Appropriate informed consent was obtained from all subjects.
Histological alignment
Immediately after the last clinical visit, punch biopsies were taken from target skin areas in three healthy subjects and three affected patients. The biopsies were split for standard vertical and horizontal histological preparation and stained with haematoxylin-eosin, for classical histological examination, or with the anti-mitochondrial antibody MTC02 ab3298 (Abcam, Cambridge, MA, USA), for alignment of the mitochondrial distribution, and were examined by a histopathologist and aligned to the corresponding MPT images.
Data preparation and analysis of the cellular mitochondrial distribution
Cell segmentation
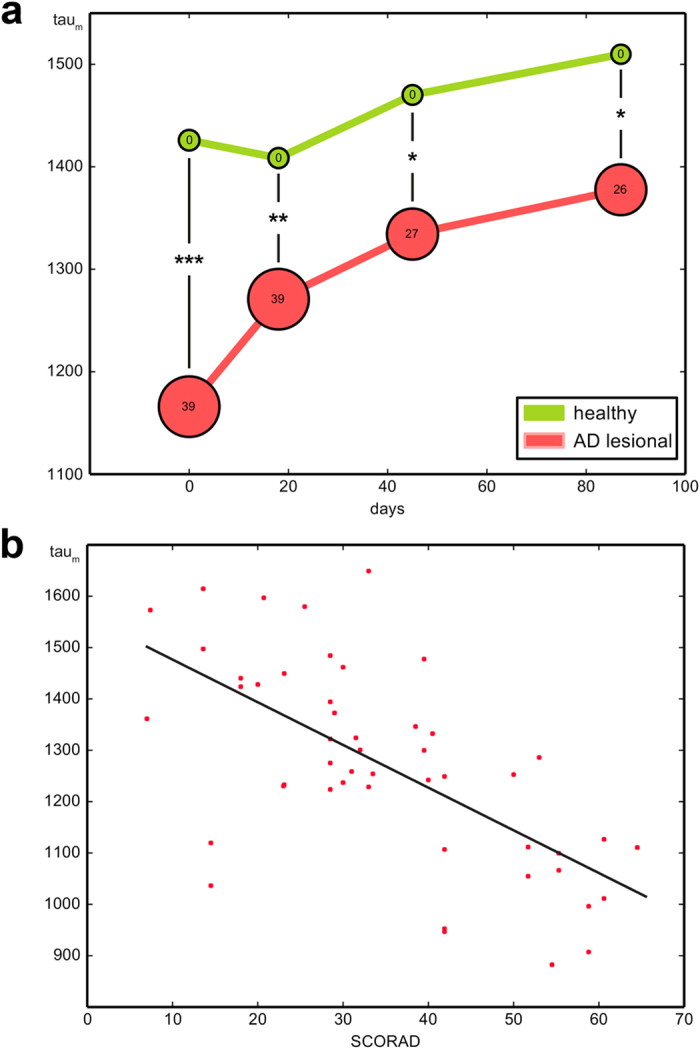
Because the mitochondrial distribution is defined per cell, the cytoplasm of different typical cells must be segmented in a preceding step. This segmentation is performed manually by defining (Software MIPAV31) two non-self-intersecting polygons: an inner polygon Pnucleus tracing the nucleus and an outer polygon Pcell tracing the cell membrane. The cytoplasm polygon is then defined by the difference Pcytoplasm = Pcell − Pnucleus, which is the region of interest (ROI) for the subsequent steps. To correct irregular illumination, we used a previously reported Gaussian filtering-based method (see Supplementary Methods and Supplementary Fig. S2)32,33.
Mitochondrial pixel classification
The mitochondrial regions of the cytoplasm are defined by a strong NADH signal resulting in a high photon count at the corresponding positions. The shading corrected photon count image Icorr used for the quantitative analysis of MD consists of a 128 × 128 matrix and each pixel I(x, y) contains the number of detected photons. To identify all pixels 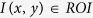 representing mitochondrial areas, an automatic thresholding algorithm is used34. The algorithm assumes that Icorr consists of two classes, foreground (high intensity) and background (low intensity) pixels, and iteratively calculates the optimal threshold by minimising the variance within each class. The detected mitochondrial pixels are coloured red in Figs 3 and 5.
representing mitochondrial areas, an automatic thresholding algorithm is used34. The algorithm assumes that Icorr consists of two classes, foreground (high intensity) and background (low intensity) pixels, and iteratively calculates the optimal threshold by minimising the variance within each class. The detected mitochondrial pixels are coloured red in Figs 3 and 5.
