Abstract
Complement sensitization of red blood cells (RBCs) can cause life-threatening hemolytic anemias. We have previously shown that complement receptor 1 (CR1) derivatives specifically the N-terminal region with decay accelerating activity (DAA) for inactivation of a key enzyme in the complement cascade can reduce complement-mediated RBC destruction in vitro and in an in vivo mouse model of hemolytic transfusion reaction. In the present study, we have modeled the N-terminal CR1 molecule based on the X-ray crystal structure of decay accelerating factor and the NMR structure of a homologous CR1 domain. Based on the homology model, we identified a 34-mer peptide encompassing the putative DAA which in vitro reduced hemolysis, C3a release and surface C3 deposition. More importantly, this peptide at 0.6 mM was effective in prolonging survival of transfused incompatible RBCs in vivo. Our results indicate that CR1-based structure–function studies may provide insights for developing structure-derived transfusion therapeutics in the future.
Keywords: Complement receptor 1, CD35, Complement inhibition, Transfusion therapy, Hemolysis, Homology modeling, Mouse model, Transfusion reactions, CR1 peptides, C3a, C3b
The complement system is an important part of the innate immune system for fighting infections and foreign molecules before an adaptive response is developed. To execute its functions, the system has to be activated. A key step in the complement activation cascade is the formation of the C3 convertase complex on the surface of target cells which cleaves C3 into C3b, the main effector of complement, and C3a, a potent anaphylatoxin that is released into the medium. C3b and its degradation products on target cells act as opsonins marking them for removal from the circulation by phagocytes. In addition, if sufficient quantities of C3b are formed on the cell surface, they can result in serial activation of complement proteins (C5–C9) that form the C5b-9 complex, also known as the membrane attack complex. Once adequate number of C5b-9 complexes form, the target cell is lysed. As expected, inappropriate complement activation can have deleterious effects. In the transfusion medicine setting, complement sensitization of red blood cells (RBCs) can result in life-threatening hemolytic transfusion reactions as well as hemolytic anemias [1]. There is thus a critical need for a therapeutically applicable complement inhibitor. To date several inhibitors have been described [2], but none of these has yet been adopted as a therapeutic agent. A strategy to develop complement inhibitors has been to manipulate naturally occurring complement regulatory proteins that act at various steps in the activation cascade and control inappropriate complement activation for therapeutic use. Complement receptor 1 (CR1) is the most versatile member of the complement regulatory proteins through its ability to bind complement split products (C3b and C4b) and possessing decay accelerating and cofactor activities that can inactivate the two critical enzymes (C3 and C5 convertases) of both the classical and alternative complement activation pathways [3–6]. A recombinant soluble form of CR1 (SCR-1) by inactivating the convertases in the complement cascade has successfully inhibited the complement activation and prevented complement-mediated tissue injury in several animal models [7,8]. More importantly, SCR-1 has been in human clinical trials for the treatment of acute respiratory distress syndrome and to reduce tissue damage in myocardial infarction and lung transplantation [9–11] with possible favorable outcomes [10].
