Abstract
Background:
Ankylosing spondylitis (AS) is the most common rheumatic condition that is slowly progressive and predominantly affects adolescents. Pathological bone formation associated with AS is an important cause of disability. The aim of the study was to investigate the possible involvement of the genes related to endochondral ossification and ectopia ossification in genetic susceptibility to AS in a Chinese Han population.
Methods:
Sixty-eight single nucleotide polymorphisms (SNPs) from 13 genes were genotyped in discovery cohorts including 300 AS patients and 180 healthy controls. The rs10019009 in dentin matrix protein 1 (DMP1) gene shown as association with AS after multiple testing corrections in discovery cohorts was replicated in a validation independent cohort of 620 AS patients and 683 healthy controls. The rs10019009 was assessed with bioinformatics including phylogenetic context, F-SNP and FastSNP functional predictions, secondary structure prediction, and molecular modeling. We performed a functional analysis of rs10019009 via reverse transcription-polymerase chain reaction, alkaline phosphatase (ALP) activity in human osteosarcoma U2OS cells.
Results:
Interestingly, the SNP rs10019009 was associated with AS in both the discovery cohort (P = 0.0012) and validation cohort (P = 0.0349), as well as overall (P = 0.0004) in genetic case–control association analysis. After a multivariate logistic regression analysis, the effect of this genetic variant was observed to be independent of linkage disequilibrium. Via bioinformatics analysis, it was found that the amino acid change of the rs10019009 led to changes of SNP function, secondary structure, tertiary conformation, and splice mode. Finally, functional analysis of rs10019009 in U2OS cells demonstrated that the risk T allele of the rs10019009 increased enzymatic activity of ALP, compared to that of the nonrisk allele (P = 0.0080).
Conclusions:
These results suggested that the DMP1 gene seems to be involved in genetic predisposition to AS, which may contribute to the ectopic mineralization or ossification in AS. In addition, DMP1 gene may be a promising intervention target for AS in the future.
Keywords: Alkaline Phosphatase, Ankylosing Spondylitis, Dentin Matrix Protein 1, Ectopic Mineralization, Polymorphisms
INTRODUCTION
Ankylosing spondylitis (AS) is a highly heritable, common rheumatic condition that primarily affects the axial skeleton and is sometimes accompanied by peripheral arthritis, enthesis and iritis, spinal deformity, and ankylosis. Pathological bone formation associated with AS is an important cause of disability. To date, susceptible gene studies of AS have primarily involved inflammation and immunization, whereas few experiments have been performed in the field of ossification. Although tumor necrosis factor-alpha (TNF-α) antagonists can provide long-term control of clinical symptoms and systemic inflammation in most AS patients, neither etanercept nor infliximab could be able to significantly slow the radiological progression of AS over a 2-year treatment period.[1] In addition, it remains unclear whether the genes other than those related to inflammation and immunization, such as ossification genes, are involved in the pathogenic mechanism of AS.
In this regard, we considered the following points: (1) ossification initiation in AS depends on inflammation, whereas ossification development in AS is independent of inflammation.[1] Ossification has its own genetic background.[2] (2) The essence of ossification in AS is predominantly endochondral ossification.[3] Histological studies have demonstrated that endochondral and direct bone formation contribute to ankylosis in spondyloarthritis.[4] (3) Maladjustment of the signaling pathway related to endochondral ossification and loss of coupling between osteoblasts and osteoclast have been identified in AS.[5,6] (4) Diarra et al.[7] demonstrated that inhibition of dickkopf-1 (DKK-1), a regulatory molecule of the Wnt pathway, reversed the bone-destructive pattern of rheumatoid arthritis to the bone-forming pattern of osteoarthritis in a mouse model. Blockade of DKK-1 induced sacroiliac joint fusion. (5) Indian hedgehog (IHH) and parathyroid hormone-related peptide participate in a feedback loop that coordinates chondrocyte proliferation and differentiation in endochondral bone development. (6) AS manifests as hydroxyapatite (HA) crystal deposition in the soft tissues of tendons and/or ligaments.[8] Dentin matrix protein 1 (DMP1) plays critical roles in the formation of collagenous mineralized tissues. Matrix Gla protein (MGP) is expressed by chondrocytes and vascular smooth muscle cells to inhibit soft tissue calcification. (7) The critical involvement of overactive bone morphogenetic protein (BMP) signaling in ectopic bone formation is well-established, and several studies have identified BMP2 and BMP4 as susceptible genes for the ossification of the posterior longitudinal ligament. (8) Following transforming growth factor-β/BMP induction, the Smad and p38 mitogen-activated protein kinase pathways converge at the runt-related transcription factor 2 (RUNX2) gene to control mesenchymal precursor cell differentiation. In this study, we aimed to assess genetic associations between candidate genes related to endochondral ossification and ectopic ossification and AS in a Chinese Han population from Shandong Province.
METHODS
Candidate gene selection strategy
Previous studies have demonstrated the selection of susceptibility genes of heterotopic ossification within the susceptible region of AS, including BMP6, RUNX2, IHH, and collagen, type XI, alpha 2 (COL11A2) genes.[9,10,11,12,13] Other important genes and transcription factors involved in signaling pathways related to selected endochondral ossification included the following: DKK1 in the Wnt signaling pathway; BMP6, BMP4, and BMP2 in the BMP signaling pathway; DMP1 and MGP in the matrix mineralization pathway; IHH in the hedgehog signaling pathway; and RUNX2- and SRY-related high mobility group-box gene 9 in transcription factors related to ossification. In addition, susceptibility genes of diseases related to ectopic ossification were chosen, including activin receptor-like kinase 2 (ACVR1) and COL11A2.[14,15] Prostaglandin-endoperoxide synthase 2, an effective target for the treatment of heterotopic ossification, was also selected. Anthrax toxin receptor 2 (ANTXR2) showed a positive locus in a genome-wide scan of other races.[12] Ultimately, we selected 13 candidate genes related to ossification to determine genetic associations with AS.
Study samples
A total of 300 AS patients (the case group) and 180 ethnically matched healthy controls (the control group) were enrolled in the discovery cohort. The validation cohort included 620 AS patients and 683 healthy controls. All study subjects were all Chinese Han from Shandong Province, China. Diagnosis of AS met the modified New York criteria.[16] As previously described,[17] information was collected systematically in the discovery and validation cohorts [Table 1].
Table 1.
Demographic characteristics of all study subjects
| Characteristics | Discovery cohort | Validation cohort | ||
|---|---|---|---|---|
| Cases group (n = 300) | Control group (n = 180) | Cases group (n = 620) | Controls group (n = 683) | |
| Gender, n | ||||
| Male | 231 (77) | 135 (75) | 440 (71) | 532 (78) |
| Female | 69 (23) | 45 (25) | 180 (29) | 150 (22) |
| Age (years), (%) mean ± SD | 29.2 ± 9.9 | 24.7 ± 6.7 | 23.2 ± 8.9 | 27.3 ± 9.3 |
| Onset type, n (%) | ||||
| Juvenile onset | 120 (40) | – | 130 (21) | – |
| Adult onset | 180 (60) | – | 490 (79) | – |
| B-27, n (%) | ||||
| Negative | 24 (8) | – | 56 (9) | – |
| Positive | 276 (92) | – | 564 (91) | – |
| Kyphosis, n (%) | ||||
| Negative | 174 (58) | – | 508 (82) | – |
| Positive | 126 (42) | – | 112 (18) | – |
| Bamboo spine, n (%) | ||||
| Negative | 201 (67) | – | 428 (69) | – |
| Positive | 99 (33) | – | 192 (31) | – |
| Radiographic grading of sacroiliitis, n (%) | ||||
| 2/3 | 126 (42) | – | 366 (59) | – |
| 4 | 174 (58) | – | 254 (41) | – |
SD: Standard deviation; –: Not applicable.
This study was approved by the Ethical Committee of Shandong Academy of Medicinal Sciences. All study subjects provided their written consent to participate in the study and to allow their biological samples to be genetically analyzed.
Single nucleotide polymorphism selection
Tag single nucleotide polymorphisms (SNPs) covering the genomic regions of selected genes (including 3000 base pairs upstream of the first exon) were selected using Tagger in Haploview 4.0 (Broad Institute of MIT and Harvard, Cambridge, Massachusetts, USA). The criterion for r2 was set at 0.8. SNPs within the promoter, coding region, splice sites, and missense and nonsense sites were preferentially selected. SNPs with a multiple testing correction of P < 0.05 and statistical power greater than 80% were subjected to replication.
Genotyping
A total of 68 SNPs in 13 genes were genotyped using the Illumina GoldenGate typing platform in the discovery cohort, and we used TaqMan-polymerase chain reaction (PCR) in the validation cohort. The probes for rs10019009 were ordered from Applied Biosystems (Applied Biosystems, Carlsbad, California, USA).
Bioinformatics analysis
Alignment of the phylogenetic context of rs10019009 was obtained from the Ensembl genome browser (http://asia.ensembl.org/Homo_sapiens/Variation/Compara_Alignments?align = 582 and db = core and g = ENSG00000152592 and r = 4%3A88571459-88585513 and t = ENST00000339673 and v = rs10019009 and vdb = variation and vf = 44284457). The function prediction of rs10019009 was performed using F-SNP (http://compbio.cs.queensu.ca/F-SNP/) and FastSNP (http://fastsnp.ibms.sinica.edu.tw/OnlineAnalysis/AgentOutput.jsp?taskid = TK59976 and objectName = rs10019009 and RiskType = 12) online. Predictions of secondary and tertiary structures were generated using the Phyre version 2 web server (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id = index).[18]
Investigation of rs10019009 function
Plasmid construction
We constructed the expression plasmid pEGFP-1-DMP1. DMP1 expression vectors containing the p.Cys69, p.Arg69, and p.Gly69 variants were generated using site-directed mutagenesis (Beyotime Biotechnology, China) with the use of wild-type (WT) human DMP1 (containing p.Ser69) complementary DNA (cDNA) as the template. All new clones were confirmed using direct sequencing. The primer sets for site-directed mutagenesis of rs10019009 are listed in Supplementary Table 1. Ser69 and Cys69 variants are WT model variants, whereas Arg69 and Gly69 are mutation model variants. We examined the four variants at position 69 to evaluate the impact of changes at this residue on the function of the entire DMP1 protein.
