Abstract
Lampreys are extant representatives of agnathans. Descriptions of lamprey development, physiology and genome have provided critical insights into early evolution of vertebrate traits. However, efficient means for genetic manipulation in agnathan species have not been developed, hindering functional studies of genes in these important Evo-Devo models. Here, we report a CRISPR/Cas system optimized for lamprey genomes and use it to disrupt genomic loci in the Northeast Chinese lamprey (Lethenteron morii) with efficiencies ranging between 84~99%. The frequencies of indels observed in the target loci of golden (gol), kctd10, wee1, soxe2, and wnt7b, estimated from direct sequencing of genomic DNA samples of injected lamprey larvae, were 68/69, 47/56, 38/39, 36/37 and 36/42, respectively. These indels often occurred in both alleles. In the CRISPR/Cas9 treatment for gol or kctd10, 38.6% or 85.3% of the targeted larvae had the respective recessive null-like phenotypes, further confirming the disruption of both loci. The kctd10 gRNA, designed against an essential functional region of Kctd10, resulted in null-like phenotypes and in-frame mutations in alleles. We suggest that the CRISPR/Cas-based approach has the potential for efficient genetic perturbation in organisms less amenable to germ line transmission based approaches.
Lampreys and hagfishes are sole surviving lineages of jawless vertebrates, or agnathans. As an out-group to the gnathostomes (vertebrates with hinged jaws), the development and evolution of agnathans have been examined extensively. These studies have provided important insights into the origin and early radiation of the vertebrate linage1,2,3. In particular, studies in lampreys have helped reveal possible developmental mechanisms of the craniate, and the origin of the features unique to vertebrate animals, such as the neural crest, the true brain, the pharyngeal skeleton, the jaw, the vertebra and the appendage2,4. In addition, lampreys have been used extensively as biomedical models for elucidation of mechanisms that govern adaptive immunity, spinal cord regeneration, locomotion, and neuronal degeneration5,6.
With a few exceptions, such as elucidation of particular gene functions through morpholino-knockdowns and RNA interference technology7,8,9, studies of lamprey development and gene function have remained largely descriptive. To define the functions of essential genes in an organism, both genome data and a means to edit the genome are indispensable. The draft genomes of two lamprey species, the sea lamprey (Petromyzon marinus) and Japanese lamprey (Lethenteron japonicum), have become available10,11. However, techniques for genetic mutagenesis in lamprey species have not been established. Established techniques that generate mutations through germ line transmission are difficult to implement in lamprey species, which develop through a lengthy (up to 20 years to reach sexual maturation) and complicated life cycle3,12,13,14. Not surprisingly, culture of a lamprey (or any agnathan) species through a complete life cycle in captivity has not been reported. Furthermore, lampreys are semelparous species; sexually mature adults spawn once and expire, making it exceedingly difficult to obtain sufficient F1 and F2 individuals even if one can cultivate lamprey to maturation. We reasoned that a method that efficiently disrupts both alleles of genomic loci in the founder (F0) generation would be amenable for studies of gene function in lamprey species. Furthermore, many species that are uniquely advantageous for addressing certain biological questions but less tractable genetically through germ line transmission based approaches should also become more useful as model systems when genome editing at the (F0) generation becomes available.
Recently, a series of designer nucleases targeting specific endogenous DNA sequences has been developed15,16. Among these systems, the CRISPR/Cas (clustered regularly interspaced short palindromic repeats/CRISPR-associated proteins) system emerged as a method of choice for its simplicity and efficiency in introducing mutations of target genes in a variety of model organisms17,18,19,20. Furthermore, CRISPR/Cas9 systems with high cleavage activities have been shown to cut both alleles of a target gene efficiently in embryonic stem cells as well as in organisms including mouse, pig and zebrafish21,22,23,24.
Here we report the development of a CRISPR/Cas9 system that targets genomic loci in lamprey larvae with high efficiencies. Since the CRISPR/Cas9 system relies on an RNA-guided nuclease system derived from prokaryotes, the Cas9 nuclease activity may be enhanced if its cDNA is optimized according to the codon usage in the target eukaryote genome. We applied the optimized CRISPR/Cas9 system in five loci, wee1, soxe2, kctd10, wnt7b and golden (gol or slc24a5). These genes were selected because they have confirmed functions in vertebrate development or give convenient and consistent readouts of phenotypes, or both7,8,25,26. In particular, the gol encodes the sodium/calcium exchanger Slc24a5 that affects pigmentation in zebrafish and human. A mutation of gol in zebrafish, golb1, results in hypopigmentation of skin melanophores and retinal pigment epithelia27. The loss of function for gol is recessive, and therefore the retinal color pattern is a convenient readout for testing the efficiency of biallelic loci disruption. kctd10 encodes the potassium channel tetramerization domain-containing 10. In zebrafish, mutation of kctd10 causes malformation of the atrioventricular canal, resulting in a stretched and string-like heart28. The loss-of-function of kctd10 is recessive, and the phenotype of the mutant is consistently recognizable. In the Northeast Chinese lamprey (Lethenteron morii), injection of the Cas9 mRNA optimized for lamprey codon usage along with the relevant gRNAs into one-cell stage embryos disrupted both alleles of each of the five endogenous genes in F0. In the two loci, gol and kctd10, that give clear phenotype readouts in the larval stage, the null-like phenotypes were observed in 38.6% and 85.3% of the founders, respectively. We conclude that the CRISPR/Cas9-based technique is an efficient means to disrupt both alleles of endogenous genes in lamprey.