Our studies were the first to demonstrate a potential for SCR-1 for inhibiting complement-mediated red cell destruction following transfusion immunization events [12,13]. We reasoned that by identifying the functional regions of CR1 (decay accelerating and cofactor activities), structure-based small synthetic molecules with more specific inhibitory activities, but lacking immunogenecity can be developed, thereby permitting the administration of lower drug doses and better efficacy. A combined in vitro and in vivo structure–function analysis of CR1 domains responsible for complement inhibition has been our approach towards the future design of such molecules. The extracellular 1930 residue long domain of CR1 can be divided into 30 short consensus repeats (SCRs), each of 59–72 amino acids (aa) with sequence homology between SCRs ranging from 60% to 90% [14]. Through structure–function analysis we identified a 254 aa domain at the N-terminus of CR1, consisting of 4 SCRs (SCRs-1–4), that has anti-hemolytic activity both in vitro and in vivo in a mouse model of complement-mediated hemolytic transfusion reaction [15]. This region mediates inhibition of the classical pathway and possesses decay acceleration of the C3 convertases [16]. Previous mutagenesis data of SCRs-1–4 have identified several critical residues for decay accelerating activities in the N-terminal domain [16–20]. For example, three positively charged amino acids Arg59, Arg60, and Lys61 at SCR-1/SCR-2 junction, Arg64, Asn65, Thr103, and Thr110 in SCR-2 as well as Gly35 in SCR-1 were found to be important for both decay accelerating activity while Phe82 was required primarily for decay accelerating activity [20]. To date, no structural data for the N-terminal domain of CR1 is available. However, the NMR structure of a homologous region (SCRs-15–17) in CR1 has been reported [21]. In addition, there is crystallographic data on the structure of the 4 SCRs of decay accelerating factor (DAF)/CD55 [22]. In the present study, we have modeled the amino terminal SCRs based on the crystal structure of DAF [22] and the NMR structure of SCRs-15–17, designed peptides based on this homology model and tested their anti-hemolytic activities. Using this rational approach, we have identified a 34-mer peptide in SCR-2 which has anti-complement activity both in vitro and in vivo.
Methods and materials
Modeling CR1 based on DAF crystal structure and SCRs-15–17 NMR structure
Homology models of the first 3 N-terminal SCRs of CR1 were built based on the X-ray crystal structure of DAF/CD55 (pdb code: 1ojw) and on the average NMR structure of the SCRs-15–16 of CR1 (CD35) (pdb code: 1gkn) using the automated software Modeller [23] within Quanta 2000 (Accelrys, San Diego, CA) running on a Silicon Graphics Octane with a dual R12000 processor (sgi, Mountain View, CA). ‘Refine 3’ option in Modeller, which uses conjugated gradient together with molecular dynamics by simulated annealing technique, was used to optimize the models. Five models for each run were developed and the model with the lowest Objective Function was selected. The resulting models were evaluated for its stereochemical properties by Procheck software [24] at 2.0Å and by Quanta Protein Health programs within Quanta 2000.
Synthetic peptides
The peptides in Fig. 2 were synthesized by Genemed Synthesis Inc. (South San Francisco, CA) and by the Biomolecular Synthesis Laboratory of the New York Blood Center. A standard solid-phase Fmoc method was used for peptide synthesis. Peptides were purified to homogeneity (purity >90%) by HPLC and identified by laser desorption mass spectrometry.
Fig. 2.
CR1 peptides. (A) Schematic representation of CR1 peptides in relation to full-length CR1 is depicted. (B) CLUSTAL W (1.82) alignment of SCR-1 and -2 of CR1 with SCR-2 and -3 of DAF. The degree of conservation is shown above the sequences (*, same residue; “:”, conserved substitution; “.”, semi-conservative substitution). The residues in DAF that have been shown to be critical for decay acceleration of the C3 convertases are underlined. The position of the predicted hydrophobic patch in SCR-2 based on the homology model is shown in boxed grey. The sequence of the peptides 1–4 (P1–4) and their position in relation to full-length SCR-1 and SCR-2 of CR1 are depicted.
In vitro CR1 functional assays
For the hemolytic assay, we used papain-treated group A RBCs. Papain digestion of RBCs was performed on washed RBCs by incubating cells with 0.05% final concentration of papain (Calbiochem, San Diego, California) in PBS at 37 °C for 20 min as previously described [25]. Hemolytic assays were performed using human sera, as source of complement, from two different group O volunteers that were preincubated for 15 min at room temperature with mock or 10 mM EDTA to inhibit all complement activation pathways or different concentrations of CR1 derivatives. The treated sera were then added to a final serum concentration of 30% to papain-treated group A RBCs (108 per ml) and incubated for 30 min at 37 °C. Hemolysis was assessed by spectrophotometric measurement of hemoglobin in the supernatants and the mock-treated samples were used as control: the amount of hemolysis in the experimental samples was expressed as a percentage of the control (taken as 100%) as previously described [12]. We assayed for complement activation by measuring the levels of C3 deposition on the RBCs using FITC-conjugated goat anti-human C3 (ICN, Aurora, Ohio) by flow cytometry. As control, we also measured levels of IgG which should not be affected by complement inhibitors using FITC-conjugated anti-human IgG. By expressing the levels of anti-C3 reactivity on buffer-treated samples as 100%, complement deposition on the remaining samples were then calculated as a percentage of the control. C3a levels were measured using an OptEIA™ Human C3a ELISA kit (BD Pahrmingen) according to the protocol outlined by the manufacturers.
