Summary
HIV-1 recruits cellular Endosomal Sorting Complexes Required for Transport (ESCRTs) to bud virions from the membrane. Disruption of the viral nucleocapsid (NC) domain integrity affects HIV-1 budding. However, the molecular mechanisms of NC’s involvement in HIV budding remain unclear. We find that NC mimics the PDZ domains of syntenin, a membrane-binding adaptor involved in cell-to-cell contact/communication, to capture the Bro1 domain of ALIX, which is an ESCRTs recruiting cellular adaptor. NC binds membranes via basic residues in either the distal or proximal zinc fingers and NC-membrane binding is essential for Bro1 capture and HIV-1 budding. Removal of RNA enhances NC membrane binding suggesting a dynamic competition between membrane lipids and RNA for same binding sites in NC. Remarkably, syntenin PDZ can substitute for NC function in HIV-1 budding. Thus, NC mimics syntenin PDZs to function as a membrane-binding adaptor critical for HIV-1 budding at microdomains of the membrane.
Introduction
Viruses use mimicry to co-opt host factors and mechanisms to perform a variety of processes necessary for replication. This mimicry is best exemplified with retroviruses’ hijacking of multiple cellular components at the membrane to perform assembly and budding, two late steps which precede release and are key for virus transmission and persistence. HIV-1 Gag uses lipid rafts as preferred assembly platforms to gain access to specific lipids that favor Gag anchorage, assembly and deformation of membrane into a particle (Dick et al., 2012; Ono et al., 2004; Ono and Freed, 2001; Ono et al., 2007; Yandrapalli et al., 2014). Gag orchestrates a dynamic and rapid membrane scission catalyzed by a subset of ESCRTs (Morita et al., 2011). While various components of lipid-rafts are believed to play important roles in Gag interactions with the membrane, direct involvement in virus budding is unknown. Moreover, it is well established that in late assembly retroviruses use adaptor proteins (e.g. ALIX) to recruit ESCRTs (Votteler and Sundquist, 2013); the nature of interplay between lipid, virus and ESCRTs and the mechanisms of mimicry involved are not yet elucidated.
Gag (Pr55Gag) is the main structural component of virus particles. It carries 4 distinct domains: the N-terminal matrix (MA) and central capsid (CA) domains are responsible for membrane binding and homo-oligomerization, respectively, and two C-terminal domains, the nucleocapsid (NC), and p6 endowed with specific late assembly functions. NC interacts with RNA, which is believed to accelerate Gag high order assembly (Burniston et al., 1999; Cimarelli et al., 2000; Muriaux et al., 2001). The highly conserved Late (L) domains in p6 are necessary for virus budding (Garrus et al., 2001; Gottlinger et al., 1991; Huang et al., 1995; VerPlank et al., 2001); L domains carry the PTAP and LYPXnL motifs and bind the cellular proteins TSG101 (Garrus et al., 2001; Martin-Serrano et al., 2001; VerPlank et al., 2001) and ALIX (Strack et al., 2003), respectively; which drive two independent HIV-1 budding pathways, and both require a subset of ESCRT-III isoforms (Morita et al., 2011), and the activity of the disassembly factor VPS4 AAA-type ATPase (Martin-Serrano et al., 2003; von Schwedler et al., 2003).
ALIX contains three distinct structural domains: an N-terminal Bro1 domain, a central V domain, and a C-terminal proline-rich domain (PRD) (Fisher et al., 2007; Kim et al., 2005; Lee et al., 2007). Bro1 has a boomerang-shaped structure, binds CHMP4 isoforms and is essential for ALIX function in HIV-1 release (Fisher et al., 2007; Usami et al., 2007). A hydrophobic surface is centered by the phenylalanine residue 105 (Phe105) at the tip of a long flexible loop protruding from the convex side of Bro1. Disruption of this interface impairs ALIX ability to promote HIV-1 release, suggesting the loop plays key functional role(s) (Sette et al., 2011a, b; Zhai et al., 2011b). The Phe105 loop inserts in endosomal membrane in lysobisphosphatidic acid (LBPA)-dependent manner (Bissig et al., 2013), a lipid found exclusively at the endosome. The ALIX V-shaped domain binds the LYPXnL motif, which is the strongest interaction between ALIX and Gag (Fisher et al., 2007; Lee et al., 2007; Zhai et al., 2008). The PRD domain connects ALIX to multiple cellular processes (Chatellard-Causse et al., 2002). Domain flexibility within ALIX domains has been reported (Fisher et al., 2007; Zhai et al., 2011a) suggesting ALIX dynamically responds to various interactors and/or effectors.
Gag harbors sequences that target virus assembly to specific micro-domains of the plasma membrane (PM) where proteins and lipids that favor virus production and transmission concentrate (Hubner et al., 2009; Jin et al., 2011; Jolly and Sattentau, 2005, 2007; Ono et al., 2004; Ono and Freed, 2001, 2005; Ono et al., 2007; Thomas et al., 2015). A bipartite signal in MA composed of myristic acid and clusters of basic residues are involved in targeting Gag to PI(4,5)P2 and cholesterol-rich microdomains (Dick et al., 2012; Ono et al., 2004; Ono et al., 2007; Saad and Muriaux, 2015). NC is also involved in Gag selective targeting to sites of virus transmission (Llewellyn et al., 2010). Gag membrane interaction is regulated by RNA (Chukkapalli et al., 2010). MA basic residues bind either RNA or membrane depending on Gag localization in the cell (Kutluay et al., 2014). Similarly, positively charged residues in NC that bind viral and cellular RNAs also interact with lipids in vitro (Kempf et al., 2015). The functional significance of NC binding to lipid is however unclear.
We previously showed that disruption of p6 access to ESCRTs or NC integrity was equally detrimental to virus budding (Bello et al., 2012; Dussupt et al., 2009; Dussupt et al., 2011) suggesting a role for NC in virus production. While the nature of NC involvement in this process is unclear, our data supported a model in which NC and p6 functionally cooperate to promote virus budding. This is best illustrated by the dual binding of ALIX to NC and its adjacent domain p6 in order to promote the severing of HIV-1 budding stalks (Dussupt et al., 2009; Popov et al., 2008). A stable NC-Bro1 interaction is required and sufficient to promote membrane scission, provided Bro1 retains access to ESCRT-III (Dussupt et al., 2009). To gain insights into the molecular mechanisms that govern NC function in this process, we sought to identify ALIX Bro1’s native cellular partner(s). We found that Bro1 binds the PDZ domains (PDZs) of syntenin, a membrane-binding cellular adaptor that localizes at cell-to-cell adhesion/contact domains and is involved in membrane dynamics and cell signaling. Striking parallels between ALIX Bro1 interactions with its viral NC or native partner syntenin PDZ(s) revealed that NC mimics PDZs to bind the membrane and co-opt ALIX Bro1/ESCRTs in cholesterol-rich domains, and each of these steps is equally important to ALIX-mediated HIV-1 budding. Like PDZs, NC binds anionic lipids and RNA regulates binding. PDZ1 substitutes for NC functionally and promotes HIV-1 budding. Our studies reveal that NC uses mimicry to function as an adaptor that binds the membrane to gain access to ALIX Bro1/ESCRTs at specific domains that favor virus assembly and transmission.
Results
Syntenin’s PDZ tandem is an ALIX Bro1 binding partner
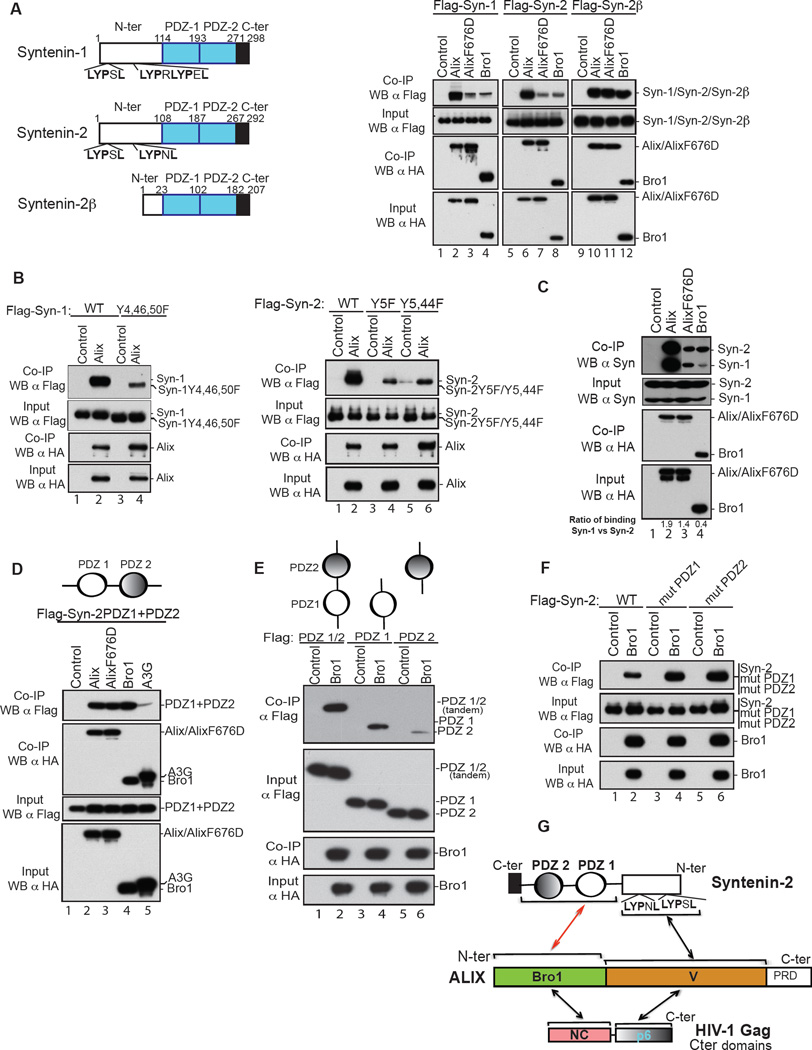
We previously reported that HIV-1 NC binds the Bro1 domain of ALIX (Dussupt et al., 2009; Popov et al., 2008) and that binding is required for ALIX-mediated HIV-1 budding (Dussupt et al., 2009). The nature of NC involvement in these processes is unknown. We opted to search for the native cellular partner of ALIX Bro1 with the reasoning that its identification should help elucidate the role of NC-Bro1 interactions in HIV-1 budding. Y2H screens captured known partners of ALIX, CHMP4B, cellular ALIX [through ALIX dimerization (Pires et al., 2009)], and an unknown binding partner syntenin-2 (syn-2), a member of syntenin family (Figure 1A, left). Syn-2 is an isoform of the recently identified ALIX V domain-binding protein, syntenin-1 (Baietti et al., 2012) and a membrane adaptor, which functions in important cellular processes at the cell membrane including signaling, cell-to-cell communication/adhesion and cancer metastasis (Beekman and Coffer, 2008; Friand et al., 2015; Philley et al., 2015).