Supplementary Table 1.
The primer sets used for site-directed mutagenesis of rs10019009
| Variant or mutant | Primer sets |
|---|---|
| Cys69 | 5’atgaagaccccagtgactgcactcagtcagaggagg 3’ (sense) |
| 5’cctcctctgactgagtgcagtcactggggtcttcat 3’ (antisense) | |
| Arg69 | 5’ atgaagaccccagtgaccgcactcagtcagaggagg 3’ (sense) |
| 5’ cctcctctgactgagtgcggtcactggggtcttcat 3’ (antisense) | |
| Gly69 | 5’ tgaagaccccagtgacggcactcagtcagaggag 3’ (sense) |
| 5’ ctcctctgactgagtgccgtcactggggtcttca 3’ (antisense) |
The triplet codon of the mutant amino acid is in bold.
Cell culture
Human osteoblastic osteosarcoma cell line U2OS was cultured in Dulbecco's Modified Eagle's Medium with high glucose (4.5 g/L) supplemented with 10% fetal calf serum (FCS). Ascorbic acid, dexamethasone, 1,25-dihydroxyvitamin D3, and β-glycerophosphate were not added to the culture medium. Cells were plated in a 24-well plate and 90% were confluent at the time of transfection (Lipofectamine 2000, Invitrogen, USA). pEGFP-1 not containing the DMP1 cDNA was used for mock transfections; non-transfected cells were used as controls. The transfection efficacy was quantified as the transfected cells/all cells in one microscope field ×100%.
Alkaline phosphatase activity
Cells were harvested 120 h after transfection, and ALP activity was assessed at 37°C using 10 mmol/L pNPP (Sigma, USA) as the substrate. The protein concentration was determined using a protein assay kit (Beyotime Biotechnology). ALP activity is expressed as nmol·min−1·mg−1. The enzymatic activity of ALP was assessed in five different experiments.
Reverse transcription-polymerase chain reaction
Total RNA was extracted using an RNeasy Mini Kit (Omega, Georgia, USA), and reverse transcription-PCR (RT-PCR) was performed. Messenger RNA (mRNA) was amplified using the primers described in Supplementary Table 2, and glyceraldehyde-3-phosphate dehydrogenase mRNA was used as an internal standard.
Supplementary Table 2.
The primer sets for genes related to ossification used in RT-PCR
| Genes | Primer sets |
|---|---|
| DMP1 | 5’AGTGCCCAAGATACCACCAG3’ (sense) |
| 5’CATTCCCTCATCGTCCAACT3’ (antisense) | |
| RUNX2 | 5’GAATGCTTCATTCGCCTCACA3’ (sense) |
| 5’TGACCTGCGGAGATTAACCAT3’ (antisense) | |
| OCN | 5’AAGAGACCAAGGCGCTACCT3’ (sense) |
| 5’GCCGATAGGCCTCCTTGAAAG3’ (antisense) | |
| BMP2 | 5’CCCCTACATGCTAGACCTGTATCG3’ (sense) |
| 5’CACTCGTTTCTGGTAGTTCTTCCA’ (antisense) | |
| BMP6 | 5’CGGGTCTCCAGTGCTTCAGATT3’ (sense) |
| 5’GCATCCACAAGCTCTTACAACC3’ (antisense) | |
| GAPDH | 5’TGAACGGGAAGCTCACTGG3’ (sense) |
| 5’TCCACCACCCTGTTGCTGTA3’ (antisense) |
In the discovery cohort, we found an association between RUNX2, BMP2, and BMP6 and AS. We explored whether the DMP1 gene promoted the expression of RUNX2, BMP2, and BMP6 genes in transfected U2OS cells to examine the downstream pathways of DMP1 and to clarify the ectopic ossification mechanisms of AS. We detected the mRNA expression of OCN in transfected U2OS cells because the OCN protein is a marker of the bone formation process. Many studies use OCN as a preliminary biomarker of the efficacy of a given drug on bone formation. RT–PCR: Reverse transcription–polymerase chain reaction; DMP1: Dentin matrix protein 1; RUNX2: Runtrelated transcription factor 2; OCN: Osteocalcin; GAPDH: Glyceraldehyde–3–phosphate dehydrogenase; mRNA: MessengerRNA.
Statistical analysis
An exact test for Hardy–Weinberg equilibrium was performed in healthy controls (P > 0.05). The allele and genotype frequencies of the AS patients and controls were compared using Fisher's exact test. We calculated the sample capacity of patients and healthy controls via PS: Power and Sample Size Calculation version 3.1.2, 2014 (http://biostat.mc.vanderbilt.edu/wiki/Main/PowerSampleSize). Permutation-based hypothesis testing (10,000 permutations) was performed for multiple testing corrections using the SHEsis permutation function (http://analysis.bio-x.cn/myAnalysis.php). We also performed linkage disequilibrium (LD) analysis in the case group using SHEsis software online, and we performed a multivariate logistic regression analysis to show whether the association was due to LD or to a true association with functionally relevant polymorphisms using SPSS version 17.0 (SPSS Inc., Chicago, IL, USA). Statistically significant differences between cells transfected with c.205A plasmids and c.205T, c.205C, or c.205G plasmids were estimated using nonparametric testing in SPSS 17.0.
RESULTS
Inclusion criteria
A total of 296 AS patients, 170 healthy controls, and 65 SNPs were retained for the final analyses in the discovery cohort after filtering (Hardy–Weinberg disequilibrium for healthy controls >0.05, minor allele frequency >0.05, call rate for individual sample >0.9, and call frequency for individual SNP >0.9). A total of 612 AS patients and 672 healthy controls were retained for the final analyses in the validation cohort after filtering.
Quality control
We randomly re-genotyped ten selected samples in the discovery cohort, and the reproducibility of the genotype was more than 96%. In the validation experimental group, we randomly replicated the ten genotyped samples in the discovery cohort, and the concordance rates were 100%. According to our experimental design and parameter settings, we had 80% power to detect SNP with odds ratio >1.5 or <0.64.
Association and linkage disequilibrium analyses
Case–control association analysis in the discovery cohort
SNP rs10019009 was significantly associated with AS (uncorrected P = 0.0012, corrected P = 0.0460, dominant P = 0.0012, and recessive P = 0.0360) with a power of 90%. However, the allele association of 4 other SNPs (rs16873348, rs35565233, rs6910759, and rs3178250) with AS disappeared after multiple test correction excluding rs10019009. An association of genotype frequencies of 3 SNPs (rs10019009, rs16873348, and rs35565233) was found [Supplementary Table 3]. Multivariate logistic regression analysis demonstrated that rs10019009 has an independent, key role compared to other loci within the DMP1 and ANTXR2 genes (P = 0.0030) [Supplementary Figure 1].
Supplementary Table 3.