Mouse transfusions
RBCs were labeled with the fluorescent dye, PKH-26 (Sigma, St. Louis, Missouri) according to manufacturer's instructions and resuspended in known amounts of peptide or buffer to give a final hematocrit of 20% in 500 μl. Eight- to ten-week old C57Bl/6 female mice were injected by the tail-vein equivalent to 10% of total mouse blood volume. Blood samples (25 μl) by retro-orbital sinus bleeding were obtained at the time points indicated after transfusion and the clearance of fluorescent RBCs was measured by flow cytometry as previously described [12]. The fraction of fluorescent RBCs of the total number of recipient RBCs studied (200,000 per sample) was determined for each animal.
Statistical analysis
The significance of differences between groups of mice was calculated using analysis of variance, Anova test and only p values less than 0.05 were considered as significant.
Results and discussion
Modeling CR1 based on DAF crystal structure and SCRs-15–17 NMR structure
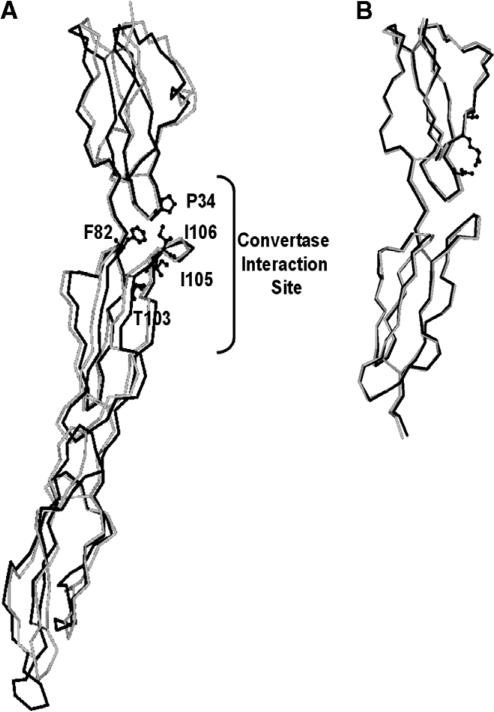
The superimposed models of the first 3 N-terminal SCRs of CR1 based on the X-ray crystal structure of DAF and NMR structure of the SCRs-15–17 of CR1 showed similar folding patterns (Fig. 1). The crystallographic data together with mutagenesis studies from DAF suggest that a strong candidate for A protein-binding site contacting the catalytic subunit of the C3 convertases is a hydrophobic patch Pro-97, Phe-148, Phe-169, Leu-171, and Ile-172 (Fig. 2) [22,26]. Equivalent residues in CR1 (Pro-34, Phe-82, Thr-103, Ile-105, and Ile-106) are conserved for the most part and by our modeling studies also appear to form a hydrophobic patch near the SCR-1–2 junction with almost all the residues present on SCR-2 (Figs. 1 and 2). In addition, based on the model it appears that the convertase contact sites are far away from the third SCR (SCR-3) (Fig. 1), indicating that SCR-3 has little or no role in direct binding although it may still have a role as a structural element.
Fig. 1.
Homology model of N-terminal domain of CR1. (A) C-α tracing of SCR-1–SCR-3 domains of CR1 (black) superimposed on the X-ray crystal structure (grey) of human complement regulator CD55. The residues, forming the hydrophobic patch and responsible for interaction with convertase, are shown as ball-and-stick model. (B) C-α tracing of SCR-1 and SCR-2 domains of CR1 (black) superimposed on the NMR structure (grey) of SCRs-15–16 of CR1.