Annuli filling analysis (AFA)
To measure the spatial distribution of mitochondrial pixels within the cytoplasm of each cell, the 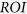 is divided into 10 circular regions (annuli) from the centre to the periphery (A0, …, A9) defined by concentric circles C0, …, C9 (see upper right of Fig. 3a–d). With
is divided into 10 circular regions (annuli) from the centre to the periphery (A0, …, A9) defined by concentric circles C0, …, C9 (see upper right of Fig. 3a–d). With  as the centroid of the inner polygon Pnucleus, the radius r0 of the innermost circle C0 is defined by the maximum distance between c and the bounding box of Pnucleus. The radius r9 of the outer circle C9 is the minimum distance between
as the centroid of the inner polygon Pnucleus, the radius r0 of the innermost circle C0 is defined by the maximum distance between c and the bounding box of Pnucleus. The radius r9 of the outer circle C9 is the minimum distance between  and the bounding box of Pcell. The radii r1, …, r8 are defined by
and the bounding box of Pcell. The radii r1, …, r8 are defined by 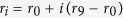 /9 with
/9 with 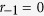 . Given the Euclidian distance
. Given the Euclidian distance 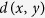 , a pixel
, a pixel 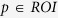 resides in annulus
resides in annulus 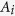 if
if 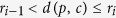 . Following this scheme, every mitochondrial pixel
. Following this scheme, every mitochondrial pixel 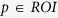 can be assigned to exactly one annulus Ai, and a relative mitochondrial count mi can be calculated per Ai by dividing the number of mitochondrial pixels by the number of all pixels in Ai.
can be assigned to exactly one annulus Ai, and a relative mitochondrial count mi can be calculated per Ai by dividing the number of mitochondrial pixels by the number of all pixels in Ai.
In the next step, the relative mitochondrial counts m0, …, m9 (black points in the graphs of Figs 3 and 5) are used to calculate a 4th degree polynomial 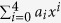 via weighted polynomial interpolation (green plot in the graphs of Figs 3 and 5). Two extra points
via weighted polynomial interpolation (green plot in the graphs of Figs 3 and 5). Two extra points 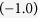 and
and 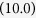 (not shown in Figs 3 and 5) with a high weight are included in the interpolation to force the polynomial maxima within the range of 0 and 9. Furthermore, higher mi values receive a higher weight to map the polynomial to the maxima. The gradient
(not shown in Figs 3 and 5) with a high weight are included in the interpolation to force the polynomial maxima within the range of 0 and 9. Furthermore, higher mi values receive a higher weight to map the polynomial to the maxima. The gradient 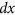 of the inflexion point (red points in Figs 3 and 5) with the smallest distance from the dominant maximum (grey points in Figs 3 and 5) describes the mitochondrial distribution. For complete mitochondrial centralisation the gradient becomes minimal (
of the inflexion point (red points in Figs 3 and 5) with the smallest distance from the dominant maximum (grey points in Figs 3 and 5) describes the mitochondrial distribution. For complete mitochondrial centralisation the gradient becomes minimal ( ; Fig. 3a). In the case of complete peripheralisation, the gradient is maximal (
; Fig. 3a). In the case of complete peripheralisation, the gradient is maximal (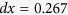 ; Fig. 3c) and a homogeneous MD is denoted by a
; Fig. 3c) and a homogeneous MD is denoted by a 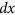 close to 0 (Fig. 3b).
close to 0 (Fig. 3b).
Radial profiles analysis (RPA)
Although the AFA method works well in cells with a relatively circular shape, problems arise in cells with extremely ragged outlines or eccentric nuclei. Thus, we have developed an alternative method based on a previously published algorithm35, which is unaffected by these problems. In general, RPA works by generating a number of radial profiles within the cytoplasm, and subsequent analysis is performed on the grey level distribution.
In the first step, l equiangular radial line segments within Pcytoplasm are generated (Fig. 4b,h, coloured lines). Each line is then sampled at  equidistant points and the bilinear interpolated grey values at these sampling points form one radial profile p. All profiles are stacked together to form a s × l matrix, which represents the unrolled length-normalised cytoplasm (Fig. 4e,f). To reduce the effect of non-mitochondrial pixels and background noise, each column profile p is thresholded separately via Otsu’s method34, depicted in Fig. 4c (before thresholding) and Fig. 4d (after thresholding).
equidistant points and the bilinear interpolated grey values at these sampling points form one radial profile p. All profiles are stacked together to form a s × l matrix, which represents the unrolled length-normalised cytoplasm (Fig. 4e,f). To reduce the effect of non-mitochondrial pixels and background noise, each column profile p is thresholded separately via Otsu’s method34, depicted in Fig. 4c (before thresholding) and Fig. 4d (after thresholding).