Case–control association analyses in the discovery cohort
| Gene | Chromosome | SNP | Allele | Genotype | P | Pc | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele | Frequency (control) | Frequency (case) | P | Pc | OR | 95% CI | Case frequency | Control frequency | |||||||||
| 1/1 | 2/2 | 1/2 | 1/1 | 2/2 | 1/2 | ||||||||||||
| PTGS2 | 1 | rs5275 | A | 0.792 | 0.783 | 0.746 | 1.000 | 0.947 | 0.681–1.317 | 0.593 | 0.027 | 0.379 | 0.615 | 0.030 | 0.355 | 0.869 | 1.000 |
| PTGS2 | 1 | rs20417 | C | 0.938 | 0.935 | 0.871 | 1.000 | 0.956 | 0.552–1.653 | 0.876 | 0.007 | 0.117 | 0.875 | 0.000 | 0.125 | 0.554 | 1.000 |
| PTGS2 | 1 | rs689466 | A | 0.550 | 0.523 | 0.434 | 1.000 | 0.899 | 0.688–1.174 | 0.251 | 0.205 | 0.544 | 0.294 | 0.194 | 0.512 | 0.608 | 1.000 |
| PTGS2 | 1 | rs12042763 | A | 0.303 | 0.253 | 0.100 | 1.000 | 0.781 | 0.581–1.049 | 0.053 | 0.546 | 0.400 | 0.082 | 0.477 | 0.441 | 0.235 | 1.000 |
| ACVR1 | 2 | rs7595478 | A | 0.259 | 0.263 | 0.878 | 1.000 | 1.024 | 0.756–1.387 | 0.064 | 0.537 | 0.399 | 0.082 | 0.565 | 0.353 | 0.524 | 1.000 |
| ACVR1 | 2 | rs10933441 | A | 0.376 | 0.383 | 0.854 | 1.000 | 1.026 | 0.780–1.350 | 0.154 | 0.389 | 0.456 | 0.118 | 0.365 | 0.518 | 0.358 | 1.000 |
| ACVR1 | 2 | rs4380178 | A | 0.188 | 0.193 | 0.849 | 1.000 | 1.034 | 0.736–1.451 | 0.013 | 0.627 | 0.360 | 0.018 | 0.641 | 0.341 | 0.869 | 1.000 |
| ACVR1 | 2 | rs10497192 | A | 0.482 | 0.450 | 0.337 | 1.000 | 0.877 | 0.672–1.146 | 0.161 | 0.261 | 0.579 | 0.235 | 0.271 | 0.494 | 0.097 | 1.000 |
| IHH | 2 | rs3100776 | A | 0.524 | 0.540 | 0.627 | 1.000 | 1.068 | 0.818–1.395 | 0.283 | 0.203 | 0.513 | 0.265 | 0.218 | 0.518 | 0.884 | 1.000 |
| IHH | 2 | rs7592246 | A | 0.746 | 0.720 | 0.395 | 1.000 | 0.877 | 0.647–1.187 | 0.493 | 0.054 | 0.453 | 0.550 | 0.059 | 0.391 | 0.423 | 1.000 |
| ANTXR2 | 4 | rs4461470 | A | 0.732 | 0.692 | 0.195 | 1.000 | 0.822 | 0.611–1.106 | 0.498 | 0.113 | 0.388 | 0.541 | 0.077 | 0.382 | 0.388 | 1.000 |
| ANTXR2 | 4 | rs4690127 | A | 0.306 | 0.298 | 0.808 | 1.000 | 0.965 | 0.722–1.289 | 0.093 | 0.497 | 0.410 | 0.082 | 0.470 | 0.447 | 0.723 | 1.000 |
| ANTXR2 | 4 | rs9992915 | A | 0.759 | 0.723 | 0.236 | 1.000 | 0.831 | 0.612–1.129 | 0.527 | 0.080 | 0.393 | 0.577 | 0.059 | 0.365 | 0.497 | 1.000 |
| ANTXR2 | 4 | rs12643227 | A | 0.662 | 0.618 | 0.184 | 1.000 | 0.828 | 0.627–1.094 | 0.397 | 0.160 | 0.443 | 0.429 | 0.106 | 0.465 | 0.265 | 1.000 |
| ANTXR2 | 4 | rs4333130 | A | 0.876 | 0.875 | 0.970 | 1.000 | 0.991 | 0.630–1.561 | 0.758 | 0.008 | 0.233 | 0.759 | 0.008 | 0.233 | 0.996 | 1.000 |
| ANTXR2 | 4 | rs7668399 | A | 0.209 | 0.211 | 0.946 | 1.000 | 1.011 | 0.729–1.403 | 0.033 | 0.612 | 0.354 | 0.065 | 0.647 | 0.288 | 0.134 | 1.000 |
| ANTXR2 | 4 | rs17509725 | A | 0.626 | 0.587 | 0.235 | 1.000 | 0.847 | 0.645–1.114 | 0.354 | 0.181 | 0.465 | 0.394 | 0.141 | 0.465 | 0.479 | 1.000 |
| ANTXR2 | 4 | rs6818557 | C | 0.638 | 0.625 | 0.686 | 1.000 | 0.945 | 0.717–1.245 | 0.373 | 0.123 | 0.503 | 0.424 | 0.147 | 0.429 | 0.300 | 1.000 |
| ANTXR2 | 4 | rs12651388 | A | 0.349 | 0.312 | 0.331 | 1.000 | 0.847 | 0.604–1.186 | 0.057 | 0.433 | 0.510 | 0.118 | 0.420 | 0.462 | 0.143 | 1.000 |
| DMP1 | 4 | rs1462367 | A | 0.879 | 0.848 | 0.199 | 1.000 | 0.772 | 0.520–1.147 | 0.707 | 0.010 | 0.283 | 0.770 | 0.012 | 0.219 | 0.311 | 1.000 |
| DMP1 | 4 | rs1462369 | A | 0.703 | 0.747 | 0.146 | 1.000 | 1.246 | 0.926–1.676 | 0.550 | 0.057 | 0.393 | 0.494 | 0.088 | 0.418 | 0.303 | 1.000 |
| DMP1 | 4 | rs10019009 | T | 0.368 | 0.477 | 0.0012# | 0.046 | 1.565 | 1.192–2.044 | 0.247 | 0.201 | 0.552 | 0.388 | 0.126 | 0.488 | 0.0028# | 0.122 |
| DMP1 | 4 | rs2627724 | A | 0.291 | 0.234 | 0.054 | 0.983 | 0.744 | 0.551–1.005 | 0.053 | 0.585 | 0.361 | 0.088 | 0.506 | 0.406 | 0.151 | 1.000 |
| BMP6 | 6 | rs7753111 | A | 0.709 | 0.681 | 0.367 | 1.000 | 0.875 | 0.655–1.170 | 0.452 | 0.090 | 0.485 | 0.494 | 0.077 | 0.429 | 0.649 | 1.000 |
| BMP6 | 6 | rs6910759 | A | 0.812 | 0.747 | 0.024* | 0.839 | 0.686 | 0.494–0.953 | 0.555 | 0.060 | 0.385 | 0.659 | 0.035 | 0.306 | 0.076 | 0.997 |
| BMP6 | 6 | rs270406 | A | 0.397 | 0.401 | 0.898 | 1.000 | 1.018 | 0.776–1.336 | 0.164 | 0.361 | 0.475 | 0.171 | 0.377 | 0.453 | 0.900 | 1.000 |
| BMP6 | 6 | rs911749 | A | 0.326 | 0.325 | 0.963 | 1.000 | 0.993 | 0.748–1.319 | 0.110 | 0.460 | 0.430 | 0.118 | 0.465 | 0.418 | 0.951 | 1.000 |
| BMP6 | 6 | rs1226104 | A | 0.524 | 0.503 | 0.552 | 1.000 | 0.922 | 0.707–1.204 | 0.238 | 0.231 | 0.532 | 0.277 | 0.229 | 0.494 | 0.621 | 1.000 |
| BMP6 | 6 | rs1358893 | A | 0.515 | 0.524 | 0.793 | 1.000 | 1.036 | 0.794–1.354 | 0.294 | 0.247 | 0.460 | 0.288 | 0.259 | 0.453 | 0.958 | 1.000 |
| BMP6 | 6 | rs1150890 | A | 0.303 | 0.321 | 0.565 | 1.000 | 1.088 | 0.816–1.451 | 0.110 | 0.468 | 0.421 | 0.100 | 0.494 | 0.406 | 0.850 | 1.000 |
| BMP6 | 6 | rs198354 | A | 0.374 | 0.401 | 0.413 | 1.000 | 1.121 | 0.852–1.475 | 0.165 | 0.364 | 0.471 | 0.141 | 0.394 | 0.465 | 0.715 | 1.000 |
| BMP6 | 6 | rs267174 | A | 0.694 | 0.680 | 0.654 | 1.000 | 0.936 | 0.702–1.248 | 0.450 | 0.090 | 0.460 | 0.458 | 0.071 | 0.471 | 0.764 | 1.000 |
| BMP6 | 6 | rs267180 | A | 0.527 | 0.572 | 0.181 | 1.000 | 1.201 | 0.919–1.570 | 0.314 | 0.171 | 0.515 | 0.272 | 0.219 | 0.509 | 0.370 | 1.000 |
| BMP6 | 6 | rs267183 | A | 0.680 | 0.677 | 0.905 | 1.000 | 0.983 | 0.739–1.307 | 0.460 | 0.107 | 0.433 | 0.438 | 0.077 | 0.485 | 0.415 | 1.000 |
| BMP6 | 6 | rs267205 | A | 0.412 | 0.405 | 0.832 | 1.000 | 0.971 | 0.741–1.273 | 0.144 | 0.334 | 0.522 | 0.171 | 0.347 | 0.482 | 0.641 | 1.000 |
| BMP6 | 6 | rs1225923 | A | 0.565 | 0.589 | 0.476 | 1.000 | 1.103 | 0.843–1.444 | 0.358 | 0.181 | 0.462 | 0.312 | 0.182 | 0.506 | 0.570 | 1.000 |
| BMP6 | 6 | rs1225933 | A | 0.617 | 0.657 | 0.217 | 1.000 | 1.191 | 0.902–1.573 | 0.428 | 0.114 | 0.458 | 0.377 | 0.144 | 0.479 | 0.461 | 1.000 |
| COL11A2 | 6 | rs2071025 | A | 0.453 | 0.396 | 0.089 | 1.000 | 0.792 | 0.605–1.037 | 0.148 | 0.356 | 0.497 | 0.177 | 0.271 | 0.553 | 0.162 | 1.000 |
| COL11A2 | 6 | rs9277934 | A | 0.247 | 0.212 | 0.236 | 1.000 | 0.820 | 0.590–1.139 | 0.068 | 0.644 | 0.288 | 0.078 | 0.584 | 0.338 | 0.469 | 1.000 |
| COL11A2 | 6 | rs9277935 | A | 0.494 | 0.550 | 0.100 | 1.000 | 1.251 | 0.958–1.635 | 0.263 | 0.163 | 0.573 | 0.225 | 0.237 | 0.539 | 0.140 | 1.000 |