Results
CRISPR/Cas9 system mediated gene targeting in lamprey
We examined three Cas9 proteins for their efficiency in altering lamprey alleles: 1) ZCas9, the Streptococcus pyogenes Cas9 optimized for zebrafish codon usage with an SV40 NLS at the 5′ end and a nucleoplasmin NLS at the 3′ end29; 2) LCas9-1 and 3) LCas9-2, both of which were optimized according to the codon usage determined from 15,790 coding sequences of the sea lamprey draft genome (Fig. S1, Table S1). LCas9-1 and LCas9-2 differed slightly in their amino acid residue sequences. The codon optimized LCas9-1 and -2 were fused to NLS at both 5′ and 3′ ends (Fig. S1). The three Cas9 cDNAs were ligated separately into a pxT7 expression vector, and transcribed individually into Cas9 mRNAs. The guide RNAs (gRNAs) against the loci of the five targeted endogenous genes of lamprey (Fig. S2 and Table S2) were designed and synthesized. The according gRNA for each examined gene was injected, along with each of the three Cas9 mRNAs simultaneously, respectively, into lamprey embryos at the one-cell stage, resulting in a total of 15 combinations of gRNA and Cas9 mRNA.
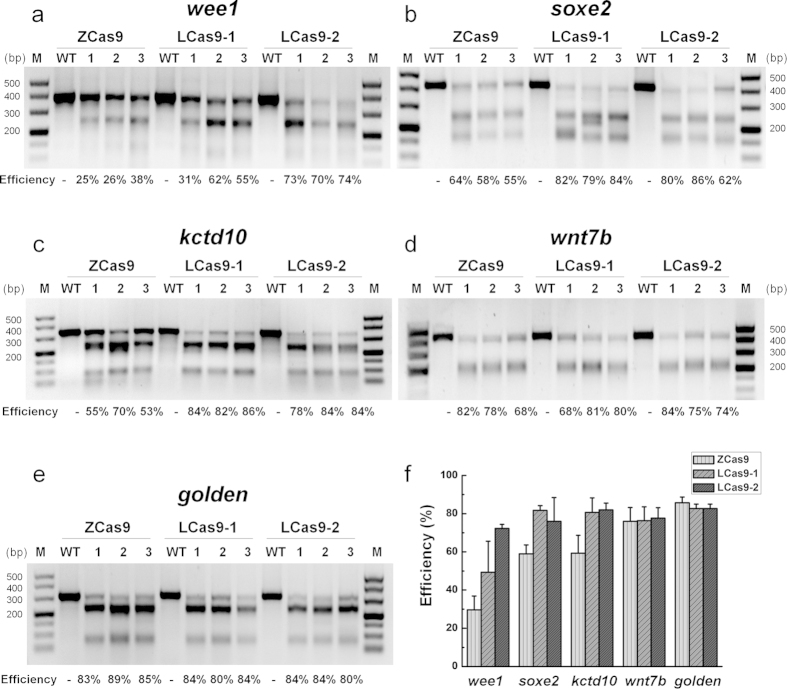
The T7 Endonuclease I (T7EI) assay30 revealed robust mutagenesis at all five loci targeted by the CRISPR/Cas9 systems (Fig. 1). All tested Cas9s cleaved the target regions of the lamprey genome in the presence of the appropriate gRNA. In most cases, the Cas9s optimized for the lamprey genome appeared to be more efficient (Fig. 1f). Approximately 60% to over 80% of the PCR products derived from the target regions of these five genes were cleaved by T7EI when LCas9-2 was used (Fig. 1). These data demonstrate that the optimized CRISPR/Cas9 systems targeted the lamprey genomic loci effectively.
Figure 1. The CRISPR/Cas9 system induced mutagenesis in five endogenous genes of lamprey.
Cas9 mRNA and gRNA against each of the five endogenous genes were injected separately into lamprey embryos at the one-cell stage. The control embryos (WT) were wild type. T7EI assays showed the mutagenesis rates in three groups of pooled injected embryos for each target site, and each group contained five randomly selected individual. For every endogenous gene and for every type of Cas9, the rates of mutagenesis ranged from 25% to 89%.
Treatment of Cas9 and gol gRNA induced null-like phenotypes in F0
To further evaluate the efficiency of the optimized CRISPR/Cas9 systems in inducing biallelic disruptions of endogenous genes in lamprey, we targeted gol loci in L. morii. Analyses through sequence alignment indicated that Slc24a5 is conserved within the vertebrate clade. The conservation of amino acid residues between lamprey and human, mouse or zebrafish are at 54%, 55% and 56% identity, respectively (Fig. S3), suggesting that Slc24a5 may have similar functions in these animals.
To target the loci of gol, Cas9 mRNAs were injected into one-cell stage lamprey embryos along with gol gRNA (Fig. S2). The gol gRNA was designed to target a DNA segment found in the coding region of lamprey gol but absent in the human, mouse or zebrafish orthologs (Fig. S3). The retina of L. morii larvae was examined for pigmentation on day 19 after the injection (Table 1). The injection largely affected the rate of null-like phenotypes (χ2 = 170.24, p < 0.0001). Among the larvae (n = 132) injected with LCas9-1 and gol gRNA, 67.4% showed hypopigmentation, of which 38 larvae (28.8% of the injected larvae) had mosaic hypopigmentation (Fig. 2b,e, and Fig. S4d–f). A total of 51 larvae (38.6% of the injected larvae) had no observable pigment in their retinas, indicating that biallelic disruption of gol occurred in the majority of the cells in these individuals (Fig. 2c,f, and Fig. S4g–S4i). LCas9-1 and LCas9-2 differed in their effects on pigmentation patterns (χ2 = 10.01, p = 0.007), with LCas9-1 inducing a higher rate of the null-phenotype and LCas9-2 inducing a higher rate of mosaic pigmentation in the retina (Table 1).
