It has been documented that there are differences in disease susceptibilities between humans and non-human primates. We investigate one of these differences in fibroblasts to examine differences in cellular adhesion between humans and chimpanzees using microscopy and gene expression and have found significant differences in both datasets. These results suggest that human and chimpanzee fibroblasts may have somewhat different adhesive properties, which could play a role in differential disease phenotypes and responses to external factors.
Keywords: human evolution, focal adhesion, fibroblast, cancer
Abstract
There are a number of documented differences between humans and our closest relatives in responses to wound healing and in disease susceptibilities, suggesting a differential cellular response to certain environmental factors. In this study, we sought to look at a specific cell type, fibroblasts, to examine differences in cellular adhesion between humans and chimpanzees in visualized cells and in gene expression. We have found significant differences in the number of focal adhesions between primary human and chimpanzee fibroblasts. Additionally, we see that adhesion related gene ontology categories are some of the most differentially expressed between human and chimpanzee in normal fibroblast cells. These results suggest that human and chimpanzee fibroblasts may have somewhat different adhesive properties, which could play a role in differential disease phenotypes and responses to external factors.
BACKGROUND
Recent efforts to expose genotypic differences between humans and our closest relatives have uncovered the question of gene expression changes and the role they play in influencing phenotype. By determining the genetic differences between humans and chimpanzees, we can learn about how we have evolved and adapted during the ∼6 million year divergence between these species. Comparative analyses can be informative about not only evolution but also in questions of different disease susceptibilities between species [1, 2]. There have been a number of studies looking at differences in gene expression between humans and chimpanzees [3–7]; however, very few studies have undertaken a comparative analysis at the level of phenotypic cell biology in these species.
Our study uses focal adhesions to illustrate how we differ from our closest relatives at the phenotypic level, and how this relates to differences in gene expression. Focal adhesions are large complexes of proteins that are usually found at the periphery of cells [8]. One of their primary functions is to facilitate cell attachment to the extra-cellular matrix (ECM), primarily through the use of proteins called integrins [8, 9]. However, focal adhesions are also involved in several other aspects of cell function including cell–cell signaling, detection of the ECM and cell movement [8–10]. Focal adhesions play an early and vital role in many signaling pathways and allow cells to respond to various stimuli [8, 9, 11–13]. In addition, focal adhesions are key factors in cell mobility as they are transported from the lagging areas of a moving cell to its leading edge in order to form a new attachment with the ECM and stabilize the cell [8, 9, 14–17]. As a result, focal adhesions are critical to normal cell function as well as a cell’s ability to react to its environment [10, 16, 18, 19].
A phenotypic difference that affects focal adhesions could impair or modify basic cellular functions; interfering with focal adhesion function can cause reduced cell motility [10] and dramatic changes can affect the phenotype to the point of cell death [15, 16, 18–20]. In addition, differences in the prevalence and availability of many of the signaling proteins associated with focal adhesions have been shown to produce phenotypes such as cardiac disease or several types of cancer [21–23], and when some of these same proteins are targeted, they can have beneficial effects in the treatment of those diseases [13, 24]. Furthermore, there are broader implications to modifications in focal adhesions as a new phenotype can have an effect at the organismal level. For example, adjusting focal adhesion phenotype could cause differences in cell sensitivity due to increased signaling or faster wound healing due to increased cell motility [8, 9]. A significant difference in focal adhesion phenotype between species could be an indicator of a change in fibroblast adherence and interaction with the environment.
In order to assay focal adhesion phenotypes, a way of determining focal adhesion location and size is required. Focal adhesions are composed of several proteins and compounds, one of which is vinculin. Vinculin is a focal adhesion-specific protein that is critical to the structure of focal adhesions [20, 25, 26]. It is primarily involved in cell attachment and motility and localizes to focal adhesions [14, 27]. It is a key protein in cell structure as it anchors F-actin to the cell membrane at focal adhesions and has no close relatives that can fulfill its function if it is not present [28, 29]. Vinculin knockout mice were shown to die in early development and have defects in their partially formed hearts and brains [28]. Vinculin can be used as a proxy for focal adhesion presence as it is required for focal adhesion function, it is specific to focal adhesions and it is irreplaceable [29, 30]. Additionally, vinculin is pervasive throughout the focal adhesions’ structures, which allow it to be used to observe their size as well [30, 31]. As such it has been used by several studies to study focal adhesion location and function [29, 30]. Here, we examine focal adhesions within human and chimpanzee primary skin fibroblasts using this protein proxy. As a primarily exploratory study, we are investigating the following question: To what extent do human and chimpanzee skin fibroblasts differ in adhesive properties?