| RUNX2 | 6 | rs16873348 | A | 0.740 | 0.665 | 0.017* | 0.743 | 0.697 | 0.520–0.940 | 0.427 | 0.097 | 0.477 | 0.539 | 0.059 | 0.402 | 0.049* | 0.973 |
| RUNX2 | 6 | rs35565233 | A | 0.324 | 0.403 | 0.016* | 0.699 | 1.411 | 1.067–1.867 | 0.127 | 0.321 | 0.552 | 0.082 | 0.435 | 0.482 | 0.032* | 0.911 |
| RUNX2 | 6 | rs6908650 | A | 0.374 | 0.353 | 0.526 | 1.000 | 0.914 | 0.694–1.206 | 0.110 | 0.405 | 0.485 | 0.106 | 0.359 | 0.535 | 0.562 | 1.000 |
| RUNX2 | 6 | rs9472487 | A | 0.471 | 0.416 | 0.103 | 1.000 | 0.800 | 0.612–1.046 | 0.142 | 0.311 | 0.547 | 0.194 | 0.253 | 0.553 | 0.214 | 1.000 |
| RUNX2 | 6 | rs7771980 | A | 0.944 | 0.910 | 0.060 | 0.989 | 0.598 | 0.349–1.028 | 0.823 | 0.003 | 0.173 | 0.894 | 0.006 | 0.100 | 0.092 | 0.999 |
| RUNX2 | 6 | rs1321075 | A | 0.212 | 0.245 | 0.247 | 1.000 | 1.208 | 0.877–1.663 | 0.043 | 0.553 | 0.403 | 0.035 | 0.612 | 0.353 | 0.465 | 1.000 |
| RUNX2 | 6 | rs2820339 | C | 0.272 | 0.243 | 0.330 | 1.000 | 0.860 | 0.635–1.165 | 0.073 | 0.587 | 0.340 | 0.077 | 0.533 | 0.391 | 0.510 | 1.000 |
| RUNX2 | 6 | rs12665622 | A | 0.791 | 0.797 | 0.841 | 1.000 | 1.034 | 0.745–1.436 | 0.640 | 0.046 | 0.313 | 0.629 | 0.047 | 0.324 | 0.973 | 1.000 |
| RUNX2 | 6 | rs9463090 | A | 0.176 | 0.173 | 0.903 | 1.000 | 0.978 | 0.690–1.388 | 0.027 | 0.680 | 0.293 | 0.024 | 0.671 | 0.306 | 0.945 | 1.000 |
| RUNX2 | 6 | rs7750470 | A | 0.718 | 0.706 | 0.698 | 1.000 | 0.943 | 0.703–1.266 | 0.485 | 0.074 | 0.442 | 0.524 | 0.088 | 0.388 | 0.510 | 1.000 |
| DKK1 | 10 | rs1528877 | A | 0.638 | 0.676 | 0.238 | 1.000 | 1.184 | 0.895–1.566 | 0.433 | 0.081 | 0.487 | 0.394 | 0.118 | 0.488 | 0.373 | 1.000 |
| DKK1 | 10 | rs1569198 | A | 0.738 | 0.775 | 0.204 | 1.000 | 1.221 | 0.897–1.663 | 0.580 | 0.030 | 0.390 | 0.553 | 0.077 | 0.371 | 0.073 | 0.996 |
| DKK1 | 10 | rs7069912 | A | 0.029 | 0.025 | 0.692 | 1.000 | 0.849 | 0.377–1.911 | 0.003 | 0.953 | 0.044 | 0.000 | 0.941 | 0.059 | 0.575 | 1.000 |
| MGP | 12 | rs4236 | A | 0.865 | 0.857 | 0.733 | 1.000 | 0.935 | 0.636–1.375 | 0.740 | 0.027 | 0.233 | 0.753 | 0.024 | 0.224 | 0.946 | 1.000 |
| BMP4 | 14 | rs17563 | A | 0.675 | 0.673 | 0.969 | 1.000 | 0.994 | 0.748–1.322 | 0.430 | 0.083 | 0.487 | 0.450 | 0.101 | 0.450 | 0.681 | 1.000 |
| BMP4 | 14 | rs762642 | A | 0.541 | 0.545 | 0.910 | 1.000 | 1.016 | 0.773–1.335 | 0.267 | 0.178 | 0.555 | 0.306 | 0.225 | 0.469 | 0.202 | 1.000 |
| SOX9 | 17 | rs2229989 | A | 0.035 | 0.052 | 0.248 | 1.000 | 1.489 | 0.754–2.940 | 0.000 | 0.897 | 0.103 | 0.000 | 0.929 | 0.071 | 0.500 | 1.000 |
| SOX9 | 17 | rs1042667 | A | 0.491 | 0.430 | 0.069 | 0.994 | 0.781 | 0.598–1.020 | 0.184 | 0.324 | 0.492 | 0.224 | 0.241 | 0.535 | 0.149 | 1.000 |
| BMP2 | 20 | rs1049007 | A | 0.238 | 0.210 | 0.316 | 1.000 | 0.850 | 0.619–1.168 | 0.033 | 0.613 | 0.353 | 0.047 | 0.571 | 0.382 | 0.572 | 1.000 |
| BMP2 | 20 | rs1005464 | A | 0.376 | 0.393 | 0.610 | 1.000 | 1.074 | 0.817–1.412 | 0.133 | 0.347 | 0.520 | 0.118 | 0.365 | 0.518 | 0.856 | 1.000 |
| BMP2 | 20 | rs235768 | A | 0.762 | 0.808 | 0.096 | 1.000 | 1.314 | 0.952–1.812 | 0.642 | 0.027 | 0.331 | 0.565 | 0.041 | 0.394 | 0.226 | 1.000 |
| BMP2 | 20 | rs3178250 | A | 0.594 | 0.525 | 0.041* | 0.956 | 0.755 | 0.577–0.988 | 0.267 | 0.217 | 0.517 | 0.359 | 0.171 | 0.471 | 0.096 | 0.999 |
| BMP2 | 20 | rs6054512 | A | 0.650 | 0.672 | 0.499 | 1.000 | 1.102 | 0.832–1.458 | 0.447 | 0.103 | 0.450 | 0.394 | 0.094 | 0.512 | 0.434 | 1.000 |
*P<0.01; #P<0.05. PC, corrected P value with permutation–based hypothesis testing (10,000 permutations). OR: Odds ratio; CI: Confidence interval; SNP: Single nucleotide polymorphism; MGP: Matrix Gla protein; RUNX2: Runt–related transcription factor 2; COL11A2: Collagen, type XI, alpha 2; DMP1: Dentin matrix protein 1; ANTXR2: Anthrax toxin receptor 2; IHH: Indian hedgehog; ACVR1: Activin receptor–like kinase 2; PTGS2: Prostaglandin–endoperoxide synthase 2.
Supplementary Figure 1.
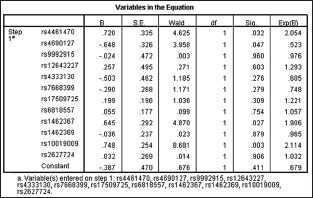
Multivariate logistic regression analyses of the loci within the dentin matrix protein 1 and anthrax toxin receptor 2 genes.
Case-control association analysis in the validation cohort
The association of rs10019009 and AS remained statistically significant in the validation cohorts [Table 2].
Table 2.
Case–control association analyses of rs10019009 in study cohort
| Study cohort | Allele | Case group, n | Control group, n | Allele | Genotype | P | Pc† | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case frequency | Control frequency | OR | 95% CI | P* | Pc† | Case frequency | Control frequency | ||||||||||
| 1/1 | 1/2 | 2/2 | 1/1 | 1/2 | 2/2 | ||||||||||||
| Discovery cohort | T | 297 | 170 | 0.477 | 0.368 | 1.56 | 1.19–2.04 | 0.0012 | 0.0460 | 0.247 | 0.552 | 0.201 | 0.388 | 0.488 | 0.124 | 0.0028 | 0.1220 |
| Validation cohort | T | 612 | 672 | 0.439 | 0.398 | 1.20 | 1.01–1.39 | 0.0349 | – | 0.338 | 0.446 | 0.216 | 0.365 | 0.474 | 0.161 | 0.0425 | – |
| Combined cohort | T | 909 | 842 | 0.451 | 0.392 | 1.28 | 1.11–1.47 | 0.0004 | – | 0.308 | 0.481 | 0.211 | 0.370 | 0.477 | 0.153 | 0.0016 | – |
*Fisher’s exact P value; †Corrected P value with permutation–based hypothesis testing (10,000 permutations). OR: Odds ratio; CI: Confidential interval.
In the discovery cohort and validation cohort, no association was found between rs10019009 and onset type (juvenile onset/adult onset), kyphosis (negative/positive), bamboo spine (negative/positive), and radiographic grading of sacroiliitis (2/3–4).
Linkage disequilibrium analysis
LD analysis demonstrated that two SNPs (rs4333130 and rs10019009) were in LD (|D’| = 0.70) in the case group [Supplementary Figure 2].
Supplementary Figure 2.
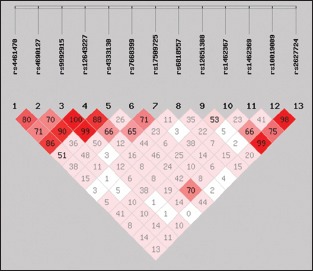
Linkage disequilibrium analysis of single nucleotide polymorphisms in the dentin matrix protein 1 and anthrax toxin receptor 2 genes in the case group. rs4333130 and rs10019009 were in linkage disequilibrium (|D’| = 0.70) in the case group.
Bioinformatics analyses
Phylogenetic context
Alignment of the rs10019009 locus in 31 mammals, from mice to humans, revealed that this site in humans exclusively displays the polymorphism c.205A>T; in contrast, this site is an A in the other thirty mammals. Nucleotide 211 in humans is a C (Gln71), an A (Lys71) in Tupaia belangeri but a G (Glu71 or Asp71) in the other 29 mammals [Figure 1].
Figure 1.