Table 1. Retina pigmentation of lamprey larvae from Cas9 and gol gRNA treatment.
| Treatment | Total Number of larvae | Normal pigment | No pigment | Mosaic pigment |
|---|---|---|---|---|
| Control | 116 | 116 | 0 (0%) | 0 (0%) |
| LCas9-1 + gol gRNA | 132 | 43 | 51 (38.6%) | 38 (28.8%) |
| LCas9-2 + gol gRNA | 109 | 33 | 25 (22.9%) | 51 (46.8%) |
| ZCas9 + gol gRNA | 90 | 32 | 25 (27.7%) | 33 (36.7%) |
Each Cas9 induced hypopigmentation in comparison to control (p < 0.0001 for each Cas9). LCas9-1 and LCas9-2 differed in inducing the pigmentation patterns (χ2 = 10.01, p = 0.007), with LCas9-1 inducing a higher rate of null-phenotypes whereas LCas9-2 inducing a higher rate of mosaic pigmentation in the retina.
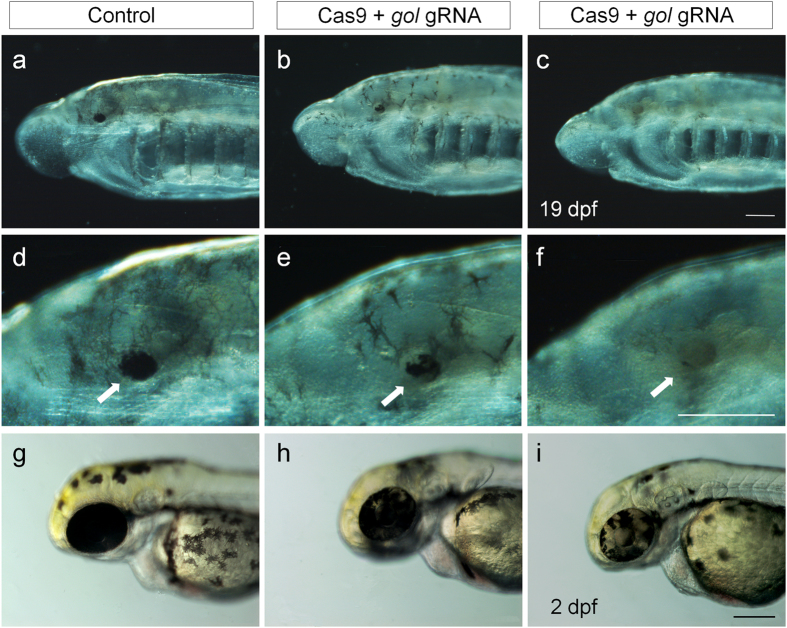
Figure 2. Retina pigmentation in lamprey and zebrafish developed from Cas9 and gol gRNA injected embryos.
(a–f) Lateral views of control (a,d) and gol gRNA injected larvae (b,c,e,f) on 19 dpf (days post fertilization). Phenotypes shown include the wild-type (arrow in (d)), mosaic hypopigmentation (arrow in (e)) and no observable pigment (arrow in (f)) in the retina. Zebrafish embryos injected with Cas9 and the appropriate gol gRNA on 2 dpf also showed mosaic pigmentation (h,i) in the retinas. Scale bars: 0.2 mm.
As a positive control and to compare the efficiency of allelic disruption, we targeted the gol loci in zebrafish using ZCas929 and an appropriate gRNA21. The injected zebrafish larvae showed the three pigmentation phenotypes similar to those obtained in lamprey larvae (Fig. 2g–i), confirming that the function of gol is highly conserved in vertebrates. Notably, the percentage of the gol null-like phenotype was drastically lower than the mosaic pigment phenotype in zebrafish compared to lamprey (Table S3). This discrepancy is most likely because CRISPR/Cas9 based biallelic disruption of gol in founders was more efficient in the lamprey than in the zebrafish.
Treatment of Cas9 disrupted both alleles of gol in virtually all cells
We sequenced the target region of the genome from injected animals to examine the exact alteration induced by the CRISPR/Cas9 systems at the lamprey gol locus. Genomic DNA extracted from five founder individuals (two with no pigment, two with mosaic pigment and one with normal pigment) were examined. Twenty independent colonies derived from each DNA sample were sequenced (Table 2 and Fig. S5). The genomic DNA sequences showed that both alleles of gol in most cells were disrupted in the null-like phenotype larvae. In one larva with no observable retinal pigments (N2), all 18 valid sequences showed indels as compared to those obtained from a wild-type lamprey, indicating that both alleles of gol were targeted (Table 2 and Fig. S5a). Seven different types of indels were found in this individual, indicating that the CRISPR/Cas9 enabled mutation occurred independently in individual cells. Most indels (14 out of 18) in this larva caused a frame shift or large fragment deletion, resulting in a null-allele. These results demonstrate that both alleles of gol in most cells of the larvae with the null-like phenotype were disrupted.
Table 2. Different alleles of gol in the injected larvae.
| Phenotype | Sample ID | Total | Wild type | In-frame indels | Frame-shift indels | Null mutation (%) |
|---|---|---|---|---|---|---|
| WT-like | W5 | 17 | 0 | 13 | 4 | 23.5 |
| Mosaic | M2 | 18 | 0 | 10 | 8 | 44.4 |
| M5 | 16 | 0 | 7 | 9 | 56.3 | |
| Null-like | N2 | 18 | 0 | 5 | 13 | 72.2 |
| N5 | 17 | 1 | 4 | 12 | 70.6 |
Total, total number of valid sequence; WT-like, wild type like.