For quantification of the mitochondrial distribution, the grey values of each column profile p are summed from the inner to the outer sampling point until 50% of the cumulative grey value of p is reached. The index 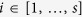 of this point is the distribution value Distr of one profile, and the normalised mean of all profiles
of this point is the distribution value Distr of one profile, and the normalised mean of all profiles 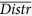 represents the mitochondrial distribution of the cell. Distinct eccentric nuclei might be indicative of tilted optical sectioning, resulting in a single-sided impaired information density. Therefore, extraordinarily long and short radial profile line outliers are filtered out using the median absolute deviation (MAD)36.
represents the mitochondrial distribution of the cell. Distinct eccentric nuclei might be indicative of tilted optical sectioning, resulting in a single-sided impaired information density. Therefore, extraordinarily long and short radial profile line outliers are filtered out using the median absolute deviation (MAD)36.
Cell culture and in vitro measurement of cell metabolism using MPT-FLIM
The human keratinocyte cell line HaCaT was cultivated as previously reported37,38. Briefly, HaCaT cells were maintained at 37 °C under 5% CO2 in RPMI 1640 medium supplemented with 10% foetal calf serum, 1% L-glutamine and 1% penicillin/streptomycin. Twenty-four hours prior to the experiment, the cells were seeded into a 0.2 μm luer slide (ibidi GmbH, Munich, Germany). Multiphoton tomography was carried out 4 hours after treatment with 5 μM rotenone or 20 mM glucose (both Sigma-Aldrich, Steinheim, Germany) or 10 nM recombinant human TNF-alpha. The experiments were performed in HEPES-buffered Ringer’s solution containing (in mmol/L): 140 NaCl, 5 KCl, 1 MgCl, 1 CaCl2, 5 glucose, and 10 HEPES (N-2-hydroxyethylpiperazin-N0-2-ethanesulfonic acid), adjusted to a pH of 7.4, at 37 °C.
Preparation and analysis of in vivo MPT-FLIM data
Because of the vast amount of obtained image and FLIM data, a specialised system for structured data storage, retrieval and analysis was needed. For this purpose, we used the Open Microscopy Environment (OME). OME consists of OME-XML27, an open standard format for the description and annotation of microscopy image data, and OMERO (OME Remote Objects)39, an open source software for the management and analysis of image data.
To enrich the existing image data with histological parameters, all images were first categorised by a dermatologist with expertise in the field of histopathology. For categorisation, a region of interest consisting only of cells from one single epidermal layer was defined for each image. Additional examples of parameters are the image quality with regard to suitability for automatic analysis and diagnosis from a histological perspective. Categorisation parameters, clinical values (e.g., medication and the SCORAD) and technical image acquisition data (e.g., excitation wavelength and laser energy) were transformed into the OME-XML format, merged with the associated image and FLIM data and then imported to the OMERO server. Various clients (OMERO.insight, OMERO.web) can be used for manual image and metadata browsing. Furthermore, the functionality of the OMERO server can be used and extended to implement algorithms for analysis. To obtain a tool for specialised and explorative data analysis and quantification, we have chosen a two-staged approach. In the first step, the user creates a subset of all images by defining various metadata constraints (e.g., image quality: high to very high; epidermal layers: Stratum granulosum and spinosum, and SCORAD: 10 to 50), and in a second step, the parameters for quantitative analysis (e.g., SCOARD vs. taum) are chosen. For rapid evaluation of results, different visualisation methods, such as box or scatter plots, are available, and the quantitative results can be exported for further analysis.
Statistical computation
Mean data from the experiments are presented with the standard error of the mean (SEM). Statistical computations were performed with SAS 9.2 (SAS Institute Inc., Cary, NC, USA).
Additional Information
How to cite this article: Huck, V. et al. From morphology to biochemical state – intravital multiphoton fluorescence lifetime imaging of inflamed human skin. Sci. Rep. 6, 22789; doi: 10.1038/srep22789 (2016).