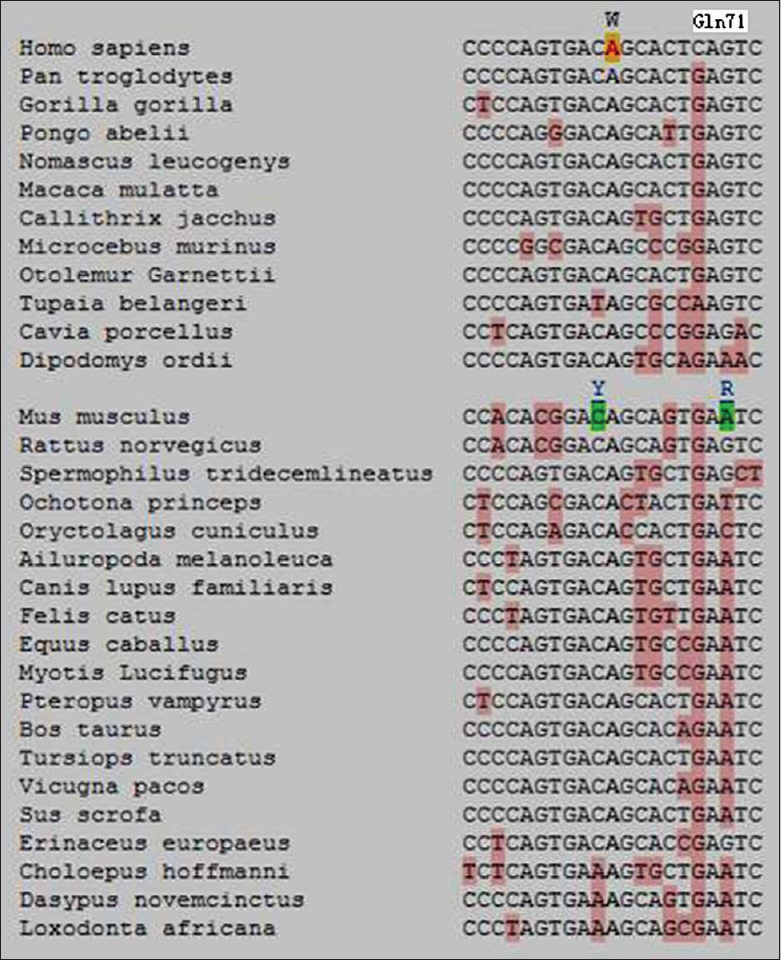
Phylogenetic context of rs10019009. The locus 205 exclusively displays polymorphism c.205A>T in humans. The locus 211 exhibits a G (Glu71 or Asp71) in the other 29 mammals but a C (Gln71) in humans.
F-single nucleotide polymorphism and fast single nucleotide polymorphism functional predictions of rs10019009
The F-SNP score was 0.274, and the functional significance scores (FSs) ranged between 0 and 1. An FS of 0 indicates that none of the tools predict a deleterious effect, whereas an FS of 1 suggests that all tools predict a deleterious effect. FS scores were deemed significant if FS ≥0.5, but it must be considered that 45% of disease-related SNPs were previously found to be <0.5.[19] According to the PolyPhen and SNPeffect predicting tools, the predicted result in the protein-coding region of DMP1 was possibly damaging and deleterious, respectively. Functional effects were predicted using ESRSearch, and the results revealed an alteration of splicing regulation at the splicing level. Golden Path showed predicted functional effects at the level of transcriptional regulation. The predicted result using fastSNP demonstrated that rs10019009, which is in the coding region, leads to a nonsynonymous and nonconservative change that affects protein structure, with an estimated risk rank = 3–4. FastSNP assigns an integer score to each SNP between 0 and 5, called the risk rank, which quantifies the likelihood that the SNP will exhibit functional effects that lead to disease phenotypes. The following possible effects of rs10019009 were identified: (1) an amino acid change to one with different structural characteristics, and (2) disruption of the exonic splicing enhancer binding site in the coding sequence, abolishing the protein domain.
Secondary structure prediction
The Phyre2 web server revealed that when the amino acid is at position 69 changes from Ser to Cys, the helix including QSEE (71–74) changes correspondingly to a helix that includes PSDCTQSEE (66–74) [Supplementary Figure 3].
Supplementary Figure 3.
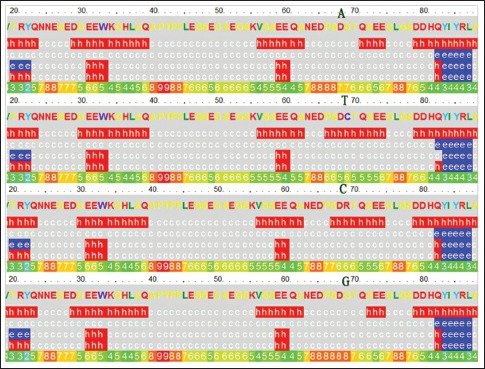
Secondary structure prediction. A, T, C, and G indicate the changes in the c.205A, c.205T, c.205C, and c.205G plasmids, respectively. Secondary structure key: e. beta strand, c.Coil, h. alpha helix.
Molecular modeling of dentin matrix protein 1
The estimated precision value of alignment of DMP1 and Borrelia burgdorferi outer surface protein A (OspA) was 100%, and the e-value was 1.5e-08. The simple way to interpret the estimated precision is as “the likelihood that the match is correct”. An 80% estimated precision means that 80% of hits with an e-value equal to or lower than that reported value are correct homologs and 20% are false positives.
The secondary and tertiary structures of DMP1 are unknown. Therefore, the X-ray-crystallographic structure of OspA (Protein Data Bank identification 2FKJ) was used as a template for modeling the tertiary structure of DMP1 according to the Phyre2 web server. The structure is displayed using the Cn3D 4.3 program (http://www.ncbi.nlm.nih.gov/Structure/CN3D/cn3dinstall.shtml) [Supplementary Figure 4].
Supplementary Figure 4.
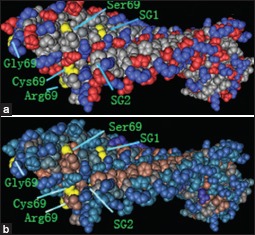
The protein structural charge and hydrophobic domains in the model of dentin matrix protein 1 and outer surface protein A (Protein Data Bank identification code 2FKJ) were visualized using Cn3D software version 4.1 (http://www.ncbi.nlm.nih.gov/Structure/CN3D/cn3d.shtml). The positions of the four amino acid substitutions of rs10019009 associated with ankylosing spondylitis susceptibility in our study are indicated. (a) Structural changes are demonstrated throughout the charged molecule after changes of the amino acid at position 69. (b) Structural changes are shown throughout the hydrophobic domain after changes of the amino acid at position 69. SG1 indicates the position of Ser109Gly110 with the change of serine and cysteine at residue 69. SG2 indicates the position of Ser109Gly110 with the change of arginine and glycine at residue 69.
Reverse transcription-polymerase chain reaction
After successful site-directed mutagenesis [Figure 2], plasmids containing c.205A, c.205T, c.205C, or c.205G were introduced into U2OS cells with a transfection efficiency of 80%. The expression of RUNX2, osteocalcin (OCN), BMP2, or BMP6 mRNA was not detected at 5 days after transfection. In addition, the expression of DMP-1 mRNA was not detected in cells containing the mock plasmid or in non-transfected cells, but the mRNA was detected in the cells harboring the c.205A, c.205T, c.205C, and c.205G plasmids at 5 days after transfection.
Figure 2.
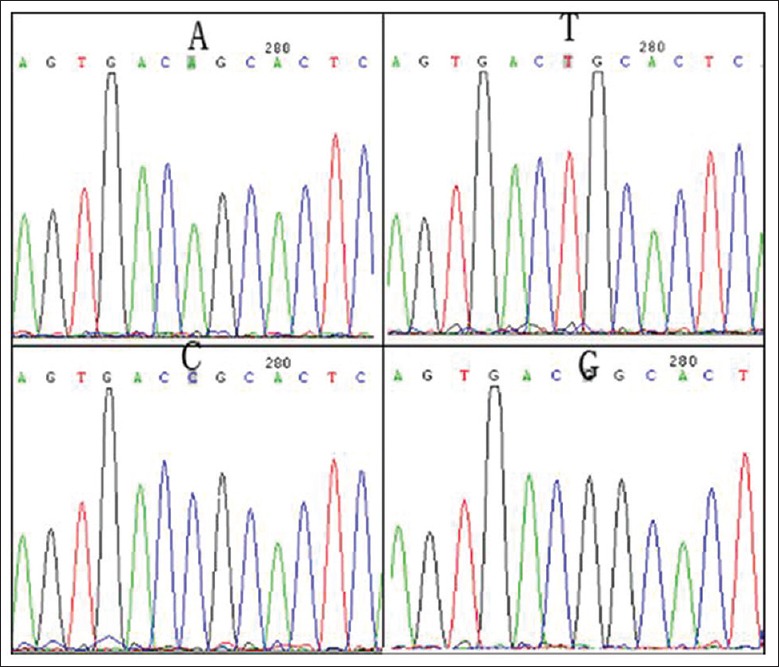
The results of site-directed mutagenesis. A, T, C, and G indicate the results detected in cells containing the c.205A (Ser69), c.205T (Cys69), c.205C (Arg69), or c.205G (Gly69) plasmid, respectively.
Alkaline phosphatase activity
ALP activity levels were low in the U2OS cells containing the mock plasmid and in the nontransfected cells. Using nonparametric testing, statistical significance was found when the relative activity of ALP of the cells transfected with the c.205A plasmid was compared to that of the cells transfected with the c.205T plasmid (P = 0.0080) [Figure 3].
Figure 3.