To our surprise, the genomic DNA sequence of injected larvae that did not display the null-like phenotype only contained mutated alleles of gol. The 34 valid sequences from the two larvae with mosaic pigmentation all contained mutated gol alleles (Table 2 and Fig. S5b). However, many deletions (10 out of 18 in one larva and 7 out of 16 in the other) caused in-frame mutations, and conserved amino acid residues were not affected in most of these deletions. Even the injected larva that had normal retinal pigmentation contained indels in all 17 valid sequences, although the majority of the mutations (13 out of 17) were in-frame deletions (Table 2 and Fig. S5c).
Collectively, among the 86 sequences obtained from the five injected lamprey larvae representing three different phenotypes, only one sequence was the wild-type. Clearly, injection of LCas9-1 and gol gRNA into lamprey embryos at the one-cell stage induced mutations in almost all the cells of the larvae, and virtually all disruptions of the genomic locus were biallelic. These results demonstrate that the optimized CRISPR/Cas9 system modified both gol alleles at an efficiency approaching 99% in F0.
Treatment of Cas9 and kctd10 gRNA induced null-like phenotypes in F0
We next evaluated the utility of the optimized CRISPR/Cas9 system in targeting the biallelic loci of genes essential for animal development. We chose kctd10 that plays an essential role in heart morphogenesis28. The lamprey Kctd10 protein shares 85~87% amino acid sequence identity with its zebrafish, mouse and human orthologs (Fig. S6). We predicted that Kctd10 may have a similar role in cardiac development in these organisms.
Lamprey embryos at the one-cell stage were injected with Cas9 mRNA and kctd10 gRNA, and their morphology observed during development. The kctd10 gRNA was designed to target a conserved region of the coding sequence (Fig. S6). The Cas9 treatments significantly affected the rate of heart malformation (χ2 = 169.61, p < 0.0001). LCas9-1 induced a higher rate of heart malformations than LCas9-2 (χ2 = 17.02, p < 0.0001) and ZCas9 (χ2 = 5.28, p = 0.022). Twenty one days after injection of larvae with LCas9-1 and kctd10 gRNA, 99 out of 116 (85.3%) showed severe pericardial edema (Table 3, Fig. 3c, and Fig. S7c,S7d). The malformed hearts had a stretched, string-like morphology and beat weakly (Fig. 3d). The zebrafish kctd10 mutant, previously identified from a spontaneous recessive mutation28, also has a string-like heart morphology and weakened heart contractility (Fig. 3g,h). Clearly, treatment of optimized Cas9 and kctd10 gRNA in lamprey resulted in a phenotype similar to that of zebrafish kctd10 mutants. These results suggest that both alleles of kctd10 in the injected lamprey larvae were disrupted, and the function of Kctd10 is conserved between lamprey and zebrafish.
Table 3. Larval heart malformations from Cas9 and kctd10 gRNA treatment.
| Treatment | Total Number of larvae | Normal larvae | Larvae with heart disorders | Mutant (%) |
|---|---|---|---|---|
| Control | 114 | 111 | 3 | 2.6 |
| LCas9-1 + kctd10 gRNA | 116 | 17 | 99 | 85.3 |
| LCas9-2 + kctd10 gRNA | 44 | 20 | 24 | 54.5 |
| ZCas9 + kctd10 gRNA | 54 | 16 | 38 | 70.4 |
Overall, treatment of Cas9 affected the phenotype (χ2 = 169.61, p < 0.0001). Treatment by each one of the three Cas9s induced more malformations than the vehicle treatment (p < 0.0001 for each comparison). LCas9-1 induced a higher rate of malformation in the heart than LCas9-2 (χ2 = 17.02, p < 0.0001) and ZCas9 (χ2 = 5.28, p = 0.022). LCas9-2 did not differ from ZCas9 in the number of induced malformations (χ2 = 2.613, p = 0.10).
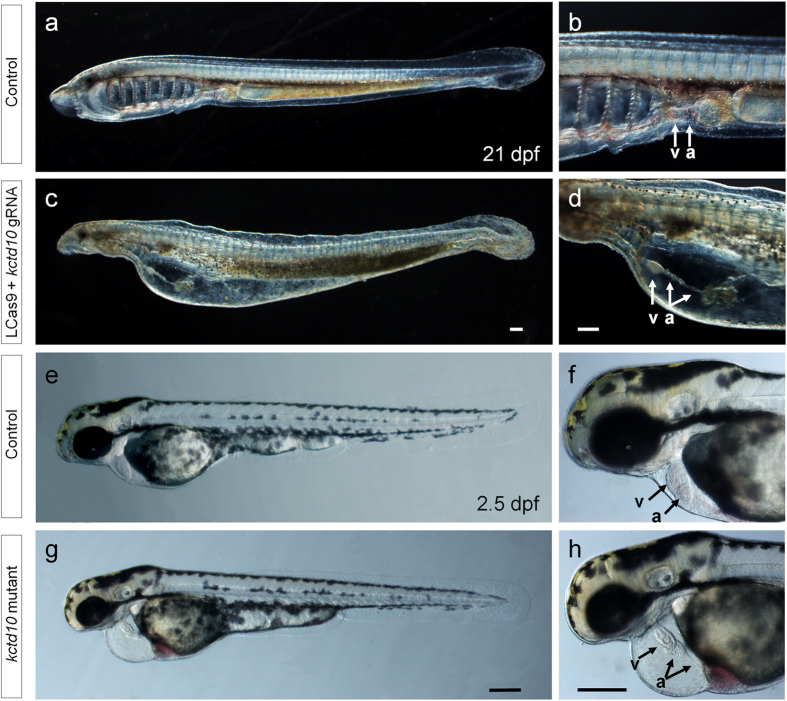
Figure 3. Heart malformations in lamprey larvae and zebrafish developed from Cas9 and kctd10 gRNA injected embryos.
