Abstract
This study aims to investigate whether the use of recombinant human cytoglobin (rhCygb) impact on hepatic fibrogenesis caused by CCl4. SD (n = 150) rats were randomly divided into three groups of normal, CCl4 model and rhCygb groups. After model establishment, rats in rhCygb groups were administered daily with rhCygb (2 mg/kg, s.c.). Histological lesions were staged according to metavir. Serum parameters including ALT, AST, HA, LN, Col III and Col IV were determined. The liver proteins were separated by 2-DE and identified. As a result, the stage of hepatic damage and liver fibrosis in rhCygb groups were significantly milder than that in CCl4 model groups. Meanwhile, rhCygb dramatically reversed serum levels of ALT and AST, and also markedly decreased the liver fibrosis markers levels of LN, HA, Col III and Col IV. In 2-DE, 33 proteins among three groups with the same changing tendency in normal and rhCygb treated groups compared with CCl4 model group were identified. GO analysis showed that several identified proteins involved in oxidative stress pathway. The study provides new insights and data for administration of rhCygb reversing CCl4-induced liver fibrosis suggesting that rhCygb might be used in the treatment of liver fibrosis.
Liver fibrosis, a pathological process common to chronic liver diseases, is caused by a range of actors, and has a highly deleterious impact on human health. It is well-established that liver fibrosis formation results from an excessive accumulation of extracellular matrix proteins including collagen, a process that occurs in most types of chronic liver disease1. Liver fibrosis is described in several stages, a normal liver is at stage 0 to 1, stage 2 represents light or minimal fibrosis, stage 3 is severe fibrosis, and stage 4 denotes advanced liver fibrosis resulting in cirrhosis, liver failure, and portal hypertension2,3. Any reversibility of advanced liver fibrosis prior to cirrhosis in patients would be of signal importance.
Because of the urgent clinical need, the development of an anti-fibrotic drug would be an auspicious moment in medical research. Up to now, several candidate treatments with anti-fibrotic properties have been developed, which variously act by blocking inflammatory pathways4,5, inhibiting profibrotic growth factors6,7,8, modulating epigenetic codes9,10, and interfering with morphogenic pathways11,12, and which have proven efficacy and tolerability in pre-clinical fibrosis models. However, they are not available to treat fibrotic diseases in clinical practice.
The Cygb protein was first discovered in 2001 by proteomic analysis in rat stellate cells, and was initially named stellate cell-activated protein (STAP)13. The protein sequence showed that Cygb was hexacoordinate hemoglobin, representing the fourth member of the globin superfamily in mammals14. Furthermore, immunohistochemical analysis revealed that Cygb was ubiquitously expressed in several organs, such as kidney and pancreas, and localized in fibroblast-like cells15. Cygb was a 21.4 kDa protein consisting of 190 amino acids and mapped at the 17q25.3 chromosomal segment. Exogenous over-expression of Cygb was soon demonstrated experimentally, both in vitro and in vivo, to protect hepatic stellate cells (HSC) from oxidative stress and suppress their differentiation to a myofibroblast-like phenotype16,17,18. Further, it was reported that Cygb had active peroxidase properties, which protected cells against oxidative damage by free radicals19,20,21. These findings prompted the hypothesis that Cygb could be effective in the prevention or treatment of liver fibrosis. In this study, therefore, we investigate the effect of rhCygb on CCl4-induced liver fibrosis in rats at various stages of the disease’s progression, from mild to severe fibrosis.
Materials and Methods
Preparation of rhCygb
Human cytoglobin cDNA was obtained from HepG2 cells by RT-PCR. A recombinant construct of pET28a-hCyt were transformed into E. coli BL21 (DE3). For expression, the bacteria cultures were induced for 4 h using 1 mM isopropyl β-d-thiogalactoside (IPTG) after the culture OD 600 has reached 0.6. The total bacteria cells were collected by centrifugation at 5,000 rpm for 10 min, resuspended in PBS, and broken down by sonication. The rhCygb was purified by sephadex G-25 and affinity chromatography which the monoclonal antibody was prepared by our laboratory. The purified protein was analyzed with SDS–PAGE and identified by mass spectrometry with ABI 4700 Proteomics Analyzer (Applied Biosystems, USA). The yields of rhCygb measured by the software BandScan 5.0 based on SDS-PAGE results. The total antioxidation capacity (T-AOC) of rhCygb was measured spectrophotometrically as described by Tang et al.22, using a T-AOC commercial kits (Jiancheng, Nanjing, China) according to the guidelines of the kit.
Ethics statement
All experimental procedures were conducted in conformity with institutional guidelines approved by the Chinese Association of Laboratory Animal Care. All experimental protocols were approved by the Institutional Animal Care and Use Committee in Southern Medical University, Guangzhou, China.
Animals
Sexually mature sprague–dawley (SD) rats (n = 150) weighing 180–220 g were obtained from the experimental animal center of Southern Medical University. Care of the animals used in this investigation was conducted according to the guidelines approved by the Chinese Association of Laboratory Animal Care. Rats were housed, five per cage, under constant temperature (20 ± 2 °C) and humidity (70%) with a 12 h light/dark cycle and with free access to chow and water.
Acute toxicity test
BALB/c mice of both sexes, weighing 18–24 g, were obtained from the experimental animal center of Southern Medical University. The acute toxicity of rhCygb was determined in vivo using the procedure approved by Chen et al.23. Uninfected mice were peritoneal injected rhCygb at doses of 100 and 500 mg/kg body weight. The animals were monitored for any toxic symptoms or mortality over a 14-day period.
Induction of fibrosis
Three degrees of liver fibrosis were induced by chronic carbon tetrachloride (CCl4) administration at three different dosage levels:
Mild model group: 2 ml/kg of 25% CCl4 in paraffin oil (Sigma Co., Milan, Italy) was administered by subcutaneous injection twice a week (Tuesday and Friday) for 10 weeks;
Medium model group: treatment in group (a) plus 50% CCl4 treatment for 3 more weeks;
Severe model group: treatment in group (b) plus 62.5% CCl4 for 2 more weeks.
Experiment design
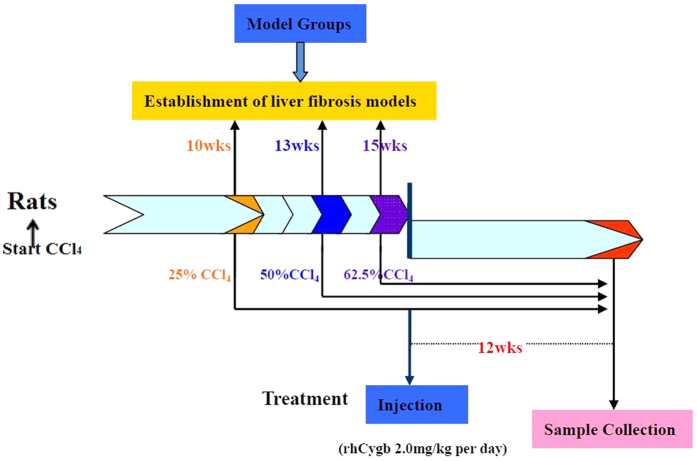
To evaluate the effect of rhCygb on liver fibrosis, 30 rats were used in each of the groups (group a, b, c). A schedule of the treatment is shown in Fig. 1. In each group, followed by CCl4 treatment, rats were further treated with saline (vehicle group), CCl4 (model group) or rhCygb (rhCygb group, 2 mg/kg body weight/day, s.c. injection) in combination with CCl4 for 12 weeks. At the end of the experiments, all rats were sacrificed within 24 h after the last injection.