Figure 1.
Syntenin PDZs is ALIX Bro1 interacting partner. (A) In all experiments below, 293T cells were transfected with expression plasmids listed before lane numbers. Clear cell lysates were incubated with anti-HA antibody-conjugated beads (in E, anti-Flag antibody-conjugated beads were used) and both input and IP complexes were subsequently analyzed by SDS-PAGE and western blot (WB) using the indicated antibodies. (Left) Domain positions in syntenins are indicated. (Right) Syntenin binds ALIX through both Bro1 and V. Flag- Syn-1, Syn-2 or Syn-2β alone (lanes 1, 5 and 9), or with HA-ALIX (lanes 2, 6 and 10), HA-ALIX F676D (lanes 3, 7 and 11), or HA-Bro1 (lanes 4, 8 and 12). (B) Syntenin proteins bind ALIX in a LYPXnL motif-independent manner. Flag- Syn-1 or Syn-2 (lane 1, right and left panels, respectively), Flag-Syn-1 Y4,46,50F (lane 2, left panel), Flag-Syn-2 Y5F (lane 3, right panel), Flag–Syn-2 Y5,44F (lane 5, left panel) alone or with HA-ALIX (lanes 2, 4, and 6 right and left panels, respectively). (C) Endogenous syntenins co-associate with ALIX and Bro1. HA-ALIX (lane 2), HA-ALIX F676D (lane 3), or HA-Bro1 (lane 4). (D) Syn-2 PDZs are sufficient to interact with ALIX Bro1. Flag-Syn-2 PDZs (PDZ1+PDZ2) alone (lane 1), or with either HA-ALIX (lane 2), HA-ALIX F676D (lane 3), HA-Bro1 (lane 4) or control HA-APOBEC3G (A3G) (lane 5). (E) ALIX Bro1-Syn-2 PDZs optimal interaction requires the entire PDZ tandem. Flag-Syn-2 PDZs (1/2) (lane 1), PDZ1 (lane 3), (PDZ2) (lane 5) alone or with HA-ALIX Bro1 (lanes 2, 4 and 6). (F) ALIX Bro1-Syn-2 interactions are PI(4,5)P2-independent. Flag-Syn-2 (lane 1), Flag-Syn-2 mutPDZ1 (K113/K167A) (lane 3) or Flag-Syn-2 mutPDZ2 (K197/K244A) (lane 5) alone or with WT HA-Bro1 (lanes 2, 4, 6). (G) Drawing of the interactions between Syn-2 and ALIX. The red arrow marks the interaction identified in this study. See also S1 and S2.
We excluded the possibility that syn-1 and syn-2 are co-factors in virus budding through RNAi depletion and virus budding assays using HIV-1 and EIAV (Figure S1). Next, mapping of syntenin-2-ALIX binding sites was performed. In addition to two LYPXnL L-domain-like motifs --similar to those found in HIV-1 p6 region and syn-1 (Grootjans et al., 1997), syn-2 harbors two PDZ domains organized in tandem (Kang et al., 2003) that target syn-2 preferentially to PI(4,5)P2-enriched domains of the PM (Mortier et al., 2005), where HIV-1 is also known to selectively assemble and transmit infection (Hubner et al., 2009; Jolly and Sattentau, 2007; Jouvenet et al., 2006). Further examination revealed a LYPXnL-independent interaction of syn-1 and syn-2 to ALIX (Figure 1A, right) and interaction was retained by syntenin-2β (Syn-2β). Despite missing the LYPXnL-containing first 85 residues, Syn-2β readily bound ALIX (Fig. 1A, lanes 9–11) as well as Bro1 alone, which recapitulated binding at the level seen with ALIX (Fig. 1A, lane 12). Syn-1 (Y4, 46, 50F) and Syn-2 (Y5, Y5, 44F), two mutants carrying disrupted LYPXnL motifs also retained binding to ALIX (Figure 1B) further confirming the existence of an ALIX binding site in syntenin. ALIX also captured the cellular syntenins (Figure 1C, lane 2) and Bro1 displayed a clear preference for endogenous syn-2 as higher amounts (> 5-times) were consistently captured (Figure 1C, lane 4), comparable to those immunoprecipitated with F676D, an ALIX mutant lacking the LYPXnL-binding site (lane 3). Collectively, the data reveal LYPXnL-independent ALIX-cellular syntenin interactions whose determinants lie within Bro1. Further mapping analysis showed that syn-2 PDZ1-PDZ2 tandem is sufficient to bind Bro1 with efficiency comparable to that observed with ALIX (Figure 1D). Deletion of either the proximal PDZ1 or distal PDZ2 domain reduced binding to Bro1, with removal of the latter causing the least adverse effect (Figure 1E). These interactions were also assessed using GST in vitro binding assays. Both recombinant Bro1 and Bro1-V captured Flag-tagged Syn-1, Syn-2 and Syn-2β in cellular extracts (Figure S2A). GST-Bro1 also bound recombinant strep-tagged syntenins (Figure S2B). Only weak interactions between recombinant PDZ-1-PDZ2 and Bro1 were observed (Figure S2C), suggesting additional cellular factor(s) might help stabilize binding in cellular extracts. Syntenin PDZ1-PDZ2 domains bind PI(4,5)P2 and mutation of lysine residues mediating binding prevented syn-2 enrichment at the PM (Mortier et al., 2005). We generated these mutants in syn-2 in either PDZ1 (mut PDZ1) or PDZ2 domain (mut PDZ2) and tested the involvement of PI(4,5)P2 binding sites in capturing ALIX Bro1. These changes had no effect on interactions with Bro1 (Figure 1F) and similar results were obtained with all other syntenins (data not shown). The data here indicate that i) syn-2 PDZ1-PDZ2 mediate interaction(s) with Bro1, ii) both PDZ1 and PDZ2 are needed for strong interactions, iii) that PDZs engage distinct residues to bind PI(4,5)P2 and Bro1 suggesting they can accommodate binding to both at the membrane, and iv) syntenin PDZ1-PDZ2 tandem is a native binding partner of ALIX Bro1 (Figure 1G).
Bro1 requires insertion in cholesterol-rich membrane to capture syntenin PDZ tandem
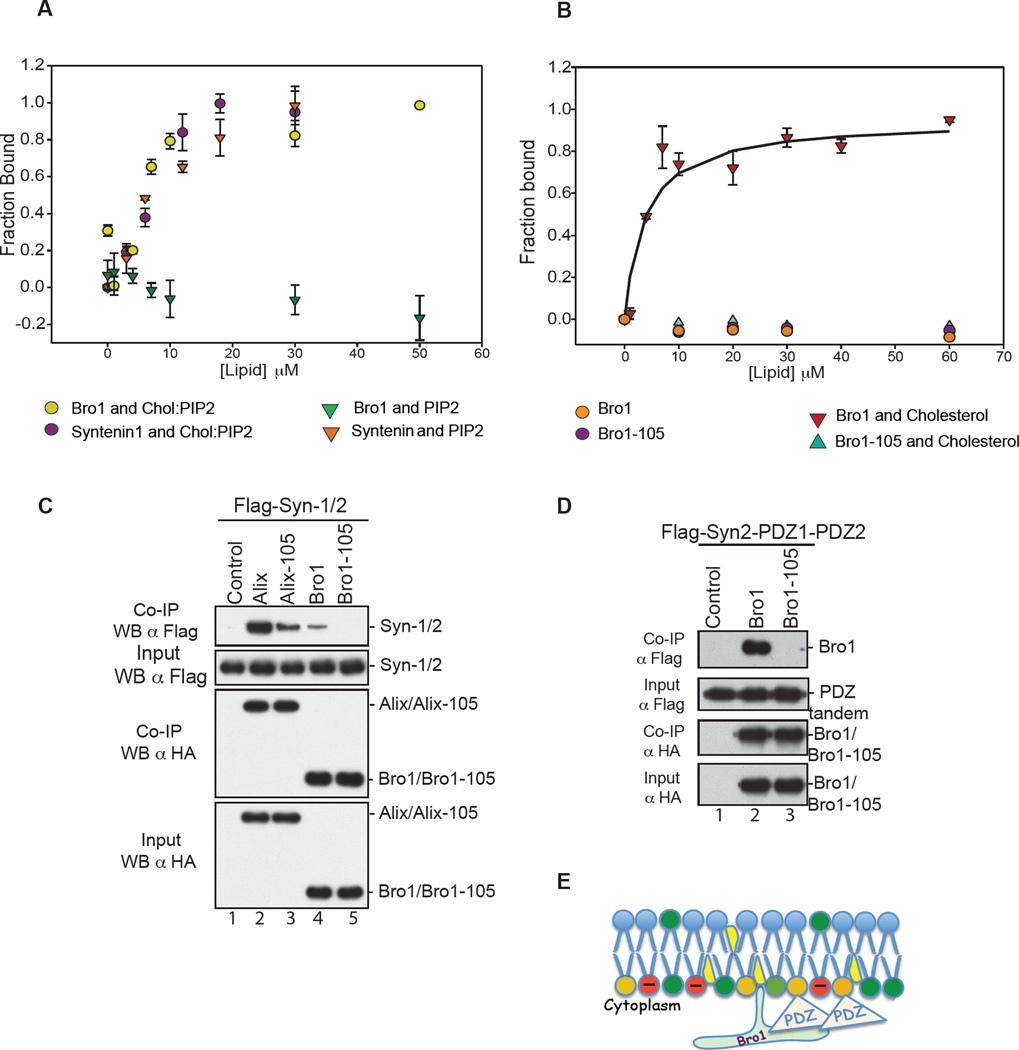
Bro1 uses the Phe105 loop to bind membrane containing lysobisphosphatidic acid (LBPA), a lipid found exclusively at the endosome (Bissig et al., 2013) while syntenin PDZ(s) bind PI(4,5)P2 at the membrane (Zimmermann et al., 2002). This suggested that Bro1 and PDZs might co-insert at the endosomal membrane to function. To assess whether Bro1 uses similar mechanisms at HIV-1 budding sites, we tested Bro1 interaction to Large Unilamellar Vesicles (LUVs) with a composition that mimics that of the PM, with or without lipids known to favor HIV-1 assembly. Bro1 and syntenin bind (LUVs) containing PI(4,5)P2 and cholesterol (Figure 2A). Conversely, omission of cholesterol prevented Bro1 binding to PI(4,5)P2-containing LUVs while syntenin binding was retained under identical conditions as expected (Figure 2A). Disruption of the lipid binding Phe105 loop in Bro1 however eliminated binding to LUVs (Figure 2B) in a manner comparable to that observed with the removal of cholesterol (2A). This data show that the Phe105 loop is the primary determinant for Bro1 binding to PM-like membrane and reveal cholesterol as a binding prerequisite. Next, we investigated whether the Phe105 loop-mediated Bro1 binding to the membrane is involved in interactions with syntenin. Flag-Syn-2 interaction with Bro1 was mapped using various HA-tagged Bro1 mutants previously described in (Sette et al., 2012; Sette et al., 2011a) (Figure S3). Disruption of Phe105 loop significantly reduced ALIX binding to syntenin (Figures 2C, compare lanes 4 and 5) and loss of binding was evident when Bro1 was tested with the isolated PDZ1-PDZ2 domains (Figure 2D, lane 3). This result is consistent with a critical role for the Phe105 loop in Bro1-PDZs binding and suggests that Bro1 Phe105 loop insertion in cholesterol-rich domains is important to establish stable interaction(s) at the membrane (Figure 2E).