(a–d) Lateral views of control (a,b) and kctd10 and gRNA injected lamprey larvae (c,d) on 21 dpf (days post fertilization). The control larvae have normal heart formation (a) and the treatment larvae have severe pericardial edema (c). (b,d) enlarged views of a and c, respectively, provide a comparison that shows the marked heart malformations in the injected larvae (d). (d) the atrium (a) stretches to a string-like structure and no blood was observed in either atrium or ventricle (v). Zebrafish larvae of kctd10 mutant had pericardial edema (g) and heart malformations (h) as compared to the wild-type larvae (e,f) on 2.5 dpf. In the mutants, part of the atrium stretches to a string-like structure and no blood was observed in either atrium or ventricle (h). Scale bars: 0.2 mm.
The Cas9 system disrupted both alleles of kctd10 in lamprey
We genotyped the kctd10 loci in six LCas9-1 injected lamprey larvae (Table 4 and Fig. S8). For the three larvae that displayed heart malformations, we obtained 20, 19 and 17 valid sequencing results, respectively. Among the kctd10 sequences, 16, 18 and 13, respectively, contained indels (Fig. S8). Some of which caused a frame shift and others (>50%) caused deletions in amino acid residues that are conserved between human, mouse, zebrafish and lamprey. Overall, the frequency of the wild-type alleles of kctd10 was very low (4/20, 1/19 and 4/17), indicating that most cells of these individuals contained only mutated alleles. In other words, both alleles of kctd10 were targeted in most cells of the larvae with heart malformations. The three injected larvae with no overt heart malformations, had higher ratios of wild-type sequences (8/18, 9/20 and 13/19; Table 4 and Fig. S8b). Taken together, these results reveal that LCas9-1 efficiently mutated both alleles of kctd10, when introduced into lamprey embryos at the one-cell stage, leading to null-like phenotypes in the founder generation.
Table 4. Different kctd10 alleles in the injected larvae.
| Phenotype | Sample ID | Total | Wild type | In-frame indels |
Frame-shift indels | Null mutation (%) | |
|---|---|---|---|---|---|---|---|
| Conserved aa not affected | Conserved aa affected | ||||||
| WT-like | W1 | 18 | 8 | 0 | 7 | 3 | 55.6 |
| W2 | 20 | 9 | 0 | 5 | 6 | 55.0 | |
| W3 | 19 | 13 | 0 | 4 | 2 | 31.6 | |
| Heart disorder | H1 | 20 | 4 | 0 | 9 | 7 | 80.0 |
| H2 | 19 | 1 | 0 | 10 | 8 | 94.7 | |
| H3 | 17 | 4 | 0 | 8 | 5 | 76.5 | |
Total, the total number of valid sequence analysed; WT-like, wild type-like; aa, amino acid.
Cas9 targeting in the loci of wee1, soxe2 and wnt7b
We also confirmed by sequencing biallelic disruption in the genomic loci of wee1, soxe2 and wnt7b, in the larvae injected with the Cas9 mRNA and appropriate gRNA. Although mutated alleles of each gene were detected by the T7EI assay (Fig. 1), no apparent mutated phenotypes were observed in the larval stage. We determined if the Cas9 treatment disrupted both alleles of these loci in the target regions (Fig. S9) using the strategy outlined for the gol and kctd10 loci genotyping.
Our sequencing results showed that LCas9-1 disrupted both alleles of wee1, soxe2 or wnt7b at very high levels of efficiency. For the target region of wee1, 39 of the 40 valid sequences contained mutations, for the soxe2 target region, 36 of the 37 sequences contained indels and for the wnt7b target region, 36 of the 42 sequences contained indels. It was evident that indels occurred in both alleles of all three genes. These results further confirmed that injection of Cas9 mRNA together with the appropriate gRNA efficiently target both alleles of endogenous genes in lamprey F0.
Discussion
Lamprey species are used extensively in development and evolution studies3. However, these species are not readily amenable to either forward genetics or reverse genetics, the traditional approaches that rely on the mutations recovered and maintained through germ line transmission. In these approaches, mutations are introduced into the F0, or founder organism. Usually, the F0 is mosaic for the mutation, and is bred with the wild-type to generate the F1, some of which are heterozygous for the mutation. In reverse genetics, it is the cross between the F1 heterozygotes that results in the homozygote mutant. In forward genetics, the heterozygotes of the same mutation are obtained in the F231,32. However, it is technically challenging and costly to culture lamprey embryos to adult stages under laboratory conditions because lamprey species develop through a lengthy and complex life cycle that demands dramatic habitat changes to support the transition between life stages12. Therefore, it is not yet practical to transmit introduced mutations from F0 to F1 in this group of animals. Nonetheless, manipulation of fertilized lamprey eggs is possible as the chorions are resistant to rupture induced by surface-tension and like zebrafish and Xenopus embryos, the one-cell stage embryo can be injected in dry dishes33. The one-cell stage during lamprey embryonic development lasts for several hours, providing a very large injection window and likely increasing the cleavage efficiency of the CRISPR/Cas9 system. In the present study, we disrupted both alleles of all five examined endogenous loci in a lamprey genome by introducing the CRISPR/Cas9 system at the one-cell stage. The efficiency of biallelic mutagenesis ranged from 84% to 99%, which is comparable to that in zebrafish and cynomolgus monkey21,34, and higher than that in pig35.