Figure 1. Schedule for the treatment and experimental tests.
Rats were injected with CCl4 for three different doses to establish mild, medium and severe models at 10, 13 and 15 weeks, 2 times per week. After model was established, rats were treated daily with saline, CCl4 or rhCygb + CCl4 for 12 weeks. Rats were sacrificed within 24 h after the last treatment.
Chemical analysis
Venous blood samples were collected for serum separation. Samples were stored at −20 °C until further analysis. Aspartate aminotransferase (AST) and alanine aminotransferase (ALT) levels were measured spectrophotometrically by Olympus/AU 5200 (Konsesyum, Alternative Biomedical Services, Dallas, TX, USA).
Serum liver fibrosis markers hyaluronic acid (HA), laminin (LN), collagen III (Col III) and collagen IV (Col IV) were determined using enzyme-linked immunosorbent assay (ELISA) kits (SIGMA chemical, Corp.).
Pathological studies
In the end of the experiments, rats liver were removed and rinsed with cold saline water. Then liver lesions were fixed, embedded, and stained with Sirius red and hematoxylin-eosin (HE)24. Their evaluation was conducted using a light microscope (Axiphot2, Carl Zeiss) blindly. Fibrosis was scored according to the Metavir scoring system reported by Bedossa et al.25. Intralobular degeneration and focal necrosis, portal inflammation and fibrosis were compared across the groups. Three representative images of each histology sample section from each rat were selected randomly and all of the rats were scored.
Protein separation
Five liver samples (approximately 30–40 mg wet) were removed from vehicle group, CCl4 model group and rhCygb treated group (medium). The samples were then frozen in liquid nitrogen and homogenized with a mortar and pestle. Proteins were prepared in lysis buffer. A total of 120 μg of protein was separated by two-dimensional electrophoresis (2-DE) according to the product manual ‘2-DE for Proteomics’ (Bio-Rad, U.S). After silver staining, each spots were cut out from the gel and digested using trypsin according to the method described by Qualtieri et al.26.
MS data analysis
Peptide mixtures of each gel spot were dissolved in 0.1% TFA, desalted, and concentrated. The samples were then mixed with the same volume of matrix (CHCA in 30% ACN/0.1% TFA), spotted on a target disk, and allowed to air-dry. Samples were analyzed using a Bruker ultrafleXtreme MALDI-TOF mass spectrometer (Bruker Daltonics, Germany). The protein database search was performed using the MASCOT search engine (http://www.matrixscience.com/)27. Mass tolerance was allowed within 150 parts/million (ppm). Proteins matching more than five peptides and with a MASCOT score higher than 60 were considered significant (p < 0.05).
Bioinformatics analysis
To identify significantly represented biological themes and functional groups in the protein list (Table 1), the gene ontology (GO) and pathway analysis were performed using the database for annotation, visualization and integrated discovery (DAVID) program (https://david.ncifcrf.gov/)27. The GO analysis was used to identify enriched biological themes by the GeneCodis tool (http://genecodis.cnb.csic.es/)27. The list of identified proteins was used as the input data when the DAVID default population background was used; We used EASE scores, which modified Fisher’s exact test P values < 0.05 and corrected for multiple testing by the Benjamin–Hochberg method.
Table 1. Identification of 33 differentially expressed proteins among normal, CCl4 model, and rhCygb groups.
| Code | Protein name | Swiss-prot Accession | MW | PI | Protein function |
|---|---|---|---|---|---|
| 5 | Cathepsin D↓ | Q6P6T6 | 45107.04 | 7.11 | Proteolysis; Extracellular matrix |
| 7 | Pregnancy-specific beta 1-glycoprotein↓ | Q9Z1D6 | 43326.17 | 8.73 | Pregnancy |
| 9 | DnaJ homolog subfamily B member 1↓ | Q6TUG0 | 40754.77 | 6.26 | Serves as a co-chaperone for HSPA5 |
| 10 | Twinfilin-1↓ | Q5RJR2 | 40293.57 | 6.65 | Regulation of actinphosphorylation; Actin cytoskeleton |
| 11 | Haptoglobin↓ | P06866 | 39051.73 | 6.51 | Acute inflammatory response; Acute inflammatory response; Response to hypoxia; Organ regeneration |
| 12 | Tubulin beta-4B chain↓ | Q6P9T8 | 50225.16 | 4.52 | Major constituent of microtubules; Structural constituent of cytoskeleton |
| 15 | Transitional endoplasmic reticulum ATPase↓ | P46462 | 89976.99 | 4.89 | ER to Golgi vesicle-mediated transport; ATP catabolic process |
| 17 | Beta-lactamase-like protein 2↓ | Q561R9 | 32748.81 | 6.29 | Hydrolase activity; Metal ion binding |
| 18 | Microtubule-actin cross-linking factor 1↓ | D3ZHV2 | 623207.64 | 5.07 | Wound healing; Epidermal cell migration; Regulation of microtubule-based process |
| 20 | GTP-binding protein SAR1b↓ | Q5HZY2 | 22509.55 | 6.02 | ER-Golgi transport; Intracellular protein transport |
| 21 | Adenine phosphoribosyltransferase↓ | P36972 | 19761.48 | 6.52 | Adenine salvage; Cellular response to insulin stimulus |
| 22 | Protein Sar1a↑ | Q6AY18 | 22498.54 | 6.68 | ER-Golgi transport; Vesicle-mediated transport |
| 25 | Long-chain-fatty-acid–CoA ligase 1↑ | P18163 | 79154.6 | 6.98 | Fatty acid metabolism; Beta-oxidation |
| 28 | Protein Eea1↑ | F1LUA1 | 162091.42 | 5.54 | Endocytosis; Vesicle fusion |