In vitro anti-complement activities of N-terminal CR1 derived peptides
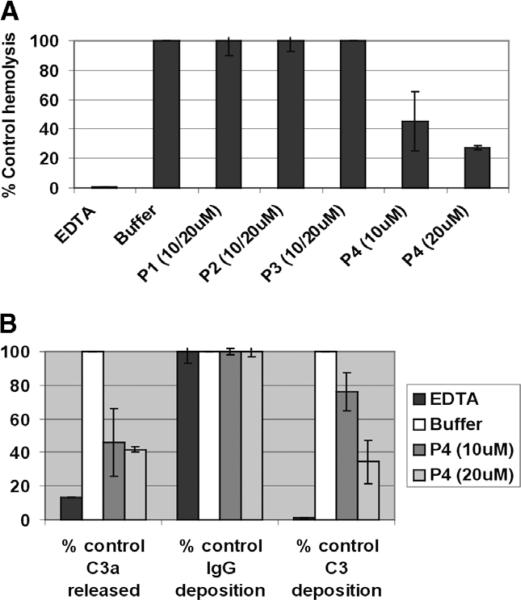
Based on the homology model, we prepared 4 peptides: the 37mer peptide 1 (P1: Gln 1–Ser 37) and 30mer peptide 2 (P2: Gln 31–Arg 60) have overlapping sequences while the 35mer peptide 3 (P3: Ser 61–Arg 95) and 34-mer peptide 4 (P4: Ser 89–Arg 120) have overlapping sequences in SCR-2. Based on the homology model, P4 is predicted to encompass the putative hydrophobic patch which based on mutagenesis is responsible for mediating decay accelerating activity (Fig. 2). We found that at 10 or 20 μM concentrations only P4 had anti-hemolytic activity in vitro (Fig. 3A). If C3 convertases were really the target of inhibition by peptide 4, the release of highly inflammatory C3a fragments and deposition of C3b opsonins should also be impaired. We found that in our classical complement activation assay, peptide 4 at either dose of 10 or 20 μM reduced release of C3a and decreased the levels of C3b deposition, but not IgG, on the cells, consistent with the complement inhibitory activity (Fig. 3B).
Fig. 3.
Anti-complement activity of CR1-derived peptides. (A) Human sera from a group O donors (=2) were pre-incubated for 15 min at room temperature with control 10 mM EDTA (for inhibition of the complement activation pathways), 10 or 20 μM peptides 1–4, before addition of human group A RBCs. Levels of hemolysis in the supernatants were measured. Data are presented as the percentage of hemolysis the buffer control from four separate experiments. (B) Surface C3 and IgG were measured on the remaining unlysed cells from P4-treated samples in A by flow cytometry. Levels of C3a released in the supernatants were measured using an ELISA kit. Data are presented as the percentage of C3 and IgG deposition and C3a release of the buffer control from four separate experiments.
Analyses of CR1-derivatives in a mouse model of complement-mediated RBC destruction
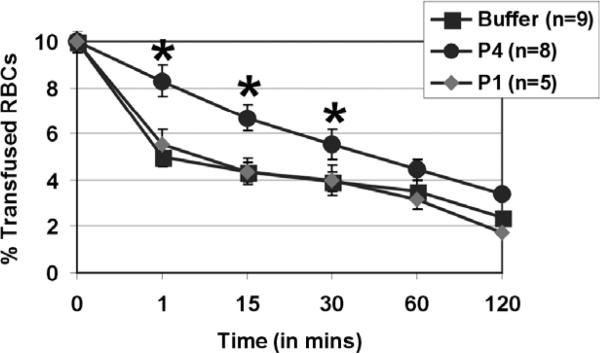
We next tested the ability of selected CR1 peptides in preventing RBCs from complement-dependent immune destruction in vivo, and in prolonging the survival of transfused human RBCs in our mouse model of transfusion reactions. In this study, we co-injected human RBCs with P1 or P4 at 0.6 mM concentration and found that only P4 prolonged transfused RBC survival in the mouse circulation compared to control buffer-treated mice for the initial times post-transfusion (p < 0.05, Fig. 4). Indeed, our data show that without treatment over 50% of transfused human red cells was destroyed in vivo by 1 min post-transfusion. However, in P4-treated group it was only after 45 min that the transfused RBC levels had reached 50% of their original numbers (Fig. 4).