For the two endogenous genes examined in detail, gol and kctd10, their disruption led to marked and expected phenotypes in F0. When the gol loci was targeted, nearly 40% of the injected larvae lacked pigment in their retina, a phenotype of the null mutation, which is similar to that in zebrafish21,36. Moreover, targeting the kctd10 loci resulted in an even higher ratio of the null-like phenotype, and up to 85% of injected larvae had a heart malformation that was virtually identical to those observed in kctd10 deficient zebrafish28. At the null-allele rates demonstrated for these two loci, the LCas9-1 based procedure is sufficiently efficient and effective in editing the lamprey genome, with practical utilities in examination of gene function in the founder generation. Although the efficiencies of biallelic disruption of the loci of wee1, soxe2 and wnt7b were also high, no marked disorders were observed in the larvae. A plausible explanation for the lack of the null phenotype is functional compensation by other genes. Another possibility is that the CRISPR/Cas9 and morpholino treatments exert different effects on the target loci, as has previously been shown in zebrafish embryos37.
A notable dichotomy in the outcomes of CRISPR/Cas9 treatment of lamprey embryos was the higher rate of frame-shifting indels in the gol locus than in the kctd10 and the dramatically higher null-phenotype rate from targeting kctd10 compared to gol. The gol locus, targeted by our optimized CRISPR/Cas9 system, led to a null-phenotype rate of approximately 40%, which is consistent with the theoretical value. In theory, the ratio of the indels that cause reading frame shift over the total indels is 2/3. Hence the probability for both alleles of a targeted locus to carry a frame shift mutation is 4/9, suggesting that 5/9 cells from an injected organism should have the wild-type or in-frame mutation alleles. To overcome this issue, we designed the kctd10 gRNA to target an essential functional domain that encodes highly conserved amino acid residues. Substitution or deletion of such amino acid residues often lead to a null mutation38. As expected, 85% of the larvae injected with Cas9 and gRNA against kctd10 showed the null-phenotype and had heart disorders, and more than 50% of the null alleles contained in-frame mutations. We suggest to improve efficiency of the CRISPR/Cas9 system to study gene function in the F0, highly conserved domains should be targeted rather than randomly selected regions.
The gRNA guided targeting of Cas9 in genomic loci appears to extend beyond the very lengthy one-cell stage of lamprey embryos. This is evident from the seven types of indels documented in genomic DNA of the target region of gol in injected individuals. If the targeting process happened only at the one-cell stage, as many as two types of indels for each targeted loci would be expected in the injected individual. We also noticed that the biallelic disruption of endogenous genes by CRISPR/Cas9 systems was more efficient in F0 lamprey than in F0 zebrafish. This could be explained by the very different durations of early embryonic development in these two animals. In lamprey embryos, the one-cell stage lasts for 5–6 hours, and each subsequent cleavage lasts for about 1 h at 18 °C (Fig. S10). In comparison, the one-cell stage in zebrafish embryos lasts for 45 minutes, and the subsequent cleavage each lasts for 15 minutes at 28.5 °C39. In future studies, it would be interesting to examine the functional dynamics of Cas9 mRNA and gRNA molecules in lamprey and zebrafish embryos through the first few stages, and infer why the Cas9 treatment results in a higher efficiency of biallelic disruptions in the lamprey founders.
In summary, we optimized a CRISPR/Cas9 system that disrupted both alleles of all five examined endogenous genes in a lamprey genome. The biallelic disruption is efficient enough to generate sufficient numbers of the null-phenotype and null-mutation in F0 for genetically tractable studies of gene functions. This method will likely enable functional studies of the genes important for the development of lamprey and for understanding early evolution of vertebrate traits. Similar approaches, with straightforward optimizations, may become available to define gene function in other organisms for which F1 and F2 generations are difficult to generate in laboratory conditions.
Material and Methods
Animals and embryos
The Northeast Chinese lampreys were obtained from the Shuifen reservoir, a tributary of Yalu River in Dandong city, Liaoning province, China. Sexually mature lamprey were transported to Shanghai Ocean University campus (Lingang, Shanghai, China), and maintained at 8 °C in tanks with aerated tap water. The eggs were striped from ovulatory females into a dish containing 300 ml autoclaved double distilled water, and the sperm from a spermiating male were expressed onto the eggs for in vitro fertilization40. As described previously28, zebrafish were raised and maintained at 28.5 °C in a re-circulating system with continuously filters. The re-circulated water was treated with UV light and aerated. Breeding was carried out in-tank to obtain the embryos, and the embryos were raised in the incubator at 28.5 °C. All handling of fishes was carried out in accordance with the guidelines on the care and use of animals for scientific purposes set up by the Institutional Animal Care and Use Committee (IACUC) of the Shanghai Ocean University (SHOU), Shanghai, China. This research was approved by the IACUC of SHOU.
Cas9 plasmids
The LCas9-1 coding sequence was from Streptococcus pyogenes MGAS1882 (YP_005388840)21. The LCas9-2 coding sequences were designed according to Liu et al.29. Both Cas9 coding sequences were codon-optimized and ordered from Sangon Biotech. The codon usage frequency of sea lamprey is listed in the Table S1. A Kozak consensus was put at the translational start and nuclear localization signal (NLS) was fused at both ends. The full length of codon-optimized Cas9 sequences with double NLS was cloned into the pXT7 vector containing T7 promoter and globin UTR. See Fig. S1 for the full sequences of the codon-optimized LCas9s.