| 29 | Cytochrome P450 2C11↑ | P08683 | 57657.6 | 7.77 | Xenobiotic metabolic process; Monooxygenase; Oxidoreductase |
| 31 | Estrogen sulfotransferase↑ | P49889 | 35734.66 | 5.52 | Utilizes 3′-phospho-5′-adenylyl sulfate (PAPS) as sulfonate donor |
| 32 | Biliverdin reductase A↑ | P46844 | 33715.58 | 6.01 | Oxidoreductase; Biliverdin reductase activity |
| 33 | Ketohexokinase↑ | Q02974 | 33298.74 | 6.65 | Carbohydrate catabolic process; Response to insulin |
| 34 | Protein Inmt↑ | D3ZNJ5 | 29989.04 | 5.81 | Thioether S-methyltransferaseactivity; Amine N-methyltransferase activity |
| 35 | Mitochondrial GTPase 1↑ | E9PTB3 | 36994.54 | 9.68 | Mitochondrial GTPase activity |
| 37 | Taste receptor type 1 member 2↑ | F8WFI0 | 62271.78 | 8.15 | Oxidoreductaseactivity; Acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor |
| 39 | Putative L-aspartate dehydrogenase↑ | G3V9Z4 | 30654.77 | 6.84 | NAD biosynthetic process; NADP catabolic process |
| 40 | Phenazine biosynthesis-like domain-containing protein↑ | Q68G31 | 31952.24 | 6.25 | Biosynthetic process |
| 41 | Carbonic anhydrase 3↑ | P14141 | 29697.82 | 7.4 | Liver response to oxidative stress; Carbonate dehydratase activity |
| 42 | Enoyl-CoA hydratase, mitochondrial↑ | P14604 | 31895.34 | 8.27 | Lipid metabolism; Fatty acid beta-oxidation |
| 43 | Protein Urad↑ | M0RCU5 | 24634.52 | 6.44 | Carboxy-lyase activity; Allantoin biosynthetic process |
| 44 | Glutathione S-transferase Yb-3↑ | P08009 | 25835.08 | 7.43 | Cellular detoxification of nitrogen compound; Xenobiotic catabolic process |
| 45 | Peroxiredoxin-2↑ | P35704 | 21941.13 | 5.28 | Removal of superoxide radicals; Response to oxidative stress; Peroxidase activity |
| 46 | ATP synthase subunit alpha↑ | F1LP05 | 59889.71 | 9.72 | ATP synthesis coupled proton transport; Lipid metabolic process |
| 47 | Glutathione peroxidase 1 ↑ | P04041 | 22463.37 | 8.05 | Oxidoreductase; Peroxidase; Cell redox homeostasis; Negative regulation of inflammatory; Response to toxic substance |
| 48 | Major urinary protein↑ | P02761 | 21008.61 | 6.13 | Negative regulation of lipid storage; Negative regulation of gluconeogenesis; Positive regulation of lipid metabolic process |
| 49 | Carboxylesterase 1D ↑ | P16303 | 62392.85 | 6.52 | Lipid degradation; Lipid metabolism; acylglycerol catabolic process; Major lipase in white adipose tissue |
| 50 | Nucleoporin GLE1↑ | Q4KLN4 | 79954.74 | 7.65 | Poly(A) + mRNA export from nucleus; Protein transport |
Modified gene set enrichment analysis (GSEA) was used to assess functional significance at the level of sets of genes as previous described by Li et al.28. All of these potential targets were compared with the “c2_all” collection of curated gene sets from the Molecular Signatures Database (GSEA Version 2.0.14), consisting of 1077 gene sets corresponding to BIOCARTA, KEGG and REACTOME biological pathways. The False Discovery Rate (FDR) q value is deemed significant value at <0.05.
Statistical analysis
All data were expressed as the mean ± standard deviation from at least 5 independent experiments. Figures were obtained from at least 5 independent experiments with similar patterns. The biochemical experimental data were analyzed by two-way ANOVA. The Kruskal-Wallis was used for comparisons. Statistical analyses were performed using SPSS software version 19.0 (SPSS Inc, Chicago, USA).
Results
Purity and activity of rhCygb
The purity of rhCygb was 95% by BandScan 5.0 software. The antioxidant activity of rhCygb is 95.58 ± 2.67 U/mg (p < 0.01) (supplemental data).
Acute toxicity results
When given as a single dose of 100 and 500 mg/kg, rhCygb produced no signs of acute toxicity in the 14-day period of observation. No death was recorded in the 100- and 500-mg/kg-treated group. All groups of mice exhibited stable temperature values throughout the period of observation.
Developments of CCl4-induced hepatic fibrosis in rats
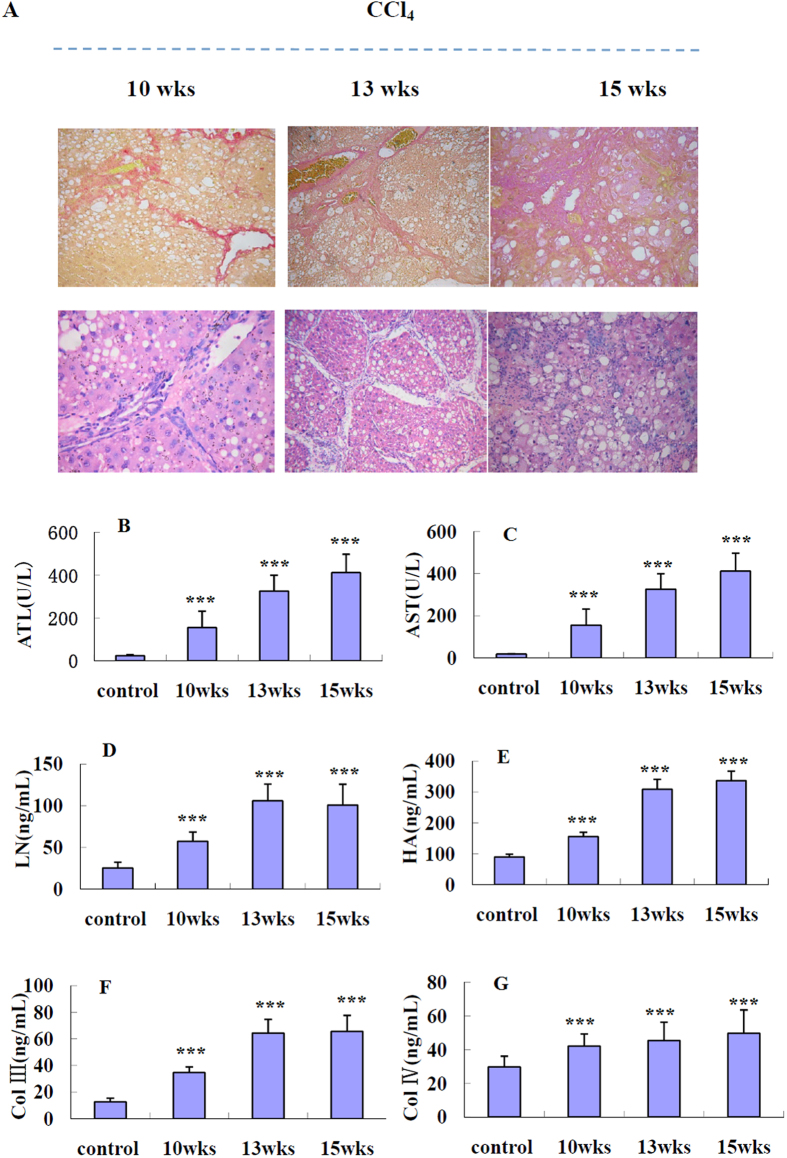
All experiments successfully induced fibrosis, as determined by the examination of liver histology and serum biochemistry (Fig. 2A–G). The mortality was 0 in all the experiment groups. According to the results of development of rat liver fibrosis model, rats treated with CCl4 were sacrificed at 3 different times, 10th week, 13th week and 15th week. At sacrifice, all rats presented marked fibrosis with significant differences in the amount of liver fibrotic tissue among groups. Hepatolobular injury was observed with centrilobular necrosis, balloon cells, and lipids accumulation in 15-weeks-model rats (Fig. 2A). The same results were obtained in an analysis of serum biochemistry (Fig. 2B–G). At the lowest dose (25% CCl4) for 10 weeks in SD rats, administration of CCl4 significantly (p < 0.01) increased the concentration of AST, ALT, LN, HA, Col III and Col IV in rat serum compared with the levels in the vehicle group (Table 2). Moreover, administration of a higher dose of CCl4 (50% CCl4) for a further 3 weeks in group b (medium) resulted in a marked prolongation of toxicity, increasing (p < 0.01) serum enzyme values about two-fold except for Col IV. However, there was no significant (p > 0.05) difference between the group b (medium) and group c (severe).