METHODOLOGY
Fibroblast staining
We examined vinculin antibody staining and cell size between representative fibroblast cell lines of the two species. Four human and four chimpanzee primary fibroblast cell lines (Coriell, Camden, NJ) were used for the experiment described here. All of the cell lines are from males of approximately the same age in each group (humans aged 22–30 years, chimpanzees aged 17–24 years) (Supplementary Table S1). Most of the human samples are from forearm samples, with one from the abdomen. Previous studies have shown that the area of the body sampled is important to note, with the upper and lower half of the body showing somewhat different changes in gene expression [32, 33]. For the chimpanzee lines, the area biopsied was not recorded by Yerkes Regional Primate Research Center although their protocols state that they should be from the ear pinna [5]. However, Shibata et al. [5] used similar cell lines in a gene expression and open chromatin analysis, and found that the fibroblast cells showed very little (<3%) overlap with the regionally affected genes from Rinn et al. [32], suggesting that the human and chimpanzee cells are from comparable regions of the body.
The fibroblast cells were expanded in Minimal Essential Medium with 10% fetal bovine serum until they were approximately 80% confluent. All cell lines were expanded until they were at similar population doubling levels and passage numbers. Cells were grown on glass coverslips in six-well plates for 24 hours (50 000 cells per well), and then fixed with a 4% formaldehyde solution. Following this, the cells underwent 3× washes with PBS before being permeabilized with 0.2% Triton-X100. The cells were then stained with a monoclonal anti-vinculin antibody (ABfinity; Thermo Fisher, Waltham, MA) and an AlexaFluor secondary (Molecular Probes; Thermo Fisher, Waltham, MA), with 3% BSA in PBS.
Cells were imaged using a Zeiss Axio Observer wide-field fluorescence microscope at Duke University’s Light Microscopy Core Facility. All cells that met the following criteria were imaged. The ∼10 clearest images from each cell line were then selected for further analysis giving a total of 60 images. Previous studies have normally had a total sample size of between 7 and 30 fibroblasts [34, 35]. The images were analyzed using the program MetaMorph. The size of the cell was determined by the actin staining. The actin image was then used to create a mask in order to determine the number and size of the focal adhesions on the exterior of the cell.
For the gene expression ontology enrichments, the raw data counts (generated using DGE-Seq [36]) were acquired from Shibata et al. [5], and normalization and differential expression were determined using edgeR [37]. These data were also generated from primary skin fibroblast lines and come from the same repository collections (Coriell, Camden, NJ), but are not from the same individual as the lines used in the analysis above. The categorical enrichments were performed using both custom software using the gene ontology (GO) databases [38].
RESULTS AND DISCUSSION
Experimental validation of differences in cell adhesion
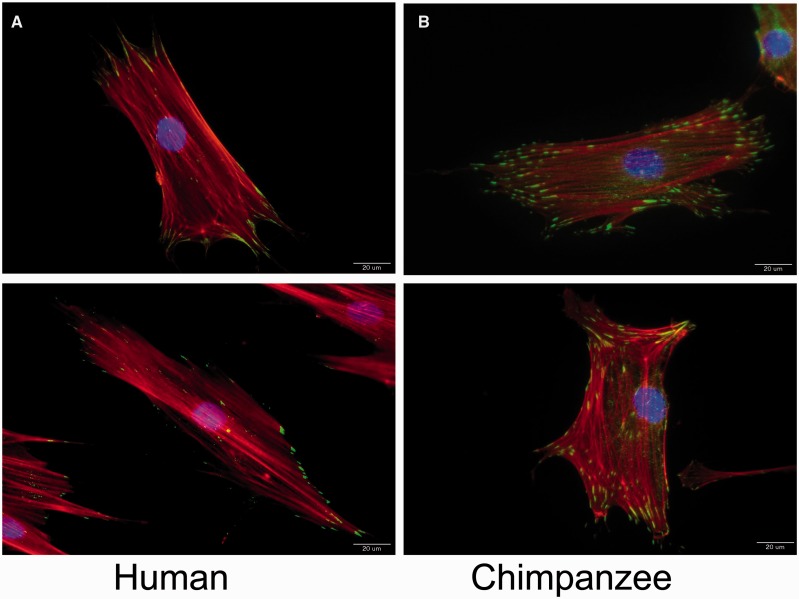