Fig. 4.
A CR1-derived 34-mer peptide prolongs the survival of transfused RBC in vivo. Fluorescently-labeled human group O RBCs equivalent to 10% of total mouse blood volume were injected intravenously into C57Bl/6 mice without (buffer, black squares, n = 9) or with peptide 1 (P1, grey diamond, n = 5) or peptide 4 (P4, black circles, n = 8) both given at 0.6 mM. At times indicated, venous blood was sampled and analyzed by flow cytometry for fraction of fluorescent RBCs. To show the clearance kinetics, injected RBCs at time 0 were taken as 10% and the remaining RBCs were calculated at different time points as the average for each group of mice with error bars depicting SEM. *p < 0.05 as compared to buffer treatment.
Conclusions
Altogether, our data indicate that a 34-mer peptide encompassing potential binding sites for mediating decay acceleration of the classical pathway C3 convertase complement activation can reduce complement-mediated RBC destruction both in vitro and in vivo.
We found that the 34-mer P4 was only effective in prolonging the survival of incompatible transfused RBCs for the first 30 min post-transfusion. This is probably due to fast renal clearance and/or enzymatic degradation of our peptide. Various approaches are used to prolong the circulating plasma half-lives of short synthetic peptides which we are testing with our candidate molecule including replacing l-analogue amino acids with d-amino acids, modification of N- and C-terminal residues or PEGylation [27]. In addition, although P4 clearly possesses anti-complement activity, we believe that other modifications such as cyclizing the peptide are most likely to increase its activity. Based on the homology model, Cys-90 in peptide 4 is disulphide-bonded with Cys-120. Future experiments are planned to test the inhibitory activity of cyclized peptide 4 having Cys-90 and -120 disulphide bonded. Although DAF has been shown to be approximately fourfold better inhibitor of the alternative pathway convertase than CR1, it is a fourfold less efficient inhibitor of the classical pathway convertase than CR1 [28]. Nevertheless, it would be informative to compare the activity of peptides encompassing the hydrophobic patch from DAF and CR1 (see Fig. 2) for prevention of the complement-mediated RBC destruction. A CR1 inhibitor (APT070) based on the N-terminal 3 SCRs that are targeted to the cell surface is in clinical trials in rheumatoid arthritis patients to explore its therapeutic potential for complement inhibition [29]. Our identification of a short peptide within this N-terminal domain may be helpful for future design of smaller peptidomimetic molecules that can replace the larger 3 SCRs in APT070. Interestingly, mutations that considerably increase decay acceleration for the classical pathway C3 convertase have been identified that map to residues within our peptide 4 [20]. By making synthetic peptides harboring such mutations we plan on testing their effectiveness in preventing complement-mediated RBC destruction using our assays.
It is important to note that a major focus of our studies has been on development of complement therapeutics for prevention of complement-mediated red cell destruction. The application for these inhibitors that we are proposing is for short-term prophylactic use as a therapeutic option in select patients with complement-mediated immune hemolysis (due to complement-fixing antibodies such as Kidd or cold agglutinins or in patients with paroxysmal nocturnal hemoglobinuria) to ameliorate the life-threatening complications.
Acknowledgments
We thank Dr. James Farmer (Biomolecular Synthesis Laboratory, NYBC) with the initial synthesis of some of the peptides and Xiaoying Zheng and Amina Mqadmi (Laboratory of Complement Biology, NYBC) for technical assistance with analysis of some of the CR1 peptides.
Footnotes
This study was supported in part by a grants (to K.Y.) from the NIH R01 HL69102, and the American Heart Association Grant-in-Aid Heritage Affiliate.