Figure 2. Examination of liver histology and serum biochemistry at different stages (10, 13, 15 weeks) in CCl4 model rats.
(A) Representative photomicrographs of HE and Sirius red staining for observing the morphology of fibrosis of rat livers at 10, 13, 15 weeks (×200); Serum levels of (B) ALT, (C) AST, (D) LN, (E) HA, (F) Col III and (G) Col IV.
Table 2. Effects of rhCygb on ALT, AST, LN, HA, Col III and Col IV of CCl4-induced liver injury SD rats (
 ± SD, n = 10).
± SD, n = 10).
| Group | ALT(U/L) | AST(U/L) | LN(ng/ml) | HA(ng/ml) | Col III(ng/ml) | Col IV(ng/ml) | |
|---|---|---|---|---|---|---|---|
| Mild model | control | 24.45 ± 4.02 | 27.37 ± 6.32 | 29.76 ± 6.32 | 112.19 ± 16.66 | 13.09 ± 2.77 | 28.28 ± 5.44 |
| CCl4 | 456.37 ± 95.34a | 588.69 ± 115.75a | 93.30 ± 11.87a | 352.64 ± 33.62a | 78.86 ± 10.18a | 50.02 ± 5.31a | |
| CCl4 + rhCygb | 48.32 ± 7.53c | 52.32 ± 5.32c | 30.37 ± 3.84c | 122.79 ± 19.96c | 19.40 ± 5.47c | 31.04 ± 5.35c | |
| Medium model | control | 33.52 ± 4.3 | 31.58 ± 6.35 | 28.45 ± 6.35 | 107.33 ± 15.26 | 15.3 ± 3.5 | 29.35 ± 5.23 |
| CCl4 | 501.46 ± 98.77a | 642.33 ± 98.77a | 113.75 ± 12.4a | 398.95 ± 48.23a | 88.23 ± 15.6a | 56.78 ± 7.89a | |
| CCl4 + rhCygb | 44.25 ± 7.6c | 45.26 ± 7.6c | 32.52 ± 7.6c | 118.56 ± 25.68c | 16.2 ± 3.3c | 30.24 ± 5.66c | |
| Severe model | control | 35.52 ± 4.3 | 37.58 ± 6.35 | 26.45 ± 6.35 | 97.33 ± 15.26 | 14.3 ± 3.5 | 26.35 ± 5.23 |
| CCl4 | 521.46 ± 98.77a | 622.33 ± 98.77a | 103.75 ± 12.4a | 378.95 ± 48.23a | 78.23 ± 14.6a | 58.78 ± 7.89a | |
| CCl4 + rhCygb | 47.25 ± 7.6c | 55.26 ± 7.6c | 28.52 ± 7.6c | 108.56 ± 25.68c | 17.2 ± 3.3c | 28.24 ± 5.66c |
aCompared with control group p < 0.01.
cCompared with CCl4 group p < 0.01.
Effects of rhCygb on hepatic histoarchitecture
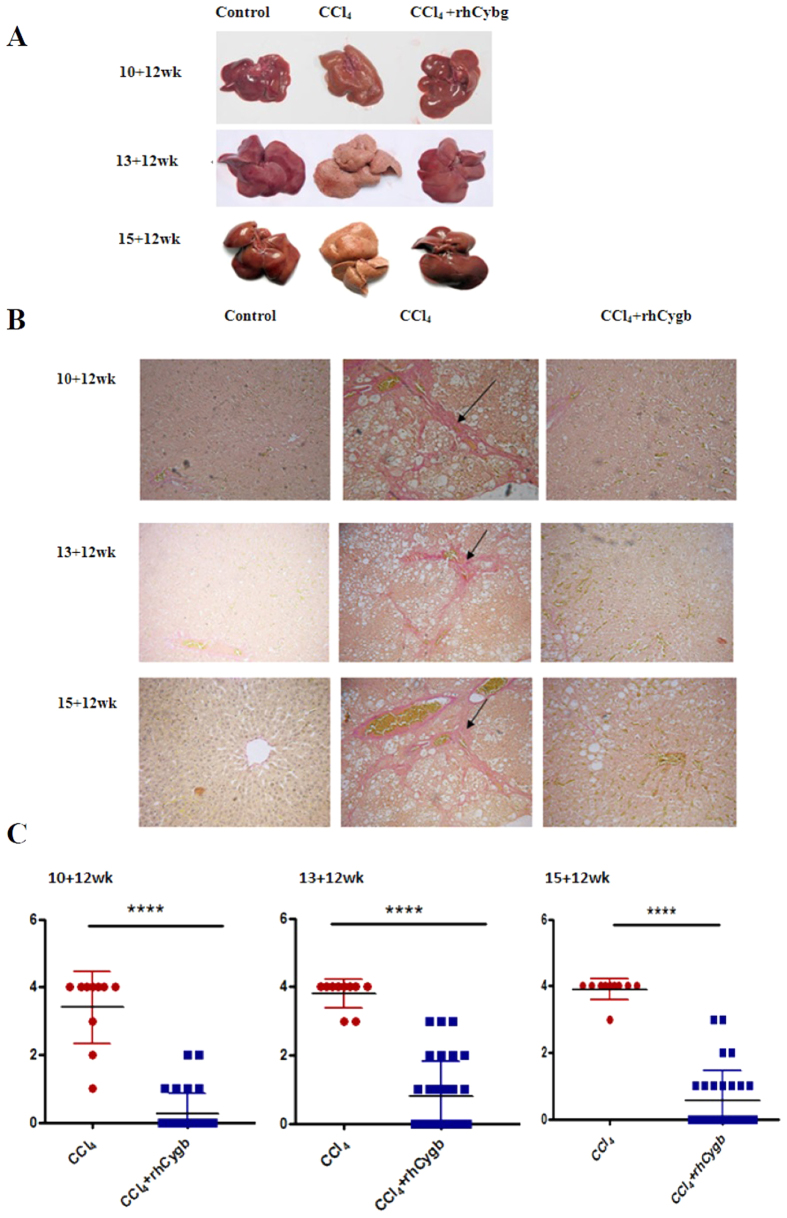
We next evaluated the effect of rhCygb on liver fibrosis following chronic CCl4 administration. Rats were treated with rhCygb (2 mg/kg/day, s.c.) in combination with CCl4 (25%, 50% or 62.5%) for 12 weeks. All of the rats survived during the experimental period. The influence of CCl4 and rhCygb on liver was depicted in Fig. 3, in which liver fibrosis was advanced for 12 more weeks. In this group, fibrotic changes caused by CCl4 reversed dramatically by rhCygb. As shown in Fig. 3A, the liver appearance of CCl4-administered rats significantly changed, especially in the medium and severe groups. These groups, after treatment with rhCygb, exhibited a clear and marked improvement in their liver, when compared with those of the CCl4-administered groups. Photomicrographs of hepatic specimens stained with Sirius red were depicted in Fig. 3B, and the scores of fibrosis variables were presented as medians in Fig. 3C. As shown in the figures, no hepatic fibrosis occurred in the control group, which showed an integrated lobular structure with central venous and hepatic cord radiation. The staging score was 0. Extensive fibrosis and ballooning of hepatocytes were observed in all the CCl4 model groups. As expected, the high CCl4 treatment caused fibrous connective tissue proliferation as well as a good deal of collagen fiber formation. Moreover, increased dosages caused even more frequent pseudo lobules to appear in model control groups. The hepatic fibrosis score in the CCl4 groups increased to 3.40 ± 0.33, 3.80 ± 0.13 and 3.90 ± 0.1, respectively. In the rhCygb treated groups, liver had little collagen fiber formation in the peripheral area, and no pseudo lobules. The positive area of Sirius red staining was significantly lower than that of the CCl4 groups. The scores were 0.27 ± 0.58 (mild group), 0.8 ± 0.19 (medium group) and 0.57 ± 0.16 (severe group), respectively. Similar observations were made in rhCygb groups which evinced statistically significant lower fibrosis scores compared to CCl4 groups.