Figure 2.
(A) ALIX Bro1 binds the membrane in PI(4,5)P2-independent manner. Comparison of membrane binding of 130 nM CPM-Bro1 ( ) and 130 nM Syn-1 (
) and 130 nM Syn-1 ( ) to LUVs containing both PI(4,5)P2 and cholesterol, (PC:PE:PS:Chol: PI(4,5)P2 at 26:33:27:9:5) (n=6), to membrane binding of CPM-Bro1 (
) to LUVs containing both PI(4,5)P2 and cholesterol, (PC:PE:PS:Chol: PI(4,5)P2 at 26:33:27:9:5) (n=6), to membrane binding of CPM-Bro1 ( ) and CPM-Syn-1 (
) and CPM-Syn-1 ( ) to membranes without cholesterol, (PC:PS:PE: PI(4,5)P2 at 28:34:28:10), (n=6). (B) Cholesterol is critical for ALIX Bro1 binding to the membrane. The fraction of protein bound to membranes was determined by the increase in fluorescence of a 130 nM CPM-Bro1 or the CPM-Bro1 105 mutant as LUVs with or without cholesterol were incrementally added. Neither CPM-Bro1 (
) to membranes without cholesterol, (PC:PS:PE: PI(4,5)P2 at 28:34:28:10), (n=6). (B) Cholesterol is critical for ALIX Bro1 binding to the membrane. The fraction of protein bound to membranes was determined by the increase in fluorescence of a 130 nM CPM-Bro1 or the CPM-Bro1 105 mutant as LUVs with or without cholesterol were incrementally added. Neither CPM-Bro1 ( ) nor CPM-Bro1-105 (
) nor CPM-Bro1-105 ( ) bound to PC:PE:PS (1:1:1) lipids (n=6), while CPM-Bro1 (
) bound to PC:PE:PS (1:1:1) lipids (n=6), while CPM-Bro1 ( ) but not CPM-Bro1-105 (
) but not CPM-Bro1-105 ( ) showed significant binding to PC:PE:PS:Cholesterol (28:34:28:10) lipids (n=9). (C) Flag- Syntenin expressed in 293T cells alone (lane 1), or with either HA-Alix (lane 2), HA-Alix-105 (lane 3), HA-Bro1 (lane 4) or HA-Bro1-105 (lane 5). (D) In parallel, cells were also transfected with Flag-Syn-2 PDZ tandem (1/2) alone (lane 1) or with HA-Bro1 (lane 2) or HA-Bro1 Phe105 (lane 3). Cell lysates were incubated with anti-HA antibody-conjugated beads. Both input and IP complexes were analyzed by SDS-PAGE and WB as indicated. (E) Model of ALIX Bro1-syn-2 PDZ tandem co-anchorage at lipid-rich domains of the membrane. PI(4,5)P2 (orange), cholesterol (yellow), anionic phospholipids PS (red), Bro1 and Phe105 loop (green), and PDZs tandem (pink). See also S3.
) showed significant binding to PC:PE:PS:Cholesterol (28:34:28:10) lipids (n=9). (C) Flag- Syntenin expressed in 293T cells alone (lane 1), or with either HA-Alix (lane 2), HA-Alix-105 (lane 3), HA-Bro1 (lane 4) or HA-Bro1-105 (lane 5). (D) In parallel, cells were also transfected with Flag-Syn-2 PDZ tandem (1/2) alone (lane 1) or with HA-Bro1 (lane 2) or HA-Bro1 Phe105 (lane 3). Cell lysates were incubated with anti-HA antibody-conjugated beads. Both input and IP complexes were analyzed by SDS-PAGE and WB as indicated. (E) Model of ALIX Bro1-syn-2 PDZ tandem co-anchorage at lipid-rich domains of the membrane. PI(4,5)P2 (orange), cholesterol (yellow), anionic phospholipids PS (red), Bro1 and Phe105 loop (green), and PDZs tandem (pink). See also S3.
NC mimics Syntenin PDZ tandem to bind membrane and promote HIV-1 budding
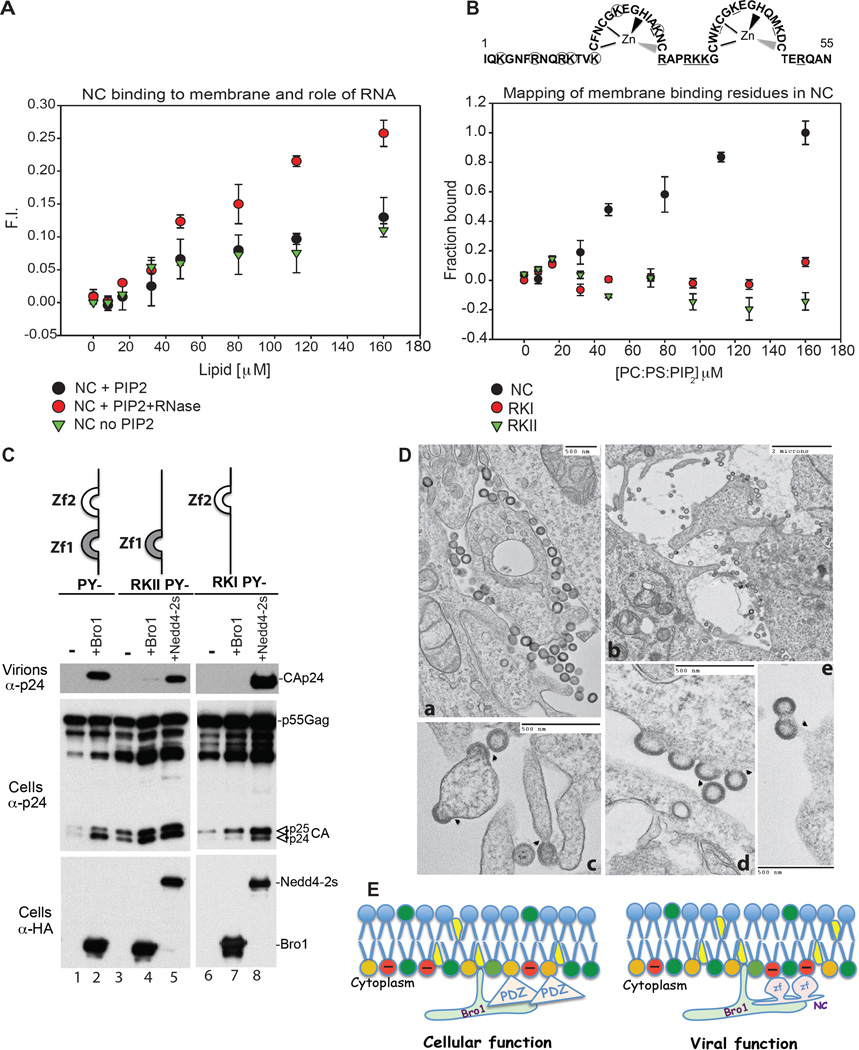
Our data suggested that syntenin engages in dual interactions with ALIX that are strikingly similar to those observed with the Gag NC and p6 domains (Dussupt et al., 2009; Fisher et al., 2007; Popov et al., 2008; Strack et al., 2003). Considering these similarities, we hypothesized that NC mimics syntenin PDZs to bind lipid and usurps Bro1. GST-NC binding to membrane was tested using PM-like LUVs . Unlike syntenin or PDZs, NC bound membranes independently of PI(4,5)P2 (Figure 3A); binding was enhanced with nuclease treatment and-removal of RNA (Figure 3A), implying lipids and RNA compete for the same binding sites within NC. Similarly, mutations of basic residues (R and K amino-acids) in either the N-terminal or C terminal region of NC (panel B) eliminated binding (Figure 3B) indicating anionic lipids mediate NC binding to membrane. Since these same residues are also known to bind RNA (Aldovini and Young, 1990), we concluded that NC membrane binding is regulated by RNA in a manner reminiscent of that observed with MA (Chukkapalli et al., 2010; Kutluay et al., 2014).
Figure 3.