RNA Synthesis
All three Cas9 plasmids were linearlized by XbaI. Capped mRNAs of Cas9s were synthesized in vitro by mMESSAGE mMACHINE T7 ULTRA kit (Ambion), and purified using RNeasy Mini Kit (Qiagen). Double strand cDNA for specific gRNA was amplified by PCR (NEB) using corresponding primers (target site specific forward primer 5′-TAATACGACTCACTATANNNNNNNNNNNNNNNNNNNNGTTTTAGAGCTAGAAATAGC-3′; common reverse primer 5′-AAAAAAAGCACCGACTCGGTGCCAC-3′). The template sequence is GTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACT TGAAAAAGTGGCACCGAGTCGGTGCTTTTTTT41. Each target sequence contains the GG at the beginning, which is required for T7 RNA polymerase starting. After gel extraction of the PCR product, gRNA was synthesized using MAXIscript T7 kit (Ambion), and digested with DNase I (Ambion) for 15 min to remove DNA. The gRNA was purified using mirVana™ miRNA Isolation Kit (Ambion).
Microinjection
The microinjection of the Northeast Chinese lamprey embryos followed the protocol of Nikitina33 with modification. Briefly, the fertilized eggs (one-cell stage) were washed with 18 °C autoclaved double distillted water for several times, placed on a microinjection dish with a nylon mesh at 18 °C, and each embryo injected with a mixture of the Cas9 mRNA (400 pg) and gRNA (80 pg). Over 200 embryos were injected for each combination of Cas9 mRNA and gRNA.
The microinjection of zebrafish embryos was performed at the one-cell stage as described previously42. Approximately 400 pg mRNA encoding Cas9 and 80 pg gRNA were injected into each embryo.
T7EI assay
Five days after injection of each combination of gRNA and Cas9 mRNA, genomic DNA was isolated from three groups of pooled injected larvae, and each group contained five randomly selected individuals. The target region was amplified with PCR using the according primer pairs (for details, see Table S2). The resulted products were denatured and annealed, and treated with T7 Endonuclease I (NEB) for 45 min at 37 °C. The digested PCR products were examined by agarose gel (2%) electrophoresis.
Sequence analysis
The genomic DNA was isolated from the injected individual, and the target regions were amplified from the genomic DNA using the according specific PCR primers. Each product was ligated into the pMD19-T TA cloning vector (Takara) and transformed into DH5α competent cells (Tiangen). For each amplicon, approximately 20 independent colonies were sequenced by Dye terminator sequencing.
Imaging
Embryos were treated with 0.02% tricaine (3-amino benzoic acid ethyl ester), mounted in 3% methyl-cellulose, and visualized either under a Stereoscopic Microscopes (Zeiss, Discovery. V20) or a Fluorescence Microscope (Zeiss, Axioimager. M2).
Additional Information
How to cite this article: Zu, Y. et al. Biallelic editing of a lamprey genome using the CRISPR/Cas9 system. Sci. Rep. 6, 23496; doi: 10.1038/srep23496 (2016).
Supplementary Material
Acknowledgments
We thank Dr. Bo Zhang (School of Life Sciences, Peking University) for providing the ZCas9 plasmid, Dr. Jingwei Xiong (Institute of Molecular Medicine, Peking University) for providing the pUC19-scaffold plasmid for gRNA synthesis. We thank Dr. Deborah Power (Universidade do Algarve) for proofreading this manuscript, Dr. An Xiao (Eastern Virginia Medical School) for useful discussions. We also thank Xueying Zhao and Jing Xie for their technical assistance and Dr. Yu-Wen Chung-Davidson (Michigan State University) for her assistance in statistical analysis. This work was supported by the National Natural Science Foundation of China (No. 31501166), the Sailing Program of Science and Technology Commission of Shanghai Municipality (15YF1405000), the Chenguang Program of the Shanghai Municipal Education Commission (14CG49), the Doctor Startup Fund of Shanghai Ocean University (A2-0302-14-300064), the SHOU&MSU Joint Research Center Grant (A1-0209-15-0806), and the Peak Discipline Program for Fisheries from the Shanghai Municipal Government.
Footnotes
Author Contributions Y.Z. and W.L. conceived the project. Y.Z. designed the experiments. X.Z. made the Cas9 mRNA and gRNA. X.D. and Y.Z. performed the microinjections, analyzed the phenotypes and captured the images. X.Z. and Z.Z. performed the T7E1 assays and sequenced the target sites. J.L., X.Z. and Q.Z. captured and maintained the lampreys. J.R. identified the sequences of target genes and optimized the codon of the LCas9. Y.Z. analyzed the data. Y.Z. and W.L. wrote the manuscript.