Figure 3. rhCygb treatment attenuates CCl4-induced rat liver fibrosis.
(A) The liver appearance of vehicle control, CCl4 model and rhCygb groups. (B) Representative photomicrographs of Sirius red staining for observing the morphology of fibrosis of rat livers from different groups (×200). Black arrows indicate fibrosis. (C) Fibrosis scores (Y axis) of CCl4 model groups (n = 10) and rhCygb treatment groups (n = 30). Fibrosis scores are based on the percentage of liver area positively stained by Sirius red; ****p < 0.001.
Indices of hepatotoxicity: liver marker enzymes
The levels of serum ALT and AST are markers of liver damage. As shown in Table 2, AST and ALT level significantly increased in mild, medium and severe CCl4 model groups compared with normal control groups, reflecting hepatocelluar damage in CCl4-induced liver fibrosis. Then rhCygb treatment significantly (p < 0.01) reversed serum ALT and AST levels in 10 + 12 weeks, 13 + 12 weeks and 15 + 12 weeks groups, to the levels almost equal to vehicle controls.
Effects of rhCygb on levels of LN, HA, Col III and Col IV
In this study, serum LN, HA, Col III and Col IV levels in CCl4 groups were higher (p < 0.01) than controls (Table 2). Compared with the CCl4 groups, rhCygb also markedly decreased (p < 0.01) liver fibrosis marker levels of LN, HA, Col III and Col IV in 10 + 12 weeks, 13 + 12 weeks and 15 + 12 weeks groups.
Differential 2-DE analysis of liver tissue proteins
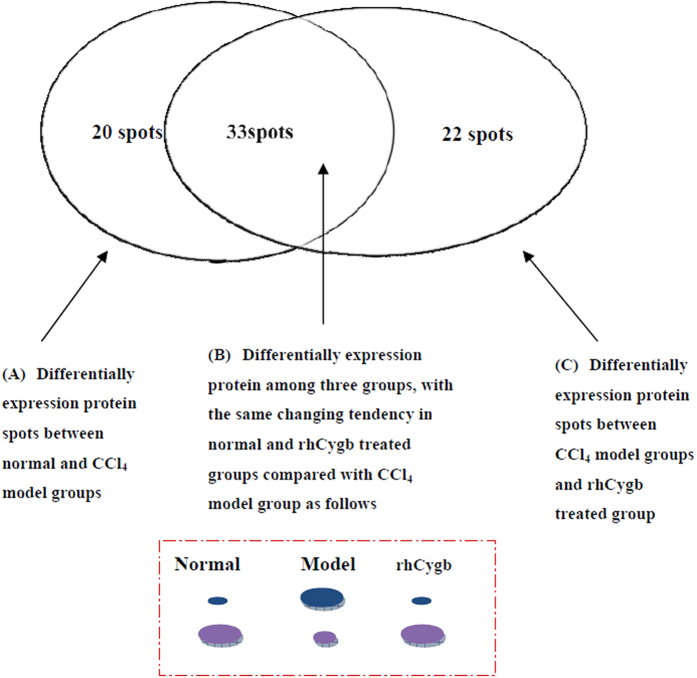
To study the influence of rhCygb on the proteome of CCl4-induced fibrotic liver, liver tissues were harvested from the (13 + 12)-weeks-old control group, the CCl4 model group, and rhCygb treatment group of rats. When examined, the 2-DE gel showed an obvious separation of liver tissue proteins, with 1312 proteins in the vehicle group, 1325 proteins in the CCl4 model group and 1259 proteins in rhCygb group. Comparing the spots of vehicle to model group or model to rhCygb group by PDquest software for image analysis, a total of 75 protein spots exhibited significant (p < 0.05) differences in protein expression between vehicle and model or between model and rhCygb group (supplement data). These 75 protein spots were grouped into three major patterns (Fig. 4): pattern A: 53 spots differentially expressed between vehicle and CCl4 group; pattern C: 55 spots differentially expressed between CCl4 model and rhCygb group; pattern B: 33 spots which overlapped pattern A and pattern C. For example, the expression of cathepsin D in the vehicle group was lower compared to model, while that in rhCygb treated group was also lower. Among the 33 proteins, the expression of 21 proteins were three times higher and that of 12 proteins was three times lower in the vehicle and rhCygb treated groups than in the CCl4 model group significantly (p < 0.05). A total of 33 peptide mass fingerprints (PMFs) were matched and identified by MALDI-TOF-MS and protein database searching, as displayed in Table 1.
Figure 4. Grouping of differentially expressed protein spots.
There were 53 differential spots between vehicle and CCl4 model groups (pattern A), 55 differential between model and rhCygb groups (pattern C), and 33 spots were overlapped between pattern A and pattern C, in which vehicle and rhCygb groups had the same tendency of differential expression compared with ones in model group (pattern B).