In order to begin to explore cell adhesion at a cellular level, we examined the differences in aspect of adhesion in human and chimpanzee primary fibroblast cells (Fig. 1). Vinculin can be used as a proxy indicator for focal adhesions, allowing for the detection of the number, size and location of the focal adhesions present within a cell [25, 26]. Vinculin is a focal adhesion-specific protein that is critical to the structure of focal adhesions [25, 26]. Additionally, transfection of vinculin cDNA into tumor cell lines expressing lower levels of the endogenous protein results in a significant suppression of their tumorigenic ability and an increase in substrate adhesiveness [39], suggesting that vinculin expression drives this change in phenotype. Here, we see that the ratio of focal adhesions to cell size between humans and chimpanzees is significantly different between species (Fig. 1), with an upregulation in the chimpanzee cells (analysis of variance, P < 0.00001). This is true even when we normalized for any differences in cell size, as measured by the actin cytoskeleton (Fig. 2). When the chimpanzee cell line with some outlier points is removed (S008975), this difference is also significant (P = 0.01476) (Supplementary Figure S1). The variance in the chimpanzee cell lines is much larger when cell lines are plotted individually (Supplementary Figure S1). Larger sample sizes will be needed in the future to determine more quantitatively what component of this variance is due to cell line or species effects. Given that the principal function of a focal adhesion is cell attachment [8], it is possible that the biological interpretation of this difference results in differential adhesive and migratory properties. It is possible that chimpanzee fibroblasts can attach to their substrate more firmly than human cells, which could indicate a difference in the way fibroblast cells move normally or in response to injury.
Figure 1.
Example images of the stained human and chimpanzee fibroblast cells. Nuclei (blue), actin (red), and vinculin, a label for focal adhesions (green), are merged in these images, with 20 μm for the scale bar
Figure 2.
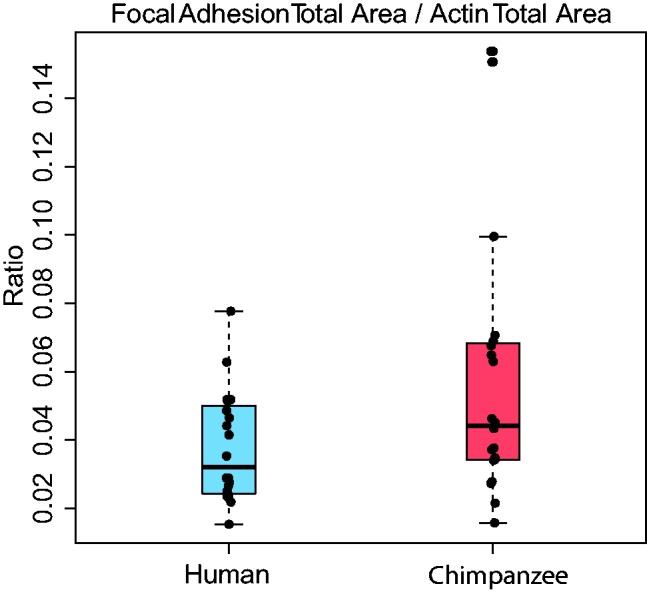
Box plot of the differences in the ratio of focal adhesion to total cell size (measured by actin staining). The human ratio is in blue and the chimpanzee in red
In the event of a disruption in tissue continuity, i.e. a wound, new cells must differentiate to replace those lost and healthy cells must migrate to seal the gaps in the tissue layer [40]. This results in a large number of focal adhesions being disassembled and reassembled at the leading edge of a migrating cell [40, 41]. Recent work has shown that the rate of wound closure will increase with a firmer wound bed [42]. Cells that are more firmly attached to their substrate might be more difficult to dislodge and may form a more robust cell layer. The efficiency with which a wound can be sealed and repaired could therefore be dramatically affected by the number of focal adhesions present in the cells forming the bed of the wound have. There have been reports of faster wound healing in wild primates than in humans [43, 44], but so far these are isolated events. Additionally, it is well documented that the gene expression response to wound healing is highly similar to gene expression changes in cancer progression [45, 46]. Epithelial cancers occur at much higher rates in humans than in non-human primates [47–50], and while most of that difference is likely due to environmental factors, there is evidence for some genetic and cellular differences as well.