Supplementary Material
Acknowledgments
We thank A. Wissel for excellent technical assistance and S. Mühl as well as F. Valentin for technical and editorial support. This work has been funded by the German federal ministry for education & research BMBF (grant nos. 13N9283 and 13N11248). It was approved by the Bundesinstitut für Arzneimittel und Medizinprodukte (BfArM) of Germany and the Ethics Committee of the Medical Association Westfalen-Lippe (Münster, Germany).
Footnotes
K.K. is the CEO of JenLab GmbH.
Author Contributions V.H. and C.M. conceived, designed and performed most of the experiments, analysed the data, contributed materials and analysis tools and wrote this manuscript. Additionally, the project supervisor S.W.S. and D.M. conceived and designed the experiments and contributed materials to this project. C.G., K.T., V.G. and T.R.U. performed the continuative examinations. C.G., V.G., V.N., K.Z., J.S.F. and M.A.F. were responsible for analysis of the data, for which C.M. developed new analytical tools. S.S., T.A.L. and K.K. contributed materials to this project.
References
- Bieber T. Atopic dermatitis. N. Engl. J. Med. 358, 1483–1494 (2008). [DOI] [PubMed] [Google Scholar]
- Cork M. J. et al. New perspectives on epidermal barrier dysfunction in atopic dermatitis: gene-environment interactions. J. Allergy Clin. Immunol. 118, 3–21; quiz 22–23 (2006). [DOI] [PubMed] [Google Scholar]
- Becker W. et al. Fluorescence lifetime imaging by time-correlated single-photon counting. Microsc. Res. Tech. 63, 58–66 (2004). [DOI] [PubMed] [Google Scholar]
- Koehler M. J. et al. Clinical application of multiphoton tomography in combination with confocal laser scanning microscopy for in vivo evaluation of skin diseases. Exp. Dermatol. 20, 589–594 (2011). [DOI] [PubMed] [Google Scholar]
- Koenig K. & Riemann I. High-resolution multiphoton tomography of human skin with subcellular spatial resolution and picosecond time resolution. J Biomed Opt 8, 432–439 (2003). [DOI] [PubMed] [Google Scholar]
- Williams R. M., Zipfel W. R. & Webb W. W. Multiphoton microscopy in biological research. Curr. Opin. Chem. Biol. 5, 603–608 (2001). [DOI] [PubMed] [Google Scholar]
- Fischer F. et al. Assessing the risk of skin damage due to femtosecond laser irradiation. J Biophotonics 1, 470–477 (2008). [DOI] [PubMed] [Google Scholar]
- Kable E. P. & Kiemer A. K. Non-invasive live-cell measurement of changes in macrophage NAD(P)H by two-photon microscopy. Immunol. Lett. 96, 33–38 (2005). [DOI] [PubMed] [Google Scholar]
- Lakowicz J. R., Szmacinski H., Nowaczyk K. & Johnson M. L. Fluorescence lifetime imaging of free and protein-bound NADH. Proc. Natl. Acad. Sci. USA 89, 1271–1275 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneckenburger H., Koenig K., Kunzi-Rapp K., Westphal-Frosch C. & Ruck A. Time-resolved in-vivo fluorescence of photosensitizing porphyrins. J. Photochem. Photobiol. B. 21, 143–147 (1993). [DOI] [PubMed] [Google Scholar]
- Skala M. C. et al. In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia. Proc. Natl. Acad. Sci. USA 104, 19494–19499 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patalay R. et al. Multiphoton multispectral fluorescence lifetime tomography for the evaluation of basal cell carcinomas. PLoS One 7, e43460 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidenari S. et al. Multiphoton laser tomography and fluorescence lifetime imaging of basal cell carcinoma: morphologic features for non-invasive diagnostics. Exp. Dermatol. 21, 831–836 (2012). [DOI] [PubMed] [Google Scholar]
- Ulrich M. et al. In vivo detection of basal cell carcinoma: comparison of a reflectance confocal microscope and a multiphoton tomograph. J Biomed Opt 18, 61229 (2013). [DOI] [PubMed] [Google Scholar]
- Paul R. J. & Schneckenburger H. Oxygen concentration and the oxidation-reduction state of yeast: determination of free/bound NADH and flavins by time-resolved spectroscopy. Naturwissenschaften 83, 32–35 (1996). [DOI] [PubMed] [Google Scholar]
- Chia T. H., Williamson A., Spencer D. D. & Levene M. J. Multiphoton fluorescence lifetime imaging of intrinsic fluorescence in human and rat brain tissue reveals spatially distinct NADH binding. Opt Express 16, 4237–4249 (2008). [DOI] [PubMed] [Google Scholar]