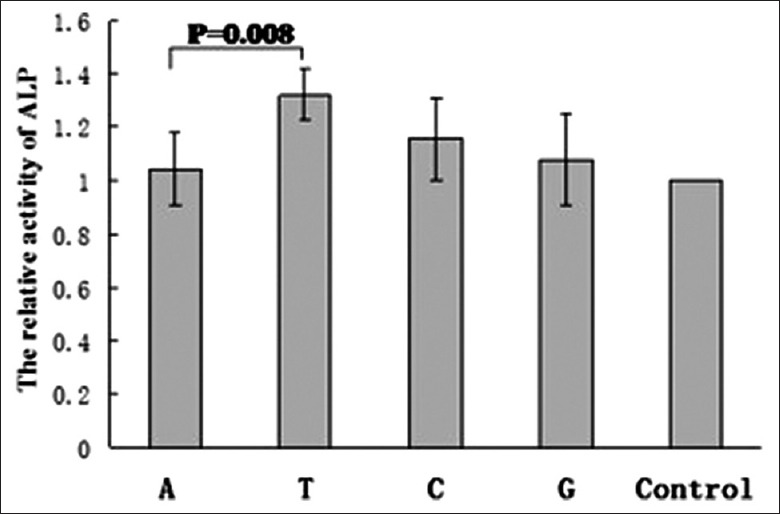
The enzymatic relative activity of alkaline phosphatase at 120 h after transfection. The relative activity of alkaline phosphatase is indicated as the ratio of values from cells transfected with the c.205A or c.205T plasmid/values from nontransfected cells. A, T, C, G, and control indicate the relative activity of alkaline phosphatase from cells transfected with c.205A, c.205T, c.205C, or c.205G or nontransfected cells, respectively (n = 5 in each group).
DISCUSSION
We performed an analysis of candidate genes related to endochondral ossification and ectopic ossification in AS in a Chinese Han population. In our case–control association analysis, the SNP rs10019009 of the DMP1 gene was found to be associated with AS in both the discovery (P = 0.0012) and the replication (P = 0.0349) cohorts as well as overall (P = 0.0004).
Recent large-scale genome-wide association studies have revealed that genetic variants within ANTXR2 are responsible for AS [Supplementary Table 4]. However, the present study found no association of rs4333130 of ANTXR2 and AS, consistent with previous findings.[20,21] Thus, polymorphisms in the ANTXR2 gene may not be major contributors to AS susceptibility in the Chinese Han population.[20] An association was found between the SNP rs10019009 in the DMP1 gene and AS in the current study; this gene is located on chromosome 4q21 and is about 7 cM downstream from ANTXR2. LD analysis demonstrated that two SNPs (rs4333130 and rs10019009) were in LD (|D’| = 0.70) in the case group, and multivariate logistic regression analysis demonstrated that rs10019009 was likely a functionally relevant SNP not due to LD. The 4q21.1 region contains a cluster of structurally and phylogenetically related genes that encode matricellular phosphoglycoproteins that function in bone formation and growth. Genome-wide association studies for AS, body height, and bone density have provided evidence that ANTXR2 and BMP3, protein kinase, cGMP-dependent, type II, RasGEF domain family, member 1B, integrin-binding sialoprotein, and matrix extracellular phosphoglycoprotein are linked in this chromosome region [Supplementary Table 4 and Supplementary Figure 5]. ANTXR2 functions as a receptor for the anthrax toxin, though little is known about the exact function of this receptor in AS or the exact function of the protein in general. In summary, the ANTXR2-DMP1 region may be a positional candidate for susceptibility to AS, and rs10019009 is a promising locus for AS predisposition.
Supplementary Table 4.
The present study and genome-wide association studies in the ANTXR2-DMP1 region
| Chromosome | Neighboring | Related | P* | OR | Study | |||
|---|---|---|---|---|---|---|---|---|
| BP | SNP | Genes | A 1 | Disease/traits | ||||
| 4q21 | 88583135 | rs10019009 | DMP1 | A | AS | 3.6×10−4 | 0.78 | The current study |
| 4q21 | 81168853 | rs4333130 | ANTXR2 | G | AS | 9.3×10−8 | 0.82 | Australo–Anglo–American Spondyloarthritis Consortium (TASC) et al. 2010[1] |
| 4q21 | 81165499 | rs4389526 | ANTXR2 | A | AS | 9.4×10−8 | 1.21 | Evans et al. 2011[2] |
| 4q21 | 82149831 | rs710841 | BMP3 PRKG2 RASGEF1B |
A | Body height | 1.9×10−8 | – | Gudbjartsson et al. 2008[3] |
| 4q21 | 82150006 | rs788867 | PRKG2/BMP3 | T | Body height | 8.9×10−28 | – | Lango Allen et al. 2010[4] |
| 4q21 | 82165790 | rs994014 | PRKG2 | T | Body height | 7.8×10−10 | – | N’Diaye et al. 2011[5] |
| 4q21 | 88732692 | rs1054627 | IBSP | A | Bone density | 4.6×10−5 1.5×10−4 7.6×10−7 |
– – – |
Styrkarsdottir et al. 2009[6] Koller et al. 2010[7] Duncan et al. 2011[8] |
| 4q21 | 88775243 | rs1471403 | MEPE | T | Bone density | 1.5×10−8 | – | Rivadeneira et al. 2009[9] |
*P. Allelic test. The 4q21.1 region contains a cluster of structurally and phylogenetically related genes encoding matricellular phosphoglycoproteins that function in bone formation and growth. Genome–wide association studies for AS, body height, and bone density revealed evidence that ANTXR2 and BMP3, PRKG2, RASGEF1B and IBSP, and MEPE are linked in this chromosome region. OR: Odds ratio; ANTXR2–DMP1: Anthrax toxin receptor 2–Dentin matrix protein 1; SNP: Single nucleotide polymorphism; PRKG2: Protein kinase, cGMPdependent, type II; RASGEF1B: RasGEF domain family, member 1B; IBSP: Integrinbinding sialoprotein; MEPE: Matrix extracellular phosphoglycoprotein; BMP3: Bone morphogenetic protein 3.
Supplementary Figure 5.
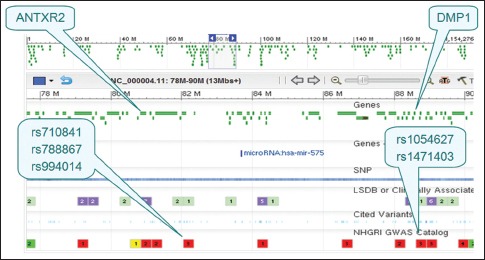
Graphics of the anthrax toxin receptor 2-dentin matrix protein 1 region. Dentin matrix protein 1 is located approximately 7 cM downstream from anthrax toxin receptor 2. rs710841, rs788867, and rs994014 are related to body height. rs1054627 and rs1471403 are related to bone density.
A previous study indicated that the sites of acidification, phosphorylation, and splice modes are important for DMP1 to function as a mineralization promoter or inhibitor.[22] Therefore, investigations of the effects of rs10019009 on DMP1 function are challenging. To this end, we performed bioinformatics analyses and preliminary functional studies of rs10019009.
We used software predictions of DMP1 secondary structure and simulations of tertiary structure and found that the amino acid change in rs10019009 leads to changes in SNP function, secondary structure, tertiary conformation, and splice mode. Some of the N-terminal fragments of DMP1 exist as a proteoglycan form, referred to as “DMP1-PG”, which acts as a mineralization inhibitor.[23] A single glycosaminoglycan chain predominantly composed of chondroitin-4-sulfate is linked to the core protein via Ser109, which is located in the SG (Ser109-Gly110) dipeptide. Sequence alignment analyses demonstrated that the Ser109-Gly110 dipeptide and its flanking regions are highly conserved in a wide range of species from caiman to Homo sapiens.[23] Further studies should be conducted to clarify the effect of rs10019009 on DMP1 function via interaction with SG (Ser109-Gly110) or RGD (arginine-glycine-aspartate) and changes in phosphorylation, splice mode, mechanical loading, charge distribution, and hydrophobic groups.
Notably, the c.205A site is a serine in mammals from mouse to gorilla, but this site does exhibit polymorphism (rs10019009, c.205A>T, p.Ser69Cys) in humans. Residue 71 is a Glu in mammals from mouse to gorilla but is a Gln in humans. Humans walk upright, and bony proliferations in the form of osteophytes and syndesmophytes may be a response-to-stress strategy of the joints. Biomechanical factors (upright walking) and microdamage (inflammation) likely play roles in “entheseal stress”, which triggers an acute inflammatory reaction and progenitor cells for new bone formation.[24] Mechanical loading intensifies DMP1 expression[25] and osteocytes, which conduct mechanical force, highly express DMP1. Therefore, we speculate that the T allele in rs10019009 with or without Gln71 and upright walking lead to the susceptibility of AS.
We found associations between RUNX2, BMP2, and BMP6 and AS in the discovery cohort. However, these associations disappeared after multiple test correction. To explore the pathways downstream of the DMP1 gene and to clarify the ectopic ossification mechanism of AS, we performed RT-PCR to further investigate whether DMP1 promotes the expression of RUNX2, BMP2, and BMP6 in transfected U2OS cells; however, we did not detect expression. DMP1 may be involved in the ectopic mineralization of AS through other mechanisms.
U2OS cells exhibit low phosphatase activity and lack the mineralization activity of human osteoblast-like cells. The main cause for U2OS cells as a cell model is whether overexpression of risk allele in rs10019009 can increase the activity of ALP and the ability of mineralization in U2OS cells. In the present study, the T allele of rs10019009 promoted the enzymatic activity of ALP compared to the nonrisk allele in U2OS cells. ALP (EC.3.1.3.1) plays a key role in mineralization and is often elevated in the peripheral blood of AS patients, especially individuals with severe disease. Indeed, Choi et al.[26] and Mitra et al.[27] found that serum ALP levels in AS patients were more elevated compared to healthy controls (P < 0.001).