We then looked at differences between human and chimpanzee fibroblasts at the level of gene expression based on the published gene expression dataset described Shibata et al. [5], which were also generated from primary skin fibroblast cell lines. We re-analyzed those differential gene expression data and found that two of the highest categories are related to cellular adhesive properties (biological adhesion, FDR = 1.07E−06 and cell adhesion, FDR = 1.84E−06) (Table 1 and Supplementary Table S2). Overall, categories involved in cellular adhesion are some of the most differentially expressed GO categories in human and chimpanzee fibroblasts (Table 1 and Supplementary Table S2).
Table 1.
Top 10 GO biological process ontology enrichments from the fibroblast RNA-Seq data in Shibata et al. [5]
| Category | P value | Total occurrence |
|---|---|---|
| Multicellular organismal process | 7.36E−09 | 1048 |
| Biological adhesion | 7.78E−07 | 409 |
| Cell adhesion | 7.78E−07 | 409 |
| Oxidation reduction | 9.32E−06 | 339 |
| Regulation of multicellular organismal process | 2.94E−05 | 358 |
| Developmental process | 3.21E−05 | 1756 |
| Cellular ion homeostasis | 4.49E−05 | 117 |
| Branching morphogenesis of a tube | 5.10E−05 | 40 |
| Morphogenesis of a branching structure | 5.84E−05 | 41 |
| Cellular chemical homeostasis | 9.57E−05 | 119 |
The ontology enrichments were performed using the DAVID Functional Annotation tool [52], and used all genes measured in the study as the background gene set.
Our results suggest that the chimpanzee fibroblast cells might be naturally more adhesive, and the human cells then possibly more prone to changes towards cancer morphologies. These results illustrate the importance of comparative studies at the cellular level. Other studies looking at cellular differences in fibroblasts have also shown differential apoptotic function in humans when compared with chimpanzees [51], also suggesting that these species-specific differences in fibroblasts may lead to differences in epithelial cancers. It will be important to understand how these cellular phenotypes functionally link changes from gene expression to organismal phenotype, and how that might assist in our understanding of susceptibilities to diseases, such as epithelial cancer.
CONCLUSION
Fibroblasts show differences in focal adhesions between humans and chimpanzees at both the genetic and cellular levels. Understanding the evolutionary history and phenotypic impact of these changes is essential in understanding the differences in fibroblast function in normal and diseased tissue. Our results suggest that human and chimpanzee fibroblasts may differ in adhesive properties, which then may play a role in differential phenotypes and responses to environmental factors.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Greg Wray and the members of the Wray and Babbitt laboratories for support and input. We also thank Lisa Pfefferle for guidance and advice throughout the project. Additionally, we thank Sam Johnson and Yasheng Gao at Duke’s Light Microscopy Core Facility.
FUNDING
Funding was provided by Duke University Primate Genomics Initiative Fellowship and the Howard Hughes Medical Institute’s Vertical Integration Partners Program for undergraduates to A.S.A. and A.C.
SUPPLEMENTARY DATA
Supplementary data is available at EMPH online.
Conflict of interest: None declared.