Functional annotation of differentially expressed proteins
We analyzed the GO enrichment of these upregulated and downregulated proteins (Table 1). Relevant Protein functions were listed in Table 3. These dysregulated proteins were predicted to be involved in different metabolic processes including the response to stimulus, oxidation-reduction processes, heat-shock protein, the catabolic process and the cytoskeleton.
Table 3. Identification of the differential expressed proteins through the use of GO analysis.
| Protein function categories | Up-regulation in rhCygb groups | Down-regulation in rhCygb groups |
|---|---|---|
| Response to stimulus | Haptoglobin; Adenine phosphoribosyltransferase | |
| Oxidation-reduction process | Peroxiredoxin-2; Glutathione S-transferase Yb-3; Glutathione peroxidase 1; Cytochrome P450 2C11; Biliverdin reductase A; Estrogen sulfotransferase; Taste receptor type 1 member 2; Putative L-aspartate dehydrogenase | |
| Heat shock protein/chaperones | DnaJ homolog subfamily B member 1; GTP-binding protein SAR1b | |
| Catabolic process | Long-chain-fatty-acid–CoA ligase 1; Enoyl-CoA hydratase, mitochondrial; Carboxylesterase 1D; ATP synthase subunit alpha; Major urinary protein; Protein Inmt; Phenazine biosynthesis-like domain-containing protein; Ketohexokinase | |
| Cytoskeleton | Tubulin beta-4B chain; Microtubule-actin cross-linking factor 1; Cathepsin D |
GSEA (Table 4) revealed that up-regulated genes in rhCygb treated group were significantly enriched in the down-regulated gene set in hepatocellular carcinoma (ranked 1st, p = 3.33 e−8, FDR = 2.27 e−4), or in the genes highly expressed in hepatocellular carcinoma with good survival (ranked 4th, p = 4.19 e−5, FDR = 2.77 e−2). FDR q-value was considered significant at < 0.05.
Table 4. GSEA result for the up-regulated genes in rhCygb treated groups vs CCl4 model group.
| Gene Set Name | Description | Genes in Overlap | p-value | FDR q-value |
|---|---|---|---|---|
| SHETH LIVER CANCER VS TXNIP LOSS PAM4 | Cluster PAM4: genes down-regulated in hepatocellular carcinoma (HCC) vs normal liver tissue from mice deficient for TXNIP [GeneID = 10628]. | 5 | 3.33 e−8 | 1.13 e−4 |
| AMBROSINI FLAVOPIRIDOL TREATMENT TP53 | Genes down-regulated by flavopiridol [PubChem = 5287969] in the HCT116 cells (colon cancer) depending on their TP53 [GeneID = 7157] status: wild-type vs loss of the gene’s function (LOF). | 3 | 2.57 e−5 | 2.03 e−2 |
| ACEVEDO_LIVER_CANCER_DN | Genes down-regulated in hepatocellular carcinoma (HCC) compared to normal liver samples | 4 | 3.97 e−5 | 2.03 e−2 |
| LEE LIVER CANCER SURVIVAL UP | Genes highly expressed in hepatocellular carcinoma with good survival. | 3 | 8.05 e−5 | 3.39 e−2 |
| HOUSTIS ROS | Genes known to modulate ROS or whose expression changes in response to ROS | 2 | 8.98 e−5 | 3.39 e−2 |
| VARELA ZMPSTE24 TARGETS DN | Top genes down-regulated in liver tissue from mice with knockout of ZMPSTE24 [GeneID = 10269]. | 2 | 8.98 e−5 | 3.39 e−2 |
| OHGUCHI LIVER HNF4A TARGETS UP | Genes up-regulated in liver samples of liver-specific knockout of HNF4A [GeneID = 3172] | 2 | 1.21 e−4 | 4.1 e−2 |
Discussion
Much experimental evidence and clinical studies has shown that the physiological functions of Cygb are associated with various diseases, such as cancer29, gastroesophageal reflux disease30, psychomotor retardation epilepsy31, and certain neurodegenerative disorders32, especially liver fibrosis. Hence, the aim of the present research was to study the mechanism of liver fibrosis and rhCygb anti-fibrotic action.
Effects of rhCygb on serum markers of fibrosis
CCl4 proved to be highly useful as an experimental model for the study of liver damage in humans33,34,35. It is usually used through intraperitoneal injection or oral administration, and with a dosage of 0.2 to 5 ml/kg liver fibrosis is achieved between 4 and 18 weeks36,37. In this study, the mild, medium and severe liver fibrosis rat models were developed through differential doses (25–62.5%) and exposure duration (10–15 weeks) of CCl4. The results of the present study revealed that continuous and increasing doses of CCl4 resulted in advanced fibrosis which caused an elevation of levels of serum marker enzymes such as ALT, AST, HA, LN, Col III and Col IV from the mild to the severe model. The serum marker enzymes ALT and AST have previously been reported as important diagnostic factors for hepatic diseases, as they are cytosolic enzymes of the hepatocyte, and increased activity in their circulation reflects cell damage and leakage38. High ALT and AST levels are associated with a higher risk of fibrosis progression. The present study demonstrates that rhCygb can reduce heightened ALT and AST, indicating that they can promote the repair of injured liver cell.
In addition, HA, LN, Col III and Col IV have been found to be ideal serum markers of hepatic fibrosis, which play an important role in detecting the degree of hepatic fibrosis39. In the present study, CCl4 model groups increased markedly compared with normal groups. However, after the treatment of rhCygb (therapy but not removing CCl4), we detected that the serum contents of HA, LN, Col III and Col IVdecreased to the level of normal groups. This may be the basic mechanism of rhCygb reversing rat hepatic fibrosis.
Effects of rhCygb on stages of fibrosis
In response to liver injury induced by whatever cause, collagen and matrix proteins are produced in much higher quantities by activated hepatic stellate cells, which ultimately leads to liver fibrosis40. Fibrosis is evaluated by using Sirius red staining and scored using the fibrosis staging system (the Metavir score). The Metavir system scores necroinflammatory activity from 0 to 3 and fibrosis from 0 to 425. In this study, evaluation of staging revealed a score of 4 in different cases in CCl4 model groups: 70% in the mild group, 80% in the medium group and 90% in the severe group. However, score 4 didn’t show up in any of the rhCygb treated groups. In time, score 0 was present in 24 rats in the mild group, 18 rats in the medium group, and 19 rats in the severe group. These data are in solid agreement with the activities of serum biomarkers. Interestingly, the stages of fibrosis were previously believed to be progressive and largely irreversible. There is now mounting experimental and clinical evidence suggesting that liver fibrosis can regress in all chronic liver diseases by withdrawing the cause of liver injury or by treatment of the disease41,42. Our paper also obtains that CCl4-induced rat liver fibrosis was markedly reversed by administration of rhCygb. He et al. reported the similar findings in their use of rhCybg in the thioacetamide (TAA)-induced rat liver fibrosis model43.