Human ALP is classified into four types: tissue nonspecific, intestinal, placenta, and germ cell. Tissue-nonspecific alkaline phosphatase (TNAP) is ubiquitously expressed in many tissues,[28] and TNAP, nucleotide pyrophosphatase/phosphodiesterase-1 (NPP1) and human progressive ankylosis (ANKH) regulate HA formation.[29] TNAP hydrolyzes inorganic pyrophosphate (PPi), inhibiting HA formation, and NPP1 generates PPi, which is transported out of cells by ANKH. Mineralization begins with HA formation in matrix vesicles; HA crystals penetrate the matrix vesicle membrane and elongate due to the effect of TNAP, and the crystals are then deposited between collagen fibrils.
Osteophyte formation requires mineralization deposition and is generally linked to endochondral bone formation, which first leads to the differentiation of hypertrophic chondrocytes and proteoglycan-rich matrix deposition prior to bone build up. Morphological studies of the small facet joint of the vertebral column suggest that endochondral ossification participates in joint fusion through the building of a bridge of hypertrophic chondrocytes that fill the joint gap.[30] Notably, the mechanisms of soft tissue calcification are similar to normal skeletal development.[31] One feature that is common to most normal mineralization mechanisms is the elaboration of matrix vesicles; in fact, matrix vesicle-like membranes are observed in several ectopic calcifications. DMP1 may be involved in the mineralization that is elaborated by matrix vesicles[32] and is one of the noncollagenous proteins considered to be key regulator in matrix mineralization deposition.[33] DMP1 nucleates HA in vitro when immobilized on type I collagen fibrils, and DMP1 mRNA is expressed in coincidence with the start of mineral deposition. Acidic proteins, such as DMP1, osteopontin, bone sialoprotein, osteonectin, OCN, and bone acidic glycoprotein 75, are expressed by odontoblasts and osteoblasts, which likely play an important role in matrix mineralization through the regulation of crystal size and morphology.[34] One unique feature of DMP1 is that it is highly hydrophilic because the principal amino acids are glutamic acid, aspartic acid, and serines, which are important in the affinity of DMP1 for Ca2+ and the ability to induce in vitro mineralization.[35] These steps initiate the nucleation process and activate the entire cascade of regulated HA crystal growth.
The Pi/PPi ratio may reflect the overall differentiation state of chondrocytes (mature vs. hypertrophic), the expression levels of TNAP, NPP1, or other proteins that affect Pi and PPi concentrations and the balance between pro- and anti-calcification factors, and this ratio may serve as an indicator of the calcification process.[36] Currently available information clearly demonstrates the centrality of TNAP in the formation of minerals in mineralizing tissues, which likely occurs through its ability to alter the Pi/PPi ratio in the premineralized matrix of these tissues. In contrast, increased inorganic phosphate in cementoblasts up-regulates DMP1, osteopontin, NPP1 and ankylosis proteins, and down-regulates TNAP expression.[37] Therefore, we hypothesized that DMP1 may affect ALP expression via the correct Pi/PPi ratio.
Anti-TNF-α inhibits the production of pro-inflammatory cytokines including interleukin-1 (IL-1), IL-6, IL-8, and granulocyte-macrophage colony stimulating factor.[38] This inhibition validates the theory that TNF-α is at the apex of the pro-inflammatory cascade in rheumatoid arthritis[39] and AS.[40] Studies of TNF-α and AS susceptibility have primarily concentrated on the promoter region of the TNF-α gene, but the results are not consistent.[41,42] TNF-α was not identified in a GWAS of AS.[9,10,11,12,13] Therefore, the genes that are involved in the pathogenesis of this disease are not necessarily disease susceptibility genes. Nonetheless, the susceptibility genes of a disease are certainly involved in the pathogenesis of the disease. If disease susceptibility genes are pathogenic manipulators, then TNF-α would be the pathogenic executor in the pathogenesis of AS, and treatment of diseases with complex traits and multigenetic diseases should target the pathogenic executor with a nonspecific but strong effect rather than the pathogenic manipulator with a specific but weak effect [Supplementary Figure 6]. As TNF-α is at the apex of the pro-inflammatory cascade, and DMP1 is a key regulator of matrix mineralization,[33] DMP1 is a promising candidate gene for AS treatment because of its central role in the matrix mineralization pathway. DMP1 is a susceptibility gene for AS, and it may be the pathogenic manipulator and executor. In summary, DMP1 is a promising intervention target.
Supplementary Figure 6.
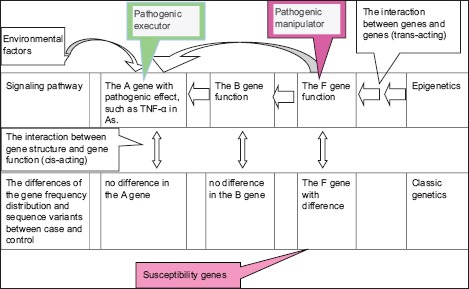
The cis-acting (gene structure and gene function) and trans-acting (the F gene → the B gene → the A gene) of genes. This figure explains why we did not find susceptibility genes from the candidate genes involved in the pathogenesis of the disease. We cannot elucidate the pathogenesis of the disease using the susceptibility genes that were detected in the genome-wide scan. However, it is likely that the susceptibility gene primarily exerts its effect via interaction between genes (trans-acting).
Supplementary information is linked to the online version of the paper on the Chinese Medical Journal website.
Financial support and sponsorship
This study was supported by grants from the National Natural Science Foundation of China (No. 81371920) and the Natural Science Foundation of Liaoning Province (No. 2013022060).
Conflicts of interest
There are no conflicts of interest.
Footnotes
Edited by: Xin Chen
REFERENCES
- 1.van der Heijde D, Salonen D, Weissman BN, Landewé R, Maksymowych WP, Kupper H, et al. Assessment of radiographic progression in the spines of patients with ankylosing spondylitis treated with adalimumab for up to 2 years. Arthritis Res Ther. 2009;11:R127. doi: 10.1186/ar2794. doi:10.1186/ar2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schett G, Rudwaleit M. Can we stop progression of ankylosing spondylitis? Best Pract Res Clin Rheumatol. 2010;24:363–71. doi: 10.1016/j.berh.2010.01.005. doi:10.1016/j.berh.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 3.Schett G. Bone formation versus bone resorption in ankylosing spondylitis. Adv Exp Med Biol. 2009;649:114–21. doi: 10.1007/978-1-4419-0298-6_8. doi:org/10.1007/978-1-4419-0298-6_8. [DOI] [PubMed] [Google Scholar]
- 4.Appel H, Kuhne M, Spiekermann S, Köhler D, Zacher J, Stein H, et al. Immunohistochemical analysis of hip arthritis in ankylosing spondylitis: Evaluation of the bone-cartilage interface and subchondral bone marrow. Arthritis Rheum. 2006;54:1805–13. doi: 10.1002/art.21907. doi:10.1002/art.21907. [DOI] [PubMed] [Google Scholar]
- 5.Lories R. The balance of tissue repair and remodeling in chronic arthritis. Nat Rev Rheumatol. 2011;7:700–7. doi: 10.1038/nrrheum.2011.156. doi:10.1038/nrrheum.2011.156. [DOI] [PubMed] [Google Scholar]
- 6.Huang W, Schwarz EM. Mechanisms of bone resorption and new bone formation in spondyloarthropathies. Curr Rheumatol Rep. 2002;4:513–7. doi: 10.1007/s11926-002-0059-0. doi:org/10.1007/s11926-002-0059-0. [DOI] [PubMed] [Google Scholar]
- 7.Diarra D, Stolina M, Polzer K, Zwerina J, Ominsky MS, Dwyer D, et al. Dickkopf-1 is a master regulator of joint remodeling. Nat Med. 2007;13:156–63. doi: 10.1038/nm1538. doi:10.1038/nm1538. [DOI] [PubMed] [Google Scholar]
- 8.Sampson HW, Davis RW, Dufner DC. Spondyloarthropathy in progressive ankylosis mice: Ultrastructural features of the intervertebral disk. Acta Anat (Basel) 1991;141:36–41. doi: 10.1159/000147096. doi:org/10.1159/000147096. [DOI] [PubMed] [Google Scholar]
- 9.Huang J, Li C, Xu H, Gu J. Novel non-HLA-susceptible regions determined by meta-analysis of four genomewide scans for ankylosing spondylitis. J Genet. 2008;87:75–81. doi: 10.1007/s12041-008-0010-x. doi:org/10.1007/s12041-008-0010-x. [DOI] [PubMed] [Google Scholar]
- 10.Carter KW, Pluzhnikov A, Timms AE, Miceli-Richard C, Bourgain C, Wordsworth BP, et al. Combined analysis of three whole genome linkage scans for Ankylosing Spondylitis. Rheumatology (Oxford) 2007;46:763–71. doi: 10.1093/rheumatology/kel443. doi:10.1093/rheumatology/kel443. [DOI] [PubMed] [Google Scholar]