REFERENCES
- 1.Olson MV, Varki A. Sequencing the chimpanzee genome: insights into human evolution and disease. Nat Rev Genet 2003;4:20–8. [DOI] [PubMed] [Google Scholar]
- 2.Varki A, Nelson DL. Genomic comparisons of humans and chimpanzees. Ann Rev Anthropol 2007;36:191–209. [Google Scholar]
- 3.Babbitt CC, Fedrigo O, Pfefferle AD. et al. Both noncoding and protein-coding RNAs contribute to gene expression evolution in the primate brain. Genome Biol Evol 2010;2010:67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blekhman R, Oshlack A, Chabot AE. et al. Gene regulation in primates evolves under tissue-specific selection pressures. PLoS Genetics 2008;4:e1000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shibata Y, Sheffield N, Fedrigo O. et al. Extensive evolutionary changes in regulatory element activity during human origins are closely associated with altered gene expression and positive selection. PLoS Genet 2012;8:e1002789.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brawand D, Soumillon M, Necsulea A. et al. The evolution of gene expression levels in mammalian organs. Nature 2011;478:343–8. [DOI] [PubMed] [Google Scholar]
- 7.Somel M, Franz H, Yan Z. et al. Transcriptional neoteny in the human brain. Proc Natl Acad Sci U S A 2009;106:5743–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burridge K, Chrzanowska-Wodnicka M. Focal adhesions, contractility, and signaling. Annu Rev Cell Dev Biol 1996;12:463–518. [DOI] [PubMed] [Google Scholar]
- 9.Bershadsky AD, Ballestrem C, Carramusa L. et al. Assembly and mechanosensory function of focal adhesions: experiments and models. Eur J Cell Biol 2006;85:165–73. [DOI] [PubMed] [Google Scholar]
- 10.Ilic D, Furuta Y, Kanazawa S. et al. Reduced cell motility and enhanced focal adhesion contact formation in cells from FAK-deficient mice. Nature 1995;377:539–44. [DOI] [PubMed] [Google Scholar]
- 11.Woods A, Couchman JR. Syndecan-4 and focal adhesion function. Curr Opin Cell Biol 2001;13:578–83. [DOI] [PubMed] [Google Scholar]
- 12.Romer LH, Birukov KG, Garcia JG. Focal adhesions: paradigm for a signaling nexus. Circ Res 2006;98:606–16. [DOI] [PubMed] [Google Scholar]
- 13.Nelson ES, Folkmann AW, Henry MD. et al. Vinculin activators target integrins from within the cell to increase melanoma sensitivity to chemotherapy. Mol Cancer Res 2011;9:712–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ezzell RM, Goldmann WH, Wang N. et al. Vinculin promotes cell spreading by mechanically coupling integrins to the cytoskeleton. Exp Cell Res 1997;231:14–26. [DOI] [PubMed] [Google Scholar]
- 15.Tamura M, Gu J, Matsumoto K. et al. Inhibition of cell migration, spreading, and focal adhesions by tumor suppressor PTEN. Science 1998;280:1614–7. [DOI] [PubMed] [Google Scholar]
- 16.Parsons JT. Focal adhesion kinase: the first ten years. J Cell Sci 2003;116:1409–16. [DOI] [PubMed] [Google Scholar]
- 17.Opazo Saez A, Zhang W, Wu Y. et al. Tension development during contractile stimulation of smooth muscle requires recruitment of paxillin and vinculin to the membrane. Am J Physiol Cell Physiol 2004;286:C433–47. [DOI] [PubMed] [Google Scholar]
- 18.Frisch SM, Vuori K, Ruoslahti E. et al. Control of adhesion-dependent cell survival by focal adhesion kinase. J Cell Biol 1996;134:793–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilmore AP, Romer LH. Inhibition of focal adhesion kinase (FAK) signaling in focal adhesions decreases cell motility and proliferation. Mol Biol Cell 1996;7:1209–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen H, Cohen DM, Choudhury DM. et al. Spatial distribution and functional significance of activated vinculin in living cells. J Cell Biol 2005;169:459–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McLean GW, Komiyama NH, Serrels B. et al. Specific deletion of focal adhesion kinase suppresses tumor formation and blocks malignant progression. Genes Dev 2004;18:2998–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hakim ZS, DiMichele LA, Rojas M. et al. FAK regulates cardiomyocyte survival following ischemia/reperfusion. J Mol Cell Cardiol 2009;46:241–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sulzmaier FJ, Jean C, Schlaepfer DD. FAK in cancer: mechanistic findings and clinical applications. Nat Rev Cancer 2014;14:598–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng Z, DiMichele LA, Hakim ZS. et al. Targeted focal adhesion kinase activation in cardiomyocytes protects the heart from ischemia/reperfusion injury. Arterioscler Thromb Vasc Biol 2012;32:924–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burridge K, Mangeat P. An interaction between vinculin and talin. Nature 1984;308:744–6. [DOI] [PubMed] [Google Scholar]