(A) NC binding to membrane is enhanced by RNAse treatment. The fraction of NC bound to lipid was determined by the increase in fluorescence of the CPM-labeled protein as LUVs were incrementally added. Binding of 130 nM CPM-NC treated with RNAse to LUVs containing PI(4,5)P2 (PC:PE:PS: PI(4,5)P2 at 31:32:32:5) ( ) showed a different pattern when compared to NC binding to LUVs with PI(4,5)P2 PC:PE:PS: PI(4,5)P2 at 31:32:32:5) without RNAse (
) showed a different pattern when compared to NC binding to LUVs with PI(4,5)P2 PC:PE:PS: PI(4,5)P2 at 31:32:32:5) without RNAse ( ) or without PI(4,5)P2 (PC:PE:PS at 1:1:1) (
) or without PI(4,5)P2 (PC:PE:PS at 1:1:1) ( ) (n=9). (B) Disruption of NC zinc finger basic residues ablates membrane binding. Substitution to A in RKI (circles) or RKII (underlined) are shown above the panel. While 130 nM CPM-WT NC was observed to bind to LUVs containing PI(4,5)P2 (PC:PE:PS: PI(4,5)P2 at 31:32:32:5) (
) (n=9). (B) Disruption of NC zinc finger basic residues ablates membrane binding. Substitution to A in RKI (circles) or RKII (underlined) are shown above the panel. While 130 nM CPM-WT NC was observed to bind to LUVs containing PI(4,5)P2 (PC:PE:PS: PI(4,5)P2 at 31:32:32:5) ( ), neither 130 nM CPM-NC RKI (
), neither 130 nM CPM-NC RKI ( ) nor 130 nM CPM-NC RKII (
) nor 130 nM CPM-NC RKII ( ) bound to the same lipid (n=9) in identical conditions. (C) Disruption of NC membrane binding ability prevents utilization of ALIX Bro1 in HIV-1 budding. 293T cells were transfected with HIV-1 PY- (lanes 1–2), RKIIPY- (3–5) or RKIPY- (lanes 6–8) either alone, with HA-Bro1 (lanes 2, 4 and 7) or HA-Nedd4.2s (lanes 5 and 8). Cells and virus protein content was analyzed by SDS-PAGE and WB. (D) TEM images of RKI (a, d, e) and RKII (b, c) reveal arrested particles at the PM. (E) Model of ALIX Bro1-HIV-1 NC co-anchorage at lipid-rich domains for viral and cellular function. PI(4,5)P2 (orange), zinc fingers (zf) (pink).
) bound to the same lipid (n=9) in identical conditions. (C) Disruption of NC membrane binding ability prevents utilization of ALIX Bro1 in HIV-1 budding. 293T cells were transfected with HIV-1 PY- (lanes 1–2), RKIIPY- (3–5) or RKIPY- (lanes 6–8) either alone, with HA-Bro1 (lanes 2, 4 and 7) or HA-Nedd4.2s (lanes 5 and 8). Cells and virus protein content was analyzed by SDS-PAGE and WB. (D) TEM images of RKI (a, d, e) and RKII (b, c) reveal arrested particles at the PM. (E) Model of ALIX Bro1-HIV-1 NC co-anchorage at lipid-rich domains for viral and cellular function. PI(4,5)P2 (orange), zinc fingers (zf) (pink).
We and colleagues previously observed that NC role in virus budding necessitates the capture of Bro1 (Dussupt et al., 2011; Popov et al., 2008) whose own membrane binding through the Phe105 loop is required for function at the PM (Bissig et al., 2013; Sette et al., 2011a; Zhai et al., 2011b). Next, we tested how disruption of NC membrane binding sites affects these functions (Figure 3C). Mutation of membrane binding basic residues in NC, in either the distal or proximal zinc fingers eliminated the ability of HIV-1 to use ALIX Bro1 and promote virus budding (Figure 3C, RKII PY-, lanes 3–4 or RKI PY-, lanes 6–7), in contrast to the control virus lacking all domains but retaining a functional NC (PY-, lanes 1–2). Despite loosing binding to the membrane, both NC mutants RKI and RKII retain the ability to fully assemble particles at the membrane (Figure 3D), where arrested budding virions accumulated. This showed that NC requires membrane binding to utilize Bro1 in virus budding necks and sever developing virions from cells. NC and Bro1 concomitantly insert in the membrane (Figure 3E, right), in a manner similar to that seen between syn-2 PDZs and Bro1 during cellular function (3E, left).
NC membrane binding is necessary for the capture of ALIX Bro1 and HIV-1 budding
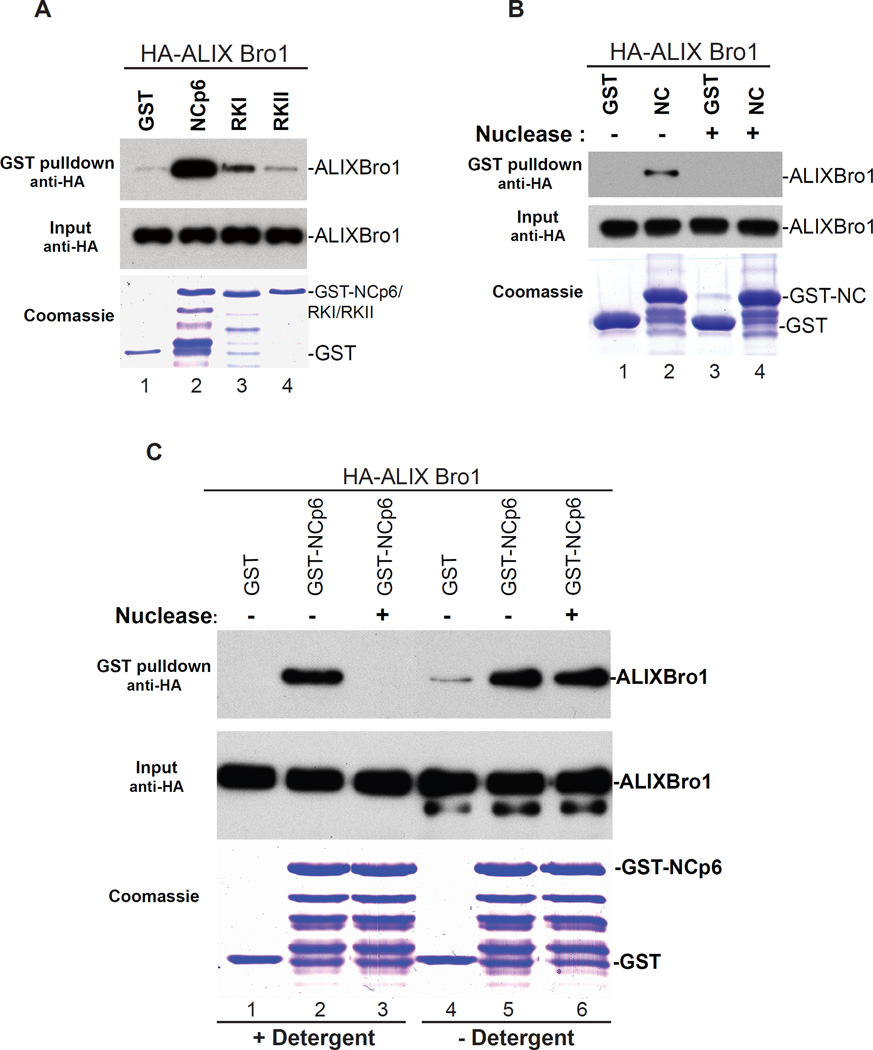
NC basic residues bind either the membrane or RNA. Mutation of NC basic residues, which disrupts zinc fingers and in turn inhibits interactions with RNA (Darlix et al., 1995; Gorelick et al., 1988; Summers et al., 1990), also ablated interactions with ALIX Bro1 to near undetectable levels (Figure 4A). Similarly, NC alone fails to capture Bro1 following RNase treatment (Figure 4B) emphasizing a role for RNA in NC-Bro1 interactions. The nature of RNA involvement in these interactions is however unclear. Since NC interacts with RNA during Gag-Gag multimerization to form large assemblages, while NC binds Bro1 late in assembly at HIV-1 budding necks, we hypothesized that NC (or a subset of NC molecules) which localizes near or at HIV-1 budding sites might dynamically trade RNA for membrane to capture ALIX Bro1. In this case, the apparent requirement for RNA in in vitro binding assays might actually be reflecting NC’s need for membrane binding in order to capture Bro1 in HIV-1 budding necks. To test this hypothesis, we assessed Bro1 binding to NC in presence or absence of natural cellular membrane and with or without RNase treatment. Removal of RNA eliminated NC-Bro1 interactions when cells were lysed and interactions assessed in membrane stripping detergent-containing buffer (Figure 4C, lanes 1–3). Conversely, NC-Bro1 complexes became insensitive to RNAse when cells were lysed mechanically in absence of detergent to preserve the cell’s natural membranes (Figure 4C, lanes 4–6). These data indicate that in the cell NC-Bro1 interactions are RNA-independent. Instead, late in assembly NC binds the membrane to recruit Bro1, thus revealing a requirement for the scission of HIV-1 budding necks.
Figure 4.
NC binding to cellular membranes is necessary for the capture of ALIX Bro1. (A) NC zinc fingers are necessary for ALIX Bro1 capture. GST alone (lane 1), GST-NC-p6 (lane 2), GST-RKI or GST-RKII (lanes 3 and 4) were purified and subsequently tested for binding with HA-ALIX Bro1- containing cellular extracts. Captured complexes and input fractions were analyzed by SDS-PAGE and WB. GST proteins were visualized by Coomassie blue. (B) RNA removal ablates NC-ALIX Bro1 interactions. Pull down assays similar to those described in (A) were conducted with GST control (lane 1) and GST-NC (lane 2) without (lanes 1 and 2) or with nuclease treatment (lanes 3 and 4). Input and eluate fractions were analyzed as described above. (C) GST pull down assays were conducted in conditions similar to those described in (B) except in the way the HA-ALIX Bro1 extracts were prepared. 293T cells were either lyzed in presence of detergent (lanes 1–3) or in hypotonic buffer and lyzed mechanically (lanes 4–6) and interactions with NC-p6-ALIX Bro1 were assessed with or without nuclease. Input and eluate fractions were analyzed as described above.