- Niesner R. et al. Selective detection of NADPH oxidase in polymorphonuclear cells by means of NAD(P)H-based fluorescence lifetime imaging. J Biophys 2008, 602639 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su G. C., Wei Y. H. & Wang H. W. NADH fluorescence as a photobiological metric in 5-aminolevlinic acid (ALA)-photodynamic therapy. Optics express 19, 21145–21154 (2011). [DOI] [PubMed] [Google Scholar]
- Vishwasrao H. D., Heikal A. A., Kasischke K. A. & Webb W. W. Conformational dependence of intracellular NADH on metabolic state revealed by associated fluorescence anisotropy. The Journal of biological chemistry 280, 25119–25126 (2005). [DOI] [PubMed] [Google Scholar]
- Seidenari S. et al. Multiphoton laser microscopy and fluorescence lifetime imaging for the evaluation of the skin. Dermatol Res Pract 2012, 810749 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig K. Robert Feulgen Prize Lecture. Laser tweezers and multiphoton microscopes in life sciences. Histochem. Cell Biol. 114, 79–92 (2000). [DOI] [PubMed] [Google Scholar]
- Heikal A. A. Intracellular coenzymes as natural biomarkers for metabolic activities and mitochondrial anomalies. Biomark Med 4, 241–263 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chance B. Pyridine nucleotide as an indicator of the oxygen requirements for energy-linked functions of mitochondria. Circ. Res. 38, I31–38 (1976). [PubMed] [Google Scholar]
- Bito T. & Nishigori C. Impact of reactive oxygen species on keratinocyte signaling pathways. J. Dermatol. Sci. 68, 3–8 (2012). [DOI] [PubMed] [Google Scholar]
- Gros E., Bussmann C., Bieber T., Forster I. & Novak N. Expression of chemokines and chemokine receptors in lesional and nonlesional upper skin of patients with atopic dermatitis. J. Allergy Clin. Immunol. 124, 753–760 e751 (2009). [DOI] [PubMed] [Google Scholar]
- Suarez-Farinas M. et al. Nonlesional atopic dermatitis skin is characterized by broad terminal differentiation defects and variable immune abnormalities. J. Allergy Clin. Immunol. 127, 954–964 e951–954 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkert M. et al. Metadata matters: access to image data in the real world. J. Cell Biol. 189, 777–782 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severity scoring of atopic dermatitis: the SCORAD index. Consensus Report of the European Task Force on Atopic Dermatitis. Dermatology 186, 23–31 (1993). [DOI] [PubMed] [Google Scholar]
- Rickham P. P. Human Experimentation. Code of Ethics of the World Medical Association. Declaration of Helsinki. Br. Med. J. 2, 177 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use. (2015) Available at: http://www.ich.org. (Accessed: 15th May 2015). [DOI] [PMC free article] [PubMed]
- McAuliffe M. J. et al. In Computer-Based Medical Systems, 2001. CBMS 2001. Proceedings. 14th IEEE Symposium on. 381–386.
- Gonzalez R. C. & Woods R. E. In Digital image processing 3rd edn, 100–101 (Prentice Hall, 2008). [Google Scholar]
- Leong F. J., Brady M. & McGee J. O. Correction of uneven illumination (vignetting) in digital microscopy images. J. Clin. Pathol. 56, 619–621 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsu N. Threshold Selection Method from Gray-Level Histograms. Ieee T Syst Man Cyb 9, 62–66 (1979). [Google Scholar]
- Sanches J. M. et al. Quantification of mutant E-cadherin using bioimaging analysis of in situ fluorescence microscopy. A new approach to CDH1 missense variants. Eur. J. Hum. Genet. 23, 1072–1079 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leys C., Ley C., Klein O., Bernard P. & Licata L. Detecting outliers: Do not use standard deviation around the mean, use absolute deviation around the median. J. Exp. Soc. Psychol. 49, 764–766 (2013). [Google Scholar]
- Boukamp P. et al. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 106, 761–771 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz J. et al. The amido black assay: a simple and quantitative multipurpose test of adhesion, proliferation, and cytotoxicity in microplate cultures of keratinocytes (HaCaT) and other cell types growing adherently or in suspension. J. Immunol. Methods 167, 1–13 (1994). [DOI] [PubMed] [Google Scholar]
- Swedlow J. R. & Eliceiri K. W. Open source bioimage informatics for cell biology. Trends Cell Biol. 19, 656–660 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.