- 26.Humphries JD, Wang P, Streuli C. et al. Vinculin controls focal adhesion formation by direct interactions with talin and actin. J Cell Biol 2007;179:1043–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geiger B. Involvement of vinculin in contact-induced cytoskeletal interactions. Cold Spring Harb Symp Quant Biol 1982;46 Pt 2:671–82. [DOI] [PubMed] [Google Scholar]
- 28.Xu W, Baribault H, Adamson ED. Vinculin knockout results in heart and brain defects during embryonic development. Development 1998;125:327–37. [DOI] [PubMed] [Google Scholar]
- 29.Goldmann WH, Ingber DE. Intact Vinculin Protein Is Required for Control of Cell Shape, Cell Mechanics, and rac-Dependent Lamellipodia Formation. Biochemical and Biophysical Research Communications 2002;290:749–55. [DOI] [PubMed] [Google Scholar]
- 30.Balaban NQ, Schwarz US, Riveline D. et al. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat Cell Biol 2001;3:466–72. [DOI] [PubMed] [Google Scholar]
- 31.Ziegler WH, Liddington RC, Critchley DR. The structure and regulation of vinculin. Trends Cell Biol 2006;16:453–60. [DOI] [PubMed] [Google Scholar]
- 32.Rinn JL, Bondre C, Gladstone HB. et al. Anatomic demarcation by positional variation in fibroblast gene expression programs. PLoS Genet 2006;2:e119.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang HY, Chi JT, Dudoit S. et al. Diversity, topographic differentiation, and positional memory in human fibroblasts. Proc Natl Acad Sci U S A 2002;99:12877–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mao H, Wang Y, Li Z. et al. Hsp72 interacts with paxillin and facilitates the reassembly of focal adhesions during recovery from ATP depletion. J Biol Chem 2004;279:15472–80. [DOI] [PubMed] [Google Scholar]
- 35.Kuo JC, Han X, Hsiao CT. et al. Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for beta-Pix in negative regulation of focal adhesion maturation. Nat Cell Biol 2011;13:383–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.t Hoen PAC, Ariyurek Y, Thygesen HH. et al. Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Research 2008;36: Article No.: e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat Genet 2000;25:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodriguez Fernandez JL, Geiger B, Salomon D. et al. Suppression of tumorigenicity in transformed cells after transfection with vinculin cDNA. J Cell Biol 1992;119:427–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin P. Wound healing–aiming for perfect skin regeneration. Science 1997;276:75–81. [DOI] [PubMed] [Google Scholar]
- 41.Gabbiani G. The myofibroblast in wound healing and fibrocontractive diseases. J Pathol 2003;200:500–3. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Wang G, Luo X. et al. Substrate stiffness regulates the proliferation, migration, and differentiation of epidermal cells. Burns 2012;38:414–20. [DOI] [PubMed] [Google Scholar]
- 43.Sullivan TP, Eaglstein WH, Davis SC. et al. The pig as a model for human wound healing. Wound Repair Regen 2001;9:66–76. [DOI] [PubMed] [Google Scholar]
- 44.Hedlund M, Tangvoranuntakul P, Takematsu H. et al. N-glycolylneuraminic acid deficiency in mice: implications for human biology and evolution. Mol Cell Biol 2007;27:4340–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang HY, Nuyten DS, Sneddon JB. et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci U S A 2005;102:3738–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang HY, Sneddon JB, Alizadeh AA. et al. Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds. PLoS Biol 2004;2:E7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Varki A. A Chimpanzee Genome Project is a biomedical imperative. Genome Res 2000;10:1065–70. [DOI] [PubMed] [Google Scholar]
- 48.McClure HM. Tumors in nonhuman primates: observations during a six-year period in the Yerkes Primate Center Colony. Am J Phys Anthropol 1973;38:425–9. [DOI] [PubMed] [Google Scholar]
- 49.Seibold HR, Wolf RH. Neoplasms and proliferative lesions in 1065 nonhuman primate necropsies. Lab Animal Sci 1973;23:533–9. [PubMed] [Google Scholar]
- 50.Beniashvili DS. An overview of the world literature on spontaneous tumors in nonhuman primates. J Med Primatol 1989;18:423–37. [PubMed] [Google Scholar]
- 51.Arora G, Mezencev R, McDonald JF. Human cells display reduced apoptotic function relative to chimpanzee cells. PLoS One 2012;7:e46182.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.