NC associates with cellular ALIX at HIV-1 budding sites at the plasma membrane
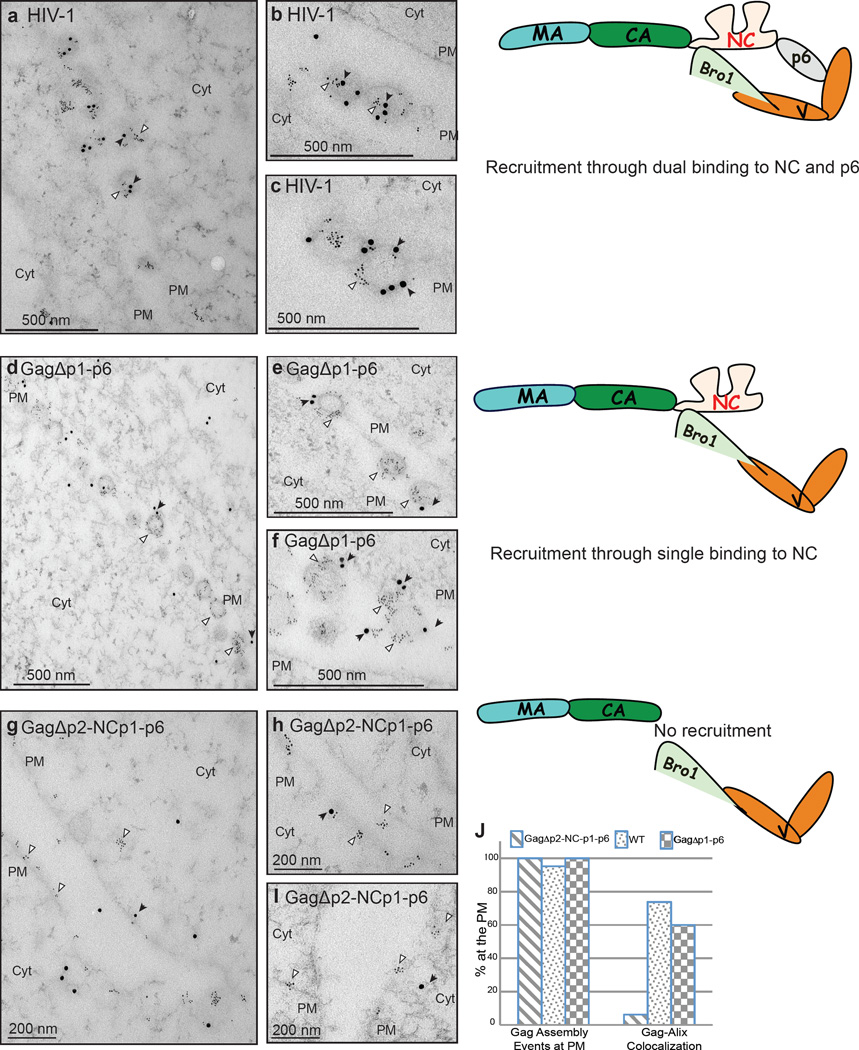
The biochemical and functional data above suggested that NC binds and co-inserts with Bro1 at the membrane where ESCRT-III is recruited to catalyze scission of HIV-1 budding stalks. However, whether NC is sufficient to associate with endogenous ALIX Bro1 in the cell at the PM is unknown. We employed double immunogold labeling to examine whether cellular ALIX associates with Gag assembly sites at the PM of HIV-1 expressing cells. Double labeling with anti-Gag and anti-Bro1 antibodies showed ALIX co-localized with HIV-1 budding sites at the PM (Figure 5, panels a, b and c). ALIX co-localization at the membrane was retained by a Gag lacking the entire p6 domain (panels d, e and f), and was lost only when a HIV-1 mutant lacking both NC and p6 was used (panels g, h and i), a finding consistent with ALIX’s inability to bind Gag in absence of the Bro1 domain-binding site in NC. Counting double-labeled Gag assembly events at the PM revealed that Gag and its p6-deleted counterpart exhibited ~10-fold more co-localization with ALIX than Gag lacking NC-p6 (Figure 5J). Thus NC is required for association with endogenous ALIX at HIV-1 budding sites and implied that NC binds Bro1 in natural conditions.
Figure 5.
Co-localization of endogenous ALIX with Gag at HIV-1 budding sites at the PM requires NC. COS-1 cells expressing WT Gag (panel a, low-power view, and panels b and c, high-power view of other fields), GagΔp2-NCp1-p6 (panels d low-power view, and panels e and f high-power view of other fields) or GagΔp1-p6 (panels g low-power view, and h and i high-power view of other fields) were processed by immunogold double labeling with an antibody to Gag (6-nm gold; indicated by white arrowheads) and cellular ALIX (15-nm gold; indicated by black arrows). Scale bars correspond to 200 nm (panels g, h, i) or 500 nm (a, b, c, d, e, f); (PM) to PM and (Cyt) to cytoplasm. Gag-ALIX colocalization was evaluated by counting gold particles of ~ 300 assembly events at the PM (J).
Syntenin PDZ1 replaces NC functionally in HIV-1 budding
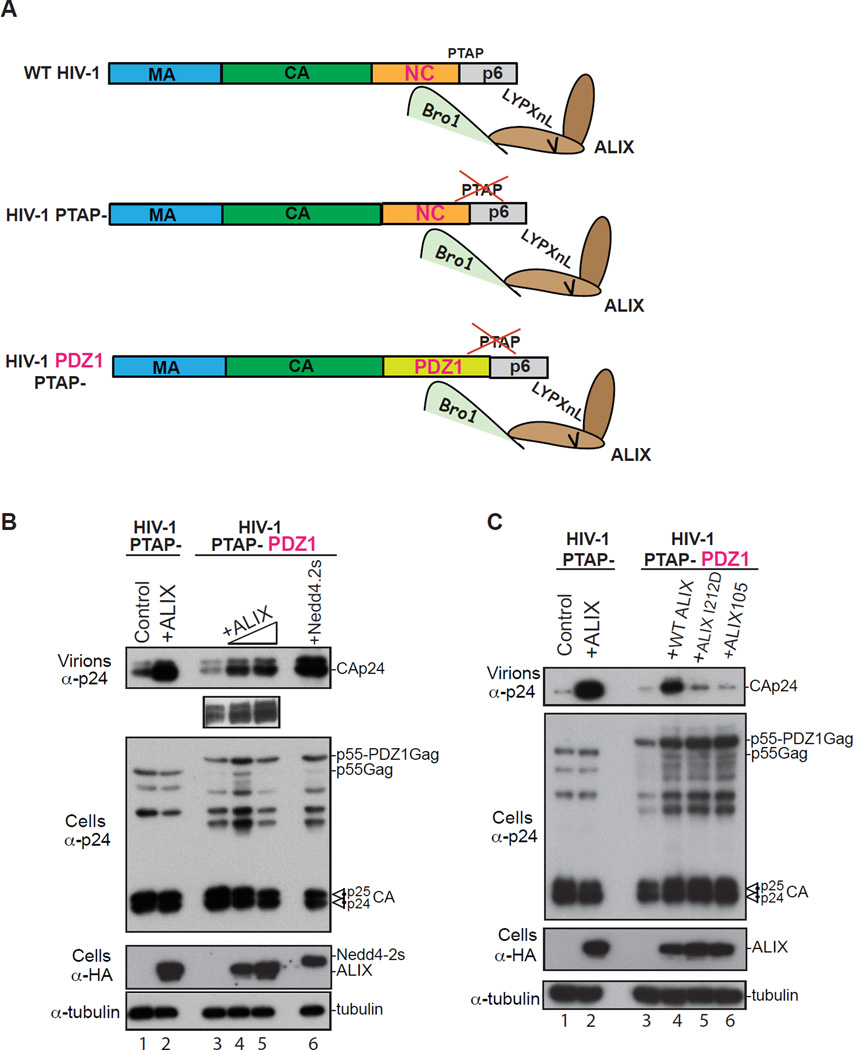
Our studies indicate that cellular syntenin PDZs and viral NC share several biochemical and functional properties that support a mimicry model. To further test this model, we examined whether syntenin PDZs can substitute for NC functionally in HIV-1 budding. An HIV-1 chimera construct where the Gag NC domain was replaced with syn-2 PDZ1, a domain that retained binding to Bro1 (Figure 1E) was constructed and tested in virus budding assays (Figure 6A). Remarkably, ectopic expression of ALIX stimulated the budding of an HIV-1 PDZ1 lacking the PTAP in a dose-dependent manner (Figure 6B, lanes 4–5 and inset) and as well as the HIV-1 PTAP- control carrying a WT NC domain (lanes 1 and 2) suggesting that ALIX Bro1 utilized syntenin PDZ1 in lieu of NC to stimulate HIV-1 budding. Ectopic expression of Nedd4.s also robustly stimulated virus budding (lane 6) suggesting that the HIV-1 Gag-PDZ1 chimera is assembly competent. Disruption of the ESCRT-III factor CHMP4 binding site or the membrane anchoring Phe105 loop in ALIX Bro1 (Figure 6C, lanes 5 and 6, respectively) significantly inhibited ALIX function in virus budding confirming that similarly to its WT counterpart, Gag-PDZ1 chimera virus requires access to ESCRTs and insertion of ALIX Bro1 at the membrane to promote scission of virus necks. These data indicate that the membrane adaptor syn-2-PDZ domain can function in lieu of HIV-1 NC within Gag and utilize Bro1/ESCRTs to promote ALIX-mediated HIV-1 budding.
Figure 6.
Syn-2 PDZ1 substitutes for NC functionally and promotes ALIX-mediated HIV-1 budding. (A) Drawing of HIV-1 constructs used in this experiment and their ability to bind ALIX. ALIX engages in dual binding with WT HIV-1 NC and p6 domains, can stimulate the release of the PTAP- mutant. HIV-1 PDZ1 chimera where NC was replaced with Syn-2 PDZ1 is shown (B) Syn-2 PDZ1 substitutes for NC functionally. 293T cells were transfected with either HIV-1 PTAP- (lane 1) or with HA-ALIX (lane 2), with HIV-1 PTAP-PDZ1 chimera alone (lane 3), with increasing amounts of HA-ALIX (lanes 4 and 5) or HA-Nedd4-2s (lane 6). Cells, virus and input were analyzed by SDS-PAGE and WB as indicated. Inset shows the WB of higher amounts of virus released (lanes 3–5). (C) Gag-PDZ1 requires access to membrane- and ESCRT-binding ALIX Bro1 to promote HIV-1 budding. Experiments identical to those described in B were performed with the addition of ALIX I212D and ALIX105 (lanes 5, 6).