References
- 1.Petz LD, Garratty G. Immune Hemolytic Anemias. Churchill Livingston; 2004. [Google Scholar]
- 2.Mollnes TE, Kirschfink M. Strategies of therapeutic complement inhibition. Mol. Immunol. 2006;43:107–121. doi: 10.1016/j.molimm.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 3.Hourcade D, Liszewski MK, Krych-Goldberg M, Atkinson JP. Functional domains, structural variations and pathogen interactions of MCP, DAF, and CR1. Immunopharmacology. 2000;49:103–116. doi: 10.1016/s0162-3109(00)80296-9. [DOI] [PubMed] [Google Scholar]
- 4.Iida K, Nussenzweig V. Complement receptor is an inhibitor of the complement cascade. J. Exp. Med. 1981;153:1138–1150. doi: 10.1084/jem.153.5.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fearon DT. Regulation of the amplification C3 convertase of human complement by an inhibitory protein isolated from human erythrocyte membrane. Proc. Natl. Acad. Sci. USA. 1979;76:5867–5871. doi: 10.1073/pnas.76.11.5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ross GD. Complement receptor type 1. Curr. Top. Microbiol. Immunol. 1992;178:31–44. doi: 10.1007/978-3-642-77014-2_3. [DOI] [PubMed] [Google Scholar]
- 7.Makrides SC. Therapeutic inhibition of the complement system. Pharmacol. Rev. 1998;50:59–87. [PubMed] [Google Scholar]
- 8.Weisman HF, Bartow T, Leppo MK, Marsh HC, Jr., Carson GR, Concino MF, Boyle MP, Roux KH, Weisfeldt ML, Fearon DT. Soluble human complement receptor type 1: In vivo inhibitor of complement suppressing post-ischemic myocardial inflammation and necrosis. Science. 1990;249:146–151. doi: 10.1126/science.2371562. [DOI] [PubMed] [Google Scholar]
- 9.Perry GJ, Eisenberg PR, Zimmerman JI, Levin J. Phase I safety trial of soluble complement receptor 1 (TP10) in acute myocardial infarction. J. Am. Coll. Cardiol. 1998;31:411A. [Google Scholar]
- 10.Zamora MR, Davis RD, Keshavjee SH, Schulman L, Levin J, Ryan U, Patterson GA. Complement inhibition attenuates human lung transplant reperfusion injury: a multicenter trial. Chest. 1999;116:46S. doi: 10.1378/chest.116.suppl_1.46s. [DOI] [PubMed] [Google Scholar]
- 11.Zimmerman JL, Dellinger RP, Straube RC, Levin JL. Phase I trial of the recombinant soluble complement receptor 1 in acute lung injury and acute respiratory distress syndrome. Crit. Care Med. 2000;28:3149–3154. doi: 10.1097/00003246-200009000-00004. [DOI] [PubMed] [Google Scholar]
- 12.Yazdanbakhsh K, Kang S, Tamasauskas D, Sung D, Scaradavou A. Complement receptor 1 inhibitors for prevention of immune-mediated red cell destruction: potential use in transfusion therapy. Blood. 2003;101:5046–5052. doi: 10.1182/blood-2002-10-3068. [DOI] [PubMed] [Google Scholar]
- 13.Yazdanbakhsh K, Scaradavou A. CR1-based inhibitors for prevention of complement-mediated immune hemolysis. Drug News Perspect. 2004;17:314–320. doi: 10.1358/dnp.2004.17.5.829035. [DOI] [PubMed] [Google Scholar]
- 14.Klickstein LB, Bartow TJ, Miletic V, Rabson LD, Smith JA, Fearon DT. Identification of distinct C3b and C4b recognition sites in the human C3b/C4b receptor (CR1, CD35) by deletion mutagenesis. J. Exp. Med. 1988;168:1699–1717. doi: 10.1084/jem.168.5.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mqadmi A, Abdullah Y, Yazdanbakhsh K. Characterization of complement receptor 1 domains for prevention of complement-mediated red cell destruction. Transfusion. 2005;45:234–244. doi: 10.1111/j.1537-2995.2004.04163.x. [DOI] [PubMed] [Google Scholar]