Effects of rhCygb on protein species
We systematically analyzed the effect of rhCygb on liver proteome in the CCl4-induced rat model. Thirty-three differentially expressed proteins appeared among the three groups, with the same alteration profile in the vehicle and rhCygb treated groups compared with the CCl4 model group, identified through comparative proteomics. Functional annotation analysis showed several identified proteins involved in oxidative stress pathways, such as the response to chemical stimuli, oxidative-reduction, and xenobiotic metabolic processes. Oxidative stress is thought to be a critical factor in the development of CCl4-induced liver fibrosis44. Previous studies have reported the effects of rhCygb on the levels of oxidative stress biomarkers and anti-oxidative capacity45. The current results show that rhCygb regulates antioxidant enzymes such as peroxiredoxin-2, glutathione S-transferase, glutathione peroxidase 1, cytochrome P450 2C11, biliverdin reductase A, estrogen sulfotransferase, and taste receptor type 1 member 2. One of these proteins, P450 2C11, an enzyme that was increased by rhCygb, plays an important role in the deactivation of estrogens via 16a-hydroxylation and catechol formation. It is particularly active in relation to the development of fatty change, hepatic fibrosis, portal hypertension, and cirrhosis46. Earlier studies have shown that P450 2C11 activity in rat models of experimental liver disease including hepatic cirrhosis is produced by CCl447. The present study also demonstrated that the level of protein P450 2C11 in the liver decreased significantly in this model of liver fibrosis at the 25-week time point. In addition, we also found that enzymes involved in fatty acid β-oxidation (Long-chain-fatty-acid-CoA ligase 1, Enoyl-CoA hydratase, Carboxylesterase 1D and ATP synthase sub-unit alpha) were down-regulated in CCl4 model group, whereas in the rhCygb-treated group they were up-regulated (as they were in the vehicle group). These findings show that impairment of fatty acid oxidation may contribute to hepatic fibrosis.
In summary, the results of the present study demonstrate that in addition to the effect of reversing liver fibrosis, rhCygb could reduce the serum levels of ALT, AST, HA, LN, Col III and Col IV, indicating that it has definite effects in reversing the formation of liver fibrosis in the course of chronic hepatitis. The differentially expressed proteins have several groups of biological functions, especially relating to oxidative stress, which are helpful in revealing the underlying mechanisms of rhCygb action against liver fibrosis. rhCygb could be a novel candidate drug in combating liver fibrosis.
Additional Information
How to cite this article: Li, Z. et al. The Effect of rhCygb on CCl4-Induced Hepatic Fibrogenesis in Rat. Sci. Rep. 6, 23508; doi: 10.1038/srep23508 (2016).
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 81200394), Guangdong Province Science and Technology Plan Project (No. 2010B031500012, No. 2013A022100027) and Guangzhou science and technology project (No. 201510010104).
Footnotes
The authors declare no competing financial interests.
Author Contributions L.Z., W.W. and C.B. performed the majority of experiments; C.G., L.X., W.P. and T.J. provided vital analysis and interpretation of data and were also involved in editing the manuscript; D.W. designed the study and wrote the manuscript.
References
- Eng F. J. & Friedman S. L. Fibrogenesis, I. New insights into hepatic stellate cell activation: the simple becomes complex. Am J Physiol 279, G7–G11 (2000). [DOI] [PubMed] [Google Scholar]
- Marcellin P., Asselah T. & Boyer N. Fibrosis and disease progression in hepatitis C. Hepatology 36, 47–56 (2002). [DOI] [PubMed] [Google Scholar]
- Ghany M. G. et al. Progression of fibrosis in early stages of chronic hepatitis C. Hepatology 32, 496A (2000). [Google Scholar]
- Riemekasten G. et al. Involvement of functional autoantibodies against vascular receptors in systemic sclerosis. Ann Rheum Dis 70, 530–536 (2011). [DOI] [PubMed] [Google Scholar]
- Rankin A. L. et al. IL-33 induces IL-13-dependent cutaneous fibrosis. J Immunol 184, 1526–1535 (2010). [DOI] [PubMed] [Google Scholar]
- Akhmetshina A. et al. Dual inhibition of c-abl and PDGF receptor signaling by dasatinib and nilotinib for the treatment of dermal fibrosis. FASEB J 22, 2214–2222 (2008). [DOI] [PubMed] [Google Scholar]
- Akhmetshina A. et al. Treatment with imatinib prevents fibrosis in different preclinical models of systemic sclerosis and induces regression of established fibrosis. Arthritis Rheum 60, 219–224 (2009). [DOI] [PubMed] [Google Scholar]
- Tashkin D. P. et al. Cyclophosphamide versus placebo in scleroderma lung disease. N Engl J Med 354, 2655–2666 (2006). [DOI] [PubMed] [Google Scholar]
- Huber L. C. et al. Trichostatin A prevents the accumulation of extracellular matrix in a mouse model of -induced skin fibrosis. Arthritis Rheum 56, 2755–2764 (2007). [DOI] [PubMed] [Google Scholar]
- Venalis P. et al. Lack of inhibitory effects of the anti-fibrotic drug imatinib on endothelial cell functions in vitro and in vivo. J Cell Mol Med 13, 4185–4191(2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann C. et al. Inhibition of glycogen synthase kinase 3 beta induces dermal fibrosis by activation of the canonical Wnt pathway. Ann Rheum Dis 70, 2191–2198 (2011). [DOI] [PubMed] [Google Scholar]
- Horn A. et al. Hedgehog signaling controls fibroblast activation and tissue fibrosis in systemic sclerosis. Arthritis Rheum 64, 2724–2733 (2012). [DOI] [PubMed] [Google Scholar]
- Kawada N. et al. Characterization of a stellate cell activation associated protein (STAP) with peroxidase activity found in rat hepatic stellate cells. J Biol Chem 276, 25318–25323 (2001). [DOI] [PubMed] [Google Scholar]
- Fago A. et al. Allosteric regulation and temperature dependence of oxygen binding in human neuroglobin and cytoglobin. Molecular mechanisms and physiological significance. J BiolChem 279, 44417–44426 (2004). [DOI] [PubMed] [Google Scholar]
- Burmester T. et al. Cytoglobin: a novel globin type ubiquitously expressed in vertebrate tissues. Mol Biol Evol 19, 416–421 (2002). [DOI] [PubMed] [Google Scholar]
- Hodges N. J. et al. Cellular protection from oxidative DNA damage by over-expression of the novel globin cytoglobin in vitro. Mutagenesis 23, 293–298 (2008). [DOI] [PubMed] [Google Scholar]