Discussion
Here we identified the native cellular partner of ALIX Bro1 as syntenin PDZs, a PI(4,5)P2-binding scaffold domains that coordinate important cell signaling pathways at the membrane (Beekman and Coffer, 2008; Friand et al., 2015; Philley et al., 2015; Wawrzyniak et al., 2013). Bro1-PDZ(s) co-association occurs in PI(4,5)P2 and cholesterol-rich membranes. In a remarkable parallel, we found that HIV-1 NC binding to anionic phospholipids is critical for the capture of Bro1, whose own anchorage at the membrane in a cholesterol-dependent manner is key for ALIX-mediated HIV-1 budding. Domain-swapping experiments showed that Bro1-binding syn-2 PDZ1 functionally substitutes for NC and efficiently facilitated ESCRTs dependent ALIX-mediated HIV-1 budding. These studies support a model in which NC mimics syn-PDZ(s) to function as a membrane adaptor that co-opts ALIX Bro1/ESCRTs and coordinates the scission of virus budding necks in microdomains of the membrane that favor HIV-1 budding and transmission (Figure 7).
Figure 7.
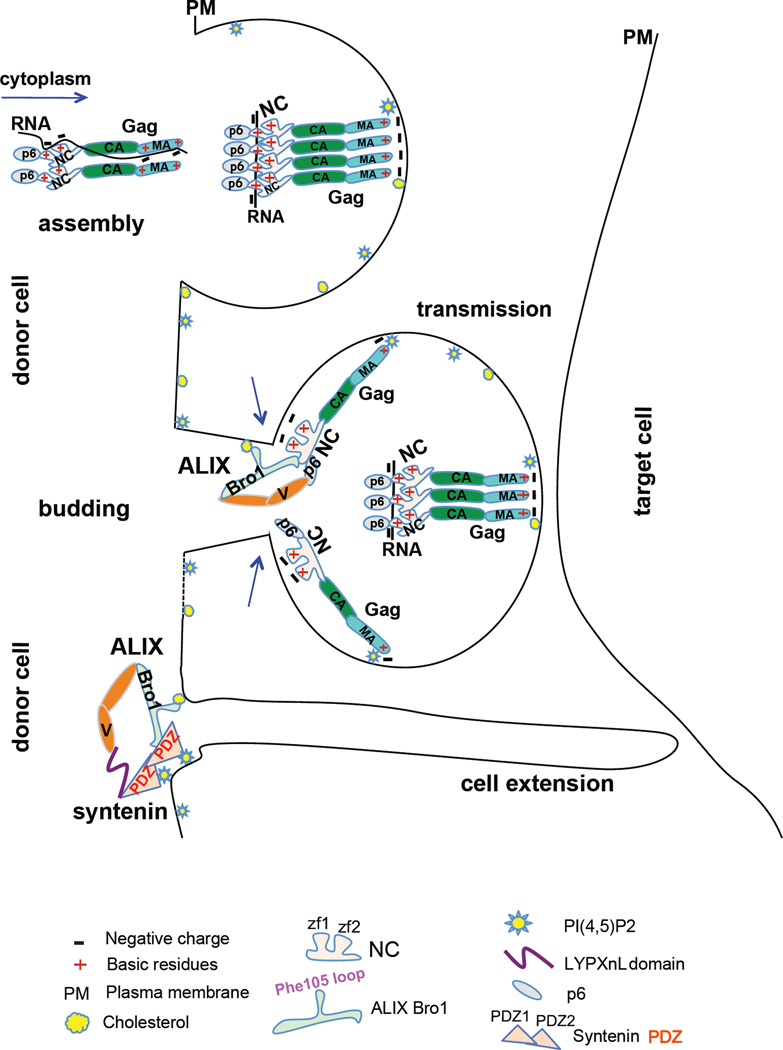
Striking parallels between NC viral function and syntenin PDZ cellular function. Gag assembly complexes associate with RNA in the cytoplasm, to stimulate Gag nucleation and prevent premature association with internal membranes. At the PM, Gag assembles in specific lipid-enriched (PI(4,5)P2 and cholesterol) domains to favor virus assembly, budding and transmission. Once a spherical particle forms, a subset of NC molecules become positioned closer to the membrane at the top of budding necks and dynamically trade RNA for lipids of the PM, where NC and ALIX Bro1 co-insert in cholesterol-rich domains. This process is known to be transient and precedes ESCRT-III recruitment and catalysis of membrane scission. In the cell, ALIX captures its cellular partner syntenin, whose PDZs associate with PI(4,5)P2-rich domains of the membrane where—like NC—it engages Bro1 to promote important signaling pathways pertinent to health and disease (i.e. cell-to-cell extension)
Several lines of evidence support the notion that NC-Bro1 interaction links Gag to the membrane and ESCRT-III in viral budding necks (Dussupt et al., 2009; Fisher et al., 2007; Lee et al., 2007; Popov et al., 2008; Sette et al., 2012): i) mutations of residues in NC that prevent Bro1 binding also halt ALIX-mediated virus budding; ii) ectopic expression of Bro1 stimulated the budding of an HIV-1 lacking all L domains, provided Gag carried a functional NC; and iii) Bro1 requires access to ESCRT-III to mediate NC-dependent HIV-1 release. In the current study, we found that ALIX engages in dual interactions with syntenin whereby LYPXnL motifs bind ALIX V domain while PDZ(s) captures Bro1 at the membrane, a mode of recruitment strikingly similar to that observed between ALIX and Gag NC-p6 domains (Figure 1). ALIX binding to syntenin LYPXnL-motifs is prominent while NC or PDZs-Bro1 interaction(s), although unambiguously detected, is discrete, owing to their transient and dynamic nature in facilitating interaction with the membrane and coordinating ESCRT-III function. We concluded that ALIX likely uses LYPXnL-V binding as a strong “anchorage point” and a lipid-binding NC/PDZ(s)-Bro1 “flexible arm” to dynamically and transiently recruit ESCRT(s) and catalyze cellular or viral membrane budding.
Our studies of ALIX Bro1-syntenin PDZ(s) interactions helped elucidate Bro1 mechanisms of function in HIV-1 budding. Membrane binding plays a key role in ALIX Bro1 ability to interact with syntenin PDZs, as disruption of the Bro1 Phe105 loop, which binds the exclusively endosomal lipid LBPA (Bissig et al., 2013) interfered with binding PDZs. Since the tip of the Phe105 loop is ultimately occupied by a lipid, we concluded that the strict requirement for the Phe105 loop in binding syn-2 PDZ(s) (Figure 2) might be due to a two step-process in which the loop first captures PDZs and second inserts in the membrane to help expose PDZs binding interface in Bro1 and establish stable interaction(s). PDZs and Bro1 require membrane-binding determinants to function in the cell (Baietti et al., 2012; Bissig et al., 2013; Zimmermann et al., 2002; Zimmermann et al., 2001), suggesting both factors likely co-anchor at the membrane to perform membrane budding (Figure 2E). Similarly, NC-mediated HIV-1 budding requires a functional Bro1 Phe105 loop (Sette et al., 2011a), and its insertion in membrane containing-cholesterol (Figure 2), a component of lipid rafts important for HIV-1 genesis (Dick et al., 2012; Ono and Freed, 2001; Ono et al., 2007) and transmission (Jolly and Sattentau, 2005). These findings unravel molecular mechanisms of ALIX Bro1 function at HIV-1 budding necks.
ALIX-Bro1 interaction with the membrane-binding syntenin PDZs was the first clue that NC might also bind membrane. Using LUVs that re-create PM-like conditions, we showed that NC binds membrane in a PI(4,5)P2-independent manner. Mutation of basic residues in NC however halted membrane binding-- indicating anionic phospholipids (PS) key involvement-- ablated NC-Bro1 interactions and in turn ALIX-mediated HIV-1 budding (Figures 3 and 4). This suggested that only a membrane-binding competent NC captures Bro1; both domains interact and co-anchor at cholesterol and PS-rich lipid raft-like domains (Pike, 2003, 2004) to promote ALIX-mediated HIV-1 budding, in line with a model in which NC mimics PDZs to capture ALIX Bro1 and perform these function(s).
ALIX Bro1 appears to recognize specific structural features in its cellular and viral binding partners. Disruption of the proximal or distal zinc finger structure in NC or either syntenin PDZ domains, adversely affected interaction with Bro1 (Figures 1 and 3) indicating Bro1 recognizes specific feature(s) in its cellular and viral partner. Alternatively, conservation of PDZ structure (Kang et al., 2003) might be necessary for the proper folding and/or exposure of the Bro1 binding interface. While syntenin PDZ1 retained ~40% binding to Bro1, strong binding was observed only with the PDZ1-PDZ2 tandem. Similarly, maintaining NC zinc fingers, which depend on interaction with RNA or cellular membrane (Figure 4), appeared to be key to binding Bro1, emphasizing the importance of maintaining a signature structure in the capture of Bro1 at sites of function (eg: plasma or endosomal membrane).
Our data suggest that NC trades RNA for membrane to capture ALIX Bro1/ESCRTs. While maintaining zinc fingers structure through binding with RNA appears to be critical for NC’s ability to capture ALIX Bro1, preserving natural cellular membranes relieved NC-Bro1 interaction from RNA dependence (Figure 4), in agreement with previous studies where NC-Bro1 binding was performed in detergent-free conditions (Popov et al., 2008). These results suggested that NC can maintain its zinc fingers structure through interactions with RNA or membrane interchangeably. Accordingly, NC binding to membrane was enhanced with RNA removal and mutation of RNA-binding basic residues also inhibited binding to PM-like membranes suggesting that RNA regulates NC binding to membrane in a manner similar to that previously described for MA (Chukkapalli et al., 2010; Kutluay et al., 2014). An attractive model would be that NC trades RNA for lipid; RNA would be actively displaced when NC is near the PM (in pinched viral neck) and/or upon binding Bro1, which itself inserts in the membrane and might be facilitating NC positioning to bind lipid. As disruption of NC membrane-binding residues halted Bro1 capture and in turn ALIX viral function (Figure 3), the data functionally tie NC ability to dynamically trade RNA for membrane to capturing Bro1/ESCRTs and promoting scission of HIV-1 budding necks.