- 16.Krych-Goldberg M, Hauhart RE, Subramanian VB, Yurcisin BM, Crimmins DL, Hourcade DE, Atkinson JP. Decay accelerating activity of complement receptor type 1 (CD35). Two active sites are required for dissociating C5 convertases. J. Biol. Chem. 1999;274:31160–31168. doi: 10.1074/jbc.274.44.31160. [DOI] [PubMed] [Google Scholar]
- 17.Krych M, Hourcade D, Atkinson JP. Sites within the complement C3b/C4b receptor important for the specificity of ligand binding. Proc. Natl. Acad. Sci. USA. 1991;88:4353–4357. doi: 10.1073/pnas.88.10.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krych M, Clemenza L, Howdeshell D, Hauhart R, Hourcade D, Atkinson JP. Analysis of the functional domains of complement receptor type 1 (C3b/C4b receptor; CD35) by substitution mutagenesis. J. Biol. Chem. 1994;269:13273–13278. [PubMed] [Google Scholar]
- 19.Krych M, Hauhart R, Atkinson JP. Structure-function analysis of the active sites of complement receptor type 1. J. Biol. Chem. 1998;273:8623–8629. doi: 10.1074/jbc.273.15.8623. [DOI] [PubMed] [Google Scholar]
- 20.Krych-Goldberg M, Hauhart RE, Porzukowiak T, Atkinson JP. Synergy between two active sites of human complement receptor type 1 (CD35) in complement regulation: implications for the structure of the classical pathway C3 convertase and generation of more potent inhibitors. J. Immunol. 2005;175:4528–4535. doi: 10.4049/jimmunol.175.7.4528. [DOI] [PubMed] [Google Scholar]
- 21.Smith BO, Mallin RL, Krych-Goldberg M, Wang X, Hauhart RE, Bromek K, Uhrin D, Atkinson JP, Barlow PN. Structure of the C3b binding site of CR1 (CD35), the immune adherence receptor. Cell. 2002;108:769–780. doi: 10.1016/s0092-8674(02)00672-4. [DOI] [PubMed] [Google Scholar]
- 22.Lukacik P, Roversi P, White J, Esser D, Smith GP, Billington J, Williams PA, Rudd PM, Wormald MR, Harvey DJ, Crispin MD, Radcliffe CM, Dwek RA, Evans DJ, Morgan BP, Smith RA, Lea SM. Complement regulation at the molecular level: the structure of decay-accelerating factor. Proc. Natl. Acad. Sci. USA. 2004;101:1279–1284. doi: 10.1073/pnas.0307200101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 24.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 25.Issitt PD. Applied Blood Group Serology. Montgomery Scientific Publications; Miami, FL: 1985. [Google Scholar]
- 26.Kuttner-Kondo LA, Mitchell L, Hourcade DE, Medof ME. Characterization of the active sites in decay-accelerating factor. J. Immunol. 2001;167:2164–2171. doi: 10.4049/jimmunol.167.4.2164. [DOI] [PubMed] [Google Scholar]
- 27.Werle M, Bernkop-Schnurch A. Strategies to improve plasma half life time of peptide and protein drugs. Amino Acids. 2006;30:351–367. doi: 10.1007/s00726-005-0289-3. [DOI] [PubMed] [Google Scholar]
- 28.Seya T, Holers VM, Atkinson JP. Purification and functional analysis of the polymorphic variants of the C3b/C4b receptor (CR1) and comparison with H, C4b-binding protein (C4bp), and decay accelerating factor (DAF) J. Immunol. 1985;135:2661–2667. [PubMed] [Google Scholar]
- 29.Smith RA. Targeting anticomplement agents. Biochem. Soc. Trans. 2002;30:1037–1041. doi: 10.1042/bst0301037. [DOI] [PubMed] [Google Scholar]