- Fordel E. et al. Neuroglobin and cytoglobin expression in mice. FEBS J 274, 1312–1317 (2007). [DOI] [PubMed] [Google Scholar]
- Xu R. et al. Cytoglobin over expression protects against damage-induced fibrosis. Mol Ther 13, 1093–1100 (2006). [DOI] [PubMed] [Google Scholar]
- Trent J. T. 3rd & Hargrove M. S. An ubiquitously expressed human hexaco-ordinate hemoglobin. J Biol Chem 277, 19538–19545 (2002). [DOI] [PubMed] [Google Scholar]
- Li H. et al. Characterization of the mechanism and magnitude of cytoglobin mediated nitrite reduction and nitric oxide generation under anaerobic conditions. J Biol Chem 287, 36623–36633 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner A. M., Cook M. R. & Gardner P. R. Nitric-oxide dioxygenase function of human cytoglobin with cellular reductants and in rat hepatocytes. J Biol Chem 285, 23850–23857 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang R. et al. Mechanisms of selenium inhibition of cell apoptosis induced by oxysterols in rat vascular smooth muscle cells. Arch Biochem Biophys 441, 16–24 (2005). [DOI] [PubMed] [Google Scholar]
- Chen K. W. & Ho W. S. Anti-oxidative and hepatoprotective effects of lithospermic acid against carbon tetrachloride-induced liver oxidative damage in vitro and in vivo. Oncol Rep 34, 673–680 (2015). [DOI] [PubMed] [Google Scholar]
- Ding H. et al. Assessment of liver fibrosis: the relationship between point shear wave elastography and quantitative histological analysis. J Gastroenterol Hepatol 30, 553–558 (2015). [DOI] [PubMed] [Google Scholar]
- Bedossa P. & Poynard T. The METAVIR cooperative study group. An algorithm for the grading of activity in chronic hepatitis C. Hepatology 24, 289–293 (1996). [DOI] [PubMed] [Google Scholar]
- Qualtieri A. et al. Two-dimensional gel electrophoresis of peripheral nerve proteins: optimized sample preparation. J Neurosci Methods 159, 125–133 (2007). [DOI] [PubMed] [Google Scholar]
- Nagasawa K. et al. Significant modulation of the hepatic proteome induced by exposure to low temperature in Xenopus laevis. Biol Open 2, 1057–1069 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. et al. Clonal expansions of cytotoxic T cells exist in the blood of patients with Waldenstrom macroglobulinemia but exhibit anergic properties and are eliminated by nucleoside analogue therapy. Blood 115, 3580–3588 (2010). [DOI] [PubMed] [Google Scholar]
- Xinarianos G. et al. Frequent genetic and epigenetic abnormalities contribute to the deregulation of cytoglobin in non-small cell lung cancer. Hum Mol Genet 15, 2038–2044 (2006). [DOI] [PubMed] [Google Scholar]
- McRonald F. E. et al. Down-regulation of the cytoglobin gene, located on 17q25, in tylosis with oesophageal cancer (TOC): evidence for trans-allele repression. Hum Mol Genet 15, 1271–1277 (2006). [DOI] [PubMed] [Google Scholar]
- Hedley-Whyte E. T. et al. Hyaline protoplasmic astrocytopathy of neocortex. J Neuropathol Exp Neurol 68, 136–147 (2009). [DOI] [PubMed] [Google Scholar]
- Powers J. M. p53-mediated apoptosis, neuroglobin over expression and globin deposits in a patient with hereditary ferritinopathy. J Neuropathol Exp Neurol 65, 716–721 (2006). [DOI] [PubMed] [Google Scholar]
- Guillaume M. et al. Recombinant human manganese superoxide dismutase reduces liver fibrosis and portal pressure in CCl4- cirrhotic rats. J Hepatol 58, 240–246 (2013). [DOI] [PubMed] [Google Scholar]
- Kataoka T. et al. Comparative study on the inhibitory effects of α-tocopherol and radon on carbon tetrachloride-induced renal damage. Ren Fail 34, 1181–1187 (2012). [DOI] [PubMed] [Google Scholar]
- Nowatzky J. et al. Inactivated Orf virus (Parapoxvirus ovis) elicits antifibrotic activity in models of liver fibrosis. Hepatol Res 43, 535–546 (2013). [DOI] [PubMed] [Google Scholar]
- Saba A. B., Onakoya O. M. & Oyagbemi A. A. Hepatoprotective and in vivo antioxidant activities of ethanolic extract of whole fruit of Lagenariabreviflora. J Basic Clin Physiol Pharmacol 23, 27–32 (2012). [DOI] [PubMed] [Google Scholar]
- Marques T. G. et al. Review of experimental models for inducing hepatic cirrhosis by bile duct ligation and carbon tetrachloride injection. Acta Cir Bras 27, 589–594 (2012). [DOI] [PubMed] [Google Scholar]
- Martinot-Peignoux M. et al. Prospective study on anti-hepatitis C virus-positive patients with persistently normal serum alanine transaminase with or without detectable serum hepatitis C virus RNA. Hepatology 34, 1000–1005 (2001). [DOI] [PubMed] [Google Scholar]
- Vaillant B. et al. Regulation of hepatic fibrosis and extracellular matrix genes by the response: new insight into the role of tissue inhibitors of matrix metalloproteinases. J Immunol 167, 7017–7026 (2001). [DOI] [PubMed] [Google Scholar]
- Beaussier M. et al. Prominent contribution of portal mesenchymal cells to liver fibrosis in ischemic and obstructive cholestatic injuries. Lab Invest 87, 292–303 (2007). [DOI] [PubMed] [Google Scholar]
- Liaw Y. F. Reversal of cirrhosis: an achievable goal of hepatitis B antiviral therapy. J Hepatol 59, 880–881 (2013). [DOI] [PubMed] [Google Scholar]
- Hui C. K. et al. Hong Kong Liver Fibrosis Study Group. Serum adiponectin is increased in advancing liver fibrosis and declines with reduction in fibrosis in chronic hepatitis B. J Hepatol 47, 191–202 (2007). [DOI] [PubMed] [Google Scholar]
- He X. et al. Cytoglobin exhibits anti-fibrosis activity on liver in vivo and in vitro. Protein J 30, 437–446 (2011). [DOI] [PubMed] [Google Scholar]
- Gutiérrez R. et al. Oxidative stress modulation by Rosmarinus officinalis in CCl4-induced liver cirrhosis. Phytother Res 24, 595–601 (2010). [DOI] [PubMed] [Google Scholar]
- Chakraborty S., John R. & Nag A. Cytoglobin in tumor hypoxia: novel insights into cancer suppression. Tumour Biol 35, 6207–6219 (2014). [DOI] [PubMed] [Google Scholar]
- Wójcikowski J., Haduch A. & Daniel W. A. Effect of antidepressant drugs on cytochrome P4502C11 (CYP2C11) in rat liver. Pharmacol Rep 65, 1247–1255 (2013). [DOI] [PubMed] [Google Scholar]
- Nomura A., Sakurai E. & Hikichi N. Effect of carbon tetrachloride-induced hepatic injury on stereoselective N-demethylation of chlorpheniramine by rat hepatic cytochrome P450 2C11 isozyme. YakugakuZasshi 118, 317–323 (1998). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.