- 11.Wellcome Trust Case Control Consortium;Australo-Anglo-American Spondylitis Consortium (TASC) Burton PR, Clayton DG, Cardon LR, Craddock N, Deloukas P, et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet. 2007;39:1329–37. doi: 10.1038/ng.2007.17. doi:10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Australo-Anglo-American Spondyloarthritis Consortium (TASC) Reveille JD, Sims AM, Danoy P, Evans DM, Leo P, et al. Genome-wide association study of ankylosing spondylitis identifies non-MHC susceptibility loci. Nat Genet. 2010;42:123–7. doi: 10.1038/ng.513. doi:10.1038/ng.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans DM, Spencer CC, Pointon JJ, Su Z, Harvey D, Kochan G, et al. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat Genet. 2011;43:761–7. doi: 10.1038/ng.873. doi:10.1038/ng.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shore EM, Xu M, Feldman GJ, Fenstermacher DA, Cho TJ, Choi IH, et al. A recurrent mutation in the BMP type I receptor ACVR1 causes inherited and sporadic fibrodysplasia ossificans progressiva. Nat Genet. 2006;38:525–7. doi: 10.1038/ng1783. doi:10.1038/ng1783. [DOI] [PubMed] [Google Scholar]
- 15.Maeda S, Koga H, Matsunaga S, Numasawa T, Ikari K, Furushima K, et al. Gender-specific haplotype association of collagen alpha2 (XI) gene in ossification of the posterior longitudinal ligament of the spine. J Hum Genet. 2001;46:1–4. doi: 10.1007/s100380170117. doi:10.1007/s100380170117. [DOI] [PubMed] [Google Scholar]
- 16.van der Linden S, Valkenburg HA, Cats A. Evaluation of diagnostic criteria for ankylosing spondylitis. A proposal for modification of the New York criteria. Arthritis Rheum. 1984;27:361–8. doi: 10.1002/art.1780270401. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Zhou X, Shan Z, Yang J, Yang Q, Cui Y, et al. The association of LRP5 gene polymorphisms with ankylosing spondylitis in a Chinese Han population. J Rheumatol. 2011;38:2616–8. doi: 10.3899/jrheum.111117. doi:10.3899/jrheum.111117. [DOI] [PubMed] [Google Scholar]
- 18.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: A case study using the Phyre server. Nat Protoc. 2009;4:363–71. doi: 10.1038/nprot.2009.2. doi:10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 19.Lee PH, Shatkay H. An integrative scoring system for ranking SNPs by their potential deleterious effects. Bioinformatics. 2009;25:1048–55. doi: 10.1093/bioinformatics/btp103. doi:10.1093/bioinformatics/btp103. [DOI] [PubMed] [Google Scholar]
- 20.Chen C, Zhang X, Wang Y. ANTXR2 and IL-1R2 polymorphisms are not associated with ankylosing spondylitis in Chinese Han population. Rheumatol Int. 2012;32:15–9. doi: 10.1007/s00296-010-1566-3. doi:10.1007/s00296-010-1566-3. [DOI] [PubMed] [Google Scholar]
- 21.Guo C, Xia Y, Yang Q, Qiu R, Zhao H, Liu Q. Association of the ANTXR2 gene polymorphism and ankylosing spondylitis in Chinese Han. Scand J Rheumatol. 2012;41:29–32. doi: 10.3109/03009742.2011.600700. doi:10.3109/03009742.2011.600700. [DOI] [PubMed] [Google Scholar]
- 22.Gericke A, Qin C, Sun Y, Redfern R, Redfern D, Fujimoto Y, et al. Different forms of DMP1 play distinct roles in mineralization. J Dent Res. 2010;89:355–9. doi: 10.1177/0022034510363250. doi:10.1177/0022034510363250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin C, Huang B, Wygant JN, McIntyre BW, McDonald CH, Cook RG, et al. A chondroitin sulfate chain attached to the bone dentin matrix protein 1 NH2-terminal fragment. J Biol Chem. 2006;281:8034–40. doi: 10.1074/jbc.M512964200. doi:10.1074/jbc.M512964200. [DOI] [PubMed] [Google Scholar]
- 24.Lories RJ, Luyten FP, de Vlam K. Progress in spondylarthritis. Mechanisms of new bone formation in spondyloarthritis. Arthritis Res Ther. 2009;11:221. doi: 10.1186/ar2642. doi:10.1186/ar2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gluhak-Heinrich J, Ye L, Bonewald LF, Feng JQ, MacDougall M, Harris SE, et al. Mechanical loading stimulates dentin matrix protein 1 (DMP1) expression in osteocytes in vivo. J Bone Miner Res. 2003;18:807–17. doi: 10.1359/jbmr.2003.18.5.807. doi:10.1359/jbmr.2003.18.5.807. [DOI] [PubMed] [Google Scholar]
- 26.Choi ST, Kim JH, Kang EJ, Lee SW, Park MC, Park YB, et al. Osteopontin might be involved in bone remodelling rather than in inflammation in ankylosing spondylitis. Rheumatology (Oxford) 2008;47:1775–9. doi: 10.1093/rheumatology/ken385. doi:10.1093/rheumatology/ken385. [DOI] [PubMed] [Google Scholar]
- 27.Mitra D, Elvins DM, Collins AJ. Biochemical markers of bone metabolism in mild ankylosing spondylitis and their relationship with bone mineral density and vertebral fractures. J Rheumatol. 1999;26:2201–4. [PubMed] [Google Scholar]
- 28.Millán JL. Alkaline phosphatases: Structure, substrate specificity and functional relatedness to other members of a large superfamily of enzymes. Purinergic Signal. 2006;2:335–41. doi: 10.1007/s11302-005-5435-6. doi:10.1007/s11302-005-5435-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hessle L, Johnson KA, Anderson HC, Narisawa S, Sali A, Goding JW, et al. Tissue-nonspecific alkaline phosphatase and plasma cell membrane glycoprotein-1 are central antagonistic regulators of bone mineralization. Proc Natl Acad Sci U S A. 2002;99:9445–9. doi: 10.1073/pnas.142063399. doi:10.1073/pnas.142063399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Appel H, Kuhne M, Spiekermann S, Ebhardt H, Grozdanovic Z, Köhler D, et al. Immunohistologic analysis of zygapophyseal joints in patients with ankylosing spondylitis. Arthritis Rheum. 2006;54:2845–51. doi: 10.1002/art.22060. doi:10.1002/art.22060. [DOI] [PubMed] [Google Scholar]
- 31.Golub EE. Biomineralization and matrix vesicles in biology and pathology. Semin Immunopathol. 2011;33:409–17. doi: 10.1007/s00281-010-0230-z. doi:10.1007/s00281-010-0230-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Massa LF, Ramachandran A, George A, Arana-Chavez VE. Developmental appearance of dentin matrix protein 1 during the early dentinogenesis in rat molars as identified by high-resolution immunocytochemistry. Histochem Cell Biol. 2005;124:197–205. doi: 10.1007/s00418-005-0009-9. doi:10.1007/s00418-005-0009-9. [DOI] [PubMed] [Google Scholar]
- 33.George A, Ramachandran A, Albazzaz M, Ravindran S. DMP1 –A key regulator in mineralized matrix formation. J Musculoskelet Neuronal Interact. 2007;7:308. [PubMed] [Google Scholar]
- 34.Linde A, Goldberg M. Dentinogenesis. Crit Rev Oral Biol Med. 1993;4:679–728. doi: 10.1177/10454411930040050301. [DOI] [PubMed] [Google Scholar]
- 35.He G, Dahl T, Veis A, George A. Nucleation of apatite crystals in vitro by self-assembled dentin matrix protein 1. Nat Mater. 2003;2:552–8. doi: 10.1038/nmat945. doi:10.1038/nmat945. [DOI] [PubMed] [Google Scholar]
- 36.Thouverey C, Bechkoff G, Pikula S, Buchet R. Inorganic pyrophosphate as a regulator of hydroxyapatite or calcium pyrophosphate dihydrate mineral deposition by matrix vesicles. Osteoarthritis Cartilage. 2009;17:64–72. doi: 10.1016/j.joca.2008.05.020. doi:10.1016/j.joca.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 37.Foster BL, Nociti FH, Jr, Swanson EC, Matsa-Dunn D, Berry JE, Cupp CJ, et al. Regulation of cementoblast gene expression by inorganic phosphate in vitro. Calcif Tissue Int. 2006;78:103–12. doi: 10.1007/s00223-005-0184-7. doi:10.1007/s00223-005-0184-7. [DOI] [PubMed] [Google Scholar]
- 38.Brennan FM, Chantry D, Jackson A, Maini R, Feldmann M. Inhibitory effect of TNF alpha antibodies on synovial cell interleukin-1 production in rheumatoid arthritis. Lancet. 1989;2:244–7. doi: 10.1016/s0140-6736(89)90430-3. doi:org/10.1016/S0140-6736(89)90430-3. [DOI] [PubMed] [Google Scholar]
- 39.Feldmann M, Maini RN. Anti-TNF alpha therapy of rheumatoid arthritis: What have we learned? Annu Rev Immunol. 2001;19:163–96. doi: 10.1146/annurev.immunol.19.1.163. doi:10.1146/annurev.immunol.19.1.163. [DOI] [PubMed] [Google Scholar]
- 40.Marzo-Ortega H, McGonagle D, O’Connor P, Hensor EM, Bennett AN, Green MJ, et al. Baseline and 1-year magnetic resonance imaging of the sacroiliac joint and lumbar spine in very early inflammatory back pain. Relationship between symptoms, HLA-B27 and disease extent and persistence. Ann Rheum Dis. 2009;68:1721–7. doi: 10.1136/ard.2008.097931. doi:10.1136/ard.2008.097931. [DOI] [PubMed] [Google Scholar]
- 41.Chung WT, Choe JY, Jang WC, Park SM, Ahn YC, Yoon IK, et al. Polymorphisms of tumor necrosis factor-a promoter region for susceptibility to HLA-B27-positive ankylosing spondylitis in Korean population. Rheumatol Int. 2011;31:1167–75. doi: 10.1007/s00296-010-1434-1. doi:10.1007/s00296-010-1434-1. [DOI] [PubMed] [Google Scholar]
- 42.Zhu X, Wang Y, Sun L, Song Y, Sun F, Tang L, et al. A novel gene variation of TNFalpha associated with ankylosing spondylitis: A reconfirmed study. Ann Rheum Dis. 2007;66:1419–22. doi: 10.1136/ard.2006.068528. doi:10.1136/ard.2006.068528. [DOI] [PMC free article] [PubMed] [Google Scholar]