References
- Delsuc F., Brinkmann H., Chourrout D. & Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature 439, 965–968 (2006). [DOI] [PubMed] [Google Scholar]
- Osorio J. & Retaux S. The lamprey in evolutionary studies. Development genes and evolution 218, 221–235 (2008). [DOI] [PubMed] [Google Scholar]
- Shimeld S. M. & Donoghue P. C. Evolutionary crossroads in developmental biology: cyclostomes (lamprey and hagfish). Development 139, 2091–2099 (2012). [DOI] [PubMed] [Google Scholar]
- Kuratani S., Kuraku S. & Murakami Y. Lamprey as an evo-devo model: lessons from comparative embryology and molecular phylogenetics. Genesis 34, 175–183 (2002). [DOI] [PubMed] [Google Scholar]
- Cohen A. H., Mackler S. A. & Selzer M. E. Functional regeneration following spinal transection demonstrated in the isolated spinal cord of the larval sea lamprey. Proc Natl Acad Sci USA 83, 2763–2766 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alder M. N. et al. Antibody responses of variable lymphocyte receptors in the lamprey. Nature immunology 9, 319–327 (2008). [DOI] [PubMed] [Google Scholar]
- McCauley D. W. & Bronner-Fraser M. Importance of SoxE in neural crest development and the evolution of the pharynx. Nature 441, 750–752 (2006). [DOI] [PubMed] [Google Scholar]
- Lakiza O., Miller S., Bunce A., Lee E. M. J. & McCauley D. W. SoxE gene duplication and development of the lamprey branchial skeleton: Insights into development and evolution of the neural crest. Developmental Biology 359, 149–161 (2011). [DOI] [PubMed] [Google Scholar]
- Heath G. et al. RNA interference technology to control pest sea lampreys-a proof-of-concept. Plos one 9, e88387 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta T. K. et al. Evidence for at least six Hox clusters in the Japanese lamprey (Lethenteron japonicum). Proc Natl Acad Sci USA 110, 16044–16049 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. J. et al. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nature Genetics 45, 415–421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beamish F. & Potter I. The biology of the anadromous Sea lamprey (Petromyzon mannus) in New Brunswick. Journal of Zoology 177, 57–72 (1975). [Google Scholar]
- Tahara Y. Normal stages of development in the lamprey, Lampetra reissneri (Dybowski). Zoological science 5, 109–118 (1988). [Google Scholar]
- Nikitina N., Bronner-Fraser M. & Sauka-Spengler T. The sea lamprey Petromyzon marinus: a model for evolutionary and developmental biology. Cold Spring Harbor protocols 2009, pdb emo113 (2009). [DOI] [PubMed] [Google Scholar]
- Gaj T., Gersbach C. A. & Barbas C. F. III ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends in biotechnol 31, 397–405 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y. et al. Making designer mutants in model organisms. Development 141, 4042–4054 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland A. E. et al. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat methods 10, 741–743 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W. Y. et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 31, 227–229 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D. L. et al. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol 31, 681–683 (2013). [DOI] [PubMed] [Google Scholar]
- Xue W. et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature 514, 380–384 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao L. E., Wente S. R. & Chen W. B. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci USA 110, 13904–13909 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Vanoli F., LaRocque J. R., Krawczyk P. M. & Jasin M. Biallelic targeting of expressed genes in mouse embryonic stem cells using the Cas9 system. Methods 69, 171–178 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemr M. & Bühler M. Single-Step Generation of Conditional Knockout Mouse Embryonic Stem Cells. Cell Reports 12, 709–716 (2015). [DOI] [PubMed] [Google Scholar]
- Wroble B. N., Finkielstein C. V. & Sible J. C. Wee1 kinase alters cyclin E/Cdk2 and promotes apoptosis during the early embryonic development of Xenopus laevis. BMC developmental biology 7, 119 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beretta C. A., Brinkmann I. & Carl M. All four zebrafish Wnt7 genes are expressed during early brain development. Gene Expr Patterns 11, 277–284 (2011). [DOI] [PubMed] [Google Scholar]
- Lamason R. L. et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science 310, 1782–1786 (2005). [DOI] [PubMed] [Google Scholar]
- Tong X. et al. Kctd10 regulates heart morphogenesis by repressing the transcriptional activity of Tbx5a in zebrafish. Nat. Commun. 5, 3153 (2014). [DOI] [PubMed] [Google Scholar]
- Liu D. et al. Efficient gene targeting in zebrafish mediated by a zebrafish-codon-optimized cas9 and evaluation of off-targeting effect. J Genet Genomics 41, 43–46 (2014). [DOI] [PubMed] [Google Scholar]
- Reyon D. et al. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol 30, 460–465 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capecchi M. R. Altering the genome by homologous recombination. Science 244, 1288–1292 (1989). [DOI] [PubMed] [Google Scholar]
- Driever W. et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development 123, 37–46 (1996). [DOI] [PubMed] [Google Scholar]
- Nikitina N., Bronner-Fraser M. & Sauka-Spengler T. Microinjection of RNA and morpholino oligos into lamprey embryos. Cold Spring Harbor protocols 2009, pdb.prot5123 (2009). [DOI] [PubMed] [Google Scholar]
- Wan H. et al. One-step generation of p53 gene biallelic mutant Cynomolgus monkey via the CRISPR/Cas system. Cell Res. 25, 258–261 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. et al. Efficient CRISPR/Cas9-mediated biallelic gene disruption and site-specific knockin after rapid selection of highly active sgRNAs in pigs. Sci. Rep. 5, 13348 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y. et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol 26, 702–708 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok F. O. et al. Reverse Genetic Screening Reveals Poor Correlation between Morpholino-Induced and Mutant Phenotypes in Zebrafish. Dev Cell 32, 97–108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablain J., Durand E. M., Yang S., Zhou Y. & Zon L. I. A CRISPR/Cas9 Vector System for Tissue-Specific Gene Disruption in Zebrafish. Dev Cell 32, 756–764 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B. & Schilling T. F. Stages of embryonic development of the zebrafish. Dev Dyn 203, 253–310 (1995). [DOI] [PubMed] [Google Scholar]
- Nikitina N., Bronner-Fraser M. & Sauka-Spengler T. Culturing lamprey embryos. Cold Spring Harbor protocols 2009, pdb prot5122 (2009). [DOI] [PubMed] [Google Scholar]
- Chang N. et al. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 23, 465–472 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu Y. et al. TALEN-mediated precise genome modification by homologous recombination in zebrafish. Nat methods 10, 329–331 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.