Our data favor a model in which NC uses mimicry to function as a membrane-binding adaptor, which coordinates HIV-1 budding. In the cell, PDZs function as scaffolds that target and cluster host factors at defined domains of the membrane, which facilitate important cell signaling including immunological synapse, cell-to-cell contact/adhesion, endosome/exosome biogenesis and cancer metastasis (Beekman and Coffer, 2008; Friand et al., 2015; Philley et al., 2015). In particular, syn-2 PDZ has the highest affinity for PI(4,5)P2 (Zimmermann et al., 2002), a PM lipid raft component that also favors virus assembly and transmission (Jolly and Sattentau, 2005; Ono et al., 2004). NC is sufficient for the recruitment of endogenous ALIX at HIV-1 budding sites at the PM, indicating that NC binds the native ALIX Bro1 (Figure 5), the natural syntenin PDZs partner. Bro1 also inserts in a cholesterol-rich lipid rafts (this study), and prefers binding syn-2 PDZ (Figure 1), suggesting the existence of a native cellular syn2-ALIX complex that plays important physiological role(s) at lipid-enriched, adhesion molecules-laden microdomains of the PM, where HIV-1 also selectively forms assembly and transmission platforms (Hubner et al., 2009). Interestingly, a role for NC in preferentially targeting Gag to such structures in T cells has been observed (Llewellyn et al., 2010) and remarkably hinged on NC’s retaining basic membrane-binding residues (this study, Figure 4). Thus NC likely participates in mechanisms of directing Gag assembly to domains that favor virus genesis and budding, and shares several properties with syn-2 PDZs (eg: binding ESCRT-adaptors). NC mimicry of PDZs membrane adaptor function(s) at HIV-1 budding necks is best illustrated with its functional replacement with syn-2 PDZ1 (Figure 6). ALIX promoted the ESCRTs-dependent release of an HIV-1 carrying syn-2 PDZ1 in lieu of NC, which is compatible with PDZ1’s ability to bind Bro1 (Figure 1), PI(4,5)P2 and to serve as a scaffolding adaptor for multiprotein complexes at specific domains of the PM (Kim and Sheng, 2004; Sheng and Sala, 2001). These three roles are analogously carried out by NC at HIV-1 budding sites, including the targeting of Gag assembly platforms (Llewellyn et al., 2010), the binding to PS-enriched lipid rafts (Pike, 2003, 2004) and co-anchoring therein with Bro1/ESCRTs (this study). In summary, we show evidence compatible with a role for NC in HIV-1 budding at the PM, where NC mimics syntenin PDZ to serve as a membrane adaptor that co-anchors with ALIX Bro1, to gain access to ESCRT-III and sever HIV-1 budding necks in specific microdomains of the membrane that also favor virus transmission (Figure 7), and possibly other cellular processes important in health and disease.
Experimental procedures
Proviral and expression vectors
syn-1 and syn-2 genes were obtained from (Open Biosystems). Syn-2β lacks syn-2 first 255 bps, and was generated by PCR. Syntenin genes were cloned in frame with Strep, 3XFlag or HA sequences. HIV-1 (pNL4-3 or Bru) and derived mutants, lacking L domains, harboring NC mutants or syn-2 PDZ1 (aa108-187) were generated by overlapping PCR as previously described in (Dussupt et al, 2009, and 2011) and (Reed et al., 2012). ALIX, Bro1 derived constructs and Nedd4.2s were previously described in (Sette et al., 2012; Sette et al., 2011a). Recombinant proteins were expressed using a modified pET30a(+)(EMD) [GB1, Strep and 6Xhis fusion tag] or pGEX-5X-2 (GE) vector.
Immunoprecipitation assays
Immunoprecipitations were conducted as previously described in (Sette et al., 2011b). 293T cells were transfected using Lipofectamine 2000. Cells were lysed in 0.5%-1% Igepal detergent buffer or mechanically in hypotonic buffer using a homogenizer (B) 48h post-transfection and lysates were incubated with HA, Flag (Sigma) or Strep (IBA) antibody-conjugated beads. After 4–6 washes, protein complexes were eluated using commercial peptides and immunoprecipitated (IP) complexes were examined by WB and antibodies indicated in figures.
Protein expression and purification and pull down assays
Strep or GST fusion proteins harboring syntenin, Bro1 and Bro1-V, NC, NC-p6 or derived mutants were expressed in BL21(DE3) pLysS E. coli (Stratagene) and purified in buffers described in Supplemental material. Interactions with Flag- or HA-fusion protein containing cell extracts were examined in pull down assays following the protocol previously described (Sette et al., 2012). Nuclease treatment was performed with 100U of Benzonase (Novagen) at 37C for 1h.
Virus release analysis
293T cells were transfected using Lipofectamine 2000 and cells were harvested 24 to 32h post-transfection. Cells and culture media were separated; released virus was pelleted by high-speed centrifugation (25000xg) at 4C and its protein content examined by WB. Buffer composition is provided in Supplemental material.
RNAi Depletion
2.5 × 106 cells/T25 were used for these experiments. siRNA duplexes (400 pmol used) against syn-1 and syn-2, respectively, were obtained from Sigma and Invitrogen. When shRNA technique was used, lentiviruses were produced by transfecting 293T cells with a mixture of p8.91, pMD.G and pGIPZ lentiviral vector (Open Biosystems) containing the appropriate shRNA sequence. All knockdowns were assessed by WB and probing for endogenous proteins.
Bro1 and NC binding to LUVs
Lipids were purchased from Avanti Polar Lipids (Alabaster, AL). Large unilamellar vesicles (100 nm) composed of POPE: POPS: POPC (1:1:1) or POPE: POPS: POPC: Cholesterol (28:34:28:10) or 5% PI(4,5)P2 were prepared as previously described (Philip and Scarlata, 2006). Binding affinity was determined by the increase in fluorescence intensity of a 130 nM CPM-labeled purified protein, as freshly extruded LUVs were incrementally added. A final concentration of RNAse at 0.1 µg/µl was used. Further details of protein-LUVs interactions are provided in Supplemental Material.
Immunogold labeling and transmission electron microscopy
Methods for virus infection followed by double immunolabeling to both Gag and endogenous ALIX have been described previously in detail in (Reed et al., 2012) and in Supplemental Material. TEM of 293T cells expressing HIV-1 or a mutant was performed as previously described in Dussupt et al., 2011.
Supplementary Material
Footnotes
Authors’ contributions
PS performed most experiments and co-analyzed data. SKO co-performed protein mapping and binding experiments. KN, VD, KC and JL performed immunogold labeling and electron microscopy experiments. SY and SS performed in vitro membrane binding assays. FB designed the project, performed PDZ chimera experiments, analyzed the data and wrote the paper.
References
- Aldovini A, Young RA. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J Virol. 1990;64:1920–1926. doi: 10.1128/jvi.64.5.1920-1926.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14:677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- Beekman JM, Coffer PJ. The ins and outs of syntenin, a multifunctional intracellular adaptor protein. J Cell Sci. 2008;121:1349–1355. doi: 10.1242/jcs.026401. [DOI] [PubMed] [Google Scholar]
- Bello NF, Dussupt V, Sette P, Rudd V, Nagashima K, Bibollet-Ruche F, Chen C, Montelaro RC, Hahn BH, Bouamr F. Budding of retroviruses utilizing divergent L domains requires nucleocapsid. J Virol. 2012;86:4182–4193. doi: 10.1128/JVI.07105-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissig C, Lenoir M, Velluz MC, Kufareva I, Abagyan R, Overduin M, Gruenberg J. Viral infection controlled by a calcium-dependent lipid-binding module in ALIX. Dev Cell. 2013;25:364–373. doi: 10.1016/j.devcel.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burniston MT, Cimarelli A, Colgan J, Curtis SP, Luban J. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J Virol. 1999;73:8527–8540. doi: 10.1128/jvi.73.10.8527-8540.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatellard-Causse C, Blot B, Cristina N, Torch S, Missotten M, Sadoul R. Alix (ALG-2-interacting protein X), a protein involved in apoptosis, binds to endophilins and induces cytoplasmic vacuolization. J Biol Chem. 2002;277:29108–29115. doi: 10.1074/jbc.M204019200. [DOI] [PubMed] [Google Scholar]
- Chukkapalli V, Oh SJ, Ono A. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc Natl Acad Sci U S A. 2010;107:1600–1605. doi: 10.1073/pnas.0908661107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimarelli A, Sandin S, Hoglund S, Luban J. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J Virol. 2000;74:3046–3057. doi: 10.1128/jvi.74.7.3046-3057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlix JL, Lapadat-Tapolsky M, de Rocquigny H, Roques BP. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J Mol Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- Dick RA, Goh SL, Feigenson GW, Vogt VM. HIV-1 Gag protein can sense the cholesterol and acyl chain environment in model membranes. Proc Natl Acad Sci U S A. 2012;109:18761–18766. doi: 10.1073/pnas.1209408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussupt V, Javid MP, Abou-Jaoude G, Jadwin JA, de La Cruz J, Nagashima K, Bouamr F. The nucleocapsid region of HIV-1 Gag cooperates with the PTAP and LYPXnL late domains to recruit the cellular machinery necessary for viral budding. PLoS Pathog. 2009;5:e1000339. doi: 10.1371/journal.ppat.1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussupt V, Sette P, Bello NF, Javid MP, Nagashima K, Bouamr F. Basic residues in the nucleocapsid domain of Gag are critical for late events of HIV-1 budding. J Virol. 2011;85:2304–2315. doi: 10.1128/JVI.01562-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RD, Chung HY, Zhai Q, Robinson H, Sundquist WI, Hill CP. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 2007;128:841–852. doi: 10.1016/j.cell.2007.01.035. [DOI] [PubMed] [Google Scholar]
- Friand V, David G, Zimmermann P. Syntenin and syndecan in the biogenesis of exosomes. Biol Cell. 2015 doi: 10.1111/boc.201500010. [DOI] [PubMed] [Google Scholar]
- Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- Gorelick RJ, Henderson LE, Hanser JP, Rein A. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a "zinc finger-like" protein sequence. Proc Natl Acad Sci U S A. 1988;85:8420–8424. doi: 10.1073/pnas.85.22.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlinger HG, Dorfman T, Sodroski JG, Haseltine WA. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci U S A. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grootjans JJ, Zimmermann P, Reekmans G, Smets A, Degeest G, Durr J, David G. Syntenin, a PDZ protein that binds syndecan cytoplasmic domains. Proc Natl Acad Sci U S A. 1997;94:13683–13688. doi: 10.1073/pnas.94.25.13683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Orenstein JM, Martin MA, Freed EO. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner W, McNerney GP, Chen P, Dale BM, Gordon RE, Chuang FY, Li XD, Asmuth DM, Huser T, Chen BK. Quantitative 3D video microscopy of HIV transfer across T cell virological synapses. Science. 2009;323:1743–1747. doi: 10.1126/science.1167525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Li F, Mothes W. Viral determinants of polarized assembly for the murine leukemia virus. J Virol. 2011;85:7672–7682. doi: 10.1128/JVI.00409-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C, Sattentau QJ. Human immunodeficiency virus type 1 virological synapse formation in T cells requires lipid raft integrity. J Virol. 2005;79:12088–12094. doi: 10.1128/JVI.79.18.12088-12094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C, Sattentau QJ. Human immunodeficiency virus type 1 assembly, budding, and cell-cell spread in T cells take place in tetraspanin-enriched plasma membrane domains. J Virol. 2007;81:7873–7884. doi: 10.1128/JVI.01845-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N, Neil SJ, Bess C, Johnson MC, Virgen CA, Simon SM, Bieniasz PD. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol. 2006;4:e435. doi: 10.1371/journal.pbio.0040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang BS, Cooper DR, Jelen F, Devedjiev Y, Derewenda U, Dauter Z, Otlewski J, Derewenda ZS. PDZ tandem of human syntenin: crystal structure and functional properties. Structure. 2003;11:459–468. doi: 10.1016/s0969-2126(03)00052-2. [DOI] [PubMed] [Google Scholar]
- Kempf N, Postupalenko V, Bora S, Didier P, Arntz Y, de Rocquigny H, Mely Y. The HIV-1 nucleocapsid protein recruits negatively charged lipids to ensure its optimal binding to lipid membranes. J Virol. 2015;89:1756–1767. doi: 10.1128/JVI.02931-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Sheng M. PDZ domain proteins of synapses. Nat Rev Neurosci. 2004;5:771–781. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- Kim J, Sitaraman S, Hierro A, Beach BM, Odorizzi G, Hurley JH. Structural basis for endosomal targeting by the Bro1 domain. Dev Cell. 2005;8:937–947. doi: 10.1016/j.devcel.2005.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutluay SB, Zang T, Blanco-Melo D, Powell C, Jannain D, Errando M, Bieniasz PD. Global changes in the RNA binding specificity of HIV-1 gag regulate virion genesis. Cell. 2014;159:1096–1109. doi: 10.1016/j.cell.2014.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Joshi A, Nagashima K, Freed EO, Hurley JH. Structural basis for viral late-domain binding to Alix. Nat Struct Mol Biol. 2007;14:194–199. doi: 10.1038/nsmb1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llewellyn GN, Hogue IB, Grover JR, Ono A. Nucleocapsid promotes localization of HIV-1 gag to uropods that participate in virological synapses between T cells. PLoS Pathog. 2010;6:e1001167. doi: 10.1371/journal.ppat.1001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J, Yarovoy A, Perez-Caballero D, Bieniasz PD. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc Natl Acad Sci U S A. 2003;100:12414–12419. doi: 10.1073/pnas.2133846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J, Zang T, Bieniasz PD. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- Morita E, Sandrin V, McCullough J, Katsuyama A, Baci Hamilton I, Sundquist WI. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe. 2011;9:235–242. doi: 10.1016/j.chom.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortier E, Wuytens G, Leenaerts I, Hannes F, Heung MY, Degeest G, David G, Zimmermann P. Nuclear speckles and nucleoli targeting by PIP2-PDZ domain interactions. EMBO J. 2005;24:2556–2565. doi: 10.1038/sj.emboj.7600722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriaux D, Mirro J, Harvin D, Rein A. RNA is a structural element in retrovirus particles. Proc Natl Acad Sci U S A. 2001;98:5246–5251. doi: 10.1073/pnas.091000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc Natl Acad Sci U S A. 2004;101:14889–14894. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Freed EO. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc Natl Acad Sci U S A. 2001;98:13925–13930. doi: 10.1073/pnas.241320298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A, Freed EO. Role of lipid rafts in virus replication. Adv Virus Res. 2005;64:311–358. doi: 10.1016/S0065-3527(05)64010-9. [DOI] [PubMed] [Google Scholar]
- Ono A, Waheed AA, Freed EO. Depletion of cellular cholesterol inhibits membrane binding and higher-order multimerization of human immunodeficiency virus type 1 Gag. Virology. 2007;360:27–35. doi: 10.1016/j.virol.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philley J, Kannan A, Dasgupta S. MDA-9/Syntenin Control. J Cell Physiol. 2015 doi: 10.1002/jcp.25136. [DOI] [PubMed] [Google Scholar]
- Pike LJ. Lipid rafts: bringing order to chaos. J Lipid Res. 2003;44:655–667. doi: 10.1194/jlr.R200021-JLR200. [DOI] [PubMed] [Google Scholar]
- Pike LJ. Lipid rafts: heterogeneity on the high seas. Biochem J. 2004;378:281–292. doi: 10.1042/BJ20031672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires R, Hartlieb B, Signor L, Schoehn G, Lata S, Roessle M, Moriscot C, Popov S, Hinz A, Jamin M, et al. A crescent-shaped ALIX dimer targets ESCRT-III CHMP4 filaments. Structure. 2009;17:843–856. doi: 10.1016/j.str.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov S, Popova E, Inoue M, Gottlinger HG. Human immunodeficiency virus type 1 Gag engages the Bro1 domain of ALIX/AIP1 through the nucleocapsid. J Virol. 2008;82:1389–1398. doi: 10.1128/JVI.01912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JC, Molter B, Geary CD, McNevin J, McElrath J, Giri S, Klein KC, Lingappa JR. HIV-1 Gag co-opts a cellular complex containing DDX6, a helicase that facilitates capsid assembly. J Cell Biol. 2012;198:439–456. doi: 10.1083/jcb.201111012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad JS, Muriaux DM. Editorial: Role of lipids in virus assembly. Front Microbiol. 2015;6:410. doi: 10.3389/fmicb.2015.00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette P, Dussupt V, Bouamr F. Identification of the HIV-1 NC binding interface in Alix Bro1 reveals a role for RNA. J Virol. 2012;86:11608–11615. doi: 10.1128/JVI.01260-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette P, Mu R, Dussupt V, Jiang J, Snyder G, Smith P, Xiao TS, Bouamr F. The Phe105 loop of Alix Bro1 domain plays a key role in HIV-1 release. Structure. 2011b;19:1485–1495. doi: 10.1016/j.str.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, Sala C. PDZ domains and the organization of supramolecular complexes. Annu Rev Neurosci. 2001;24:1–29. doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- Summers MF, South TL, Kim B, Hare DR. High-resolution structure of an HIV zinc fingerlike domain via a new NMR-based distance geometry approach. Biochemistry. 1990;29:329–340. doi: 10.1021/bi00454a005. [DOI] [PubMed] [Google Scholar]
- Thomas A, Mariani-Floderer C, Lopez-Huertas MR, Gros N, Hamard-Peron E, Favard C, Ohlmann T, Alcami J, Muriaux D. Involvement of the Rac1-IRSp53-Wave2-Arp2/3 Signaling Pathway in HIV-1 Gag Particle Release in CD4 T Cells. J Virol. 2015;89:8162–8181. doi: 10.1128/JVI.00469-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami Y, Popov S, Gottlinger HG. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J Virol. 2007;81:6614–6622. doi: 10.1128/JVI.00314-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VerPlank L, Bouamr F, LaGrassa TJ, Agresta B, Kikonyogo A, Leis J, Carter CA. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag) Proc Natl Acad Sci U S A. 2001;98:7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schwedler UK, Stuchell M, Muller B, Ward DM, Chung HY, Morita E, Wang HE, Davis T, He GP, Cimbora DM, et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- Votteler J, Sundquist WI. Virus budding and the ESCRT pathway. Cell Host Microbe. 2013;14:232–241. doi: 10.1016/j.chom.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrzyniak AM, Kashyap R, Zimmermann P. Phosphoinositides and PDZ domain scaffolds. Adv Exp Med Biol. 2013;991:41–57. doi: 10.1007/978-94-007-6331-9_4. [DOI] [PubMed] [Google Scholar]
- Yandrapalli N, Muriaux D, Favard C. Lipid domains in HIV-1 assembly. Front Microbiol. 2014;5:220. doi: 10.3389/fmicb.2014.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q, Fisher RD, Chung HY, Myszka DG, Sundquist WI, Hill CP. Structural and functional studies of ALIX interactions with YPX(n)L late domains of HIV-1 and EIAV. Nat Struct Mol Biol. 2008;15:43–49. doi: 10.1038/nsmb1319. [DOI] [PubMed] [Google Scholar]
- Zhai Q, Landesman MB, Chung HY, Dierkers A, Jeffries CM, Trewhella J, Hill CP, Sundquist WI. Activation of the retroviral budding factor ALIX. J Virol. 2011a;85:9222–9226. doi: 10.1128/JVI.02653-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q, Landesman MB, Robinson H, Sundquist WI, Hill CP. Structure of the Bro1 domain protein BROX and functional analyses of the ALIX Bro1 domain in HIV-1 budding. PLoS One. 2011b;6:e27466. doi: 10.1371/journal.pone.0027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Meerschaert K, Reekmans G, Leenaerts I, Small JV, Vandekerckhove J, David G, Gettemans J. PIP(2)-PDZ domain binding controls the association of syntenin with the plasma membrane. Mol Cell. 2002;9:1215–1225. doi: 10.1016/s1097-2765(02)00549-x. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Tomatis D, Rosas M, Grootjans J, Leenaerts I, Degeest G, Reekmans G, Coomans C, David G. Characterization of syntenin, a syndecan-binding PDZ protein, as a component of cell adhesion sites and microfilaments. Mol Biol Cell. 2001;12:339–350. doi: 10.1091/mbc.12.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.